Introduction
Ovarian cancer (OC) is one of the most lethal
gynecological malignancies in the world (1). Early detection of OC is difficult for
lack of specific diagnostic markers. Therefore, most patients are
in advanced stage when they are diagnosed (2). Although adjuvant chemotherapy has
improved the prognosis of patients, the survival rate of patients
with OC remains low due to late diagnosis and resistance to
chemotherapy (2,3). Therefore, reasonable and effective
treatment is the key to improve the survival rate and prolong the
survival time of patients with OC. Its occurrence and development
involve a variety of genetics and environmental factors (4). As important biomolecules, lncRNAs play
an important role in the communication and exchange between cells,
and the occurrence, development, metastasis and resistance of
tumors (5). In recent years, some
progress has been made in the mechanism and treatment of lncRNAs in
OC. As research continues, the function of lncRNAs will further be
clarified, and it has broad application prospects in the early
diagnosis and treatment of OC.
lncRNAs are non-coding RNA molecules, that do not
have protein-coding functions, and participate in biological
processes in the form of RNAs (6).
With the development of technology, an increasing number of studies
have revealed that lncRNAs can not only maintain the normal
physiological functions of cells, but also play an important role
in tumor progression (5). For
example, PVT1 could promote gallbladder cancer progression by
mediating the miR-143/HK2 axis (7).
Luo et al (8) have suggested
that NEAT1 could sponge miR-34a to regulate SIRT1 expression, and
activate Wnt/β-catenin signaling, thus accelerating colorectal
cancer progression. Sun et al (9) have reported XIST could suppress renal
cell carcinoma progression by regulating the miR-106b-5p/p21 axis.
It has also been reported that various lncRNAs are dysregulated in
OC, and their expression levels are related to OC progression,
prognosis, patient survival, and response to treatment, such as
H19, MALAT1, HOTAIR, NEAT1, MEG3, XIST, and MALAT1 (10,11).
However, there are few studies on small nucleolar RNA host gene 3
(SNHG3) in OC.
SNHG3 which is located on 1q36.1 and is also named
host gene of U17 (U17HG), is a member of SNHG family (12). In OC, SNHG3 expression has been
revealed to be markedly increased, and significantly related to the
malignant degree and poor prognosis of OC (13). It has been revealed that SNHG3
participated in energy metabolism in OC by regulating miRNAs and
EIF4AIII (14). However, its
potential mechanisms in OC remain to be explored. Thus, the role of
SNHG3 in OC was explored, and the signaling pathways that SNHG3
regulated or participated in OC were identified, in order to
clarify the regulation mode of SNHG3 in OC.
Materials and methods
Tissue samples
In the present study, 40 patients with OC and 19
patients with benign OC admitted to Jinan Maternal and Child Health
Care Hospital (Jinan, China) from July, 2018 to September, 2019. OC
patients were between the ages of 45 and 65 years, with an average
age of 51.8±7.6 years. The age of benign OC patients were ranged
from 40 to 55 years, with an average age of 48.8±8.6 years.
Inclusion criteria included: i) Not receiving any treatment before
surgery. ii) Confirmed by pathological examination. iii) The
patient underwent follow-up observations. Exclusion criteria
included: i) Patients with other malignant tumors. ii) Severe
abnormal liver function. iii) Serious lung diseases. iv) Cannot
cooperate with the treatment. v) Patients with mental illness or
immune disease. This study was approved by the Medical Ethics
Committee of Jinan Maternal and Child Health Care Hospital and all
subjects signed written informed consent.
Cell culture
Human OC cell lines A2780, SKOV3, OVCAR3 and OV90,
and human ovarian epithelial cell line HOSE which were purchased
from the American Type Culture Collection (ATCC) were cultured in
RPMI-1640 medium containing 10% fetal bovine serum (FBS), 1%
penicillin, and 1% streptomycin in a cell incubator at 37°C and 5%
CO2. Cell passage was performed when the degree of cell
fusion reached 80%.
Cell transfection
Small interfering RNA targeting SNHG3 (si-SNHG3) and
control [si-negative contol (NC)], Notch1 overexpression vector
(pc-Notch1), miR-139-5p mimic (mimic) and control (miR-NC),
miR-139-5p inhibitor (inhibitor) and control (anti-miR-NC) were
synthesized by Shanghai GenePharma Co., Ltd. Transfections of
siRNA, miRNA mimic/inhibitor as well as their negative controls (50
nM) were performed by Lipofectamine 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's instructions.
After 24 h of incubation at 37°C, the cells were harvested for
further analysis. The siRNA, miRNA mimic and inhibitor as well as
their negative control sequences were as follows: si-SNHG3,
5′-CCAGAATTGCTTGCCTCAT-3′; si-NC, 5′-CCAGACTGCAGGTTTGAC-3′; mimic,
5′-UCUACAGUGCACGUGUCUCCAG-3′; miR-NC, 5′-UCUCCGAACGUGUCACGU-3′;
inhibitor, 5′-ACUGGAGACACGUGCACUGUAGA-3′; anti-miR-NC,
5′-CAGUACUUUUGUGUAGUACAA-3′.
Dual luciferase reporter assay
The binding sites between SNHG3 and miR-139-5p, and
the target genes of miR-139-5p were predicted using starBase 3.0
(http://starbase.sysu.edu.cn/index.php) (15) and TargetScan 7.2 database (www.targetscan.org) (16). The pmirGLO vector (v) was used to
construct vectors [wild-type (SNHG3-wt and Notch1-wt) and mutant
type (SNHG3-mut and Notch1-mut)] for luciferase reporter assays.
Cells were seeded into a 24-well plate. After the cell density in
the well reached ~80%, miR-139-5p mimic or miR-NC (GenePharma Co.,
Ltd, Shanghai, China) was transfected into OC cells with luciferase
reporter vectors using Lipofectamine 2000 (Invitrogen; Thermo
Fisher Scientific, Inc.). After 48-h transfection, fluorescence
activity was assessed according to the instructions of the dual
luciferase detection system (Promega Corp.). The luciferase
activity was normalized to that of Renilla luciferase
activity. The experiment was repeated three times
independently.
Cell Counting Kit-8 (CCK-8) assay
A CCK-8 assay (Dojindo Molecular Technologies, Inc.)
was used to detect cell proliferation according to the
manufacturer's instructions. The transfected cells were inoculated
into 96 wells with 2×104 cells/well. After cell
attachment, they were cultured for 1, 2, 3 and 4 days, and 10 µl of
CCK-8 solution was added. Finally, the absorbance of each well at
450 nm was measured using a microplate reader (Bio-Rad
Laboratories, Inc.). The experiment was conducted independently in
triplicate.
Wound-healing assay
The transfected OC cells were placed into a 6-well
plate in a CO2 incubator for 24 h. After the cells were
completely attached to the wall, a straight line perpendicular to
the cell surface was gently drawn in the petri dish with the tip of
a 10-µl pipette. The non-adherent cells were washed off. Image-Pro
Plus software (Media Cybernetics, Inc.) was used to measure the
scratch width. Cell scratch width was analyzed and the scratch
healing rate was calculated. Scratch healing rate = (scratch width
at 0 h - scratch width at 24 h)/scratch width at 0 h × 100%. The
experiment was repeated three times.
Reverse transciption-quantitative
(RT-q)PCR
Total RNA was extracted from cells and tissues using
TRIzol Reagent (Invitrogen; Thermo Fisher Scientific, Inc.). The
Nano-Drop ND1000 spectrophotometer (NanoDrop; Thermo Fisher
Scientific, Inc.) was used to detect the ratio of D260/D280 of RNA
solution, and calculate the concentration and purity of RNA. The
ratio of D260/D280 between 1.8 and 2.1 was used for the next
experiment. Total RNA (1 µg) was reverse transcribed into cDNA
using the MMLV reverse transcription kit (C&M Biolabs). Using
this cDNA as a template, the expression of RNAs in the
aforementioned samples was detected using SYBR Green Master Mix
(Takara Bio, Inc.) on a BioRad CFX96™ system (Bio-Rad Laboratories,
Inc.). The thermocycling conditions were as follows: initiation at
94°C for 30 sec, amplification for 32 cycles at 95°C for 5 sec,
60°C for 30 sec, and 72°C for 30 sec. The relative expression
levels of RNAs were represented by a 2−∆∆Cq value
(17) and normalized by GAPDH and
U6. Primers are presented in Table
I. Data were obtained from three independent experiments.
 | Table I.Primer sequences for real-time
fluorescence quantification PCR. |
Table I.
Primer sequences for real-time
fluorescence quantification PCR.
| Gene name | Primer sequences
(5′-3′) |
|---|
| SNHG3 | F
TTCAAGCGATTCTCGTGCC |
|
| R
AAGATTGTCAAACCCTCCCTGT |
| miR-139-5p | F
TCTACAGTGCACGTGTC |
|
| R
GAATACCTCGGACCCTGC |
| Notch1 | F
TGTTAATGAGTGCATCTCCAA |
|
| R
CATTCGTAGCCATCAATCTTGTCC |
| GAPDH | F
GCACCGTCAAGGCTGAGAAC |
|
| R
TGGTGAAGACGCCAGTGGA |
| U6 | F
TCCGATCGTGAAGCGTTC |
|
| R
GTGCAGGGTCCGAGGT |
Western blotting
Cells were lysed in ice-cold RIPA buffer (Beyotime
Institute of Biotechnology) containing protease and phosphatase
inhibitors after transfection. The lysed proteins were quantified
with BCA. After equal amounts of proteins (50 µg) were separated by
10% SDS-PAGE, they were transferred to PVDF membranes. Primary
antibodies [Notch1 (dilution, 1:1,000; cat. no. sc-6014; Santa Cruz
Biotechnology Inc.) and GAPDH (dilution, 1:1,000; cat. no. 5174;
Cell Siganling Technology, Inc.)] were incubated overnight at 4°C
after being blocked with 5% skimmed milk for 1 h at room
temperature. Then membranes were incubated with a secondary
antibody conjugated to HRP (dilution, 1:5,000; cat. no. sc-2004;
Santa Cruz Biotechnology Inc.). Finally, an enhanced
chemiluminescence (ECL) kit (EMD Millipore) was used to detect
protein expression. Densitometric analysis was performed by Image
Lab software (version 5.2.1; Bio-Rad, Laboratories, Inc.). The
analysis was performed independently in triplicate.
Statistical analysis
All values were presented as the mean ± SD. GraphPad
Prism 5.0 (GraphPad Software, Inc.) statistical software was used
to analyze the data with unpaired Student's t-test or ANOVA
(parametric) with Tukey's post hoc test. P<0.05 was considered
to indicate a statistically significant difference.
Results
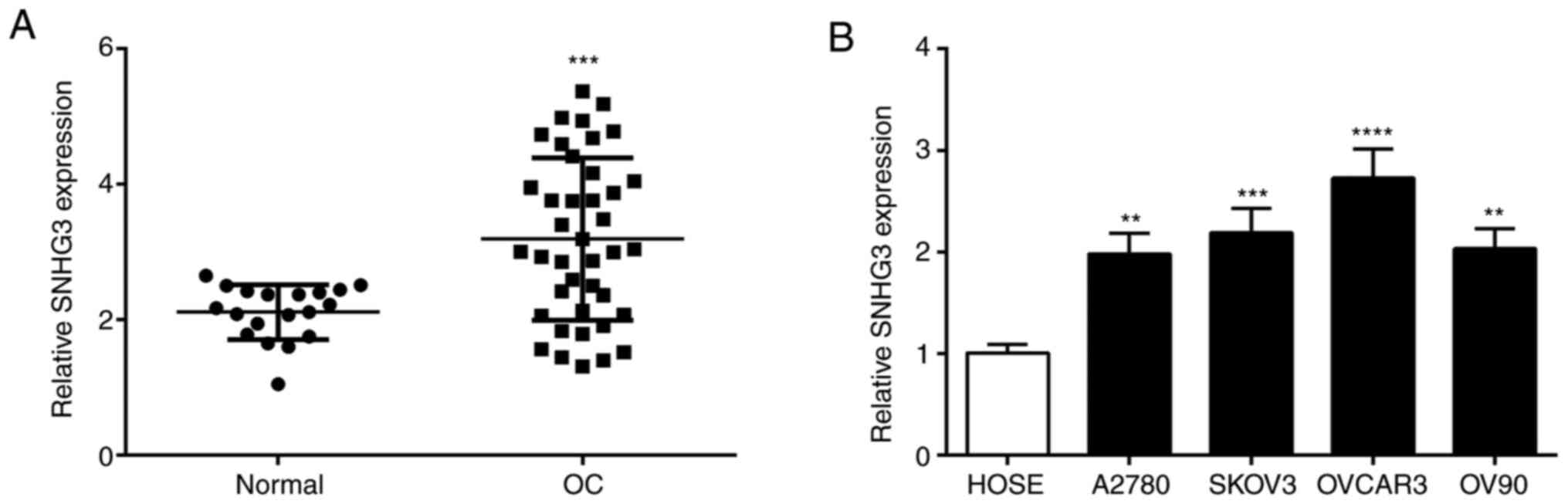
SNHG3 expression in OC
The expression level of SNHG3 in OC tissues was
significantly higher than normal tissues (Fig. 1A). Further examination of SNHG3
expression in OC cells revealed a significant increase of SNHG3
expression in different OC cell lines vs. HOSE cells (Fig. 1B). SNHG3 expression was higher in the
OVCAR3 cell line than in the other cell lines (Fig. 1B). Based on the aforementioned
experimental results, the OVCAR3 cell line was selected for further
biological functional experiments.
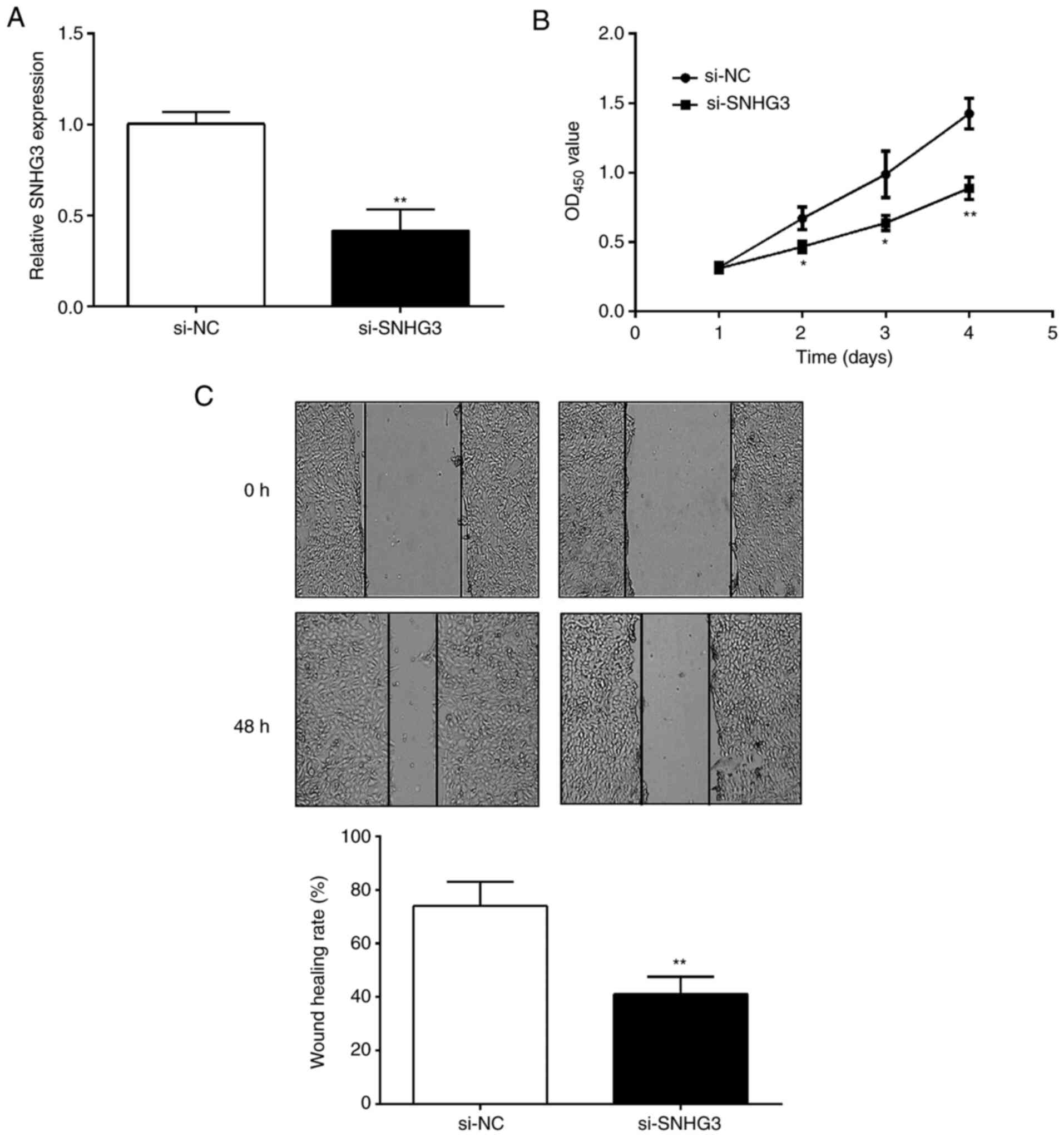
SNHG3 regulates OC cell proliferation
and migration
OC cells were collected after 48-h transfection.
Compared with the si-NC group, si-SNHG3 could effectively decrease
the expression of SNHG3 in OVCAR3 cells (Fig. 2A). Subsequently, CCK-8 and
wound-healing assays were performed to detect the effects of SNHG3
on cell proliferation and migration. The CCK-8 assay revealed that
cell proliferation ability was significantly inhibited after
downregulation of the expression level of SNHG3 in OVCAR3 cells
(Fig. 2B). Similarly, the scratch
healing rate of the si-SNHG3 group was significantly lower than in
the si-NC group (Fig. 2C),
indicating that OC cells exhibited a decreased migration ability
after SNHG3 knockdown.
SNHG3 directly targets miR-139-5p
We carried out bioinformatics analysis through the
starBase platform to predict miRNAs that could target SNHG3. The
software predicted that miR-139-5p binds to SNHG3 (Fig. 3A). OVCAR3 cells were transfected with
miR-139-5p mimic or miR-NC, miR-139-5p inhibitor or anti-miR-NC.
RT-qPCR analysis verified that miR-139-5p expression was
significantly upregulated in miR-139-5p mimic-transfected cells,
but significantly downregulated in the miR-139-5p
inhibitor-transfected cells (Fig.
3B). Dual luciferase reporter gene experiments were used to
demonstrate this prediction, and the SNHG3-wt+mimic group had a
significantly reduced luciferase activity compared with the
SNHG3-wt+miR-NC group (Fig. 3C).
Moreover, the SNHG3-mut+mimic group had no change in luciferase
activity compared with the SNHG3-mut+miR-NC group (Fig. 3C). Through this experiment, it was
demonstrated that miR-139-5p binds to SNHG3.
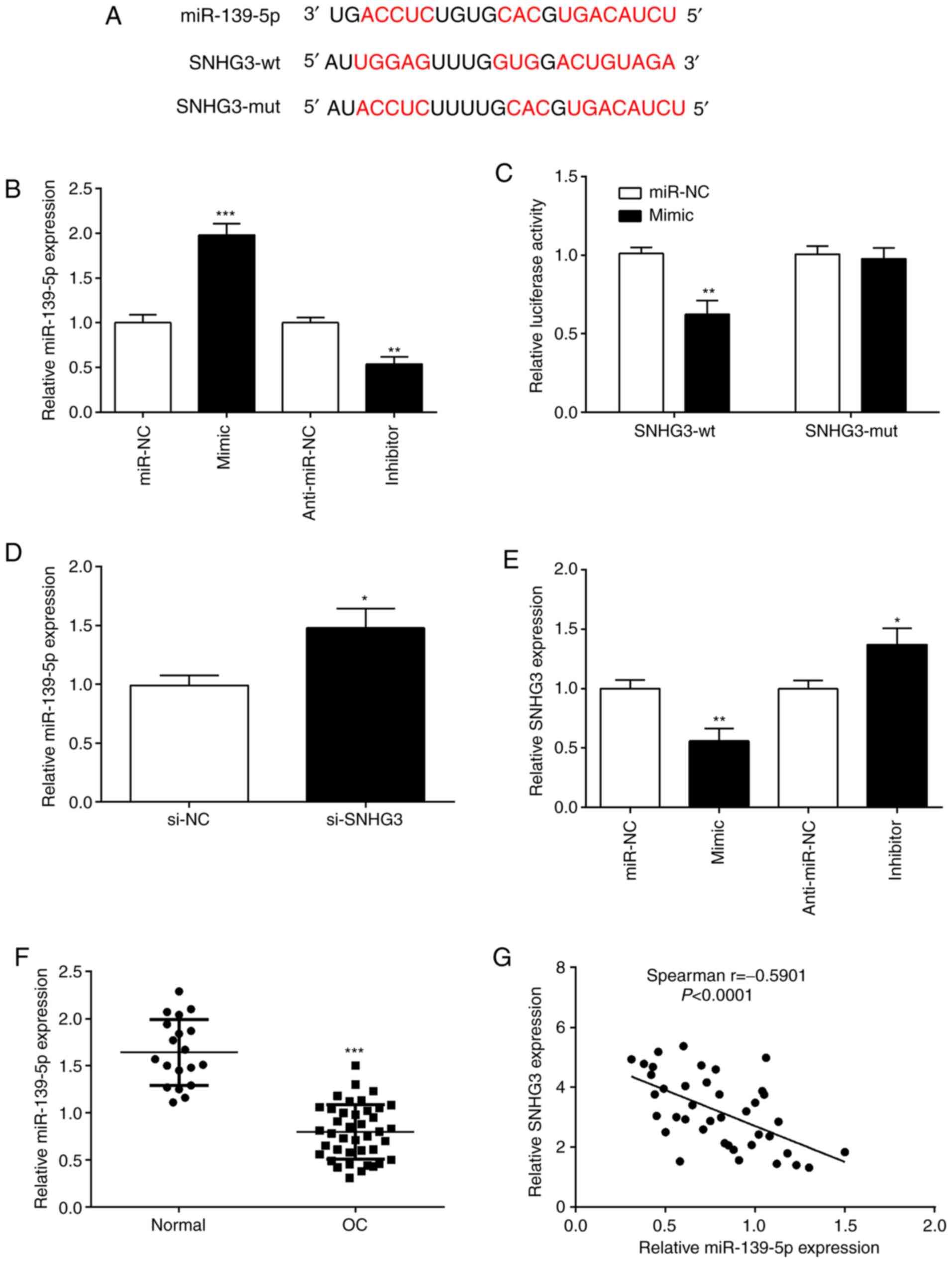 | Figure 3.SNHG3 directly binds to miR-139-5p.
(A) Predicted binding sites between miR-139-5p and SNHG3 were
predicted using starBase website. (B) OVCAR3 cells were transfected
with miR-139-5p mimic and miR-139-5p inhibitor, respectively.
RT-qPCR was performed to assess miR-139-5p expression. (C) A
luciferase reporter assay demonstrated that SNHG3 targeted
miR-139-5p. (D) RT-qPCR analysis of miR-139-5p expression in OVCAR3
cells after si-SNHG3 transfection. (E) SNHG3 expression was
quantified by RT-qPCR, in OVCAR3 cells after miR-139-5p inhibition
or overexpression. (F) Expression of miR-139-5p was determined by
RT-qPCR in tissue samples. (G) Correlation between SNHG3 and
miR-139-5p in OC tissues was evaluated by Spearman correlation
analysis. *P<0.05, **P<0.01 and ***P<0.001, compared with
miR-NC, anti-miR-NC, normal or the si-NC group. SNHG3, small
nucleolar RNA host gene 3; miR, microRNA; si-SNHG3, small
interfering RNA targeting SNHG3; OC, ovarian cancer; RT-qPCR,
reverse transciption-quantitative PCR; NC, negative control. |
In order to understand whether SNHG3 interacts with
miR-139-5p, we overexpressed or knocked down SNHG3 or miR-139-5p,
and then examined their expression. The downregulation of SNHG3
expression increased the expression of miR-139-5p, indicating that
SNHG3 inhibition increased endogenous miR-139-5p expression
(Fig. 3D). The results indicated
that SNHG3 negatively regulated miR-139-5p expression. When
miR-139-5p was overexpressed, SNHG3 expression was significantly
reduced, and it was also observed that when miR-139-5p was
inhibited, SNHG3 was upregulated (Fig.
3E). This demonstrated an inverse relationship between SNHG3
and miR-139-5p. Additionally, miR-139-5p expression in OC tissues
was significantly lower than normal group (Fig. 3F). Moreover, miR-139-5p and SNHG3
expression had a significant negative correlation in OC tissues
(Fig. 3G). Such observations proved
that miR-139-5p and SNHG3 has a negative regulatory relationship in
OC.
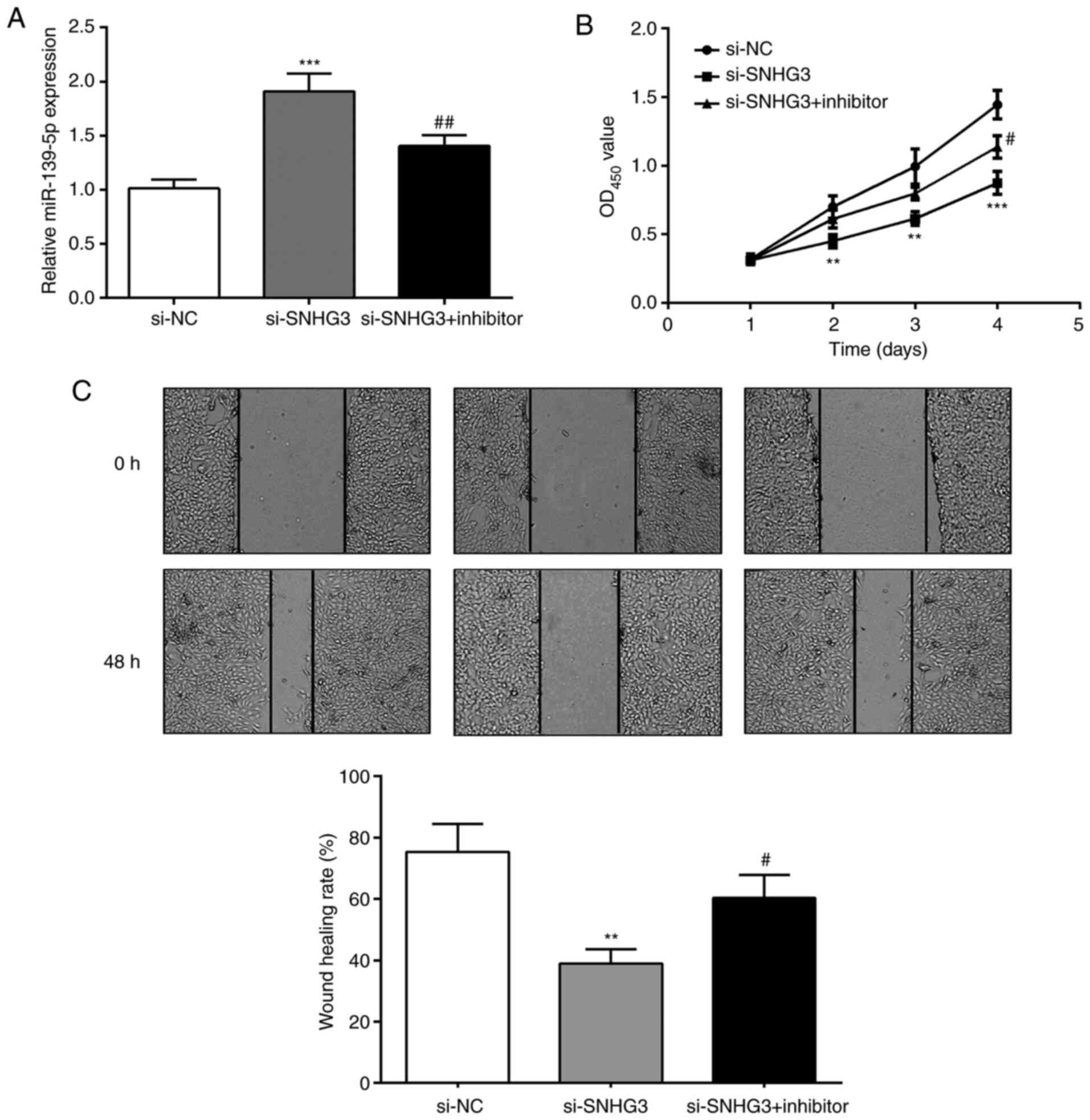
miR-139-5p regulates the suppressive
effects of SNHG3 on OC cells
Considering the relationship of SNHG3 and
miR-139-5p, SNHG3 and miR-139-5p expression were inhibited to
detect OVCAR3 cell proliferation and invasion capabilities
(Fig. 4A). The proliferation ability
of cells in the si-SNHG3 group was significantly reduced compared
with the si-SNHG3+inhibitor group (Fig.
4B). The wound-healing assay revealed that compared with the
si-SNHG3 group, the scratch healing rate of si-SNHG3+inhibitor
group was significantly increased at 48 h (Fig. 4C). The results indicated that the
suppression of si-SNHG3 on OVCAR3 cell proliferation and migration
ability could be reversed by inhibiting miR-139-5p.
Notch1 is a target of miR-139-5p
Then, bioinformatic online analysis (TargetScanHuman
7.2) predicted targets of miR-139-5p. The results revealed that
binding sites exist between Notch1 and miR-139-5p (Fig. 5A). A luciferase reporter experiment
revealed that compared with the Notch1-wt+miR-NC group, the
luciferase activity of the Notch1-wt+mimic group was significantly
reduced (Fig. 5B). However, in the
Notch1-mut group, no effect on luciferase activity was observed
(Fig. 5B). It was thus revealed that
miR-139-5p could bind to Notch1.
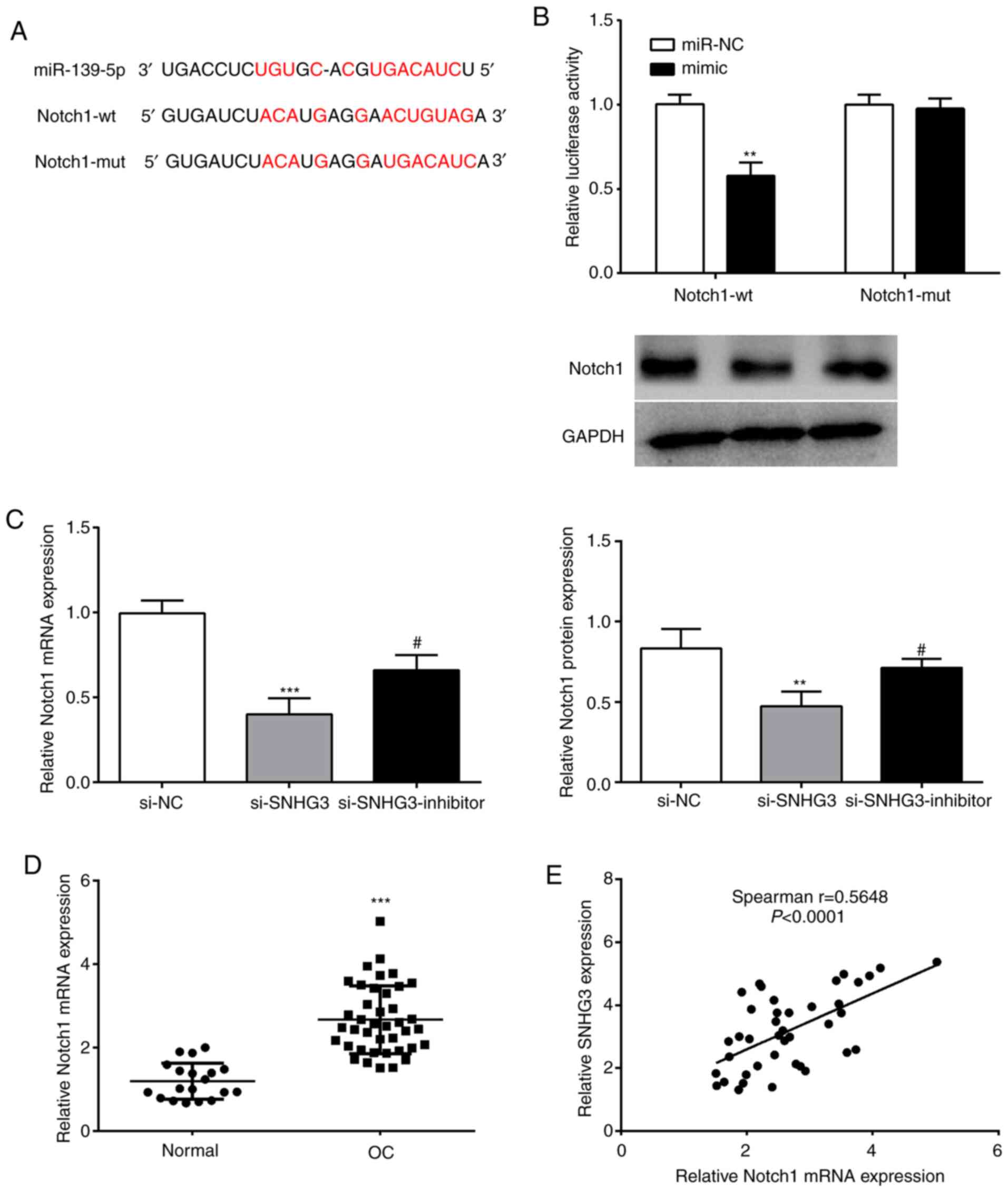 | Figure 5.miR-139-5p directly targets Notch1 in
OVCAR3 cells. (A) Putative target sites between miR-139-5p and
Notch1 were obtained by bioinformatics analysis. (B) A luciferase
reporter assay was used to confirm the binding sites of SNHG3 and
miR-139-5p. (C) RT-qPCR and western blot assays were conducted to
detect Notch1 expression in OVCAR3 cells after SNHG3 and miR-139-5p
inhibition. (D) RT-qPCR was applied to analyze Notch1 mRNA
expression in OC tissues. (E) Spearman's correlation analysis
revealed the correlation between SNHG3 and Notch1 in OC tissues.
**P<0.01 and ***P<0.001, compared with miR-NC, si-NC, or the
normal group; #P<0.05. compared with si-SNHG3. miR,
microRNA; Notch 1, Notch homolog 1, translocation-associated
(Drosophila); SNHG3, small nucleolar RNA host gene 3;
si-SNHG3, small interfering RNA targeting SNHG3; OC, ovarian
cancer; RT-qPCR, reverse transciption-quantitative PCR; NC,
negative control. |
In order to further verify the regulatory
relationship between SNHG3 and miR-139-5p on Notch1, SNHG3 and
miR-139-5p expression were interfered in OVCAR3 cells to analyze
their effects on Notch1. Notch1 mRNA and protein levels of the
si-SNHG3 group were significantly reduced vs. the si-NC group, and
miR-139-5p inhibitor reversed this effect (Fig. 5C). Moreover, an evident upregulation
of Notch1 (Fig. 5D) as well as a
significant positive correlation with SNHG3 expression in OC
tissues were observed (Fig. 5E).
Collectively, it was theorized that SNHG3 may regulate Notch1
expression through miR-139-5p in OC.
SNHG3/miR-139-5p regulates cell
proliferation and migration through Notch1
To explore whether SNHG3/miR-139-5p regulates OVCAR3
cell proliferation and migration via Notch1, Notch1 was
overexpressed in OVCAR3 cells. The results revealed that Notch1
expression was increased by pc-Notch1 but decreased by si-SNHG3 or
miR-139-5p mimic co-transfection (Fig.
6A). CCK-8 assay and wound-healing assays revealed that Notch1
overexpression promoted OVCAR3 cell proliferation and migration,
which was partially abolished by si-SNHG3 or miR-139-5p mimic
co-transfection (Fig. 6B and C).
This indicated that SNHG3 could promote the proliferation and
migration of OC by regulating miR-139-5p and Notch1.
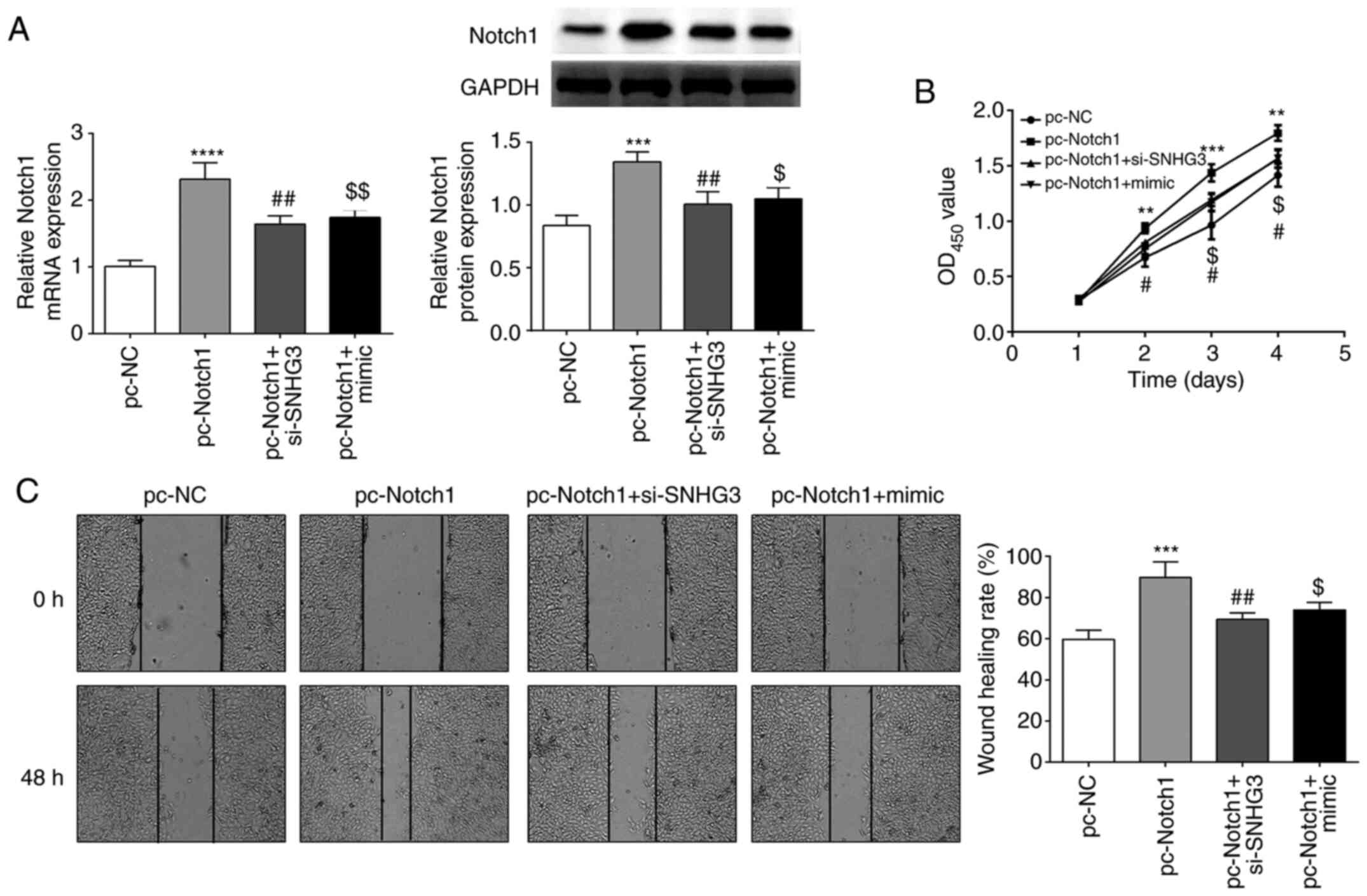 | Figure 6.Notch1 overexpression overturns the
suppressive effect of SNHG3 knockdown or miR-139-5p overexpression
on cell proliferation and migration in OVCAR3 cells. (A) Notch1
expression in OVCAR3 cells. (B and C) Cell proliferation and
migration were detected by CCK-8 and wound healing assays.
**P<0.01, ***P<0.001 and ****P<0.0001 compared with pc-NC;
#P<0.05, ##P<0.01compared with
pc-Notch1; $P<0.05, $$P<0.001 compared
with the pc-Notch1 +si-SNHG3 group. Notch 1, Notch homolog 1,
translocation-associated (Drosophila); SNHG3, small
nucleolar RNA host gene 3; miR, microRNA; si-SNHG3, small
interfering RNA targeting SNHG3; CCK-8, Cell Counting Kit-8; NC,
negative control. |
Discussion
OC is a malignant tumor of the reproductive system
that affects the health and life of women (3). It includes numerous histological
subtypes according to risk factors, origination, molecular
compositions, pathogenesis and treatments, of which epithelial OC
accounts for ~85–90%; the most common is HGSOC which accounts for
~70% of ovarian malignancies (1,4).
Although there has been great progress in OC prevention and
treatment in recent years, most patients with advanced OC will
relapse even if the treatment reaches complete remission (18). Therefore, it is particularly
important to investigate the pathogenesis of OC.
The progression of OC is a complex process, which is
regulated by various cytokines and signaling pathways (19). Previous studies have generally
revealed that numerous lncRNAs with dysregulated expression in OC
are regarded as potential therapeutic targets (11), including PVT1 (7), NEAT1 (8), and XIST (9). SNHG3 has been reported as an oncogene
in OC and its overexpression can promote cell proliferation and
invasion while knockdown of SNHG3 has the opposite effect (13). In the present study it was also
detected that SNHG3 was upregulated in OC and its knockdown
suppressed cell proliferation and migration. This was consistent
with previous research and suggested that SNHG3 plays a role as a
carcinogen (11). Notably, it was
also confirmed that SNHG3 could bind to miR-139-5p.
As a promising cancer biomarker, miR-139-5p has
various roles in different types of tumors (20). Yang et al (21) indicated that miR-139-5p expression
was decreased in prostate cancer, and was proposed to play an
anticancerous function by mediating SOX5. A previous study also
revealed that the expression of miR-139-5p was decreased in oral
squamous carcinoma cells, and had an inhibitory role in the
progression of tumorigenesis through the regulation of HOXA9
(22). However, miR-139-5p was
demonstrated to be upregulated in adrenocortical cancer and play a
promoting role in cell migration and invasion by mediating NDRG4
expression (23). In OC, low
expression of miR-139-5p has been confirmed in previous studies
(24,25). In the present study it was observed
that miR-139-5p expression was significantly decreased in OC.
Additionally, SNHG3 could bind to miR-139-5p and a negative
regulatory relationship was revealed of their expression.
Furthermore, miR-139-5p inhibition abolished the suppressive
effects of SNHG3 on OC cell proliferation and migration. The
results indicated that SNHG3 could promote OC progression by
inhibiting miR-139-5p.
Studies have revealed that lncRNAs, as carcinogenic
or tumor suppressor genes, mainly regulate the expression of miRNA
target genes by competitively binding miRNAs, and participate in
the occurrence and development of malignant tumors (26). In the present study it was confirmed
that Notch1 was a target gene of miR-139-5p. Notch1, as an
important part of Notch signaling, plays an oncogenic role in
multiple cancers (27). Its high
expression has been revealed to lead to poor overall survival of OC
patients (28). It was observed that
Notch1 was upregulated and facilitated cell proliferation and
migration of OC progression. SNHG3 inhibition or miR-139-5p
overexpression suppressed Notch1 and its promoting function on OC
cell proliferation and migration. The aforementioned results
proposed that SNHG3 regulated Notch1 and miR-139-5p in OC
progression.
There are some shortcomings in our research. For
example, additional OC tissue samples should be collected to
further explore the correlation between SNHG3 and
clinicopathological features in OC. Moreover, two or more OC cell
lines, other experiments (such as cell cloning, Transwell assay and
RNA immunoprecipitation) and animal experiments should be
introduced to investigate the function of SNHG3 in OC. Furthermore,
it is possible there are other miRNAs or target genes/signaling
pathways that are regulated by SNHG3. Hence, more research is
warranted to fully understand the potential molecular mechanism of
SNHG3 in OC.
In summary, SNHG3 expression significantly increased
and its knockdown reduced cell proliferation and migration
abilities in OC. Concurrently, the mechanism of SNHG3 was explored
and preliminarily researched. The results demonstrated that SNHG3
can regulate the expression of Notch1 and miR-139-5p, and thus play
a role in the development of OC. This provides a scientific basis
for further exploration of the mechanism of OC progression and
finding new targets for OC treatment.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
LZ, GL and HW conceived and designed the study. LZ,
XW, YZ and XH were responsible for the collection and analysis of
the experimental data. GL and XW interpreted the data and drafted
the manuscript. LZ and HW revised the manuscript critically for
important intellectual content. All authors read and approved the
final manuscript.
Ethics approval and consent to
participate
The study was approved by the Ethics Committee of
Jinan Maternal and Child Health Care Hospital. Signed written
informed consents were obtained from the patients and/or
guardians.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Torre LA, Trabert B, DeSantis CE, Miller
KD, Samimi G, Runowicz CD, Gaudet MM, Jemal A and Siegel RL:
Ovarian cancer statistics 2018. CA Cancer J Clin. 68:284–296. 2018.
View Article : Google Scholar
|
|
2
|
Li X and Wang X: The emerging roles and
therapeutic potential of exosomes in epithelial ovarian cancer. Mol
Cancer. 16:922017. View Article : Google Scholar
|
|
3
|
Moufarrij S, Dandapani M, Arthofer E,
Gomez S, Srivastava A, Lopez-Acevedo M, Villagra A and Chiappinelli
KB: Epigenetic therapy for ovarian cancer: Promise and progress.
Clin Epigenetics. 11:72019. View Article : Google Scholar
|
|
4
|
Reid BM, Permuth JB and Sellers TA:
Epidemiology of ovarian cancer: A review. Cancer Biol Med. 14:9–32.
2017. View Article : Google Scholar
|
|
5
|
Peng WX, Koirala P and Mo YY:
lncRNA-Mediated regulation of cell signaling in cancer. Oncogene.
36:5661–5667. 2017. View Article : Google Scholar
|
|
6
|
Quinn JJ and Chang HY: Unique features of
long non-coding RNA biogenesis and function. Nat Rev Genet.
17:47–62. 2016. View Article : Google Scholar
|
|
7
|
Chen J, Yu Y, Li H, Hu Q, Chen X, He Y,
Xue C, Ren F, Ren Z, Li J, et al: Long non-coding RNA PVT1 promotes
tumor progression by regulating the miR-143/HK2 axis in gallbladder
cancer. Mol Cancer. 18:332019. View Article : Google Scholar
|
|
8
|
Luo Y, Chen JJ, Lv Q, Qin J, Huang YZ, Yu
MH and Zhong M: Long non-coding RNA NEAT1 promotes colorectal
cancer progression by competitively binding miR-34a with SIRT1 and
enhancing the wnt/β-catenin signaling pathway. Cancer Lett.
440:11–22. 2019. View Article : Google Scholar
|
|
9
|
Sun K, Jia Z, Duan R, Yan Z, Jin Z, Yan L,
Li Q and Yang J: Long non-coding RNA XIST regulates miR-106b-5p/P21
axis to suppress tumor progression in renal cell carcinoma. Biochem
Biophys Res Commun. 510:416–420. 2019. View Article : Google Scholar
|
|
10
|
Tripathi MK, Doxtater K, Keramatnia F,
Zacheaus C, Yallapu MM, Jaggi M and Chauhan SC: Role of lncRNAs in
ovarian cancer: Defining new biomarkers for therapeutic purposes.
Drug Discov Today. 23:1635–1643. 2018. View Article : Google Scholar
|
|
11
|
Nikpayam E, Tasharrofi B, Sarrafzadeh S
and Ghafouri-Fard S: The role of long non-coding RNAs in ovarian
cancer. Iran Biomed J. 21:3–15. 2017. View Article : Google Scholar
|
|
12
|
Zhang T, Cao C, Wu D and Liu L: SNHG3
correlates with malignant status and poor prognosis in
hepatocellular carcinoma. Tumour Biol. 37:2379–2385. 2016.
View Article : Google Scholar
|
|
13
|
Hong L, Chen W, Wu D and Wang Y:
Upregulation of SNHG3 expression associated with poor prognosis and
enhances malignant progression of ovarian cancer. Cancer Biomark.
22:367–374. 2018. View Article : Google Scholar
|
|
14
|
Li N and Zhan X and Zhan X: The lncRNA
SNHG3 regulates energy metabolism of ovarian cancer by an analysis
of mitochondrial proteomes. Gynecol Oncol. 150:343–354. 2018.
View Article : Google Scholar
|
|
15
|
Li JH, Liu S, Zhou H, Qu LH and Yang JH:
starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA
interaction networks from large-scale CLIP-Seq data. Nucleic Acids
Res. 42:D92–D97. 2014. View Article : Google Scholar
|
|
16
|
Agarwal V, Bell GW, Nam JW and Bartel DP:
Predicting effective microRNA target sites in mammalian mRNAs, Bell
GW, Jin WN and David PB: Predicting effective microRNA target sites
in mammalian mRNAs. eLife. 4:e050052015. View Article : Google Scholar
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2-ΔΔCq method. Methods. 25:402–408. 2001. View Article : Google Scholar
|
|
18
|
Matulonis UA, Sood AK, Fallowfield L,
Howitt BE, Sehouli J and Karlan BY: Ovarian cancer. Nat Rev Dis
Primers. 2:160612016. View Article : Google Scholar
|
|
19
|
Kossaï M, Leary A, Scoazec JY and Genestie
C: Ovarian cancer: A heterogeneous disease. Pathobiology. 85:41–49.
2017. View Article : Google Scholar
|
|
20
|
Zhang HD, Jiang LH, Sun DW, Li J and Tang
JH: miR-139-5p: Promising biomarker for cancer. Tumor Biol.
36:1355–1365. 2015. View Article : Google Scholar
|
|
21
|
Yang B, Zhang W, Sun D, Wei X, Ding Y, Ma
Y and Wang Z: Downregulation of miR-139-5p promotes prostate cancer
progression through regulation of SOX5. Biomed Pharmacother.
109:2128–2135. 2019. View Article : Google Scholar
|
|
22
|
Wang K, Jin J, Ma T and Zhai H: miR-139-5p
inhibits the tumorigenesis and progression of oral squamous
carcinoma cells by targeting HOXA9. J Cell Mol Med. 21:3730–3740.
2017. View Article : Google Scholar
|
|
23
|
Agosta C, Laugier J, Guyon L, Denis J,
Bertherat J, Libé R, Boisson B, Sturm N, Feige JJ, Chabre O and
Cherradi N: miR-483-5p and miR-139-5p promote aggressiveness by
targeting N-myc downstream-regulated gene family members in
adrenocortical cancer. Int J Cancer. 143:944–957. 2018. View Article : Google Scholar
|
|
24
|
Wang Y, Li J, Xu C and Zhang X:
MicroRNA-139-5p inhibits cell proliferation and invasion by
targeting RHO-associated coiled-coil-containing protein kinase 2 in
ovarian cancer. Oncol Res. 26:411–420. 2018. View Article : Google Scholar
|
|
25
|
Liu X, Li Y, Wen J, Qi T and Wang Y: Long
non-coding RNA TTN-AS1 promotes tumorigenesis of ovarian cancer
through modulating the miR-139-5p/ROCK2 axis. Biomed Pharmacother.
125:1098822020. View Article : Google Scholar
|
|
26
|
Huang Y: The novel regulatory role of lnc
RNA-mi RNA-mRNA axis in cardiovascular diseases. J Cell Mol Med.
22:5768–5775. 2018. View Article : Google Scholar
|
|
27
|
Brzozowa-Zasada M, Piecuch A, Dittfeld A,
Mielańczyk Ł, Michalski M, Wyrobiec G, Harabin-Słowińska M, Kurek J
and Wojnicz R: Notch signalling pathway as an oncogenic factor
involved in cancer development. Contemp Oncol (Pozn). 20:267–272.
2016.
|
|
28
|
Alniaimi AN, Demorest-Hayes K, Alexander
VM, Seo S, Yang D and Rose S: Increased notch1 expression is
associated with poor overall survival in patients with ovarian
cancer. Int J Gynecol Cancer. 25:208–213. 2015. View Article : Google Scholar
|