Introduction
Lung cancer is the most common cause of
cancer-associated mortality worldwide (1,2). The
10-year survival rate of patients following diagnosis across all
stages of lung cancer is <7% (3).
Despite advancements in diagnosis, classification and therapy, the
overall survival rate of patients with lung cancer remains poor
(4). Non-small cell lung cancer
(NSCLC) accounts for 85% of all lung cancers (5). Patients with advanced or metastatic
NSCLC have poor survival outcomes, thus highlighting the need for
more effective therapies (6).
Although the diagnosis and treatment of NSCLC are continuously
being improved, patient prognosis remains unfavorable (7). Currently, the 5-year overall survival
rate is only 15% (8). Thus, it
remains critical to identify novel effective biomarkers for
accurate early diagnosis and improved prognosis of patients with
NSCLC.
Long non-coding RNAs (lncRNAs) are a novel class of
non-coding RNAs, usually defined as RNA molecules >200
nucleotides in length (9). lncRNAs
function as major regulators for gene expression, and thus play key
roles in several biological functions and disease processes,
including cancer (10,11). The lncRNA, long intragenic
non-protein-coding RNA p53-induced transcript (LINC-PINT), is
abnormally expressed in several tumors, including gastric cancer,
renal cell carcinoma and glioblastoma, and exhibits certain
diagnostic and prognostic values (12–15). In
NSCLC, LINC-PINT has been demonstrated to act as a tumor suppressor
by sponging microRNA (miRNA/miR)-208a-3p and regulating programmed
cell death 4 (PDCD4) (16). Wang
et al (17) reported that
LINC-PINT plays an important role in NSCLC by sponging miR-543 and
inducing PTEN. However, the clinical value of LINC-PINT in the
diagnosis and prognosis of NSCLC remains unclear.
Thus, the present study aimed to investigate the
clinical significance of LINC-PINT in patients with NSCLC. The
diagnostic and prognostic values of LINC-PINT were also assessed
via the receiver operating characteristic (ROC) curve, and
Kaplan-Meier and Cox regression analyses.
Materials and methods
Patients and tissue collection
A total of 122 patients who were pathologically
diagnosed with NSCLC and received resection surgery between March
2011 and June 2014 in Zibo Central Hospital were enrolled in the
present study. The patients included 53 women and 69 men with a
mean age of 61.7±13.2 years (age range, 38–84 years old). All
patients were included following the inclusion criteria: i) Tumor
tissues were histopathologically diagnosed with NSCLC; ii) Cases
had complete demographic and clinical data; iii) Cases signed
informed consent for the use of clinical samples and data. In
addition, the exclusion criteria for patient recruitment were as
follows: i) Patients with a history of other types of cancer; ii)
Cases aged <18 years; iii) Pregnant or lactating women; iv)
Cases received preoperative antitumor therapy. In addition, 62 age
(mean age, 60.8±13.8 years; age range, 37–82 years) and sex (25
women and 37 men) matched healthy individuals willing to
participate in the present study during this period were enrolled
to serve as controls. Blood samples were collected from all
participants and immediately centrifuged at 1,500 × g for 10 min at
4°C for serum extraction.
NSCLC tissues and adjacent normal tissues (at least
3 cm from the edge of tumor) were extracted from the patients
during resection surgery and frozen in liquid nitrogen at −80°C.
Demographic and clinicopathological characteristics, and the 5-year
follow-up survey (range, 0–60 months), monthly phone calls were
made for each patient and collected survival information of the
patients for subsequent analyses. Cases that died from external
events were excluded. The present study was approved the Ethics
Committee of Zibo Central Hospital (Zibo, China; approval no.
ZCHh-110824), and written informed consent was provided by all
participants prior to the study start.
Bioinformatics analysis based on The
Cancer Genome Atlas (TCGA) database
LINC-PINT expression in NSCLC and its association
with survival prognosis was assessed using the Gene Expression
Profiling Interactive Analysis (GEPIA) database (http://gepia.cancer-pku.cn/index.html)
(18), based on TCGA database
(https://cancergenome.nih.gov).
Reverse transcription-quantitative
(RT-q)PCR
Total RNA was extracted from fresh tissue and serum
samples using the GenElute Total RNA Purification kit
(Sigma-Aldrich; Merck KGaA; cat. no. RNB100). The concentration and
quality were assessed using the NanoDrop 2000 (Thermo Fisher
Scientific, Inc.), in which RNA with an absorbance ratio of optical
density (OD) 260/OD 280 results close to 2.0 were used for
subsequent RT. RT was performed using the Applied Biosystems
High-Capacity cDNA Reverse Transcription kit (Thermo Fisher
Scientific, Inc.; cat. no. 43-688-13), and the resulting cDNA was
stored at −20°C. cDNA was subsequently used as the template for
qPCR, which was performed using the SYBR-Green I Master Mix kit
(Invitrogen; Thermo Fisher Scientific, Inc.; cat. no. 4334973) and
the 7500 Real-Time PCR System (Applied Biosystems; Thermo Fisher
Scientific, Inc.). The following thermocycling conditions were
used: 95°C for 10 min, followed by 40 cycles of 95°C for 30 sec,
58°C for 20 sec and 72°C for 30 sec. The primer sequences were as
follows: LINC-PINT forward, 5′-CGTGGGAGCCCCTTTAAGTT-3′ and reverse,
5′-GGGAGGTGGCGTAGTTTCTC-3′; GAPDH forward
5′-CCTCTGACTTCAACAGCGACAC-3′ and reverse,
5′-TGGTCCAGGGGTCTTACTCC-3′. Relative expression levels were
calculated using the 2−ΔΔCq method (19) and normalized to the internal
reference gene GAPDH. Each analysis was repeated at least three
times.
Statistical analysis
Statistical analysis was performed using SPSS 21.0
software (IBM Corp.) and GraphPad 7.0 software (GraphPad Software,
Inc.). Data are presented as the mean ± standard deviation. Paired
Student's t-test was used to compare the difference in LINC-PINT
expression between NSCLC tissues and adjacent normal tissues, while
unpaired Student's t-test was used to compare serum LINC-PINT
expression between patients with NSCLC and healthy individuals, and
perform expression analysis of LINC-PINT using the GEPIA database.
The χ2 test was used to assess the association between
LINC-PINT expression and the clinicopathological characteristics of
patients with NSCLC. ROC curves were used to determine the
diagnostic value of LINC-PINT, while Kaplan-Meier and Cox
regression analyses were performed to determine the prognostic
value of LINC-PINT in NSCLC. P<0.05 was considered to indicate a
statistically significant difference.
Results
LINC-PINT expression in NSCLC based on
TCGA database
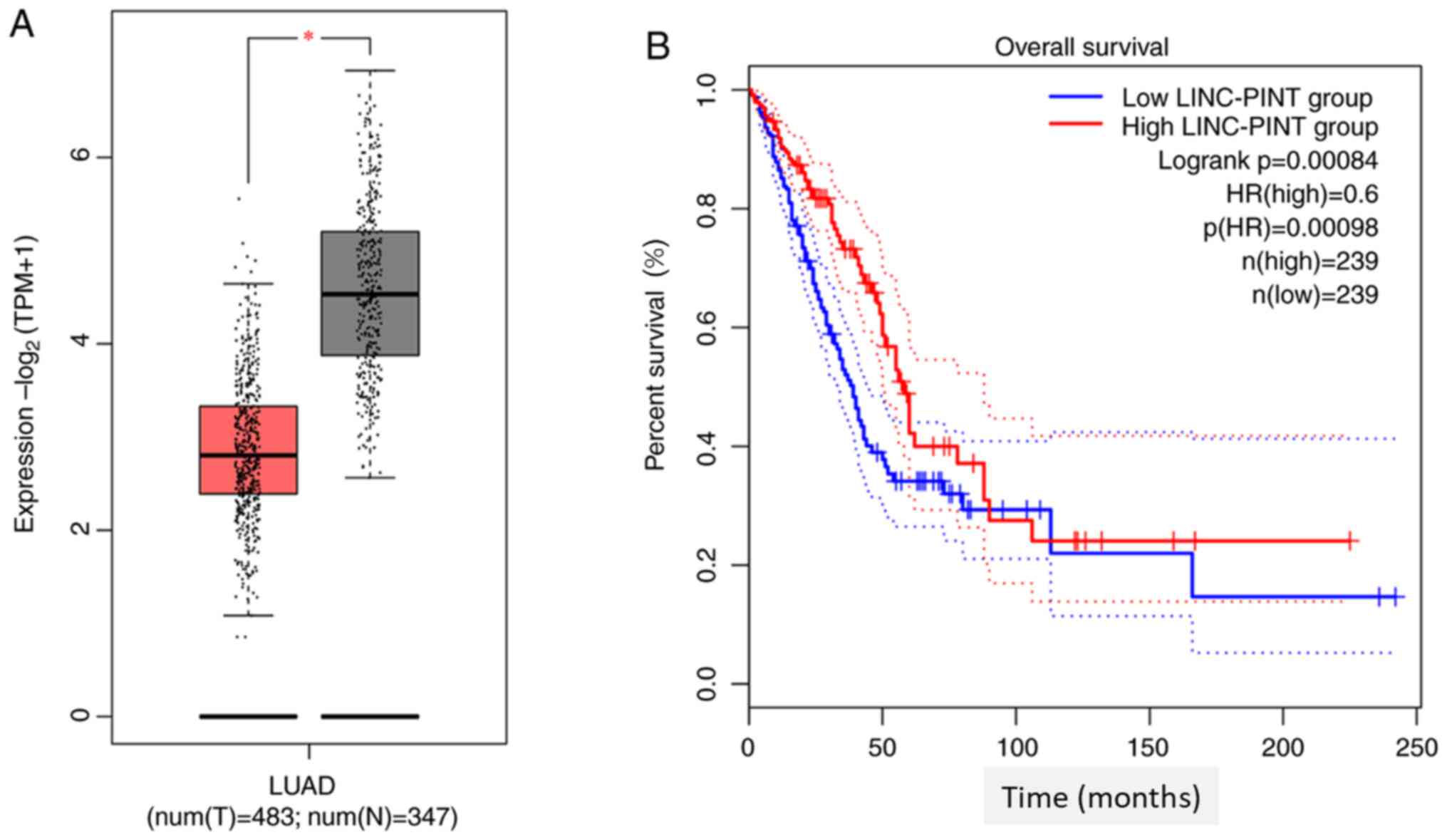
Data mining TCGA database using the GEPIA database
demonstrated that LINC-PINT expression is significantly
downregulated in NSCLC tissues compared with normal tissue
(P<0.05; Fig. 1A). Kaplan-Meier
survival analysis demonstrated that patients with low LINC-PINT
expression had a shorter overall survival time than those with high
LINC-PINT expression (Fig. 1B). In
addition, the survival curve plotted by GEPIA demonstrated that low
LINC-PINT expression was significantly associated with poor
prognosis of patients with NSCLC (P=0.00084).
LINC-PINT expression in NSCLC
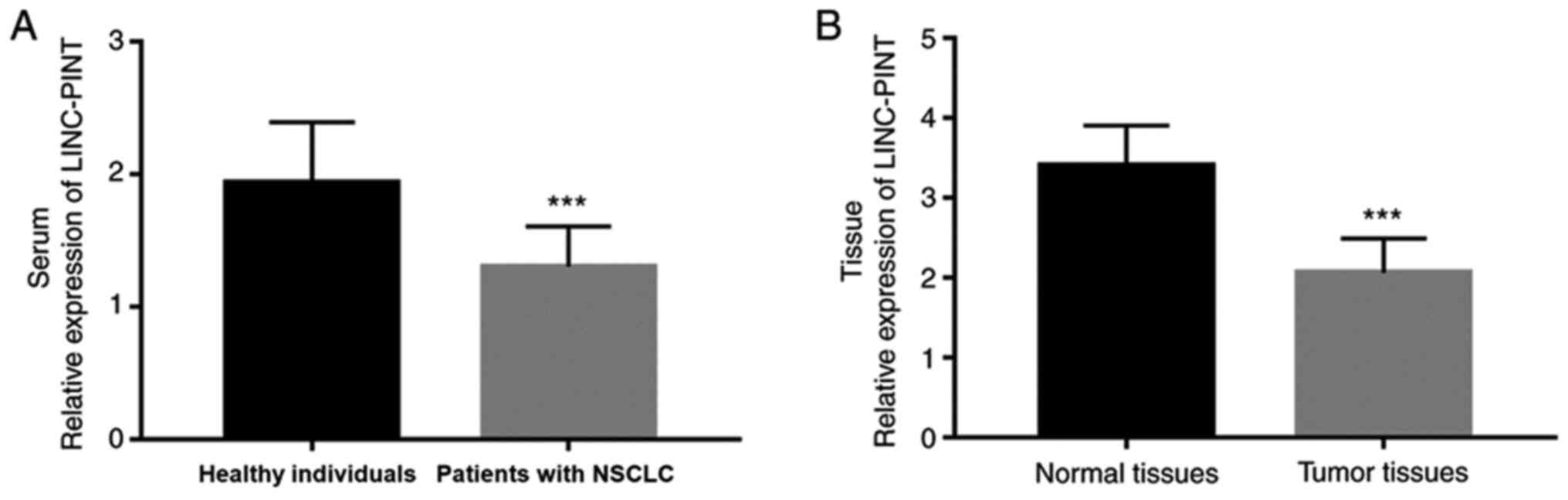
To further determine the role of LINC-PINT in NSCLC,
RT-qPCR analysis was performed to detect LINC-PINT expression in
NSCLC tissue and serum samples. The results demonstrated that serum
LINC-PINT expression was significantly downregulated in patients
with NSCLC compared with the healthy individuals (P<0.001;
Fig. 2A). Similarly, LINC-PINT
expression was significantly downregulated in NSCLC tissues
compared with adjacent normal tissues (P<0.001; Fig. 2B). These experimental results are
consistent with the analysis results from TCGA database.
Diagnostic value of serum LINC-PINT in
patients with NSCLC
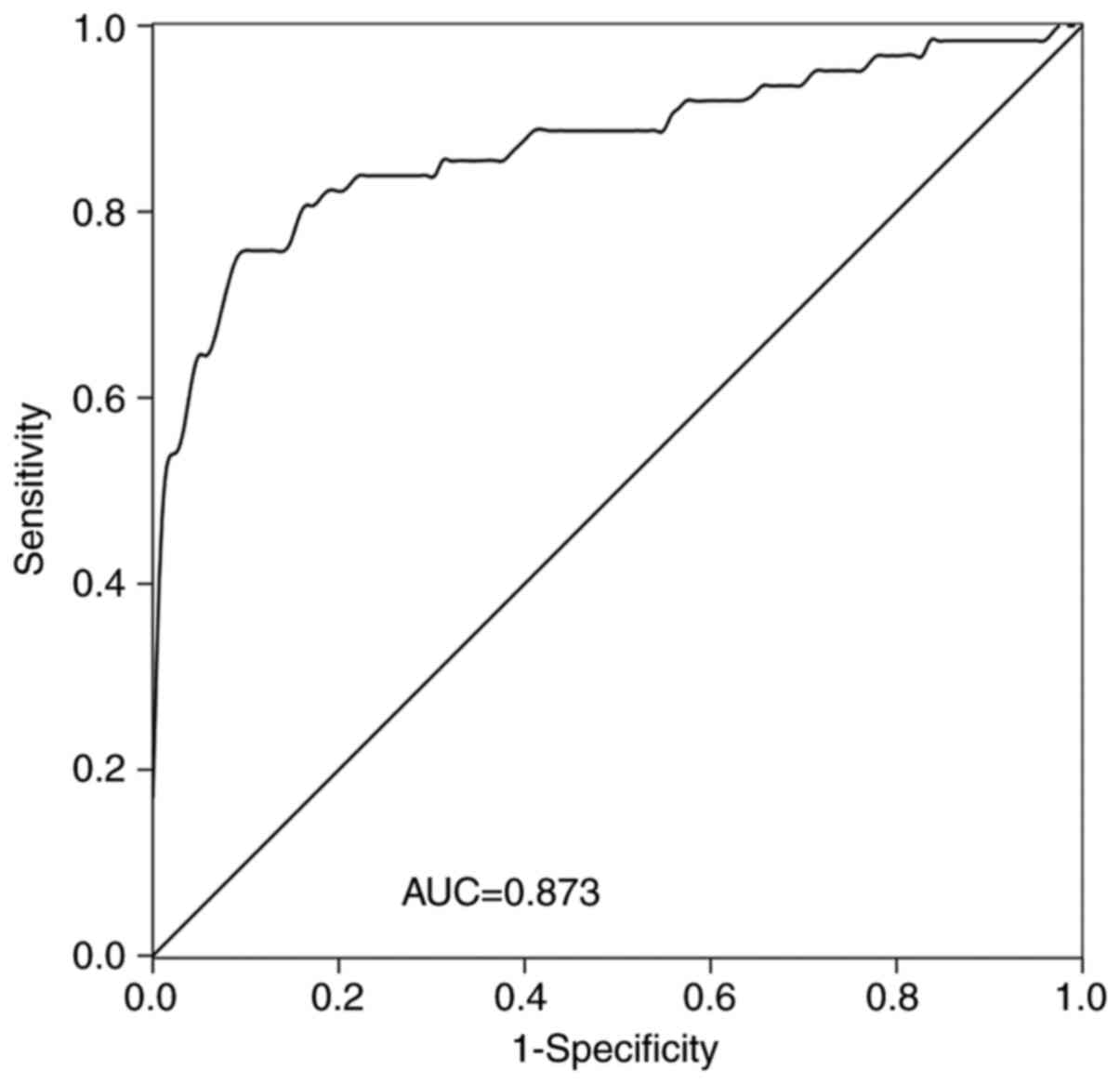
The diagnostic value of LINC-PINT in patients with
NSCLC was assessed. A ROC curve was established (Fig. 3), which demonstrated that LINC-PINT
had high diagnostic value, with an area under the curve (AUC) value
of 0.873, sensitivity of 90.9% and specificity of 75.8%. The ideal
cut-off value was 1.236.
Association between LINC-PINT
expression and the clinicopathological characteristics of patients
with NSCLC
As presented in Table
I, LINC-PINT expression was significantly associated with lymph
node metastasis (P=0.019), differentiation (P=0.028),
tumor-node-metastasis (TNM) stage (20) (P=0.020) and tumor size (P=0.027).
Conversely, no significant associations were observed between
LINC-PINT expression and age, sex and smoking history (all
P>0.05).
 | Table I.Association between LINC-PINT
expression and the clinicopathological characteristics of patients
with non-small cell lung cancer (n=122). |
Table I.
Association between LINC-PINT
expression and the clinicopathological characteristics of patients
with non-small cell lung cancer (n=122).
|
|
| LINC-PINT
expression |
|
|---|
|
|
|
|
|
|---|
| Characteristic | Number of patients,
n | Low (n=64) | High (n=58) | P-value |
|---|
| Age, years |
|
|
| 0.961 |
| ≤60 | 46 | 24 | 22 |
|
|
>60 | 76 | 40 | 36 |
|
| Sex |
|
|
| 0.943 |
|
Female | 53 | 28 | 25 |
|
|
Male | 69 | 36 | 33 |
|
| Smoking
history |
|
|
| 0.639 |
|
Never | 52 | 26 | 26 |
|
|
Ever | 70 | 38 | 32 |
|
| Tumor size, cm |
|
|
| 0.027 |
| ≤3 | 65 | 28 | 37 |
|
|
>3 | 57 | 36 | 21 |
|
|
Differentiation |
|
|
| 0.028 |
|
Well/moderate | 63 | 27 | 36 |
|
|
Poor | 59 | 37 | 22 |
|
| Lymph node
metastasis |
|
|
| 0.019 |
|
Negative | 60 | 25 | 35 |
|
|
Positive | 62 | 39 | 23 |
|
| TNM stage |
|
|
| 0.020 |
|
I–II | 56 | 23 | 33 |
|
|
III–IV | 66 | 41 | 25 |
|
Prognostic value of LINC-PINT in
patients with NSCLC
Due to the ectopic expression of LINC-PINT in NSCLC
(16), its prognostic value in
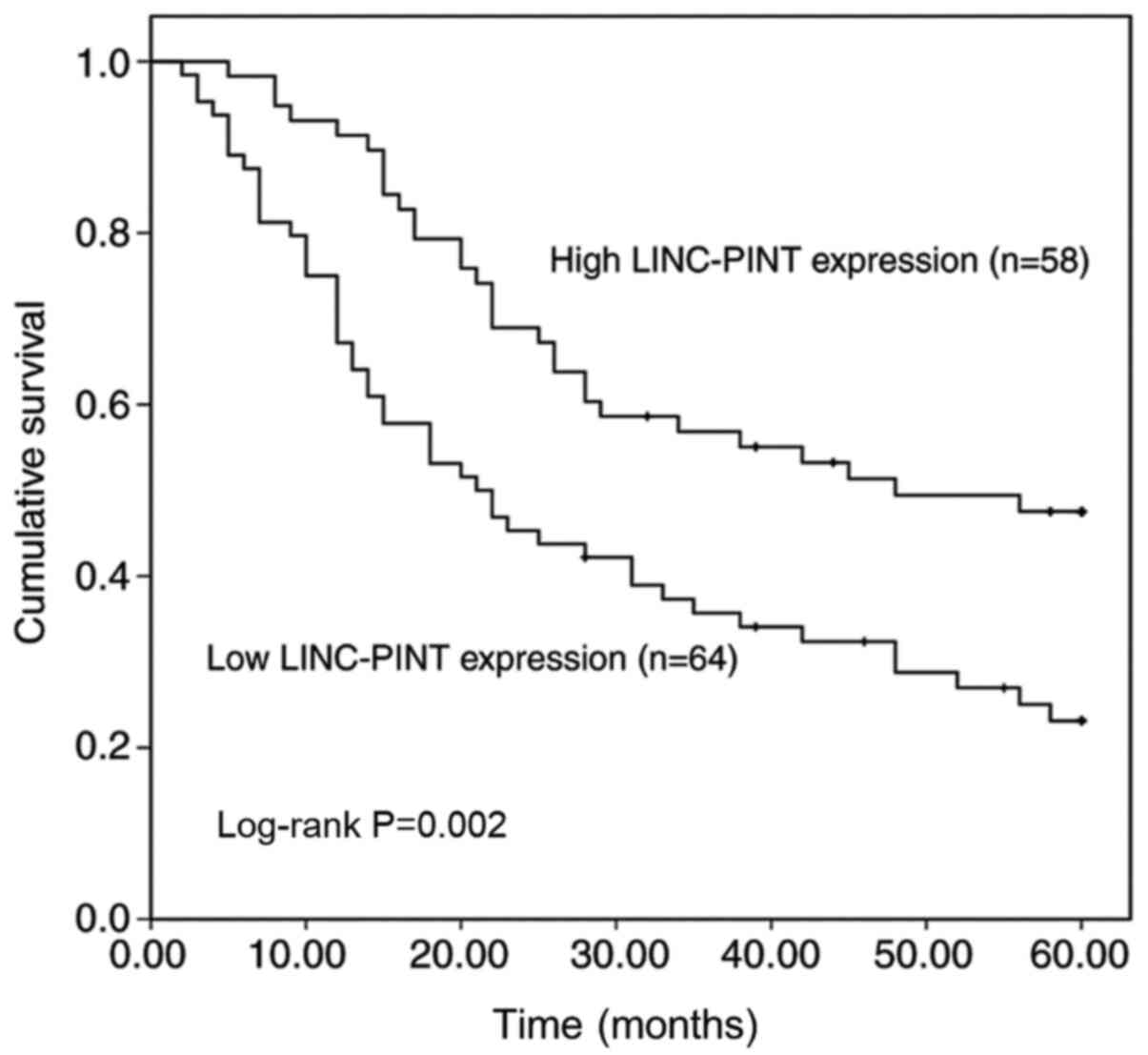
patients with NSCLC was assessed. Kaplan-Meier survival analysis
was performed to assess the association between LINC-PINT
expression and overall survival of patients with NSCLC (Fig. 4). The results demonstrated that
patients with high LINC-PINT expression had a significantly longer
overall survival time than those with low LINC-PINT expression
(P=0.002). Furthermore, the univariate and multivariate Cox
regression analysis demonstrated that LINC-PINT [hazard ratio (HR),
2.628; 95% confidence interval (CI), 1.589–4.348; P<0.001] and
TNM stage (HR, 1.810; 95% CI, 1.091–3.004; P=0.022) were two
independent prognostic factors for the survival of patients with
NSCLC (Table II).
 | Table II.Cox regression analysis of patients
with non-small cell lung cancer. |
Table II.
Cox regression analysis of patients
with non-small cell lung cancer.
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|---|
| Variable | HR | 95% CI | P-value | HR | 95% CI | P-value |
|---|
| LINC-PINT | 2.845 | 1.629–4.555 | <0.001 | 2.628 | 1.589–4.348 | <0.001 |
| Age, years | 1.141 | 0.761–1.674 | 0.499 | 1.167 | 0.721–1.888 | 0.529 |
| Sex | 1.411 | 0.857–2.166 | 0.285 | 1.479 | 0.915–2.390 | 0.110 |
| Smoking | 1.418 | 0.869–2.087 | 0.221 | 1.323 | 0.833–2.102 | 0.236 |
| Tumor size | 1.396 | 0.925–1.968 | 0.104 | 1.146 | 0.714–1.838 | 0.573 |
|
Differentiation | 1.401 | 0.991–2.120 | 0.059 | 1.358 | 0.832–2.218 | 0.221 |
| Lymph node
metastasis | 1.446 | 1.089–2.047 | 0.037 | 1.316 | 0.819–2.115 | 0.257 |
| TNM stage | 2.041 | 1.351–3.184 | 0.010 | 1.810 | 1.091–3.004 | 0.022 |
Discussion
Lung cancer is the most common malignant tumor
worldwide, with the highest mortality rate (17,21).
NSCLC is the main type of lung cancer, which accounts for ~85% of
all lung cancer cases (22), and
~30% of patients have metastatic disease at diagnosis (23). NSCLC has slower proliferation and
division of cancer cells, and relatively late spread and metastasis
compared with small cell carcinoma (24). Thus, despite advancements in
treatment, the prognosis of patients with NSCLC remains poor, and
the 5-year overall survival rate does not exceed 16% (25). Accurate biomarkers are useful in
predicting the diagnosis and prognosis of different diseases,
including NSCLC. Previous studies have proposed several biomarkers
for NSCLC (26–28). Among these, lncRNAs offer a new
direction and have attracted notable attention.
Several types of lncRNAs have been studied in NSCLC.
For example, Zhang et al (29) demonstrated that lncRNA FENDRR
inhibits the progression of NSCLC by binding to miR-761 and
regulating TIMP2 expression. In addition, lncRNA FEZF1-AS1 can act
as a tumor promoting regulator in NSCLC and may provide a target
for the treatment of NSCLC (30). It
has been demonstrated that MALAT1 can alter chemoresistance of
NSCLC cells by targeting miR-197-3p and regulating p120-ctn
expression, which may assist in improving chemotherapies for NSCLC
(31). Collectively, these results
suggest that lncRNAs play important roles in the development and
progression of NSCLC. Recently, lncRNA LINC-PINT has been
extensively studied. It has been suggested that LINC-PINT may
mediate cancer cell proliferation, invasion and migration in
osteosarcoma by binding to miRNA-21 (32). Furthermore, Zhang et al
(16) demonstrated that LINC-PINT
mediates inhibition of cell proliferation, cell cycle, and cell
migration and invasion in NSCLC via the miR-208a-3p/PDCD4 axis.
However, the clinicopathological characteristics of LINC-PINT in
NSCLC remain unclear.
In the present study, TCGA data mining and RT-qPCR
analyses demonstrated that LINC-PINT expression was significantly
downregulated in NSCLC tissues compared with normal tissues, which
was consistent with the findings by Wang et al (17). Thus, it was predicted that LINC-PINT
may be involved in the progression of NSCLC. To further investigate
its role in the development of NSCLC, the association between
LINC-PINT expression and the clinicopathological characteristics of
patients with NSCLC was assessed. The results demonstrated that
LINC-PINT expression in NSCLC was significantly associated with
lymph node metastasis, differentiation, TNM stage and tumor
size.
The clinical significance of LINC-PINT in NSCLC was
further investigated. The results demonstrated that abnormal
LINC-PINT expression was associated with the diagnosis or prognosis
of patients with NSCLC. lncRNAs are considered ideal diagnostic
tools for different human diseases due to their specific expression
and stability in blood samples (11). For example, decreased serum
lncRNA-D16366 levels serve as a non-invasive diagnostic biomarker
in patients with hepatocellular carcinoma (33), and enhanced serum lncRNA-XLOC_009167
levels may serve as a biomarker for the diagnosis of patients with
lung cancer (34). The results of
the present study demonstrated that downregulated LINC-PINT
expression increased diagnostic accuracy in patients with NSCLC.
Previous studies have investigated the diagnostic value of some
lncRNAs and a study by Xie et al (35), which investigated circulating lncRNAs
for NSCLC diagnosis, reported that SOX2OT, ANRIL, CEA, CYFRA211 and
SCCA may serve as candidate diagnostic biomarkers. In addition, the
combined diagnostic accuracy of the lncRNAs exhibited an AUC value
of 0.853. The results of the present study demonstrated that the
AUC value of LINC-PINT was 0.873, suggesting that LINC-PINT may be
a potential diagnostic biomarker for patients with NSCLC. The
prognostic value of LINC-PINT in NSCLC was also assessed in the
present study. Cancer prognosis relies on the TNM system, which
requires medical imaging support such as CT, magnetic resonance and
bone scan (36). The TNM method not
only consumes manpower and material resources, but also has a
long-time cycle (37), thus, there
is an urgent requirement to identify and develop novel prognostic
biomarkers. lncRNAs have been used as biomarkers in different types
of cancer (38). In the present
study, the prognostic value of LINC-PINT was assessed based on the
5-year follow-up survival information of patients with NSCLC.
Kaplan-Meier survival analysis demonstrated that patients with low
LINC-PINT expression had a shorter overall survival time than those
with high LINC-PINT expression. In addition, multivariate Cox
regression analysis confirmed that LINC-PINT expression can
effectively be used to predict the prognosis of patients with
NSCLC.
The biological function of LINC-PINT has been
investigated in NSCLC progression. For example, Wang et al
(17) demonstrated that LINC-PINT
can inhibit the cell proliferation and cell colony formation of
NSCLC cells, and it was concluded that LINC-PINT plays an important
biological role in NSCLC by sponging miR-543 and inducing PTEN
expression. Although this study provides evidence for the clinical
value of LINC-PINT in the diagnosis and prognosis of patients with
NSCLC, the miRNA that may be regulated by LINC-PINT in NSCLC was
not investigated in the present study. Considering the regulatory
association between LINC-PINT and miRNA in NSCLC, the clinical
significance of LINC-PINT may be improved by co-analyzing the
expression changes in the miRNAs. Thus, further studies are
required to confirm and develop the clinical application potential
of LINC-PINT, with a larger study population and analyses of
related miRNAs.
In conclusion, the results of the present study
demonstrated that lncRNA LINC-PINT expression is downregulated in
NSCLC tissue and serum samples. Furthermore, serum LINC-PINT may
serve as a candidate diagnostic biomarker to distinguish patients
with NSCLC from healthy individuals, and low LINC-PINT expression
in tumor tissues may predict poor prognosis of patients with
NSCLC.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
All data generated or analyzed during the present
study are included in this published article.
Authors' contributions
CZ and JT contributed to the conception of the work,
bioinformatics analysis, data analysis and interpretation,
manuscript writing and revision, and confirmed the authenticity of
all the raw data. CG and JL collected the clinical samples and data
and performed the experiments. All authors have read and approved
the final manuscript.
Ethics approval and consent to
participate
The present study was approved the Ethics Committee
of Zibo Central Hospital (Zibo, China; approval no. ZCHh-110824),
and written informed consent was provided by all participants prior
to the study start.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Nanavaty P, Alvarez MS and Alberts WM:
Lung cancer screening: Advantages, controversies, and applications.
Cancer Control. 21:9–14. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Nasim F, Sabath BF and Eapen GA: Lung
cancer. Med Clin North Am. 103:463–473. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Cheung CHY and Juan HF: Quantitative
proteomics in lung cancer. J Biomed Sci. 24:372017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
de Sousa VML and Carvalho L: Heterogeneity
in lung cancer. Pathobiology. 85:96–107. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Herbst RS, Morgensztern D and Boshoff C:
The biology and management of non-small cell lung cancer. Nature.
553:446–454. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Syrigos KN, Saif MW, Karapanagiotou EM,
Oikonomopoulos G and De Marinis F: The need for third-line
treatment in non-small cell lung cancer: An overview of new
options. Anticancer Res. 31:649–659. 2011.PubMed/NCBI
|
|
7
|
Richard PJ and Rengan R: Oligometastatic
non-small-cell lung cancer: Current treatment strategies. Lung
Cancer (Auckl). 7:129–140. 2016.PubMed/NCBI
|
|
8
|
Hirsch FR, Suda K, Wiens J and Bunn PA Jr:
New and emerging targeted treatments in advanced non-small-cell
lung cancer. Lancet. 388:1012–1024. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wang J, Su Z, Lu S, Fu W, Liu Z, Jiang X
and Ta S: LncRNA HOXA-AS2 and its molecular mechanisms in human
cancer. Clin Chim Acta. 485:229–233. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Peng WX, Koirala P and Mo YY:
LncRNA-Mediated regulation of cell signaling in cancer. Oncogene.
36:5661–5667. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bhan A, Soleimani M and Mandal SS: Long
noncoding RNA and cancer: A new paradigm. Cancer Res. 77:3965–3981.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Feng H, Zhang J, Shi Y, Wang L, Zhang C
and Wu L: Long noncoding RNA LINC-PINT is inhibited in gastric
cancer and predicts poor survival. J Cell Biochem. 120:9594–9600.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Duan J, Ma X, Shi J, Xuan Y, Wang H, Li P,
Zhang Y, Fan Y, Gong H, Ma X, et al: Long noncoding RNA LINC-PINT
promotes proliferation through EZH2 and predicts poor prognosis in
clear cell renal cell carcinoma. Onco Targets Ther. 12:4729–4740.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hong L, Wang H, Wang J, Wei S, Zhang F,
Han J, Liu Y, Ma M, Liu C, Xu Y and Jiang D: LncRNA PTCSC3 inhibits
tumor growth and cancer cell stemness in gastric cancer by
interacting with lncRNA linc-pint. Cancer Manag Res.
11:10393–10399. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang M, Zhao K, Xu X, Yang Y, Yan S, Wei
P, Liu H, Xu J, Xiao F, Zhou H, et al: A peptide encoded by
circular form of LINC-PINT suppresses oncogenic transcriptional
elongation in glioblastoma. Nat Commun. 9:44752018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhang L, Hu J, Li J, Yang Q, Hao M and Bu
L: Long noncoding RNA LINC-PINT inhibits non-small cell lung cancer
progression through sponging miR-218-5p/PDCD4. Artif Cells Nanomed
Biotechnol. 47:1595–1602. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang S, Jiang W, Zhang X, Lu Z, Geng Q,
Wang W, Li N and Cai X: LINC-PINT alleviates lung cancer
progression via sponging miR-543 and inducing PTEN. Cancer Med.
9:1999–2009. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Tang Z, Li C, Kang B, Gao G, Li C and
Zhang Z: GEPIA: A web server for cancer and normal gene expression
profiling and interactive analyses. Nucleic Acids Res. 45:W98–W102.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chassagnon G, Bennani S and Revel MP: New
TNM classification of non-small cell lung cancer. Rev Pneumol Clin.
73:34–39. 2017.(In French). View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Collins LG, Haines C, Perkel R and Enck
RE: Lung cancer: Diagnosis and management. Am Fam Physician.
75:56–63. 2007.PubMed/NCBI
|
|
22
|
Ettinger DS, Wood DE, Aisner DL, Akerley
W, Bauman J, Chirieac LR, D'Amico TA, DeCamp MM, Dilling TJ,
Dobelbower M, et al: Non-Small cell lung cancer, version 5.2017,
NCCN clinical practice guidelines in oncology. J Natl Compr Cancer
Netw. 15:504–535. 2017. View Article : Google Scholar
|
|
23
|
Gong HY, Wang Y, Han G and Song QB:
Radiotherapy for oligometastatic tumor improved the prognosis of
patients with non-small cell lung cancer (NSCLC). Thorac Cancer.
10:1136–1140. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Jonna S and Subramaniam DS: Molecular
diagnostics and targeted therapies in non-small cell lung cancer
(NSCLC): An update. Discov Med. 27:167–170. 2019.PubMed/NCBI
|
|
25
|
Chen G, Umelo IA, Lv S, Teugels E, Fostier
K, Kronenberger P, Dewaele A, Sadones J, Geers C and De Grève J:
MiR-146a inhibits cell growth, cell migration and induces apoptosis
in non-small cell lung cancer cells. PLoS One. 8:e603172013.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Camidge DR, Doebele RC and Kerr KM:
Comparing and contrasting predictive biomarkers for immunotherapy
and targeted therapy of NSCLC. Nat Rev Clin Oncol. 16:341–355.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Gao P, Wang H, Yu J, Zhang J, Yang Z, Liu
M, Niu Y, Wei X, Wang W, Li H, et al: MiR-3607-3p suppresses
non-small cell lung cancer (NSCLC) by targeting TGFBR1 and CCNE2.
PLoS Genet. 14:e10077902018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wang SY, Li Y, Jiang YS and Li RZ:
Investigation of serum miR-411 as a diagnosis and prognosis
biomarker for non-small cell lung cancer. Eur Rev Med Pharmacol
Sci. 21:4092–4097. 2017.PubMed/NCBI
|
|
29
|
Zhang G, Wang Q, Zhang X, Ding Z and Liu
R: LncRNA FENDRR suppresses the progression of NSCLC via regulating
miR-761/TIMP2 axis. Biomed Pharmacother. 118:1093092019. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
He R, Zhang FH and Shen N: LncRNA
FEZF1-AS1 enhances epithelial-mesenchymal transition (EMT) through
suppressing E-cadherin and regulating WNT pathway in non-small cell
lung cancer (NSCLC). Biomed Pharmacother. 95:331–338. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yang T, Li H, Chen T, Ren H, Shi P and
Chen M: LncRNA MALAT1 depressed chemo-sensitivity of NSCLC cells
through directly functioning on miR-197-3p/p120 catenin axis. Mol
Cells. 42:270–283. 2019.PubMed/NCBI
|
|
32
|
Liu W: LncRNA LINC-PINT inhibits cancer
cell proliferation, invasion, and migration in osteosarcoma by
downregulating miRNA-21. Cancer Biother Radiopharm. 34:258–263.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Chao Y and Zhou D: lncRNA-D16366 is a
potential biomarker for diagnosis and prognosis of hepatocellular
carcinoma. Med Sci Monit. 25:6581–6586. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Jiang N, Meng X, Mi H, Chi Y, Li S, Jin Z,
Tian H, He J, Shen W, Tian H, et al: Circulating lncRNA XLOC_009167
serves as a diagnostic biomarker to predict lung cancer. Clin Chim
Acta. 486:26–33. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Xie Y, Zhang Y, Du L, Jiang X, Yan S, Duan
W, Li J, Zhan Y, Wang L, Zhang S, et al: Circulating long noncoding
RNA act as potential novel biomarkers for diagnosis and prognosis
of non-small cell lung cancer. Mol Oncol. 12:648–658. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Huang SH and O'Sullivan B: Overview of the
8th edition TNM classification for head and neck cancer. Curr Treat
Options Oncol. 18:402017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Saffarzadeh AG, Blasberg JD and Beyond
TNM: Searching for new patient-centric prognostic indicators in
NSCLC. Ann Surg Oncol. 25:3425–3426. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zhou M, Zhang Z, Zhao H, Bao S, Cheng L
and Sun J: An immune-related six-lncRNA signature to improve
prognosis prediction of glioblastoma multiforme. Mol Neurobiol.
55:3684–3697. 2018.PubMed/NCBI
|