Introduction
Lung cancer remains one of the most fatal diseases
in the world, and small cell lung cancer (SCLC) accounts for
>15% of lung cancer cases (1).
The majority of SCLC cases that depend on chemotherapy treatment
for surgical resection are less responsive, but almost all cases
tend to exhibit subsequent chemoresistance, which results in
treatment failure. Therefore, investigating the mechanism of SCLC
pathogenesis is crucial.
Non-coding RNAs have no protein-coding potential,
and account for >90% of transcripts. Among them, microRNAs
(miRNAs or miRs) with a length of 19–25 nucleotides (nt) have been
extensively studied. Thousands of miRNAs regulate ~30% of
protein-coding genes (2). In
addition, long non-coding RNAs (lncRNAs), which are >200 nt in
length, play important regulatory roles in tumorigenesis and tumor
progression (3). Previous studies
have shown that lncRNAs can regulate the expression of certain
oncogenes or cancer suppressor genes by interacting with the
lncRNA-miRNA axis, thus affecting the genesis and development of
cancer. Furthermore, competitive endogenous RNA (ceRNA) has been
confirmed to form an important regulatory network of non-coding
RNAs (4,5). Although certain research has been
conducted on lncRNA-mediated sponge regulation in several cancer
types (6,7), including SCLC, additional studies on
this topic are required.
The lncRNA HOXA transcript at the distal tip
(HOTTIP), which lies at the 5′ end of the HOXA cluster, has been
confirmed to be a key regulator in several cancer types, such as
gastric cancer, pancreatic ductal adenocarcinoma, colorectal and
ovarian cancer (8). According to
previous studies on HOTTIP in tumors, miRNAs usually act as ceRNAs
of other genes by targeting HOTTIP. Due to the heterogeneity of
different tumors, different miRNAs are usually connected with
HOTTIP in different types of cancer (9,10). Our
previous studies suggested that HOTTIP participates in the
pathogenesis and chemotherapy resistance of SCLC by sponging
miR-574-5p and miR-216a-5p (11,12). In
the present study, the role of miR-574-5p in the EMT of SCLC and
its regulation via HOTTIP sponging was explored. In addition, the
proliferation- and migration-promoting functions of miR-574-5p in
SCLC, and its direct negative regulation of vimentin (VIM)
expression, were investigated. Furthermore, the role of the
HOTTIP/miR-574-5p/VIM axis in the EMT of SCLC was confirmed, and a
therapeutic strategy to regulate the function of oncogenes was
suggested.
Materials and methods
Cell culture
The SCLC cell lines H146, H446, H69 and H69AR cells
were obtained from the American Type Culture Collection (ATCC), and
were cultured in Gibco RPMI-1640 medium (Thermo Fisher Scientific,
Inc.) containing 10 or 20% fetal bovine serum (Gibco; Thermo Fisher
Scientific, Inc.) at 37°C and 5% CO2. Different from the
adherent H69AR cell line, the H146 cell line is a type of
semi-suspended and semi-adherent cell line, which makes it more
suitable for the observation of cell morphological changes during
the EMT process. Since H69AR and H69 cells grow vigorously and
exhibit fully adherent growth characteristics according to cell
growth characteristics described in ATCC, they are suitable for
cell collection, as well as for RNA and protein extraction.
According to the needs of the present study, H69, H69AR and H146
SCLC cells were selected for further experiments. Furthermore, the
293T cell line (ATCC), which was were cultured in RPMI-1640 medium
containing 10% fetal bovine serum at 37°C and 5% CO2,
was used for the dual luciferase reporter assay.
RNA interference (RNAi) and
transfection
HOTTIP small interfering (si)RNA and miR-574-5p
inhibitors or mimics (Tables I and
II) were obtained from Suzhou
GenePharma Co., Ltd. The synthesized HOTTIP and miR-574-5p
interference sequences or plasmids as well as their corresponding
NC sequences were transfected into SCLC cells, and the cells with
transient down-regulation of HOTTIP and miRNA were obtained.
Briefly, 8–10×105 cells were inoculated and cultured in
a 6-well plate at 37°C in a 5% CO2 incubator. The next
day, whe the confluence 70–80%, Lipofectamine® 2000
transfection reagent (Thermo Fisher Scientific, Inc.) was used to
transfect 10 nM plasmid or 40 nM siRNA into 1×106 cells
at 37°C for 4–6 h. Subsequently, the medium containing the
transfection reagent was discarded and the complete medium was
replaced. After 24–48 h, RNA or protein was extracted for
subsequent experiments. The lentiviral method was used for stable
transfection; the most effective interference sequence of HOTTIP
(si-h-HOTTIP-1, confirmed by RT-qPCR) was packaged in an LV3
lentiviral vector (Shanghai GenePharma Co.) for target cell
infection experiment, LV3-NC was used as a control, and SCLC cell
lines stably silencing HOTTIP were used for further
experiments.
 | Table I.Primers used for reverse
transcription-quantitative PCR analysis. |
Table I.
Primers used for reverse
transcription-quantitative PCR analysis.
| Gene | Primer sequence
(5′-3′) |
|---|
| HOTTIP | Forward:
CCTAAAGCCACGCTTCTTTG |
|
| Reverse:
TGCAGGCTGGAGATCCTACT |
| GAPDH | Forward:
GGGCTGCTTTTAACTCTG |
|
| Reverse:
TGGCAGGTTTTTCTAGACGG |
| Vimentin | Forward:
AGTCCACTGAGTACCGGAGAC |
|
| Reverse:
CATTTCACGCATCTGGCGTTC |
| E-cadherin | Forward:
ATTTTTCCCTCGACACCCGAT |
|
| Reverse:
TCCCAGGCGTAGACCAAGA |
 | Table II.miR-574-5p mimic/inhibitor/antagomir
and HOTTIP siRNA sequences. |
Table II.
miR-574-5p mimic/inhibitor/antagomir
and HOTTIP siRNA sequences.
| Molecule | Sequences
(5′-3′) |
|---|
| miR-574-5p
mimic |
UGAGUGUGUGUGUGUGAGUGUGUACACUCACACACACACACUCAUU |
| miR-574-5p
inhibitor |
ACACACUCACACACACACACUCA |
| Mimics NC |
UCUACUCUUUCUAGGAGGUUGUGA |
| Inhibitor NC |
UCUACUCUUUCUAGGAGGUUGUGA |
| HOTTIP siRNA |
GCUGCUUUAGAGCCACAUAdTdT |
| siRNA NC |
UUCUCCGAACGUGUCACGUUU |
Reverse transcription-quantitative PCR
(RT-qPCR)
RT-qPCR was performed according to the
manufacturer's instructions of the TB Green® Premix Ex
Taq™ (Tli RNaseH Plus) (Takara Bio, Inc.), and was used to detect
the mRNA expression levels of the related genes in SCLC cells. The
extraction of total RNA from cells was performed using
TRIzol® reagent (Thermo Fisher Scientific, Inc.). The
primers are listed in Table I.
SuperScript III Reverse Transcriptase kit (Thermo Fisher
Scientific, Inc.) was used to perform the reverse transcription to
synthesize cDNA through the following thermal conditions: 30°C for
10 min, 42°C for 30 min, 99°C for 5 min, 5°C for 5 min and −20°C
for storage. qPCR was performed under the following conditions:
95°C for 30 sec; 40 cycles of 95° for 5 sec and 60° for 30 sec; and
4°C for storage. The mRNA expression was normalized to the
expression of GAPDH and calculated using the 2−∆∆Cq
method. GAPDH was used as a control gene for normalization.
Western blotting and
immunofluorescence (IF) staining
Western blotting was performed as previously
described (8). Briefly, the samples
were collected from SCLC cells using RIPA buffer (Beyotime
Institute of Biotechnology). The concentration of proteins was
measured using the BCA method (Beyotime Institute of
Biotechnology). The proteins (40 µg/lane) were separated by 10%
SDS-PAGE and transferred to PVDF membranes (Thermo Fisher
Scientific, Inc.). The membranes were blocked with 5% non-fat milk
powder for 1.5 h at room temperature. Subsequently, the membranes
were incubated with anti-VIM (cat. no. 5741S; dilution, 1:1,000),
anti-E-cadherin (cat. no. 8834S; dilution, 1:1,000) and the loading
control anti-GAPDH (cat. no. 2118P; dilution, 1:1,000) (all Cell
Signaling Technology, Inc.) primary antibodies at 4°C overnight.
The next day, the membranes were washed with PBS with 0.2% Tween-20
(PBST) three times and probed with Goat Anti-Rabbit
peroxidase-conjugated secondary antibodies (cat. no. A21020;
dilution, 1:2,000; Abbkine, Inc.) for 1.5 h at room temperature.
The immunoreactive bands were washed with PBST. Finally, the signal
was monitored and visualized using an enhanced chemiluminescence
assay (EMD Millipore) and the Odyssey Infrared Imaging system
(LI-COR Biosciences). GAPDH was used as an internal control. The
intensity of the bands was analyzed by ImageJ software (version
1.8.0; National Institutes of Health).
For IF staining, H146 and H146/miR-574-5p and the
corresponding control cells were fixed with 4% paraformaldehyde at
room temperature for 20 min and blocked with PBS containing 5% goat
serum (cat. no. 16210-064; Gibco; Thermo Fisher Scientific, Inc.).
The cells were then washed three times for 5 min with PBS and
permeabilized with 0.3% Triton X-100 for 1 h at room temperature.
An additional washing step with PBS was performed three times for 5
min, and the cells were incubated with 5% BSA for 1 h at room
temperature. The samples were incubated overnight at 4°C with a
primary antibody against VIM (cat. no. 5741S; 1:100; Cell Signaling
Technology, Inc.). The samples were subsequently washed with PBS
three times for 5 min and incubated with the secondary antibody
(FITC-Labeled Anti-Rabbit IgG; cat. no. CST. 5151p; Cell Signaling
Technology, Inc.), and the nuclei were stained with DAPI (Thermo
Fisher Scientific, Inc.). An Olympus IX73 microscope was used to
capture the fluorescence images. For the quantitative analysis of
the intensity of fluorescence expression, the immunofluorescence
images were quantitatively analyzed using ImageJ software.
In vitro proliferation assay
According to a standard protocol, plate colony
formation experiments were conducted. Single-cell suspension was
prepared, and 500 cells were inoculated in a 3.5-cm dish with a
diameter of 3.5 cm. The drug concentration was determined according
to the IC50 in the treatment group on the next day.
Following culture at 5% CO2, 37°C and saturated humidity
for 14–21 days, when there were visible clones in the culture dish,
the culture was terminated, the supernatant was discarded, and the
cells were washed with PBS twice, fixed with pure methanol at room
temperature for 10 min and dyed with 0.4% crystal violet at room
temperature for 10 min. The number of colonies with >50 cells
was counted under an inverted phase-contrast microscope (CX41,
Olympus Corporation). The experiment was repeated three times to
obtain mean values (13).
In vitro migration assay
For the wound healing assay, H146 and H146/si-HOTTIP
as well as H146/si-HOTTIP + miR-574-5p cells (5×105 per
well) were incubated in 6-well plates for 24 h. When the cells were
~100% confluent, the monolayer was scratched with a 200-µl pipette
tip. The cells were washed with PBS, and fresh serum-free medium
was added. The cells were cultured at 37°C with 5% CO2
and the wound was allowed to heal. Images of the wound were
captured at 0 and 24 h. ImageJ software was used to evaluate the
wound area, and the wound closure rate was calculated using the
following formula: Closure rate=(original wound area/final wound
area)/original wound area ×20.
A Boyden chamber was used for an in vitro
migration assay according to the manufacturer's protocol (BD
Biosciences) and previously described techniques (14). Transwell chambers (8-µm pore size;
Costar, Inc.) were used to assess the migratory ability of H146 and
H146/si-HOTTIP as well as H146/si-HOTTIP + miR-574-5p cells. For
the migration assays, 5×104 cells were added into the
upper chamber. For the invasion assays, Matrigel was dissolved
overnight at 4°C, diluted with serum-free medium at a ratio of 1:3,
and added at 50 µl/well to the top chamber of a Transwell insert.
The plate was air-dried in an incubator for 4–5 h at 37°C in a cell
incubator. Subsequently, 1×105 cells were added into the
upper chamber precoated with Matrigel (BD Bioscience). A total of
500 µl medium with 15% FBS was placed into the lower chamber. The
plate was maintained at 37°C in a 5% CO2 incubator for
48 h. The cells on the lower surface of the membrane were then
fixed with 4% paraformaldehyde (25°C for 10 min) and stained with
0.5% crystal violet (25°C for 30 min). The images were captured in
three randomly selected fields under an inverted phase-contrast
microscope (Olympus, Japan) at ×20 magnification.
Dual luciferase reporter assay
The dual luciferase reporter assay was conducted by
Suzhou GenePharma Co., Ltd. Detailed information regarding the
methodology is provided on the supplier's website. The putative
binding site for miR-574-5p and HOTTIP was predicted using the
bioinformatics tool starBase (http://starbase.sysu.edu.cn/index.php). The wild-type
(WT) and mutated (MUT) HOTTIP 3′-untranslated region (UTR)
sequences were designed and constructed. The mature miR-574-5p and
its negative control (NC) were co-transfected with HOTTIP 3′-UTR-WT
and HOTTIP 3′-UTR-MUT into cells. The fluorescent enzyme activity
of the sample was detected by Dual-Luciferase® Reporter
Assay System (Promega Corporation). After 48 h, the cells were
harvested and detected for luciferase activity. Luciferase
co-transfection was used as a standard control.
Statistical analysis
All experiments were conducted in triplicate. The
data are expressed as the mean ± SD. All statistical analyses were
performed using GraphPad Prism 7 software (GraphPad Software,
Inc.). Statistical significance was analyzed by unpaired Student's
t-test, one-way ANOVA and two-way ANOVA. Bonferroni's correction
was applied as a post hoc test after ANOVA. P<0.05 was
considered to indicate a statistically significant difference.
Results
lncRNA HOTTIP may be involved in the
EMT of SCLC
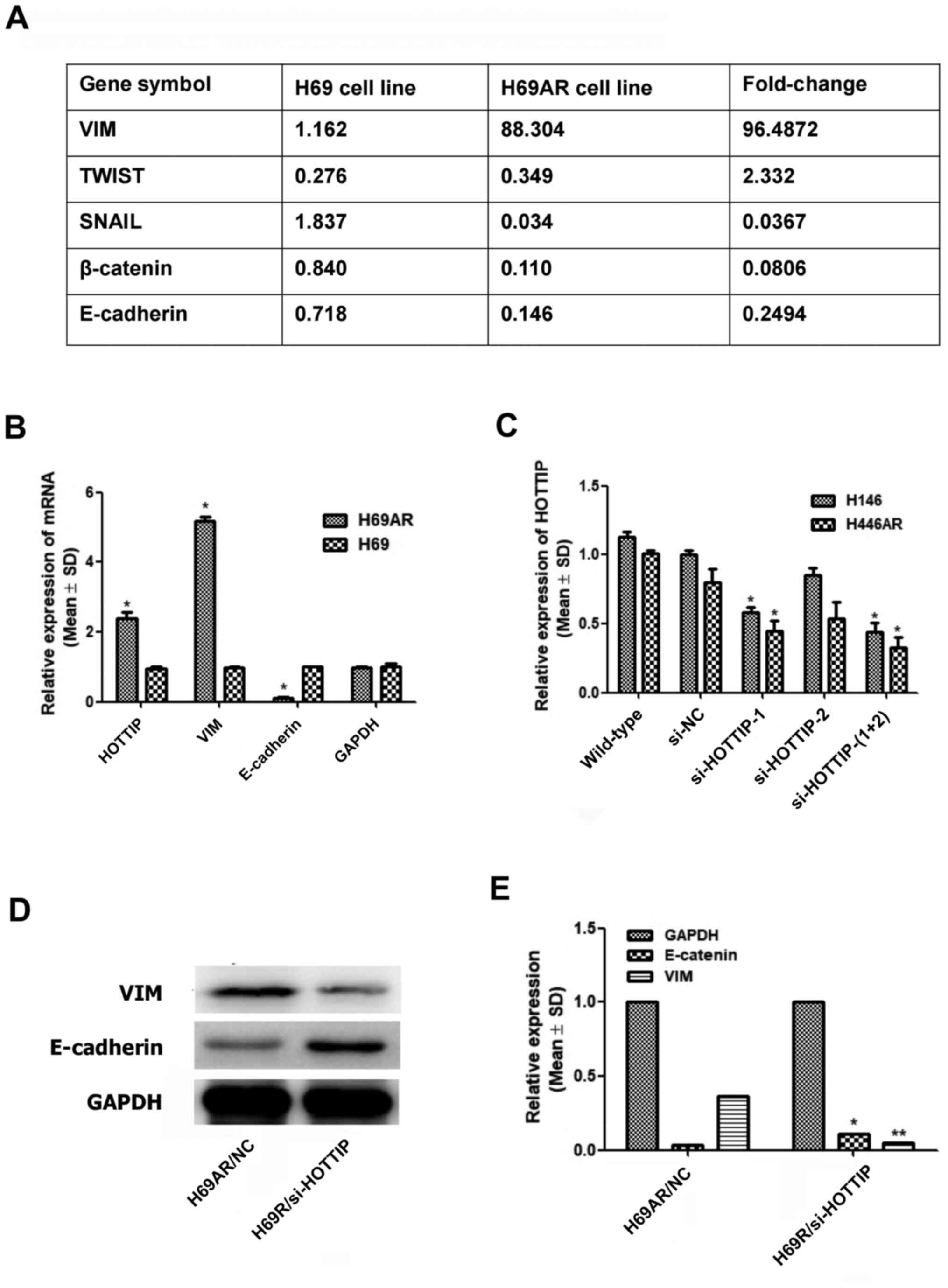
The present study suggested that several EMT markers
showed differential expression between the H69 and H69AR cell lines
(Fig. 1A). The present study
investigated the expression of two key genes in EMT, VIM and
E-cadherin, as well as that of HOTTIP, in the H69 and H69AR cell
lines. It was found that VIM was highly expressed in H69AR cells,
whereas E-cadherin was significantly decreased in H69AR cells
(Fig. 1B), and part of
HOTTIP-related data has been published (12). Notably, the transfection effect of
HOTTIP RNAi sequences as well as that of lentivirus-packaged HOTTIP
RNAi sequences were verified by immunofluorescence assay and
RT-qPCR; following the establishment of four cell lines with stable
and low expression of the HOTTIP gene, these cell lines were used
for subsequent in vitro and in vivo experiments
(Fig. 1C) (12). To analyze whether there was a
difference between the expression of HOTTIP and EMT markers, VIM
and E-cadherin, western blotting was used for protein detection
(Fig. 1D and E). The results
suggested that HOTTIP knockdown could decrease VIM expression but
increase E-cadherin expression; thus, HOTTIP may be involved in the
EMT of SCLC.
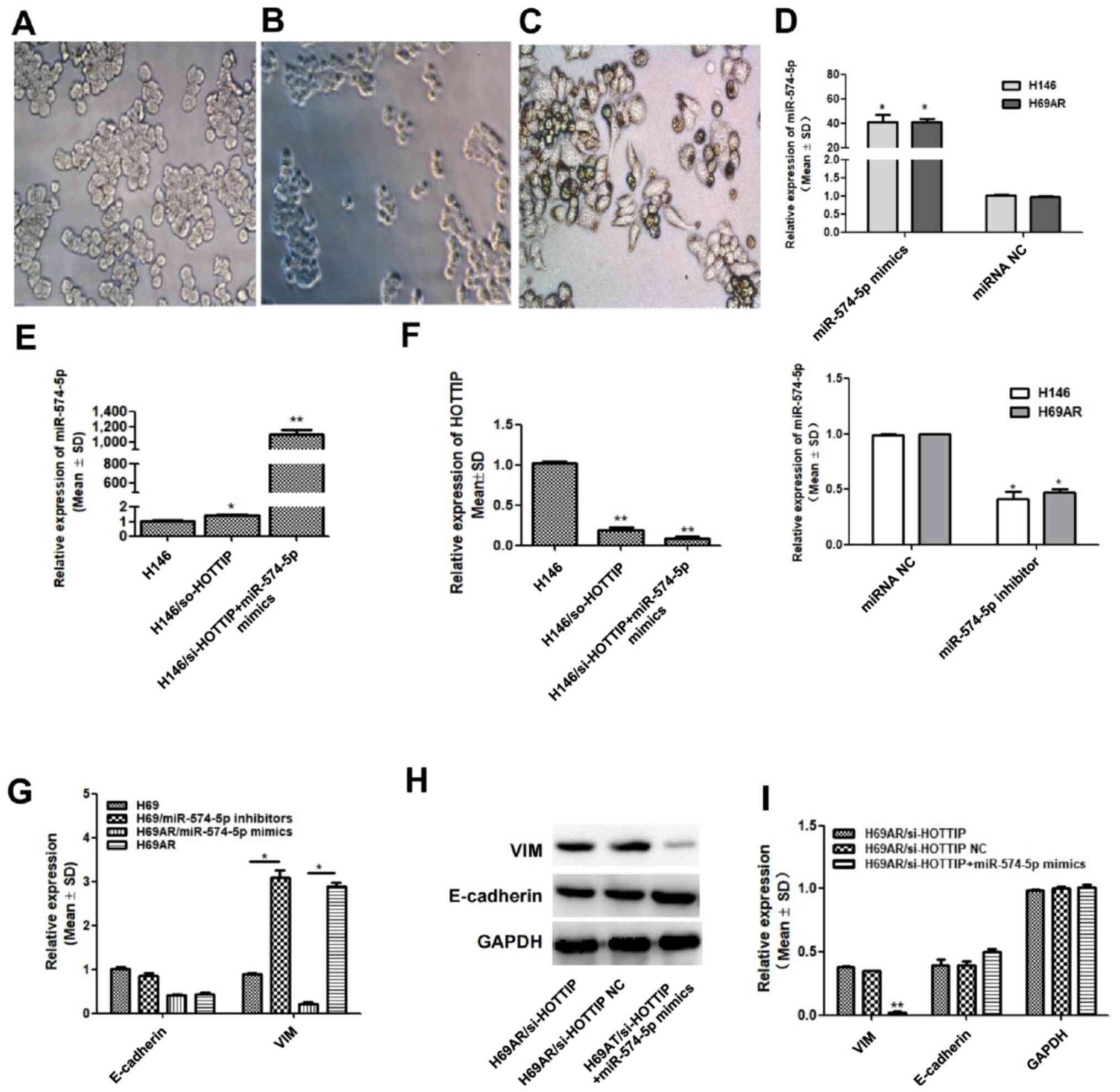
HOTTIP may participate in EMT by
binding to miR-574-5p
As described in a previous study (12), H146/si-HOTTIP cells were established
by lentiviral vector transfection (Fig.
2B). By subsequent transfection with miR-574-5p mimics,
H146/si-HOTTIP + miR-574-5p cells were established (Fig. 2C), and their cell morphology was
observed under a microscope. In contrast to H146 cells (Fig. 2A), which had an ovular and
semi-adherent morphology, the cells in the H146/si-HOTTIP group
were smaller and agglomerated into spheres (Fig. 2B), whereas the cells in the
H146/si-HOTTIP + miR-574-5p group exhibited a spindle, fusiform and
fibroblast-like morphology (Fig.
2C). The transfection efficiency of miR-574-5p mimics and
inhibitor in H146 and H69AR cells was verified (Fig. 2D). The present study next intended to
explore the mechanism of the aforementioned morphological changes
and to confirm successful transfection. RT-qPCR was used to verify
the effects of transfection, and it was found that HOTTIP-knockdown
did not affect miR-574-5p expression (Fig. 2E), whereas transfection with
miR-574-5p mimics further reduced HOTTIP expression in
H146/si-HOTTIP cells (Fig. 2F).
Based on the aforementioned results, in order to
determine whether HOTTIP induced EMT by binding to miR-574-5p, the
expression of VIM and E-cadherin in H69AR and H69 cells was
determined following transfection with miR-574-5p mimics. It was
found that, compared with that of the corresponding H69AR group,
VIM expression in the H69AR/miR-574-5p mimics group was
significantly decreased, whereas it was significantly increased in
the H69AR/miR-574-5p inhibitor group (Fig. 2G). As the H69AR/miRNA-NC cells were
contaminated and RNA could not be extracted, the wild-type group
were used as negative control for this experiment. However, changes
in E-cadherin expression following transfection were not obvious
(Fig. 2G). Therefore, the induction
of EMT by HOTTIP sponging of miR-574-5p may be achieved mainly by
the regulation of VIM at the mRNA level. Correspondingly, analysis
at the protein level by western blotting revealed that the
expression of E-cadherin changed only slightly, while VIM
expression was significantly decreased in the H69AR/si-HOTTIP +
miR-574-5p mimic group compared with that of the H69AR/si-HOTTIP
group (Fig. 2H and I). Thus,
miR-574-5p regulated the expression of HOTTIP, which has been
confirmed in a previous study (12).
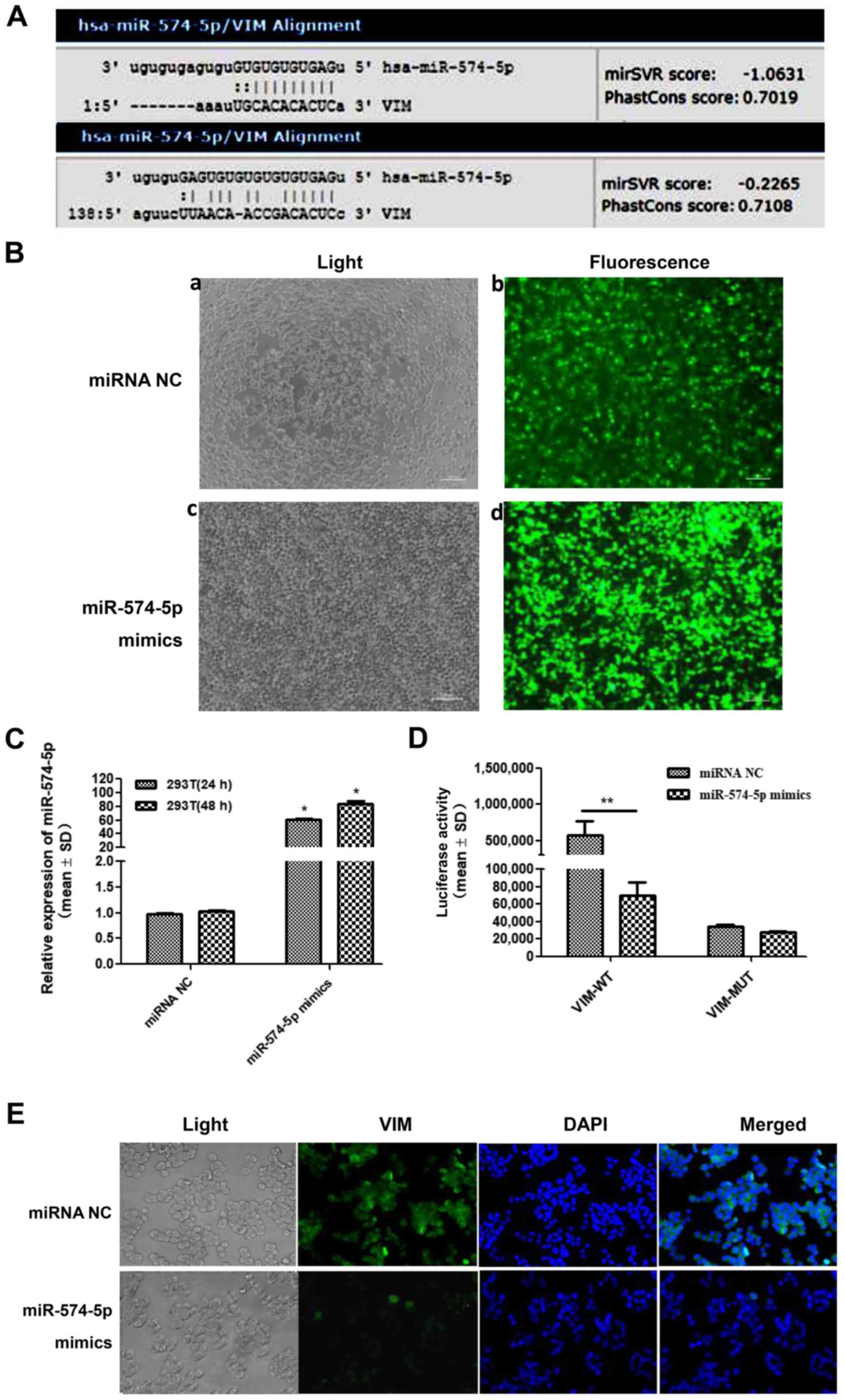
VIM is predicted and confirmed to be a
target gene of miR-574-5p
By bioinformatics prediction, two pairable base
regions were found in the sequences of miR-574-5p and VIM (Fig. 3A). To evaluate whether their binding
was effective, a dual luciferase reporter assay was performed.
Fig. 3B shows fluorescence images of
plasmid and oligo transfections, which indicated that miR-574-5p
mimics and NC were successfully transfected into 293T cells
(Fig. 3B), and RT-qPCR demonstrated
that the miR-574-5p level was increased in 293T cells following
transfection with miR-574-5p mimics (Fig. 3C). It was found that, in 293T cells,
miR-574-5p could target and regulate the expression of VIM
(Fig. 3D). Subsequent IF staining
confirmed the regulation of VIM protein by miR-574-5p. VIM
expression (green fluorescence) in the cytoplasm and nucleus of
H146/NC cells differed after transfection with miR-574-5p mimics
(Fig. 3E).
HOTTIP may affect colony formation and
cell migration by binding to miR-574-5p
To further investigate the effects of HOTTIP binding
to miR-574-5p on cell proliferation and invasion, plate
colony-forming experiments were carried out. It was found that the
colony formation ability of cells was significantly reduced in the
H146/si-HOTTIP group compared with that of the H146 group. However,
after transfection of H146/si-HOTTIP cells with miR-574-5p mimics,
the colony formation ability was restored (Fig. 4A). Fig.
4B shows the quantitative results of the cell colony formation
assay.
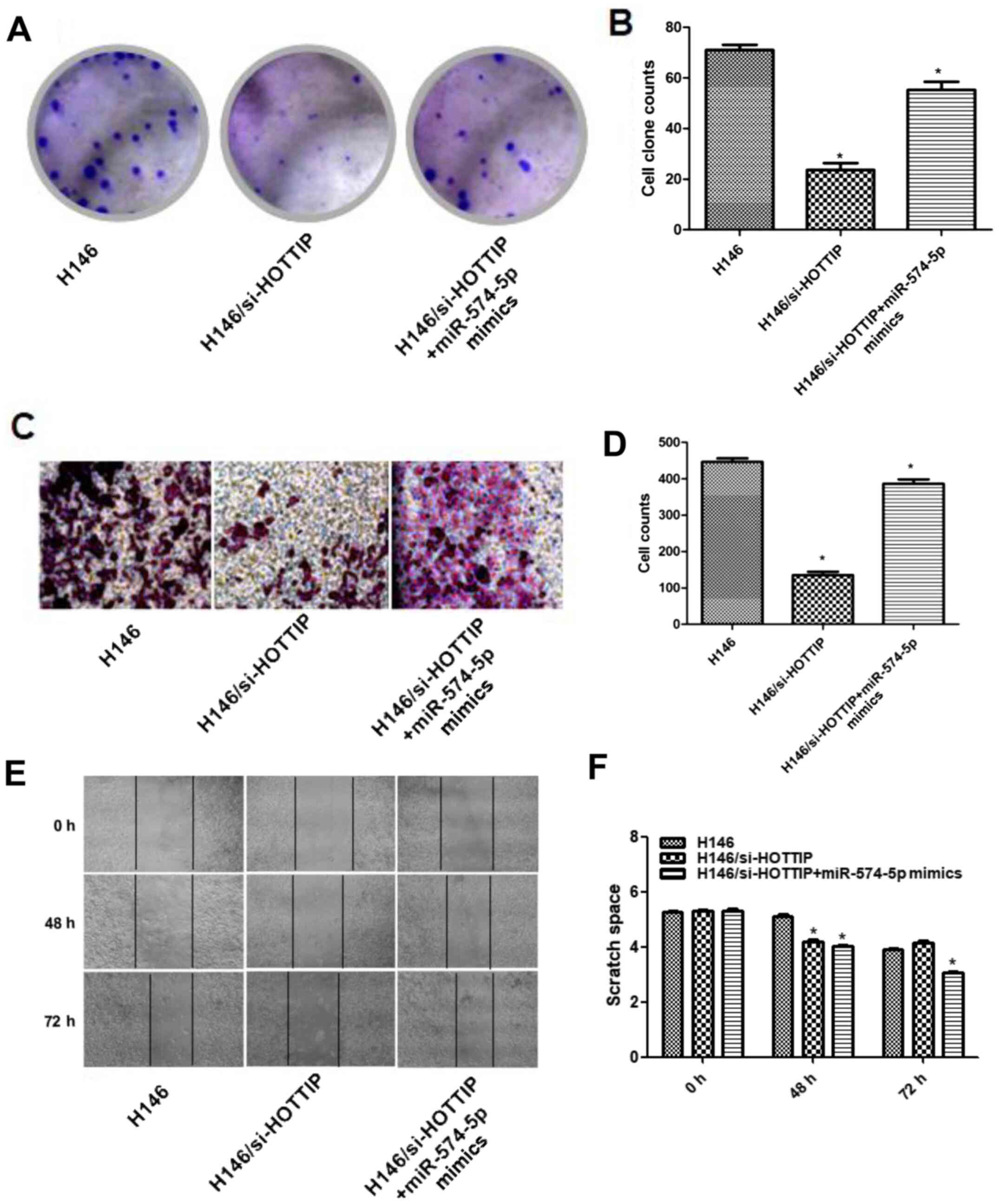 | Figure 4.HOTTIP affects colony formation and
cell migration abilities by regulating miR-574-5p. (A)
Representative images of the colony formation assay (magnification,
×20). (B) Differences in colony formation ability between H146,
H146/si-HOTTIP and H146/si-HOTTIP + miR-574-5p mimics cells after
transfection with miR-574-5p mimics (*P<0.05 vs. H146). (C)
Representative Transwell assay images (magnification, ×20). (D)
Differences of cell migration ability between H146, H146/si-HOTTIP
and H146/si-HOTTIP+miR-574-5p mimics cells, as determined by
transwell assay (*P<0.05 vs. H146). (E) Representative wound
healing assay images (magnification, ×20). (F) Differences in cell
migration ability between H146, H146/si-HOTTIP and
H146/si-HOTTIP+miR-574-5p mimics cells, as determined by wound
healing assay (*P<0.05 vs. H146 at 48 or 72 h). miR, microRNA;
HOTTIP, HOXA transcript at the distal tip; si, small
interfering. |
In a Transwell migration assay, the migration
ability of cells in the H146/si-HOTTIP group was significantly
reduced compared with that of cells in the non-transfected group,
but it was restored by subsequent transfection with miR-574-5p
mimics in the H146/si-HOTTIP + miR-574-5p group (Fig. 4C). Fig.
4D shows the quantitative results of cell migration.
Next, a wound healing experiment was conducted to
verify the aforementioned results. At 48 h, the wound healing
ability of the two treatment groups was poorer compared with that
of the control group. However, at 72 h, the H146/si-HOTTIP +
miR-574-5p group exhibited the highest wound closure rate (Fig. 4E). Fig.
4F demonstrates the quantitative results of the wound healing
assays.
Discssion
Since the prognosis of SCLC is markedly poor, the
study of the pathogenesis and development of SCLC may lead to the
identification of new biomarkers and thus aid early detection and
treatment. The ceRNA network is a key regulatory mechanism in the
pathogenesis and development of numerous tumors, and is a
regulatory network involving lncRNAs, miRNAs and their target
genes. Our previous study indicated that HOTTIP participates in the
pathogenesis, development and chemoresistance of SCLC by sponging
miR-574-5p (12). The current study
explored whether HOTTIP may play a key role in the EMT of SCLC by
sponging miR-574-5p and enhancing VIM expression.
Multiple studies have suggested that HOTTIP acts as
an oncogene in esophageal squamous cell carcinoma by inducing EMT
via the HOTTIP-miR-30b-HOXA13 axis, in which HOXA13 is a downstream
factor of HOTTIP that has been demonstrated to be directly
regulated by HOTTIP (15,16). HOTTIP has also been reported to
promote EMT in breast cancer and tongue squamous carcinoma through
the ceRNA network or other mechanisms (17,18).
Previous studies have suggested that HOTTIP and c-Myc may form a
positive feedback regulatory loop in osteosarcoma, whereas
salinomycin may reduce the development of EMT-mediated multidrug
resistance by changing the expression of HOTTIP in gastric cancer
cells (19,20). In addition, HOTTIP has been reported
to modulate the properties of cancer stem cells via EMT in
pancreatic cancer and hepatocellular carcinoma (21,22).
Previous studies on human gastric carcinoma cells
have shown that miR-574-3p regulates EMT, as well as cisplatin
resistance, by targeting zinc finger E-box-binding homeobox 1
(23); however, the association
between miR-574-5p and EMT in cancer remains unclear. The present
findings enrich the knowledge on the induction of EMT mediated by
non-coding RNA to facilitate tumor metastasis.
Our previous study evaluated HOTTIP, miR-574-5p and
the prognosis of patients with SCLC (12). The present study further revealed
that HOTTIP may induce the EMT of SCLC by sponging miR-574-5p, and
demonstrated that miR-574-5p regulates VIM directly. However,
HOTTIP-knockdown also reduced the expression of E-cadherin, another
key marker of EMT, although there may not be an obvious regulatory
association between miR-574-5p and E-cadherin. HOTTIP may affects
the expression of E-cadherin through other mechanisms, such as the
Wnt-β-catenin signaling pathway (17,24) or
modifications by salinomycin (19)
as reported in other types of cancer.
There are certain limitations in the present study,
including the absence of in vivo experiments and the use of
only one cell line for certain experiments, and no non-cancerous
cell lines used as a negative control. In-depth studies at the
tissue level were not conducted, since a suitable model has not
been established to analyze the EMT phenomenon in tumor tissues.
Future studies should be conducted to confirm the results of the
present study in additional cell lines, to verify the association
between key molecules and tumor metastasis in vivo, and to
further explore the signaling pathways involved in HOTTIP and
miR-574-5p.
In conclusion, the present study showed that HOTTIP
may participate in EMT by sponging miR-574-5p. In addition, by
using bioinformatics technology and a dual luciferase reporter
assay, it was confirmed that miR-574-5p probably inhibits the
expression of VIM, a key molecule of EMT, through direct target
binding. Future studies may focus on the regulation of SCLC
metastasis by HOTTIP.
Acknowledgements
The authors would like to thank Dr Linlang Guo
(Department of Pathology, Zhujiang Hospital of Southern Medical
University) for the designing guidance in the present study. The
dual luciferase reporter assay was performed by Suzhou GenePharma
Co., Ltd.
Funding
The present study was partly supported by the
National Natural Science Foundation of China (grant no. 81702285),
Funds for the Construction of Basic Medical Disciplines in
Guangdong Medical University (grant no. 4SG19047G), Innovation
experiment project of Guangdong Medical University in 2020 (grant
no. ZZDS007) and Key projects of Guangdong Medical University
(grant no. GDMUZ201808).
Availability of data and materials
The datasets used and/or analyzed in the current
study are available from the corresponding author upon reasonable
request.
Authors' contributions
YS, YY and SG conceived and designed the
experiments. JH and YG conceived the study. RY and CH performed the
in vitro assays. ML performed the immunofluorescence
staining. JH and YG confirm the authenticity of all the raw data.
All authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Rudin CM, Brambilla E, Faivre-Finn C and
Sage J: Small-cell lung cancer. Nat Rev Dis Primers. 7:32021.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Huarte M: The emerging role of lncRNAs in
cancer. Nat Med. 21:1253–1261. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Gomes AQ, Nolasco S and Soares H:
Non-coding RNAs: Multi-tasking molecules in the cell. Int J Mol
Sci. 14:16010–16039. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhang J, Zhang P, Wang L, Piao PL and Ma
L: Long non-coding RNA HOTAIR in carcinogenesis and metastasis.
Acta Biochim Biophys Sin (Shanghai). 46:1–5. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Cai B, Song XQ, Cai JP and Zhang S:
HOTAIR: A cancer-related long non-coding RNA. Neoplasma.
61:379–391. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Qi X, Zhang DH, Wu N, Xiao JH, Wang X and
Ma W: ceRNA in cancer: Possible functions and clinical
implications. J Med Genet. 52:710–718. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Tam C, Wong JH, Tsui SKW, Zuo T, Chan TF
and Ng TB: LncRNAs with miRNAs in regulation of gastric, liver, and
colorectal cancers: Updates in recent years. Appl Microbiol
Biotechnol. 103:4649–4677. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Liu XH, Sun M, Nie FQ, Ge YB, Zhang EB,
Yin DD, Kong R, Xia R, Lu KH, Li JH, et al: Lnc RNA HOTAIR
functions as a competing endogenous RNA to regulate HER2 expression
by sponging miR-331-3p in gastric cancer. Mol Cancer. 13:922014.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhang S, Wang W, Liu G, Xie S, Li Q, Li Y
and Lin Z: Long non-coding RNA HOTTIP promotes hypoxia-induced
epithelial-mesenchymal transition of malignant glioma by regulating
the miR-101/ZEB1 axis. Biomed Pharmacother. 95:711–720. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yang B, Gao G, Wang Z, Sun D, Wei X, Ma Y
and Ding Y: Long non-coding RNA HOTTIP promotes prostate cancer
cells proliferation and migration by sponging miR-216a-5p. Biosci
Rep. 38:BSR201805662018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sun Y, Hu B, Wang Q, Ye M, Qiu Q, Zhou Y,
Zeng F, Zhang X, Guo Y and Guo L: Long non-coding RNA HOTTIP
promotes BCL-2 expression and induces chemoresistance in small cell
lung cancer by sponging miR-216a. Cell Death Dis. 9:852018.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Sun Y, Zhou Y, Bai Y, Wang Q, Bao J, Luo
Y, Guo Y and Guo L: A long non-coding RNA HOTTIP expression is
associated with disease progression and predicts outcome in small
cell lung cancer patients. Mol Cancer. 16:1622017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Niu Y, Ma F, Huang W, Fang S, Li M, Wei T
and Guo L: Long non-coding RNA TUG1 is involved in cell growth and
chemoresistance of small cell lung cancer by regulating LIMK2b via
EZH2. Mol Cancer. 16:52017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Su F, Li H, Yan C, Jia B, Zhang Y and Chen
X: Depleting MEKK1 expression inhibits the ability of invasion and
migration of human pancreatic cancer cells. J Cancer Res Clin
Oncol. 135:1655–1663. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chen X, Han H, Li Y, Zhang Q, Mo K and
Chen S: Upregulation of long noncoding RNA HOTTIP promotes
metastasis of esophageal squamous cell carcinoma via induction of
EMT. Oncotarget. 7:84480–84485. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lin C, Wang Y, Wang Y, Zhang S, Yu L, Guo
C and Xu H: Transcriptional and posttranscriptional regulation of
HOXA13 by lncRNA HOTTIP facilitates tumorigenesis and metastasis in
esophageal squamous carcinoma cells. Oncogene. 36:5392–5406. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Han S, Jin X, Liu Z, Xing F, Han Y, Yu X,
He G and Qiu F: The long noncoding RNA HOTTIP promotes breast
cancer cell migration, invasiveness, and epithelial-mesenchymal
transition via the Wnt-β-catenin signaling pathway. Biochem Cell
Biol. 97:655–664. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Mu M, Li Y, Zhan Y, Li X and Zhang B:
Knockdown of HOXA transcript at the distal tip suppresses the
growth and invasion and induces apoptosis of oral tongue squamous
carcinoma cells. Onco Targets Ther. 11:8033–8044. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Mao Z, Wu Y, Zhou J and Xing C:
Salinomycin reduces epithelial-mesenchymal transition-mediated
multidrug resistance by modifying long noncoding RNA HOTTIP
expression in gastric cancer cells. Anticancer Drugs. 30:892–899.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Tang Y and Ji F: lncRNA HOTTIP facilitates
osteosarcoma cell migration, invasion and epithelial-mesenchymal
transition by forming a positive feedback loop with c-Myc. Oncol
Lett. 18:1649–1659. 2019.PubMed/NCBI
|
|
21
|
Fu Z, Chen C, Zhou Q, Wang Y, Zhao Y, Zhao
X, Li W, Zheng S, Ye H, Wang L, et al: LncRNA HOTTIP modulates
cancer stem cell properties in human pancreatic cancer by
regulating HOXA9. Cancer Lett. 410:68–81. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Castro-Oropeza R, Melendez-Zajgla J,
Maldonado V and Vazquez-Santillan K: The emerging role of lncRNAs
in the regulation of cancer stem cells. Cell Oncol (Dordr).
41:585–603. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wang M, Zhang R, Zhang S, Xu R and Yang Q:
MicroRNA-574-3p regulates epithelial mesenchymal transition and
cisplatin resistance via targeting ZEB1 in human gastric carcinoma
cells. Gene. 700:110–119. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Xiong L, Tang Y, Tang J, Liu Z and Wang X:
Downregulation of lncRNA HOTTIP suppresses the proliferation,
migration, and invasion of oral tongue squamous cell carcinoma by
regulation of HMGA2-mediated Wnt/β-catenin pathway. Cancer Biother
Radiopharm. 35:720–730. 2020. View Article : Google Scholar : PubMed/NCBI
|