Introduction
Breast cancer is the second most common type of
cancer worldwide (1), and the most
common type of cancer in women, with an estimated 2.09 million new
cases (24.2% of all cases of cancer in women), and 0.6 million
cancer-related deaths annually (2).
The prognosis for breast cancer patients is generally good;
however, patients with late-stage tumors (stages III and IV) have
significantly shorter overall survival (OS) (3). This is due to the fact that late-stage
breast cancers are often resistant or less responsive to
conventional medical approaches, such as conventional surgery,
chemotherapy and radiotherapy, and exhibit a high rate of both
recurrence and metastasis (3). Thus,
novel pharmacological approaches are required to manage late-stage
cancer.
Breast cancer can be classified, based on gene
expression patterns (PAM 50), into at least six subtypes:
Normal-like, luminal A, luminal B, HER2-enriched, claudin-low and
basal-like (4–7). Based on receptor expression status,
breast cancer can also be classified immunohistochemically as
estrogen receptor (ER)-positive and/or progesterone receptor
(PgR)-positive type, HER2-positive type, and triple-negative type
(ER-negative, PgR-negative, HER2-negative; TNBC). TNBC has the
poorest prognosis amongst the different breast cancer subtypes, and
70–80% of TNBCs are basal-like breast cancer (8).
Metabolic reprogramming leading to increased
glycolysis, termed the Warburg effect, is a characteristic feature
of cancer cells (9). This
enhancement of glycolysis in cancer cells contributes to their
proliferation, migration, survival and drug resistance (10). In addition, glyoxalase 1 (GLO 1),
which catalyzes the conversion of methylglyoxal (MG), a toxic
byproduct of glycolysis, to non-toxic S-D-lactoylglutathione, is
upregulated in several types of malignancy, including lung,
stomach, colon, liver, prostate, oropharyngeal, skin and breast
cancer (11–21). GLO 1 is essential for the survival of
aldehyde dehydrogenase 1 (ALDH1)-positive breast cancer stem cells,
and operates in a caspase-3-dependent manner (21). In addition, HER2/neu signaling
regulates GLO 1 expression in HER2-positive tissues and cell lines
(22). However, the signal
transduction mechanisms of GLO 1 in breast cancer remain
unclear.
It is well established that the majority of cancer
cells are derived from epithelial cells, and defects in cell
polarity are a characteristic feature of cancer cells (23). One of the atypical protein kinase C
(aPKC) isotypes, PKCλ/ι, is known to be involved in cellular
responses that include determination of cell polarity, as well as
cell proliferation, survival, chemotaxis and migration (24–26).
PKCλ is overexpressed in several types of cancer, including breast
cancer (27–42), and is known to be involved in cancer
progression, contributing to poor clinical outcomes (32–42). In
TNBC cells, TGFβ and IL1β induce PKCλ phosphorylation and promote
PKCλ-dependent proliferation, invasiveness and metastasis by
inducing NF-κB p65 nuclear translocation (30). c-Met and PKCλ are cooperatively
involved in cellular viability and tumor formation in basal-like
breast cancer cells (42). PKCλ is
also essential for the survival of ALDH1-positive breast cancer
stem cells in a caspase-3-dependent manner (41,42).
EGF, platelet-derived growth factor and insulin collectively
activate PKCλ via PI3-kinase (43–45), and
PKCλ subsequently binds to and regulates p70 S6 kinase (46). PKCλ also activates the
Rac1-Pak-Mek1/2-Erk1/2 signaling pathway, which is associated with
lung cancer cell proliferation and tumorigenicity (47). PKCλ phosphorylates FoxO1 and
modulates the DNA-binding ability of c-Myc, promoting cellular
proliferation in angiosarcoma (48).
Glucose transporter 1 (GLUT1) facilitates glucose transport and its
expression is increased in several types of cancer (including
breast cancer), where it is involved in enhanced glycolysis and
cancer progression (49,50). PKCλ regulates the translocation of
GLUT1 from intracellular vesicles to the plasma membrane in 3T3-L1
adipocytes (51). However, the role
of PKCλ in the enhanced glycolysis seen in cancer cells remains
unclear.
In the present study, the association between the
levels of GLO 1 and PKCλ expression in human breast cancer was
investigated, and their impact on the prognoses of patients with
late-stage breast cancer was assessed.
Materials and methods
Immunohistochemistry (IHC)
Specimens used for IHC were prepared at the Kanagawa
Cancer Center Research Institute from archives of surgically
removed and formalin-fixed, paraffin-embedded breast cancer tissues
in the Pathology Department. With the approval of the Research
Ethics Committee, these prepared specimens were used in the present
study through the Kanagawa Cancer Research and Information
Association, which has since been dissolved and its duties
transferred to the Kanagawa Cancer Center Research Institute
Biospecimen Center (approval no. 3-2009). The clinicopathological
data of the patients from whom the samples were obtained are
summarized in Table SI. TNM stage
data is lacking 27% because the data is already anonymized. The
research protocol used was also approved by the Institutional
Ethics Committees of Tokyo University of Science (approval nos.
13003, 15006 and 16038), and all patients provided consent for the
use of their tissue samples for research purposes.
IHC was performed as previously described (29,31–35,39,40).
Briefly, 4-µm thick paraffin embedded sections were deparaffinized,
rehydrated in a descending series of ethanol solutions and
autoclaved (120°C for 20 min) in 10 mmol/l citrate buffer (pH 6.0)
for antigen retrieval. The semi-serially prepared sections
(adjacent sections) were then immersed in 0.3% hydrogen peroxide at
room temperature for 30 min to quench the intrinsic peroxidase
activity before incubation with a primary antibody at 4°C
overnight. The antibodies used in the present study were: Mouse
anti-PKCι mAb (1:250; cat. no. 610176; BD Biosciences), mouse
anti-GLO 1 mAb (1:2,000; cat. no. NBP1-19015; Novus Biologicals,
Inc.), mouse IgG2b κ Isotype Control (eBMG2b; 1:500; cat. no.
14-4732-82; eBioscience; Thermo Fisher Scientific, Inc.) and mouse
IgG1 κ Isotype Control (P3.6.2.8.1) (1:1,000; cat. no. 14-4714-82;
eBioscience; Thermo Fisher Scientific, Inc.). The labeled antigens
were visualized using a Histo Fine kit (Nichirei) and DAB plus
(Dako; Agilent Technologies, Inc.). The sections were
counterstained with hematoxylin. The antibodies used for double
staining were: Mouse anti-PKCι mAb (1:50), rabbit anti-GLO 1 pAb
(1:200; cat. no. A1932; ABclonal, Inc.), mouse IgG2b κ Isotype
Control and normal rabbit IgG (1:952; cat. no. PM035; MBL). The
labeled antigens were visualized using a Histo Fine alkaline
phosphatase kit and DAB plus. The sections were counterstained with
hematoxylin.
IHC scoring
To evaluate the expression of GLO 1 and PKCλ
proteins using IHC, ImageJ version 1.51u was used (National
Institutes of Health) with the IHC Profiler plugin (52). The scoring system was based on the
classification calculated from the IHC Profiler (+3, high-positive;
+2, positive; +1, low-positive; and 0, negative). Signal intensity
of GLO 1 was classified into color density as follows; +3, High
positive; +2, Positive; and +1, Low positive. Signal intensity of
PKCλ was classified into color density as follows: +3, High
positive; +2, Positive; +1, Low positive; and 0, Negative. Signal
intensities were categorized as high (+3 or +2) or low (+1 or 0).
H-scores of the scatter plot data were based on calculated values
from the IHC Profiler.
Analysis of gene expression in the
breast cancer dataset from the molecular taxonomy of breast cancer
international consortium (METABRIC)
Gene expression data was downloaded from cBioportal
and analyzed as previously described (21,41,42,53,54).
Briefly, the Molecular Taxonomy of Breast Cancer International
Consortium (METABRIC) dataset (55,56) was
downloaded from cBioPortal (cbioportal.org/; last entry, 25th November 2019)
(57,58). The clinicopathological data of the
patients are summarized in Table
SII. The median age at the time of diagnosis was 61.8 years
(age range, 21.9–96.3 years). Gene expression levels were
classified as high if they were in the top 25% of Z-scores; or
otherwise, they were classed as low.
Analysis of gene expression in the
breast cancer dataset from The Cancer Genome Atlas (TCGA)
Briefly, gene expression microarray datasets from
TCGA were downloaded from Oncomine (oncomine.org; Compendia
Bioscience, 28th January 2021) (59,60), and
the breast cancer dataset (n=459) was obtained. Levels of GLO 1
(reporter, A_32_P53822) and PKCλ (reporter, A_23_P18392) mRNA
expression are presented using log2 median-centered
ratio boxplots for normal vs. cancerous tissues.
Cell culture
The MCF-10A human normal-like (non-transformed)
mammary epithelial cell line and the MDA-MB-157 and MDA-MB-468
human basal-like breast cancer cell lines were obtained from the
American Type Culture Collection. MCF-10A cells were grown in
mammary epithelial cell growth medium (MEGM; Lonza Group, Ltd.)
according to instructions from ATCC. The cancer cell lines were
cultured as previously described (21,41,42,53).
Mycoplasma testing was performed on all the cell lines used.
Inhibitory compounds
3-(1,3-Benzothiazol-2-yl)-4- (4-methoxyphenyl)
but-3-enoic acid (TLSC702) was purchased from Namiki Shoji Co.,
Ltd. and dissolved in DMSO. Aurothiomalate (ATM) was purchased from
Calbiochem (Merck KGaA) and dissolved in water.
Immunoblot analysis
Immunoblotting was performed as previously described
(21,41,42,53). The
primary antibodies used were: Mouse anti-PKCι mAb (1:5,000; cat.
no. 610176; BD Biosciences), mouse anti-GLO 1 mAb (1:2,000; cat.
no. sc-133144, Santa Cruz Biotechnology, Inc.) and mouse
anti-β-actin mAb (1:20,000; cat. no. 60008-1-Ig, ProteinTech Group
Inc.). The secondary antibody used was a goat anti-mouse IgG
horseradish peroxidase-conjugate (1:5,000; cat. no. 7076S; Cell
Signaling Technology, Inc.).
WST-8 assay
WST-8 assays were performed according to the
manufacturer's protocol, and as previously described (21,42,53).
Briefly, cells (5×103/well) were seeded into 96-well
plates (Sigma-Aldrich; Merck KGaA) and incubated for 24 h.
Inhibitors were then added to the culture medium, and the cells
were incubated for an additional 3 days, after which cell viability
was assessed using a Cell Counting Kit-8 assay (Dojindo Molecular
Technologies, Inc.). The formazan dye formed was measured using
Sunrise Remote (Tecan Group, Ltd.) at 450 nm. Assays with MCF-10A
cells were performed in MEGM supplemented with 10% FBS. Assays
using the cancer cell lines were performed in DMEM supplemented
with 10% FBS. Numerical values of the test groups were expressed
relative to the control cell (no drug).
Tumor-sphere culture
Tumor-spheres were grown as previously described
(21,41,42,53).
Briefly, cells (1×103/well) were cultured in 96-well
ultralow attachment plates (Greiner Bio-One) and treated with
inhibitors for 6 days. Images were taken through an inverted
microscope (Leica Microsystems, Inc.), and the numbers of
tumor-spheres ≥50 µm in diameter were counted. Numerical values of
the test groups are shown relative to the untreated cells. Cell
Titer-Glo® luminescence assays (Promega Corporation)
were performed according to the manufacturer's protocol, and as
previously described (21,53). Values for test groups are shown
relative to cells in the absence of the drug.
Statistical analysis
For correlation between protein GLO 1 and PKCλ
expression, statistical significance was calculated using the
χ2-test with Yates' correction. H-scores of the scatter
plot data were based on calculated values from the IHC Profiler.
Spearman's rank correlation coefficients (r) and P-value are
indicated. In the analysis of gene expression, Pearson's
correlation coefficients (r) and P-value are indicated. P-values
were calculated using a test for non-correlation. In the analysis
of gene expression, survival curves were plotted using the
Kaplan-Meier method, and P-values were calculated using the
Gehan-Breslow generalized Wilcoxon test to weight early death
points. Multiplicity was adjusted using the Holm's method for
post-hoc analysis. A multivariable Cox regression model was used to
evaluate the effect of gene expression and to estimate the adjusted
hazard ratios (HRs) with age as a confounding factor. P-values for
comparison of gene expression are presented using the
Kruskal-Wallis test with the Steel-Dwass test. Statistical analysis
was performed using BellCurve for Excel version 2.11 (SSRI). Data
for the WST-8 assay is presented as the mean ± standard deviation
of three independent experiments. Differences between groups were
compared using Tukey's test. Data for the tumor-sphere assay is
presented as the mean ± standard error of the mean of three
independent experiments. Data for the Cell Titer-Glo®
luminescence assay is presented as the mean ± standard deviation of
three independent experiments. Statistical significance was
calculated using one-way ANOVA followed by Dunnett's test. For any
of the analyses above, α-level was fixed at 0.05, and P<0.05 was
considered to indicate a statistically significant difference.
Results
GLO 1 expression is positively
correlated with PKCλ expression in breast cancer
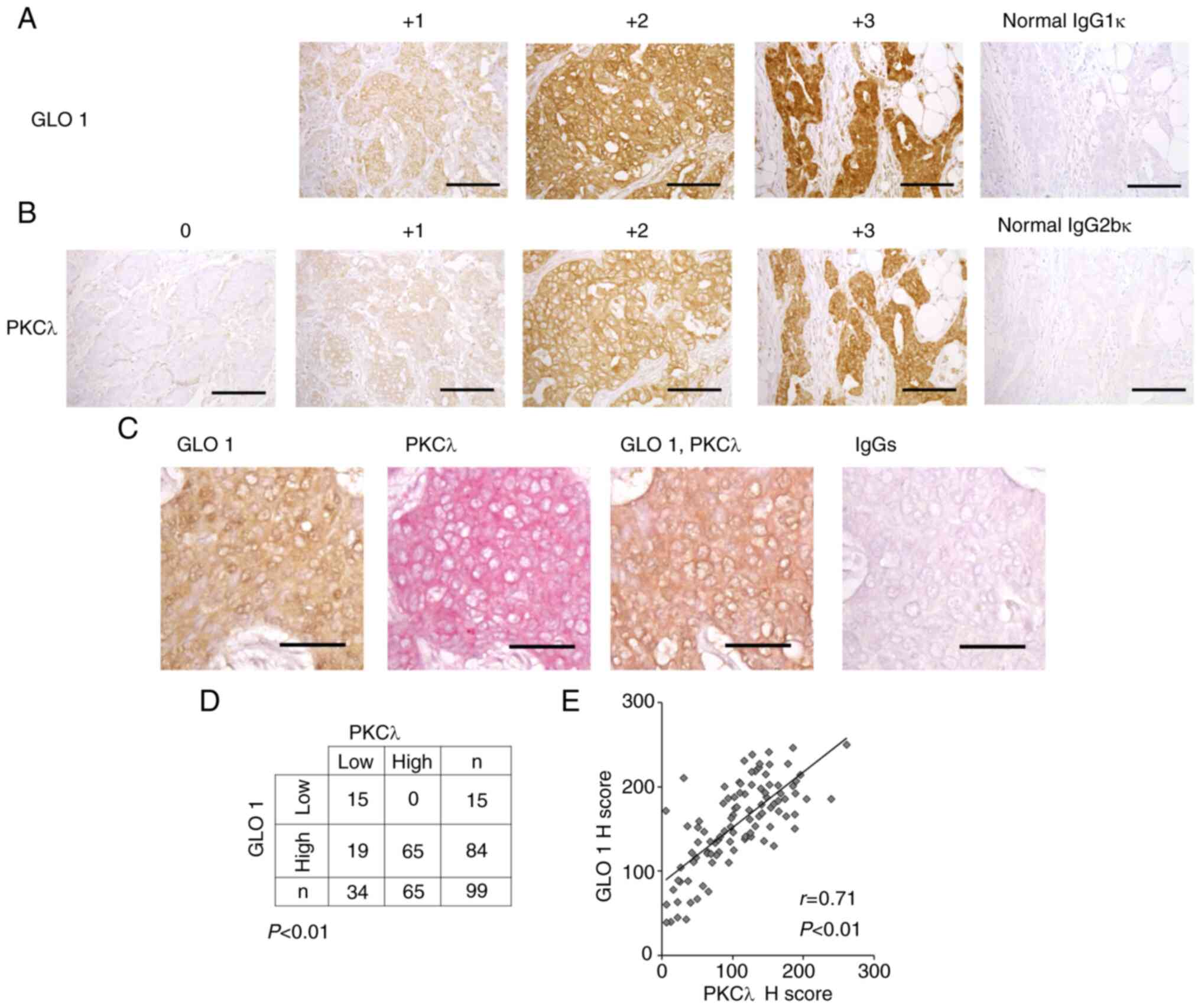
GLO 1 protein expression in both the cytosol and
nucleus of breast cancer cells was detected, and the results were
consistent with earlier observations (Fig. 1A) (20). There were no 0 image samples for GLO
1 protein. GLO 1 protein expression was detected in all breast
cancer samples, and the results were consistent with a previous
report (20). PKCλ was also
localized in the cytosol and nucleus of breast cancer cells
(Fig. 1B) as reported in our
previous study (29). Double IHC
staining showed that GLO 1 and PKCλ were colocalized in breast
cancer cells (Fig. 1C). To evaluate
the relationship between GLO 1 and PKCλ, their signal intensities
were quantified (Fig. 1A and B).
High expression of GLO 1 was significantly correlated with high
expression of PKCλ in breast cancer tissue (Fig. 1D, P<0.01, χ2-test;
Fig. 1E, r=0.71, P<0.01).
GLO 1 expression is correlated with
PKCλ expression at the mRNA level in human breast cancer
To further examine the relationship between GLO
1 and PKCλ at the mRNA level, mRNA expression data from
the METABRIC breast cancer dataset (n=1,904) was downloaded from
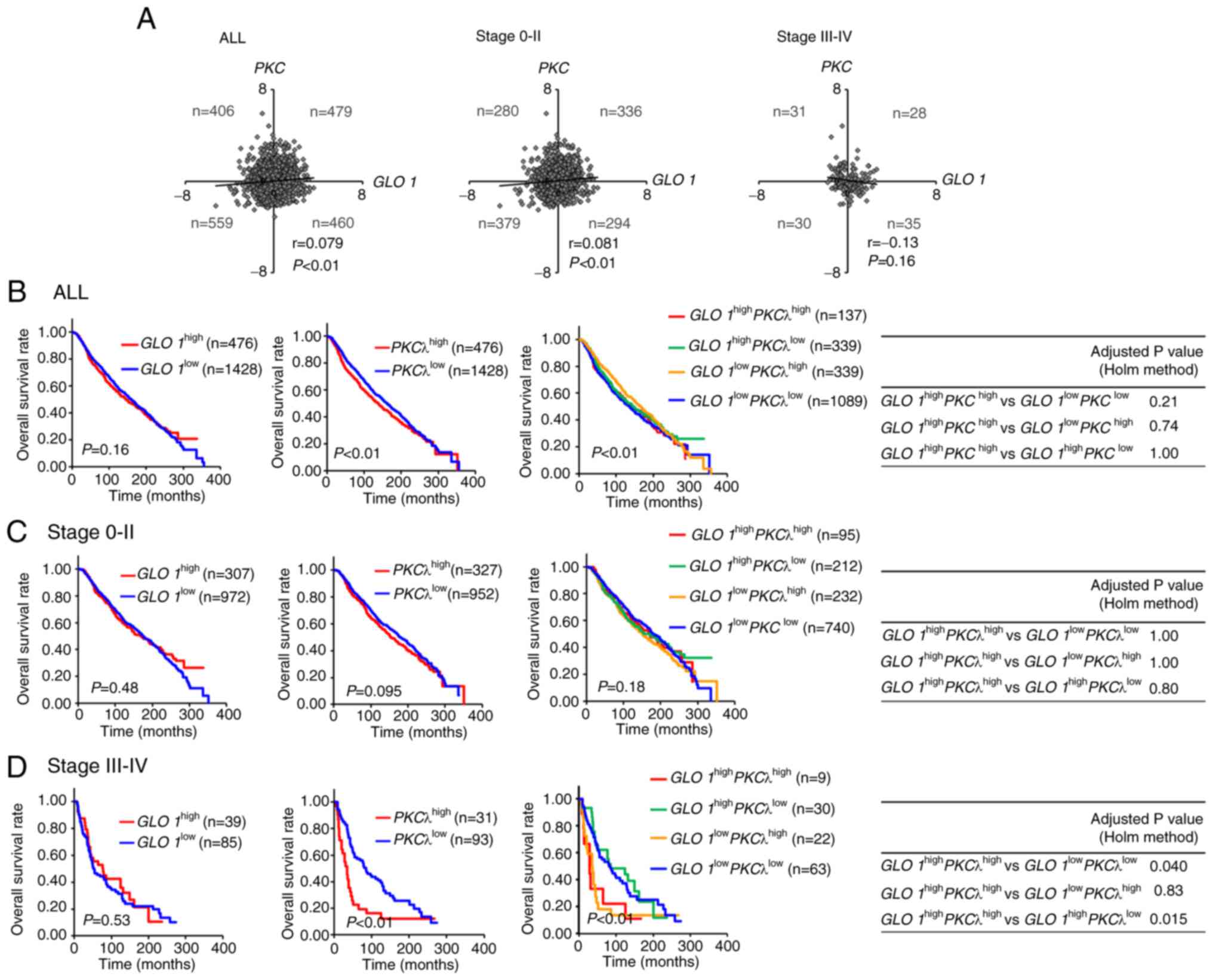
cBioPortal (Fig. 2A). In our
previous study, it was shown that GLO 1 expression is higher
in breast cancer compared with normal tissue samples (21). In addition, GLO 1 gene expression was
enhanced at all breast cancer tumor stages compared with normal
tissue samples. Similarly, PKCλ expression was significantly
higher in breast cancer tissues and at all tumor stages compared
with the normal tissues (41)
(Fig. S1). Scatter plot analysis
indicated that GLO 1 expression was weakly correlated with
PKCλ expression in all patients and in patients with
early-stage (stage 0-II) tumors (Fig.
2A), but was not correlated with PKCλ expression in
patients with late-stage (stage III–IV) tumors (Fig. 2A). As shown in Table SIII, none of the clinicopathological
parameters were correlated with high GLO 1 and PKCλ
gene expression. Nonetheless, these results, together with the IHC
findings, suggest that GLO 1 and PKCλ may be cooperatively involved
in certain cases of breast cancer.
High GLO 1 and PKCλ expression is
correlated with a poorer prognosis in patients with stage III–IV
tumors
Next, we examined weather high GLO 1 and PKCλ
expression is correlated with prognosis (Figs. 2B-D and S2; Tables I
and SIV). Examination of the
prognosis of patients with GLO 1high and
PKCλhigh breast cancer revealed that GLO
1high was not associated with a poorer OS amongst
all patients (Fig. 2B) (21). It was previously shown that
overexpression of PKCλ and its signaling promotes TNBC growth and
metastasis (30). Consistent with
this finding, patients classed as PKCλhigh had
worse OS (Fig. 2B; P<0.01)
(42). In our previous study, it was
shown that patients with stage III or IV cervical cancer with high
PKCλ expression had a worse clinical outcome (32). Thus, tumors were classified into
early-stage (stage 0-II) and late-stage (stage III–IV) tumors, and
the difference in survival was assessed using Kaplan-Meier
analysis. Patients with early-stage tumors (GLO
1high, PKCλhigh or GLO
1high PKCλhigh) exhibited similar
clinical outcomes to those expressing lower levels of these genes
(Fig. 2C), as did those with
late-stage tumors (GLO 1high; Fig. 2D). However, patients with late-stage
tumors (PKCλhigh) had a worse OS (Fig. 2D) (42). Furthermore, patients with GLO
1high PKCλhigh tumors had poorer
prognoses than patients with GLO 1high
PKCλlow (adjusted by Holm's method; P=0.015) and
GLO 1low PKCλlow (P=0.040), but
not GLO 1low PKCλhigh (P=0.83)
tumors. Multivariable Cox regression analysis revealed that GLO
1high PKCλhigh was associated with
poorer clinical outcomes than GLO 1low
PKCλlow (Table I;
HR=2.36; 95% CI, 1.08–5.16; P=0.03) and GLO 1high
PKCλlow (Table I;
HR=3.25, 95% CI, 1.26–8.35; P=0.01) in those with late-stage
tumors. Furthermore, normal-like breast cancer patients classified
as GLO 1high PKCλhigh also
exhibited a worse prognosis (Fig.
S2; adjusted by Holm's method; P=0.016 and Table SIV; HR=9.11, 95% CI, 2.07–40.15;
P<0.01). Thus, high PKCλ expression, regardless of GLO 1
expression level, contributes to poor clinical outcome. Of note,
GLO 1 protein (Fig. 1) and mRNA
(Fig. S1) expression was
considerably higher in breast cancer, reflecting enhanced
glycolysis. Therefore, GLO 1 and PKCλ may function cooperatively to
promote cancer progression, and contribute to worse clinical
outcomes in patients with late-stage breast cancer.
 | Table I.Multivariable Cox regression analysis
of the association between GLO 1 and PKCλ expression
and breast cancer in all patients, and in patients stratified by
stage (0-II and III–IV). |
Table I.
Multivariable Cox regression analysis
of the association between GLO 1 and PKCλ expression
and breast cancer in all patients, and in patients stratified by
stage (0-II and III–IV).
| Comparison | Hazard
ratioa (95% confidence
interval) | P-value |
|---|
| All |
|
|
| GLO
1high vs. GLO 1low | 1.06
(0.93–1.22) | 0.39 |
|
PKCλhigh vs.
PKCλlow | 1.20
(1.05–1.38) | <0.01 |
| GLO
1high PKCλhigh vs. GLO
1low PKCλlow | 1.18
(0.94–1.49) | 0.15 |
| GLO
1high PKCλhigh vs. GLO
1low PKCλhigh | 0.95
(0.74–1.22) | 0.69 |
| GLO
1high PKCλhigh vs. GLO
1high PKCλlow | 1.08
(0.84–1.40) | 0.54 |
| Stage 0-II |
|
|
| GLO
1high vs. GLO 1low | 0.99
(0.83–1.18) | 0.92 |
|
PKCλhigh vs.
PKCλlow | 1.14
(0.97–1.35) | 0.12 |
| GLO
1high PKCλhigh vs. GLO
1low PKCλlow | 1.03
(0.77–1.37) | 0.86 |
| GLO
1high PKCλhigh vs. GLO
1low PKCλhigh | 0.85
(0.62–1.17) | 0.32 |
| GLO
1high PKCλhigh vs. GLO
1high PKCλlow | 0.98
(0.71–1.36) | 0.91 |
| Stage III–IV |
|
|
| GLO
1high vs. GLO 1low | 0.89
(0.57–1.39) | 0.62 |
|
PKCλhigh vs.
PKCλlow | 2.23
(1.41–3.54) | <0.01 |
| GLO
1high PKCλhigh vs. GLO
1low PKCλlow | 2.36
(1.08–5.16) | 0.03 |
| GLO
1high PKCλhigh vs. GLO
1low PKCλhigh | 1.04
(0.44–2.43) | 0.93 |
| GLO
1high PKCλhigh vs. GLO
1high PKCλlow | 3.25
(1.26–8.35) | 0.01 |
TLSC702 and ATM suppresses breast
cancer cell viability
Amongst patients with basal-like breast tumors, the
proportion of patients classed as GLO 1high
PKCλhigh was larger than the proportion classed
as GLO 1low PKCλlow,
irrespective of whether they had early- or late-stage tumors
(Fig. S3). All patients with breast
cancer classed as GLO 1high
PKCλhigh primarily had luminal B type breast
cancer (43.1%, 59/137), whereas those with GLO
1low PKCλlow primarily had luminal
A type breast cancer (42.3%, 459/1,084) (Fig. S3). Similarly, amongst patients with
early-stage lesions, those classed as GLO 1high
PKCλhigh primarily had luminal B breast cancer
(48.4%, 46/95), and those with GLO 1low
PKCλlow primarily had luminal A breast cancer
(45.2%, 333/737). However, amongst patients with late-stage
lesions, those classed as GLO 1high
PKCλhigh exhibited higher incidences of luminal A
or basal-like type breast cancer (luminal A, 33.3%, 3/9;
basal-like, 33.3%, 3/9) compared with patients classed as GLO
1low PKCλlow (luminal A, 28.6%,
18/63; basal-like, 6.3%, 4/63). As shown in Fig. S4, 38% (76/199) of patients with
basal-like type cancer were classed as GLO 1high
PKCλhigh. In our previous study, it was shown
that GLO 1 expression is upregulated in basal-like breast
cancer, and that inhibition of GLO 1 suppresses basal-like breast
cancer cell viability (21). To
further clarify the effects of inhibiting GLO 1 and PKCλ in
basal-like breast cancer cells, MCF-10A human normal-like
(non-transformed) mammary epithelial cells were compared with
MDA-MB-157 and MDA-MB-468 human basal-like breast cancer cells
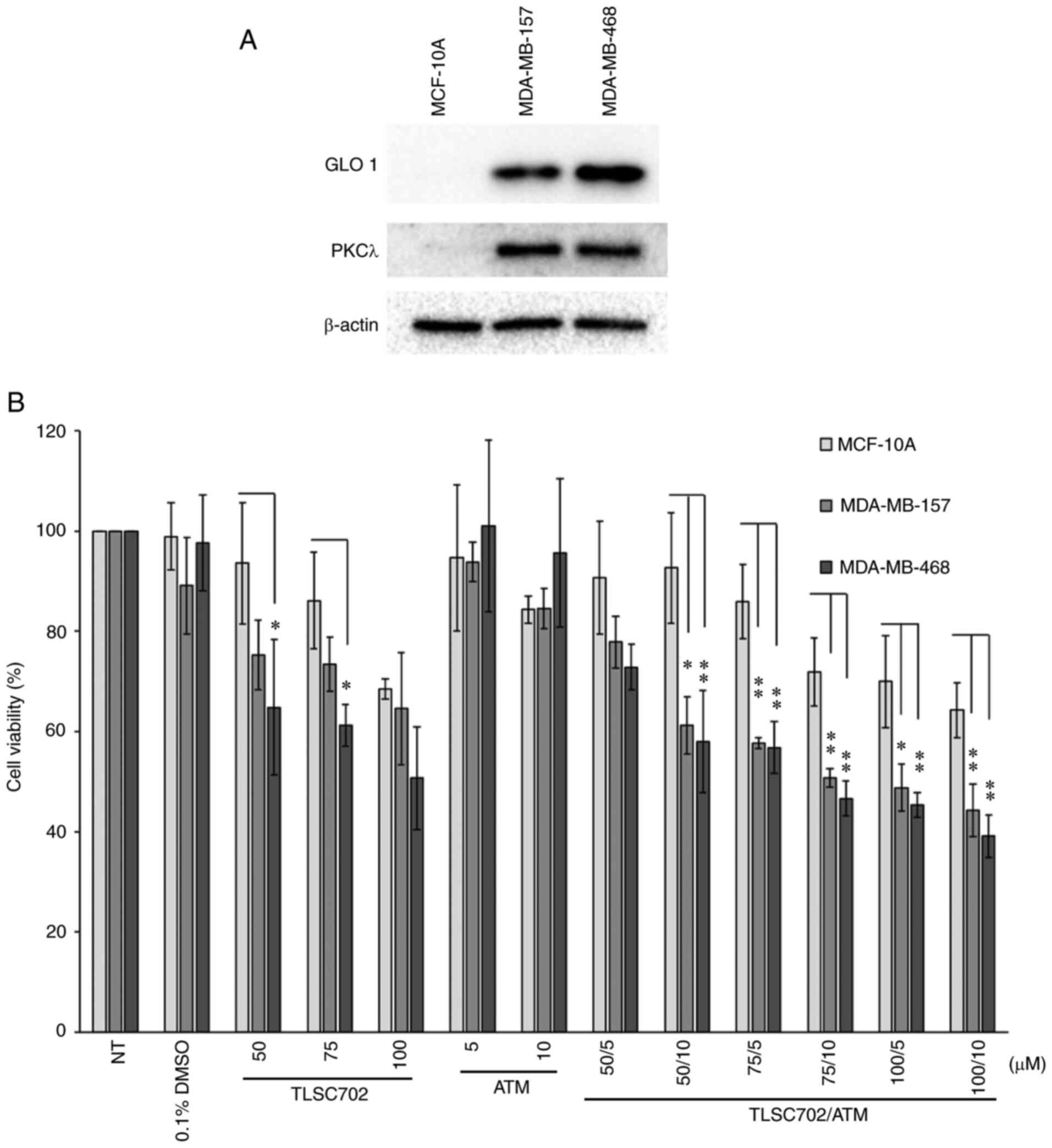
(Fig. 3) (21). Expression levels of both GLO 1 and
PKCλ were higher in MDA-MB-157 and MDA-MB-468 cells than in MCF-10A
cells (Fig. 3A). TLSC702 has
previously been shown to inhibit GLO 1 activity and induce MG
accumulation and apoptosis in cancer cells (21,61).
Based on the inhibitory effects of TLSC702 on the viability of
MCF-10A, MDA-MB-157 and MDA-MB-468 cells, concentrations of 50, 75
and 100 µM TLSC702 were used in the present study (21). To inhibit PKCλ, ATM was used (5 and
10 µM), which interferes with the PB1-PB1 domain interactions
between PKCλ and Par6 and induces apoptosis (62,63). As
a result of the inhibitory effect of 0.5, 1, 5 and 10 µM ATM on the
colony formation ability of MDA-MB-157 cells, it was found that 5
and 10 µM ATM markedly suppressed colony formation compared with
the untreated control group (unpublished data). The inhibitor
concentrations used in the present study were based on these
findings. WST-8 assays showed that MCF-10A cells were less
sensitive to GLO 1 inhibition than the two cancer cell lines,
consistent with our previous study (Fig.
3B; TLSC702 50 and 75 µM) (21).
By contrast, inhibition of PKCλ using ATM did not significantly
affect the cell viability of any of the three cell types assessed
(Fig. 3B). In addition, the
combination of TLSC702 and ATM decreased the viability of the two
cancer cell lines, which was reduced to a greater degree than that
of the control cells (TLSC702/ATM, 50/10, 75/5, 75/10, 100/5 and
100/10 µM; Fig. 3B).
TLSC702 and ATM suppresses
tumor-sphere formation in basal-like breast cancer cells
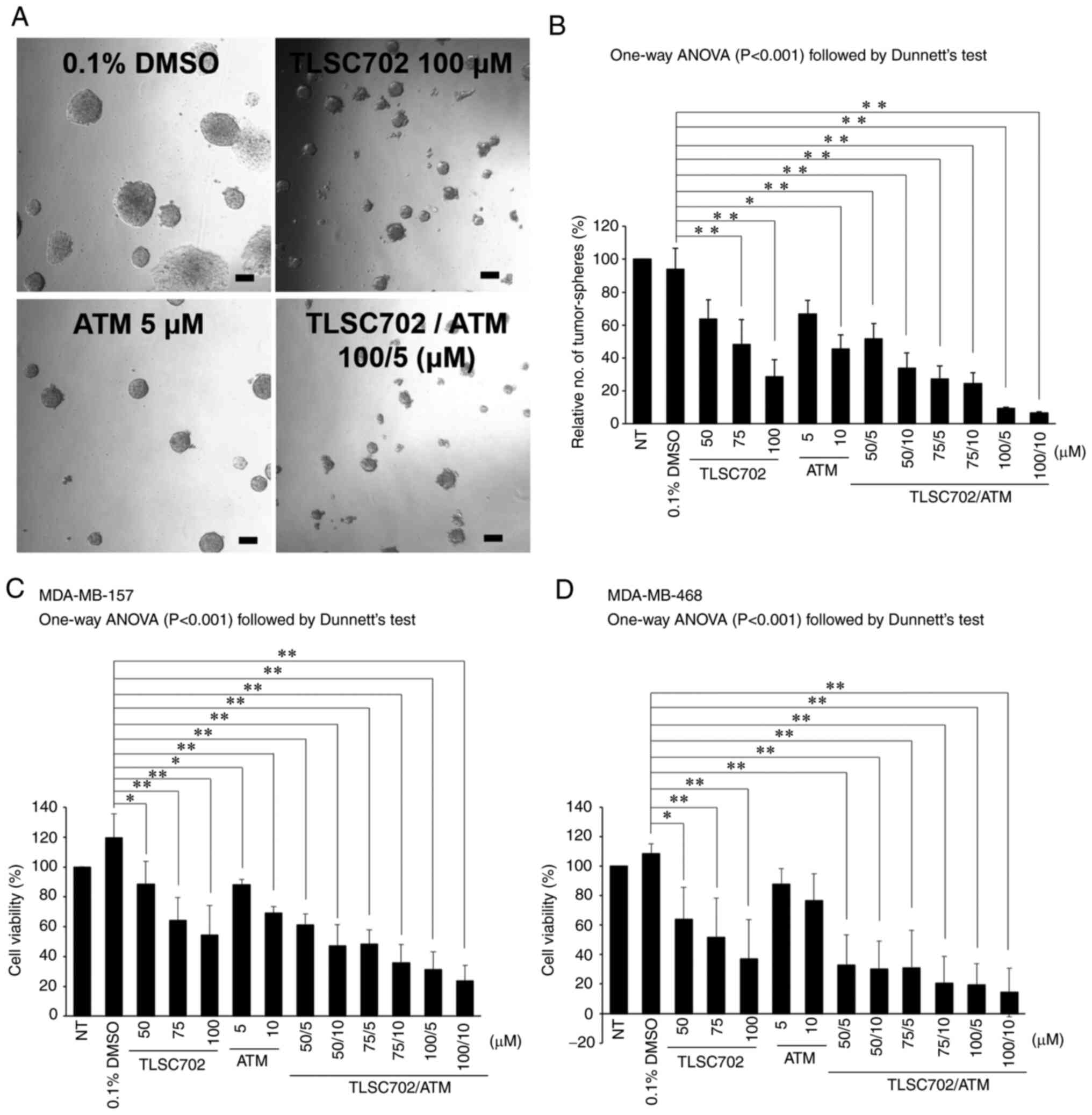
In vitro tumor-sphere formation was assessed
using MDA-MB-157 and MDA-MB-468 cells to determine the roles of GLO
1 and PKCλ in the tumorigenicity of basal-like breast cancer cells.
As shown in Fig. 4A and B, treatment
with TLSC702 or ATM reduced the relative numbers of tumor-spheres
≥50 µm in diameter. Furthermore, a combination of TLSC702 and ATM
inhibited the formation of tumor-spheres ≥50 µm in diameter
(Fig. 4A and B; P<0.05). Cell
Titer-Glo® assays also showed that the combination of
TLSC702 and ATM suppressed the viability of MDA-MB-157 and
MDA-MB-468 cells (Fig. 4C and D;
P<0.05). Taken together, these results along with the results of
the viability assays suggest that GLO 1 and PKCλ are cooperatively
involved in cancer progression and survival of basal-like breast
cancer cells.
Discussion
In the present study, it was shown that high GLO 1
expression was correlated with high PKCλ expression at the protein
and mRNA levels in breast cancer and that patients with breast
cancer classed as GLO 1high
PKCλhigh had poorer clinical outcomes for
late-stage tumors. GLO 1 and PKCλ exhibit low frequencies of gene
amplification and mutations in with breast cancer (21,41). It
thus appears that in breast cancer, higher GLO 1 and PKCλ mRNA
expression and activation reflect higher transcriptional activity
rather than gene amplification or mutations. In stage III–IV
patients, the IHC value (based on tissue acquisition) is relatively
low, and disease burden is approximated using imaging methods, such
as CT or PET. However, the present cohort data lacks CT/PET
confirmation. Therefore, GLO 1 and PKCλ may not be suitable
prognostic factors in stage III–IV patients. However, late-stage
breast cancer cases are often resistant or less responsive to
conventional medical approaches (3).
In the present study, patients with stage III–IV tumors with high
GLO 1 and PKCλ expression exhibited poorer overall survival
compared with patients expressing lower levels of these genes.
Therefore, GLO 1 and PKCλ may potentially serve as effective
therapeutic targets for late-stage human breast cancer. However,
there is a lack of in vivo studies of breast cancer using
TLSC702 and ATM, thus the use of both inhibitors requires further
investigation in vivo.
The prognosis of patients with stage 0-II breast
cancer with GLO 1high PKCλhigh
did not differ significantly from those with GLO
1low PKCλlow. Notably, amongst
patients with stage III–IV tumors, those with GLO
1high PKCλhigh status exhibited
poorer prognoses. To support increased glycolysis, glycolytic
enzymes are upregulated in cancer cells. In breast cancer, one such
enzyme is GLO 1 (19–21), and MG, an intermediate metabolite of
glycolysis, induces GLO 1 expression (64). In the present study, GLO 1 protein
expression was detected in all breast cancer samples. These results
are in line with a previous report (20). Fig.
S1 also showed that GLO 1 gene expression is enhanced at all
tumor stages of breast cancer in comparison with normal tissues.
Thus, GLO 1 mRNA and protein expression is considerably high in
breast cancer, reflecting an increased level of glycolysis.
Therefore, it is considered that there is no difference in
prognosis between GLO 1high and GLO
1low even when classified by high and low expression
in breast cancers in which GLO 1 expression was essentially high.
This is in line with the results that we previously analyzed using
same gene expression data set, which the prognosis of patients with
GLO 1high was not associated with a poorer OS
amongst breast cancer patients (21). Thus, the results of the current study
suggest that both GLO 1 and PKCλ may be cooperatively involved in
breast cancer progression, and contribute to poor prognosis.
Basal-like breast cancer, the majority of cases of
which are TNBC, has the poorest clinical outcomes amongst all
breast cancer subtypes (6). Notably,
TNBC cells show higher GLO 1 expression levels, higher GLO 1
activity and lower accumulation of a MG-arginine adduct,
Arg-pyrimidine (64). Notably, PKCλ
is upregulated in patients with TNBC (30), and it was confirmed that the fraction
of stage III–IV basal-like breast cancer cases with GLO
1high PKCλhigh were enriched
compared with all other breast cancer subtypes. Thirty eight
percent of patients (76/199) with basal-like type cancer were
classed as GLO 1high PKCλhigh.
This result suggests that GLO 1 and PKCλ are cooperatively involved
in the progression of basal-like breast cancer. Moreover, the GLO 1
inhibitor TLSC702 and the PKCλ inhibitor ATM suppressed the
viability of MDA-MB-157 and MDA-MB-468 basal-like breast cancer
cells and tumor-sphere formation using these cells. The Par6-PKCλ
complex interacts with epithelial cell transforming sequence 2 to
activate Rac1 during cancer cell proliferation (65), and PKCλ activates the
Rac1-Pak-Mek1/2-Erk1/2 signaling pathway in lung cancer cell growth
and tumorigenicity (47). PKCλ
modulates c-Myc via FoxO1 DNA-binding ability and contributes to
cell growth of angiosarcoma (48).
MG, which induces expression and activity of GLO 1, also induces
phosphorylation of Erk1/2 (66). GLO
1 modulates cell viability and tumor formation in ALDH1-positive
breast cancer stem cells (CSCs) (21). Moreover, PKCλ also modulates cell
viability, Caspase 3-dependent apoptosis and tumor formation in an
Akt independent manner in ALDH1-positive breast CSCs (41,42). The
results of the present study suggested that GLO 1 is also
functionally associated with PKCλ in the progression of
ALDH1-positive breast CSCs.
PKCλ is essential for cancer cell survival of
ALDH1-positive breast CSCs by maintaining low levels of ROS
(41). ALDH1 serves a role in the
detoxification of toxic aldehyde intermediaries generated by
ROS-induced peroxidation of intracellular lipids (67). Conversely, GLO 1 detoxifies MG, a
cytotoxic byproduct of glycolysis that induces apoptosis (68). Inhibition of GLO 1 reduces cell
viability and induces apoptosis in ALDH1-positive breast CSCs
(21). In the present study, the
combination of TLSC702 and ATM suppressed viability of basal-like
breast cancer cells. Therefore, GLO 1 and PKCλ may be involved in
cell viability by maintaining lower intracellular ROS levels and/or
detoxification of MG. However, the detailed relationship between
GLO 1 and PKCλ in ALDH1-positive breast CSCs remains to be
determined.
GLUT1 facilitates glucose transport, and its
expression is increased in several types of cancers, including
basal-like breast cancer, where it is involved in cancer
progression (49,50). PKCλ is involved in GLUT1
translocation from intracellular vesicles to the plasma membrane in
3T3-L1 adipocytes (51), where it
increases glucose accumulation and promotes cell growth via
upregulation of GLUT1 (69).
Furthermore, PKCλ is involved in insulin-dependent glucose-uptake
by GLUT4 translocation to the plasma membrane in 3T3-L1 adipocytes
(44,45). In addition, GAPDH is phosphorylated
by PKCλ and then interacts directly with the PKCλ regulatory domain
to promote microtubule nucleation. (70,71).
GLO 1 is also overexpressed in basal-like breast cancer
(21). Given the high levels of
GLO 1 and PKCλ expression in several basal-like
breast cancer types, it may be that PKCλ regulates glucose uptake
by GLUT1, leading to increased glycolysis catalyzed in part by GLO
1.
Expression of PKCζ, another aPKC isotype, is also
upregulated in breast cancer tissues compared with normal tissues
derived from the same patient (41).
c-Myc is reportedly phosphorylated by PKCζ in prostate cancer
(72), and c-Myc is known to
directly transactivate glucose metabolic genes, including GLUT1,
PFK-1, GAPDH and enolase, and to increase glucose uptake (73). Thus, c-Myc phosphorylation by PKCζ
may contribute to the regulation of glycolysis in breast cancer. In
addition, PKCζ phosphorylates Nrf2, which regulates
glucose-6-phosphate dehydrogenase (G6PD) gene expression (74). G6PD dehydrogenizes
glucose-6-phosphate, an intermediate of glycolysis, and is the rate
limiting enzyme in the pentose phosphate pathway (PPP). PPP
generates glyceraldehyde 3 phosphate (GAP), an intermediate
metabolite of glycolysis, and GAP is converted into MG.
Furthermore, despite nutrient stress, PKCζ directly phosphorylates
and inhibits the enzymatic activity of PHGDH, which suppresses
metabolic reprogramming of glycolytic intermediates (75). This suggests that both PKCλ and PKCζ
are associated with glycolysis, directly and indirectly, at
different stages in breast cancer development/progression.
Expression levels of GLO 1 and PKCλ mRNA are
correlated in ER- and/or PgR-positive and luminal B type breast
cancer. Given that luminal B type breast cancer exhibits expression
of ER and/or PgR, the correlation between GLO 1 and
PKCλ in luminal B tumors may be related to the ER and/or PgR
positivity of those tumors. The luminal B subtype is associated
with poorer clinical outcomes compared with the luminal A subtype
(76). These results therefore
suggest that high expression of GLO 1 and PKCλ may be
contributed to cancer progression in luminal B.
Kaplan-Meier and multivariable Cox regression
analyses showed that normal-like breast cancer patients classified
as GLO 1high PKCλhigh also
exhibited a worse prognosis. Earlier studies reported that patients
with normal-like tumors had a better prognosis compared with
patients with other breast cancer subtypes (6). However, unlike other breast cancer
subtypes, which have well-described molecular characteristics, the
significant features of the normal-like subtype are largely unknown
(77), and the roles of GLO 1 and
PKCλ remain to be determined.
In conclusion, the levels of GLO 1 and
PKCλ expression were shown to be correlated with breast
cancer. Patients with late-stage tumors who were classed as GLO
1high PKCλhigh had a poorer
prognosis and accounted for a large percentage of cases of
basal-like breast cancer. In addition, TLSC702, a GLO 1 inhibitor,
and ATM, a PKCλ inhibitor, reduced both cell viability and
tumor-sphere formation in basal-like breast cancer cells. It thus
appears that GLO 1 and PKCλ are cooperatively involved in cancer
progression and contribute to poorer clinical outcomes in
late-stage breast cancer patients. It is therefore suggested that
GLO 1 and PKCλ are potentially effective therapeutic targets for
treatment of late-stage breast cancer.
Supplementary Material
Supporting Data
Acknowledgements
The authors would like to thank Dr Babita Shashni
(Department of Materials Science, Graduate School of Pure and
Applied Sciences, University of Tsukuba, Ibaraki, Japan) for
proofreading the article. The authors would also like to thank Dr
Yoshiyasu Nakamura (Molecular Pathology and Genetics Division,
Kanagawa Cancer Center Research Institute, Kanagawa, Japan) for
technical support for immunohistochemistry.
Funding
This work was supported by a Grant-in-Aid for
Scientific Research (C) of JSPS (grant no. 20K07207), a
Grant-in-Aid for JSPS Research Fellows (grant no. 20J11980), JSPS
KAKENHI Grant Number JP 16H06277 (CoBiA), the MEXT's Promotion Plan
for the Platform of Human Resource Development for Cancer project,
2012–2017, Translational Research Center, Research Institute for
Science and Technology, Tokyo University of Science (S1411013) and
Nagai Memorial Research Scholarship from the Pharmaceutical Society
of Japan.
Availability of data and materials
The datasets used and/or analyzed during the
current study are available from the corresponding author on
reasonable request.
Authors' contributions
HM, AO, ST and YW performed the experiments. HM, AO
and KA confirmed the authenticity of all the raw data. HM and AO
analyzed the data using IHC Profiler. HM, AO, ST, CO, YNo, YW, YMa,
TS, KeS and KYa performed the bioinformatics analysis. RT, KYo, TH,
KaS, HI, YMi, YNa, SIT and SO contributed to acquisition of data,
and analysis and interpretation of data. HM and KA conceived the
study. HM drafted the manuscript. HM, AO, KaS, HI, SO and KA
contributed to discussion and review of the manuscript. All authors
read and approved the final manuscript.
Ethics approval and consent to
participate
The experimental protocol of the present study was
reviewed and approved by the Institutional Review Board of Kanagawa
Cancer Center Hospital (approval no. 3-2009; Kanagawa, Japan).
Written informed consent for participation was obtained from all
patients.
Patient consent for publication
Written informed consent was obtained from all
patients for publication.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
World Cancer Research Fund (WCRF), .
Breast cancer statistics. Breast cancer is the most common cancer
in women worldwide. WCRF, London. 2018.https://www.wcrf.org/dietandcancer/cancer-trends/breast-cancer-statistics
|
|
2
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Maughan KL, Lutterbie MA and Ham PS:
Treatment of breast cancer. Am Fam Physician. 81:1339–1346.
2010.PubMed/NCBI
|
|
4
|
van de Vijver MJ, He YD, van't Veer LJ,
Dai H, Hart AA, Voskuil DW, Schreiber GJ, Peterse JL, Roberts C,
Marton MJ, et al: A gene-expression signature as a predictor of
survival in breast cancer. N Engl J Med. 347:1999–2009. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Perou CM, Sørlie T, Eisen MB, van de Rijn
M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA,
et al: Molecular portraits of human breast tumours. Nature.
406:747–752. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Sørlie T, Perou CM, Tibshirani R, Aas T,
Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey
SS, et al: Gene expression patterns of breast carcinomas
distinguish tumor subclasses with clinical implications. Proc Natl
Acad Sci USA. 98:10869–10874. 2001. View Article : Google Scholar
|
|
7
|
Herschkowitz JI, Simin K, Weigman VJ,
Mikaelian I, Usary J, Hu Z, Rasmussen KE, Jones LP, Assefnia S,
Chandrasekharan S, et al: Identification of conserved gene
expression features between murine mammary carcinoma models and
human breast tumors. Genome Biol. 8:R762007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Badve S, Dabbs DJ, Schnitt SJ, Baehner FL,
Decker T, Eusebi V, Fox SB, Ichihara S, Jacquemier J, Lakhani SR,
et al: Basal-like and triple-negative breast cancers: A critical
review with an emphasis on the implications for pathologists and
oncologists. Mod Pathol. 24:157–167. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Warburg O, Wind F and Negelein E: The
metabolism of tumors in the body. J Gen Physiol. 8:519–530. 1927.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Gatenby RA and Gillies RJ: Why do cancers
have high aerobic glycolysis? Nat Rev Cancer. 4:891–899. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sakamoto H, Mashima T, Sato S, Hashimoto
Y, Yamori T and Tsuruo T: Selective activation of apoptosis program
by S-p-bromobenzylglutathione cyclopentyl diester in glyoxalase
I-overexpressing human lung cancer cells. Clin Cancer Res.
7:2513–2518. 2001.PubMed/NCBI
|
|
12
|
Cheng WL, Tsai MM, Tsai CY, Huang YH, Chen
CY, Chi HC, Tseng YH, Chao IW, Lin WC, Wu SM, et al: Glyoxalase-I
is a novel prognosis factor associated with gastric cancer
progression. PLoS One. 7:e343522012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ranganathan S and Tew KD: Analysis of
glyoxalase-I from normal and tumor tissue from human colon. Biochim
Biophys Acta. 1182:311–316. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhang S, Liang X, Zheng X, Huang H, Chen
X, Wu K, Wang B and Ma S: Glo1 genetic amplification as a potential
therapeutic target in hepatocellular carcinoma. Int J Clin Exp
Pathol. 7:2079–2090. 2014.PubMed/NCBI
|
|
15
|
Baunacke M, Horn LC, Trettner S, Engel KM,
Hemdan NY, Wiechmann V, Stolzenburg JU, Bigl M and Birkenmeier G:
Exploring glyoxalase 1 expression in prostate cancer tissues:
Targeting the enzyme by ethyl pyruvate defangs some
malignancy-associated properties. Prostate. 74:48–60. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kreycy N, Gotzian C, Fleming T,
Flechtenmacher C, Grabe N, Plinkert P, Hess J and Zaoui K:
Glyoxalase 1 expression is associated with an unfavorable prognosis
of oropharyngeal squamous cell carcinoma. BMC Cancer. 17:3822017.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zou XY, Ding D, Zhan N, Liu XM, Pan C and
Xia YM: Glyoxalase I is differentially expressed in cutaneous
neoplasms and contributes to the progression of squamous cell
carcinoma. J Invest Dermatol. 135:589–598. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Bair WB III, Cabello CM, Uchida K, Bause
AS and Wondrak GT: GLO1 overexpression in human malignant melanoma.
Melanoma Res. 20:85–96. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Rulli A, Carli L, Romani R, Baroni T,
Giovannini E, Rosi G and Talesa V: Expression of glyoxalase I and
II in normal and breast cancer tissues. Breast Cancer Res Treat.
66:67–72. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Peng HT, Chen J, Liu TY, Wu YQ, Lin XH,
Lai YH and Huang YF: Up-regulation of the tumor promoter
Glyoxalase-1 indicates poor prognosis in breast cancer. Int J Clin
Exp Pathol. 10:10852–10862. 2017.PubMed/NCBI
|
|
21
|
Tamori S, Nozaki Y, Motomura H, Nakane H,
Katayama R, Onaga C, Kikuchi E, Shimada N, Suzuki Y, Noike M, et
al: Glyoxalase 1 gene is highly expressed in basal-like human
breast cancers and contributes to survival of ALDH1-positive breast
cancer stem cells. Oncotarget. 9:36515–36529. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhang D, Tai LK, Wong LL, Chiu LL, Sethi
SK and Koay ES: Proteomic study reveals that proteins involved in
metabolic and detoxification pathways are highly expressed in
HER-2/neu-positive breast cancer. Mol Cell Proteomics. 4:1686–1696.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Gandalovičová A, Vomastek T, Rosel D and
Brábek J: Cell polarity signaling in the plasticity of cancer cell
invasiveness. Oncotarget. 7:25022–25049. 2016. View Article : Google Scholar
|
|
24
|
Akimoto K, Mizuno K, Osada S, Hirai S,
Tanuma S, Suzuki K and Ohno S: A new member of the third class in
the protein kinase C family, PKC lambda, expressed dominantly in an
undifferentiated mouse embryonal carcinoma cell line and also in
many tissues and cells. J Biol Chem. 269:12677–12683. 1994.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Suzuki A, Akimoto K and Ohno S: Protein
kinase C lambda/iota (PKClambda/iota): A PKC isotype essential for
the development of multicellular organisms. J Biochem. 133:9–16.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ohno S: Intercellular junctions and
cellular polarity: The PAR-aPKC complex, a conserved core cassette
playing fundamental roles in cell polarity. Curr Opin Cell Biol.
13:641–648. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Parker PJ, Justilien V, Riou P, Linch M
and Fields AP: Atypical protein kinase Cι as a human oncogene and
therapeutic target. Biochem Pharmacol. 88:1–11. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Murray NR, Jamieson L, Yu W, Zhang J,
Gökmen-Polar Y, Sier D, Anastasiadis P, Gatalica Z, Thompson EA and
Fields AP: Protein kinase Ciota is required for Ras transformation
and colon carcinogenesis in vivo. J Cell Biol. 164:797–802. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kojima Y, Akimoto K, Nagashima Y, Ishiguro
H, Shirai S, Chishima T, Ichikawa Y, Ishikawa T, Sasaki T, Kubota
Y, et al: The overexpression and altered localization of the
atypical protein kinase C lambda/iota in breast cancer correlates
with the pathologic type of these tumors. Hum Pathol. 39:824–831.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Paul A, Gunewardena S, Stecklein SR, Saha
B, Parelkar N, Danley M, Rajendran G, Home P, Ray S, Jokar I, et
al: PKCλ/ι signaling promotes triple-negative breast cancer growth
and metastasis. Cell Death Differ. 21:1469–1481. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Mizushima T, Asai-Sato M, Akimoto K,
Nagashima Y, Taguri M, Sasaki K, Nakaya MA, Asano R, Tokinaga A,
Kiyono T, et al: Aberrant expression of the cell polarity regulator
aPKCλ/ι is associated with disease progression in cervical
intraepithelial Neoplasia (CIN): A possible marker for predicting
CIN prognosis. Int J Gynecol Pathol. 35:106–117. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Tokinaga-Uchiyama A, Mizushima T, Akimoto
K, Nagashima Y, Sasaki K, Nakaya MA, Ohashi K, Kubota K, Maruyama
Y, Kato H, et al: Aberrant nuclear localization of aPKCλ/ι is
associated with poorer prognosis in uterine cervical cancer.
Gynecol Pathol. 38:301–309. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Baba J, Kioi M, Akimoto K, Nagashima Y,
Taguri M, Inayama Y, Aoki I, Ohno S, Mitsudo K and Tohnai I:
Atypical protein kinase Cλ/ι expression is associated with
malignancy of oral squamous cell carcinoma. Anticancer Res.
38:6291–6297. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Ishiguro H, Akimoto K, Nagashima Y, Kagawa
E, Sasaki T, Sano JY, Takagawa R, Fujinami K, Sasaki K, Aoki I, et
al: Coexpression of aPKCλ/ι and IL-6 in prostate cancer tissue
correlates with biochemical recurrence. Cancer Sci. 102:1576–1581.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ishiguro H, Akimoto K, Nagashima Y, Kojima
Y, Sasaki T, Ishiguro-Imagawa Y, Nakaigawa N, Ohno S, Kubota Y and
Uemura H: aPKClambda/iota promotes growth of prostate cancer cells
in an autocrine manner through transcriptional activation of
interleukin-6. Proc Natl Acad Sci USA. 106:16369–16374. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Regala RP, Weems C, Jamieson L, Khoor A,
Edell ES, Lohse CM and Fields AP: Atypical protein kinase C iota is
an oncogene in human non-small cell lung cancer. Cancer Res.
65:8905–8911. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Eder AM, Sui X, Rosen DG, Nolden LK, Cheng
KW, Lahad JP, Kango-Singh M, Lu KH, Warneke CL, Atkinson EN, et al:
Atypical PKCiota contributes to poor prognosis through loss of
apical-basal polarity and cyclin E overexpression in ovarian
cancer. Proc Natl Acad Sci USA. 102:12519–12524. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Scotti ML, Bamlet WR, Smyrk TC, Fields AP
and Murray NR: Protein kinase Ciota is required for pancreatic
cancer cell transformed growth and tumorigenesis. Cancer Res.
70:2064–2074. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Kato S, Akimoto K, Nagashima Y, Ishiguro
H, Kubota K, Kobayashi N, Hosono K, Watanabe S, Sekino Y, Sato T,
et al: aPKCλ/ι is a beneficial prognostic marker for pancreatic
neoplasms. Pancreatology. 13:360–368. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Takagawa R, Akimoto K, Ichikawa Y, Akiyama
H, Kojima Y, Ishiguro H, Inayama Y, Aoki I, Kunisaki C, Endo I, et
al: High expression of atypical protein kinase C lambda/iota in
gastric cancer as a prognostic factor for recurrence. Ann Surg
Oncol. 17:81–88. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Nozaki Y, Motomura H, Tamori S, Kimura Y,
Onaga C, Kanai S, Ishihara Y, Ozaki A, Hara Y, Harada Y, et al:
High PKCλ expression is required for ALDH1-positive cancer
stem cell function and indicates a poor clinical outcome in
late-stage breast cancer patients. PLoS One. 15:e02357472020.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Motomura H, Nozaki Y, Onaga C, Ozaki A,
Tamori S, Shiina TA, Kanai S, Ohira C, Hara Y, Harada Y, et al:
High expression of c-Met, PKCλ and ALDH1A3 predicts a poor
prognosis in late-stage breast cancer. Anticancer Res. 40:35–52.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Akimoto K, Takahashi R, Moriya S, Nishioka
N, Takayanagi J, Kimura K, Fukui Y, Osada SI, Mizuno K, Hirai SI,
et al: EGF or PDGF receptors activate atypical PKClambda through
phosphatidylinositol 3-kinase. EMBO J. 15:788–798. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Kotani K, Ogawa W, Matsumoto M, Kitamura
T, Sakaue H, Hino Y, Miyake K, Sano W, Akimoto K, Ohno S and Kasuga
M: Requirement of atypical protein kinase clambda for insulin
stimulation of glucose uptake but not for Akt activation in 3T3-L1
adipocytes. Mol Cell Biol. 18:6971–6982. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Farese RV: Function and dysfunction of
aPKC isoforms for glucose transport in insulin-sensitive and
insulin-resistant states. Am J Physiol Endocrinol Metab.
283:E1–E11. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Akimoto K, Nakaya M, Yamanaka T, Tanaka J,
Matsuda S, Weng QP, Avruch J and Ohno S: Atypical protein kinase
Clambda binds and regulates p70 S6 kinase. Biochem J. 335:417–424.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Regala RP, Weems C, Jamieson L, Copland
JA, Thompson EA and Fields AP: Atypical protein kinase Ciota plays
a critical role in human lung cancer cell growth and
tumorigenicity. J Biol Chem. 280:31109–31115. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Riddell M, Nakayama A, Hikita T,
Mirzapourshafiyi F, Kawamura T and Pasha A: aPKC controls
endothelial growth by modulating c-Myc via FoxO1 DNA-binding
ability. Nat Commun. 9:53572018. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Szablewski L: Expression of glucose
transporters in cancers. Biochim Biophys Acta. 1835:164–169.
2013.PubMed/NCBI
|
|
50
|
Hussein YR, Bandyopadhyay S, Semaan A,
Ahmed Q, Albashiti B, Jazaerly T, Nahleh Z and Ali-Fehmi R: Glut-1
expression correlates with basal-like breast cancer. Transl Oncol.
4:321–327. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Bosch RR, Bazuine M, Span PN, Willems PH,
Olthaar AJ, van Rennes H, Maassen JA, Tack CJ, Hermus AR and Sweep
CG: Regulation of GLUT1-mediated glucose uptake by
PKClambda-PKCbeta(II) interactions in 3T3-L1 adipocytes. Biochem J.
384:349–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Varghese F, Bukhari AB, Malhotra R and De
A: IHC Profiler: An open source plugin for the quantitative
evaluation and automated scoring of immunohistochemistry images of
human tissue samples. PLoS One. 9:e968012014. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Nozaki Y, Tamori S, Inada M, Katayama R,
Nakane H, Minamishima O, Onodera Y, Abe M, Shiina S, Tamura K, et
al: Correlation between c-Met and ALDH1 contributes to the survival
and tumor-sphere formation of ALDH1 positive breast cancer stem
cells and predicts poor clinical outcome in breast cancer. Genes
Cancer. 8:628–639. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Sato K and Akimoto K: Expression levels of
KMT2C and SLC20A1 identified by information-theoretical analysis
are powerful prognostic biomarkers in estrogen receptor-positive
breast cancer. Clin Breast Cancer. 17:e135–e142. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Curtis C, Shah SP, Chin SF, Turashvili G,
Rueda OM, Dunning MJ, Speed D, Lynch AG, Samarajiwa S, Yuan Y, et
al: The genomic and transcriptomic architecture of 2,000 breast
tumours reveals novel subgroups. Nature. 486:346–352. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Pereira B, Chin SF, Rueda OM, Vollan HK,
Provenzano E, Bardwell HA, Pugh M, Jones L, Russell R, Sammut SJ,
et al: The somatic mutation profiles of 2,433 breast cancers refine
their genomic and transcriptomic landscapes. Nat Commun.
7:114792016. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Cerami E, Gao J, Dogrusoz U, Gross BE,
Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et
al: The cBio cancer genomics portal: An open platform for exploring
multidimensional cancer genomics data. Cancer Dicov. 2:401–404.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Gao J, Aksoy BA, Dogrusoz U, Dresdner G,
Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al:
Integrative analysis of complex cancer genomics and clinical
profiles using the cBioPortal. Sci Signal. 6:pl12013. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Cancer Genome Atlas Network, .
Comprehensive molecular portraits of human breast tumours. Nature.
490:61–70. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Rhodes DR, Yu J, Shanker K, Deshpande N,
Varambally R, Ghosh D, Barrette T, Pandey A and Chinnaiyan AM:
ONCOMINE: A cancer microarray database and integrated data-mining
platform. Neoplasia. 6:1–6. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Takasawa R, Tao A, Saeki K, Shionozaki N,
Tanaka R, Uchiro H, Takahashi S, Yoshimori A and Tanuma S:
Discovery of a new type inhibitor of human glyoxalase I by
myricetin-based 4-point pharmacophore. Bioorg. Med Chem Lett.
21:4337–4342. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Stallings-Mann M, Jamieson L, Regala RP,
Weems C, Murray NR and Fields AP: A novel small-molecule inhibitor
of protein kinase Ciota blocks transformed growth of non-small-cell
lung cancer cells. Cancer Res. 66:1767–1774. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Trani M, Sorrentino A, Busch C and
Landström M: Pro-apoptotic effect of aurothiomalate in prostate
cancer cells. Cell Cycle. 8:306–313. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Chiavarina B, Nokin MJ, Durieux F, Bianchi
E, Turtoi A, Peulen O, Peixoto P, Irigaray P, Uchida K, Belpomme D,
et al: Triple negative tumors accumulate significantly less
methylglyoxal specific adducts than other human breast cancer
subtypes. Oncotarget. 5:5472–5482. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Justilien V and Fields AP: Ect2 links the
PKCiota-Par6alpha complex to Rac1 activation and cellular
transformation. Oncogene. 28:3597–3607. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Guo Y, Zhang Y, Yang X, Lu P, Yan X, Xiao
F, Zhou H, Wen C, Shi M, Lu J and Meng QH: Effects of methylglyoxal
and glyoxalase I inhibition on breast cancer cells proliferation,
invasion, and apoptosis through modulation of MAPKs, MMP9, and
Bcl-2. Cancer Biol Ther. 17:169–180. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Singh S, Brocker C, Koppaka V, Chen Y,
Jackson BC and Matsumoto A: Aldehyde dehydrogenases in cellular
responses to oxidative/electrophilic stress. Free Radic Biol Med.
56:89–101. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Kang Y, Edwards LG and Thornalley PJ:
Effect of methylglyoxal on human leukaemia 60 cell growth:
Modification of DNA G1 growth arrest and induction of apoptosis.
Leuk Res. 20:97–405. 1996. View Article : Google Scholar
|
|
69
|
Liu L, Lei B, Wang L, Chang C, Yang H, Liu
J, Huang G and Xie W: Protein kinase C-iota-mediated glycolysis
promotes non-small-cell lung cancer progression. Onco Targets Ther.
12:5835–5848. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Tisdale EJ: Glyceraldehyde-3-phosphate
dehydrogenase is phosphorylated by protein kinase Ciota/lambda and
plays a role in microtubule dynamics in the early secretory
pathway. J Biol Chem. 277:3334–3341. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Tisdale EJ: Rab2 interacts directly with
atypical protein kinase C (aPKC) iota/lambda and inhibits
aPKCiota/lambda-dependent glyceraldehyde-3-phosphate dehydrogenase
phosphorylation. J Biol Chem. 278:52524–52530. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Kim JY, Valencia T, Abu-Baker S, Linares
J, Lee SJ, Yajima T, Chen J, Eroshkin A, Castilla EA, Brill LM, et
al: c-Myc phosphorylation by PKCζ represses prostate tumorigenesis.
Proc Natl Acad Sci USA. 110:6418–6423. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Osthus RC, Shim H, Kim S, Li Q, Reddy R,
Mukherjee M, Xu Y, Wonsey D, Lee LA and Dang CV: Deregulation of
glucose transporter 1 and glycolytic gene expression by c-Myc. J
Biol Chem. 275:21797–21800. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Gjyshi O, Bottero V, Veettil MV, Dutta S,
Singh VV and Chikoti L: Kaposi's sarcoma-associated herpesvirus
induces Nrf2 during de novo infection of endothelial cells to
create a microenvironment conducive to infection. PLoS Pathog.
10:e10044602014. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Ma L, Tao Y, Duran A, Llado V, Galvez A,
Barger JF, Castilla EA, Chen J, Yajima T, Porollo A, et al: Control
of nutrient stress-induced metabolic reprogramming by PKCζ in
tumorigenesis. Cell. 152:599–611. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Franchet C, Duprez-Paumier R and
Lacroix-Triki M: Molecular taxonomy of luminal breast cancer in
2015. Bull Cancer. 102 (6 Suppl 1):S34–S46. 2015.(In French).
View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Liu Z, Zhang XS and Zhang S: Breast tumor
subgroups reveal diverse clinical prognostic power. Sci Rep.
4:40022014. View Article : Google Scholar : PubMed/NCBI
|