Introduction
Colorectal cancer (CRC) is one of the most common
malignancies worldwide (1). In 2017,
~46.9 per 100,000 population were newly diagnosed with CRC and
there are 17.7 per 100,000 CRC-associated deaths annually in the
United States (2). Unhealthy diet,
obesity, smoking and genetics may contribute to CRC, which has been
a main cause of tumor-related morbidities in China (3). Notably, biomarkers for the prognosis of
advanced CRC remain limited. Therefore, it is urgent to elucidate
the molecular mechanisms involved in the regulation of CRC
development.
Ubiquitin-conjugating enzyme E2 (UBE2) T, a member
of the E2 family, serves important roles in modulating cell
proliferation, apoptosis and signal transduction (4–6). For
instance, UBE2T induces hepatocellular carcinoma growth by
enhancing cell cycle progression (6). Additionally, UBE2T has been
demonstrated to regulate the Fanconi anemia signaling pathway by
modulating Fanconi anemia group D2 protein (FANCD2)
monoubiquitination, which is a key step in the DNA damage signaling
pathway (7). The upregulation of
UBE2T has been reported in multiple cancer types, and is associated
with the regulation of cancer cell proliferation and metastasis
(8–12), including in gastric (11) and prostate cancer (12). However, the functional roles of UBE2T
in CRC remain unclear and require further investigation.
The present study analyzed The Cancer Genome Atlas
(TCGA) dataset to identify differentially expressed genes (DEGs) in
CRC. Moreover, protein-protein interaction network analysis was
performed to identify hub regulators in the progression of CRC.
Finally, loss of function assays using short hairpin RNA (shRNA/sh)
was performed to confirm the potential functions of hub gene UBE2T
in CRC. The present results indicated that UBE2T may be a novel key
regulator of CRC progression.
Materials and methods
Analysis of public CRC RNA-Seq
data
The RNA-Seq data from TCGA (http://cancergenome.nih.gov/) was utilized to compare
gene expression between CRC and non-tumor tissues. The genes with
|log2 [fold change (FC)]|>1.0 and P-value <0.05 used as the
threshold values.
Gene Ontology (GO) and Kyoto
Encyclopedia of Genes and Genomes (KEGG) enrichment analyses of
DEGs
The Database for Annotation, Visualization and
Integrated Discovery (DAVID; version 6.8; http://david.ncifcrf.gov/summary.jsp) provides a
comprehensive set of functional annotation tools for investigators
to understand the biological meaning behind a number of different
genes (13,14). GO functional annotation and KEGG
analysis of DEGs was performed and visualized using the ImageGP
database (http://www.ehbio.com/ImageGP/index.php/Home/Index/index.html).
PPI network construction and key
module identification
The PPI network was built using the Search Tool for
the Retrieval of Interacting Genes (STRING; version 10.0) online
database (15). Cytoscape (version
3.6.1) is a bioinformatics software for the visualization of
molecular interaction networks (16). The Molecular Complex Detection
(MCODE) plug-in of Cytoscape was used to find closely connected
regions in a network (17). The PPI
network was visualized using Cytoscape and the most significant
module was identified using MCODE. The selection criteria were as
follows: Degree cut-off, 2; node score cut-off, 0.2; max depth,
100; and k-score, 2.
Cell culture
The RKO cell line was purchased from American Type
Culture Collection and cultured in DMEM (Gibco; Thermo Fisher
Scientific, Inc.) supplemented with 10% FBS (HyClone; Cytiva) at
37°C under a 5% CO2 atmosphere as previously described
(18).
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was extracted using a TRIzol®
kit (Takara Biotechnology Co., Ltd.) from RKO cells. RT was
conducted using the PrimeScript RT Reagent kit (Takara
Biotechnology Co., Ltd.). The temperature and duration of RT were:
37°C For 30 sec, followed by 85°C for 5 sec and 4°C for 10 min.
qPCR was performed using SYBR Premix Ex Taq II (Takara
Biotechnology Co., Ltd.) according to the manufacturer's protocol.
The thermocycling conditions used were as follows: 95°C For 30 sec,
followed by 40 cycles of 95°C for 5 sec and 60°C for 40 sec. The
following primer pairs were used for qPCR: UBE2T forward,
5′-ATCCCTCAACATCGCAACTGT-3′ and reverse,
5′-CAGCCTCTGGTAGATTATCAAGC-3′; and GAPDH forward,
5′-TGACTTCAACAGCGACACCCA-3′ and reverse,
5′-CACCCTGTTGCTGTAGCCAAA-3′. GAPDH was used as endogenous control.
The relative expression of target genes was calculated using the
2−ΔΔCq method (19).
Western blotting
The western blot assay was performed according to a
previous report (20). Total protein
was extracted from RKO cells using RIPA lysis buffer (Beyotime
Institute of Biotechnology). The concentration of proteins was
determined using the Bradford method. Cell lysates (30 µg/lane)
were separated using 10% SDS-PAGE and transferred to PVDF membranes
(EMD Millipore). Then, the membrane was blocked with 5% skimmed
milk for 1 h at room temperature. Next, the membrane was incubated
with primary antibodies at 4°C overnight. The antibodies used in
the present study included GAPDH (1:5,000; cat no. 10494-1-AP;
ProteinTech Group, Inc.), β-actin (1:1,000; cat. no. ab8227;
Abcam), Bcl-2 (1:1,000; cat. no. sc-492; Santa Cruz Biotechnology,
Inc.), cleaved caspase-3 (1:500; cat. no. 9664; Cell Signaling
Technology, Inc.) and UBE2T (1:2,000; cat. no. 10105-2-AP;
ProteinTech Group, Inc.). After washing with TBST (0.1% Tween)
three times, the membranes were incubated with horseradish
peroxidase-conjugated anti-rabbit IgG secondary antibody (1:3,000;
cat. no. 7074; Cell Signaling Technology, Inc.) at room temperature
for 1 h. Finally, the membrane was visualized using an Immobilon™
Western Chemiluminescent HRP substrate (EMD Millipore).
Lentivirus vector construction for RNA
interference
Recombinant lentiviral vectors were constructed as
previously described (21). shRNA
against UBE2T (shUBE2T; 5′-GCAACTGTGTTGACCTCTATT-3′) and shRNA
negative control (shNC; 5′-TTCTCCGAACGTGTCACGT-3′) were purchased
from Shanghai GeneChem Co., Ltd. The shRNAs were annealed and
ligated into the linearized GV115 lentiviral vector (Shanghai
GeneChem Co., Ltd.). Next, 10 µg pGV115-shControl
(Ctrl)/pGV115-shUBE2T were transfected into 293T cells (Sangon
Biotech, Co., Ltd.) with the pHelper system (Shanghai GeneChem Co.,
Ltd.) to produce lentiviral particles. Lentiviruses were harvested
after 48 h. Next, RKO cells were infected and cultured in RPMI-1640
medium with lentiviruses at a multiplicity of infection of 10 for
48 h at 37°C. At 48 h after transfection, the transfection
efficiency was determined using reverse transcription-quantitative
(RT-q)PCR and western blotting.
Cell proliferation analysis
An adherent cell cytometry system,
Celigo®, was used to detect the proliferation of CRC
cells as previously described (21).
Colony formation assays
A total of 1,000 RKO cells were seeded into 6-well
plates and cultured with DMEM (Gibco; Thermo Fisher Scientific,
Inc.) at 37°C with 5% CO2 for 2 weeks. Subsequently, the
colonies were washed with 1 ml PBS and fixed with 1 ml 4%
paraformaldehyde (Beyotime Institute of Biotechnology) for 15 min
at room temperature. Next, the colonies were stained with 500 µl
Giemsa (Sigma-Aldrich; Merck KGaA) for 15–20 min at room
temperature. Finally, the colonies were observed under an inverted
light microscope (magnification, ×200; IX71; Olympus Corporation)
and counted using ImageJ software (version 4.0; National Institutes
of Health).
Cell cycle and apoptosis assays
At 2 days after transfection with shUBE2T or shNC
lentivirus, RKO cells were collected, washed twice with PBS and
resuspended in staining buffer containing 0.03% Triton X-100.
Subsequently, 5 µl Annexin V-allophycocyanin (eBioscience; Thermo
Fisher Scientific, Inc.) was added to the cells to detect cell
apoptosis, while 50 ng/ml PI (Sigma-Aldrich; Merck KGaA) was added
to the cells to analyze the cell cycle. Finally, the effects of
UBE2T knockdown on the cell cycle and apoptosis were detected using
a FACSCalibur instrument (BD Biosciences). The cell cycle was
analyzed using the FACSCalibur flow cytometer (BD Biosciences).
Statistical analysis
Statistical analysis was performed using GraphPad
Prism v6.0 software (GraphPad Software, Inc.). All assays were
performed three times and data are presented as the mean ± standard
deviation. Unpaired Student's t-test was used to compare
differences between two groups P<0.05 was considered to indicate
a statistically significant difference.
Results
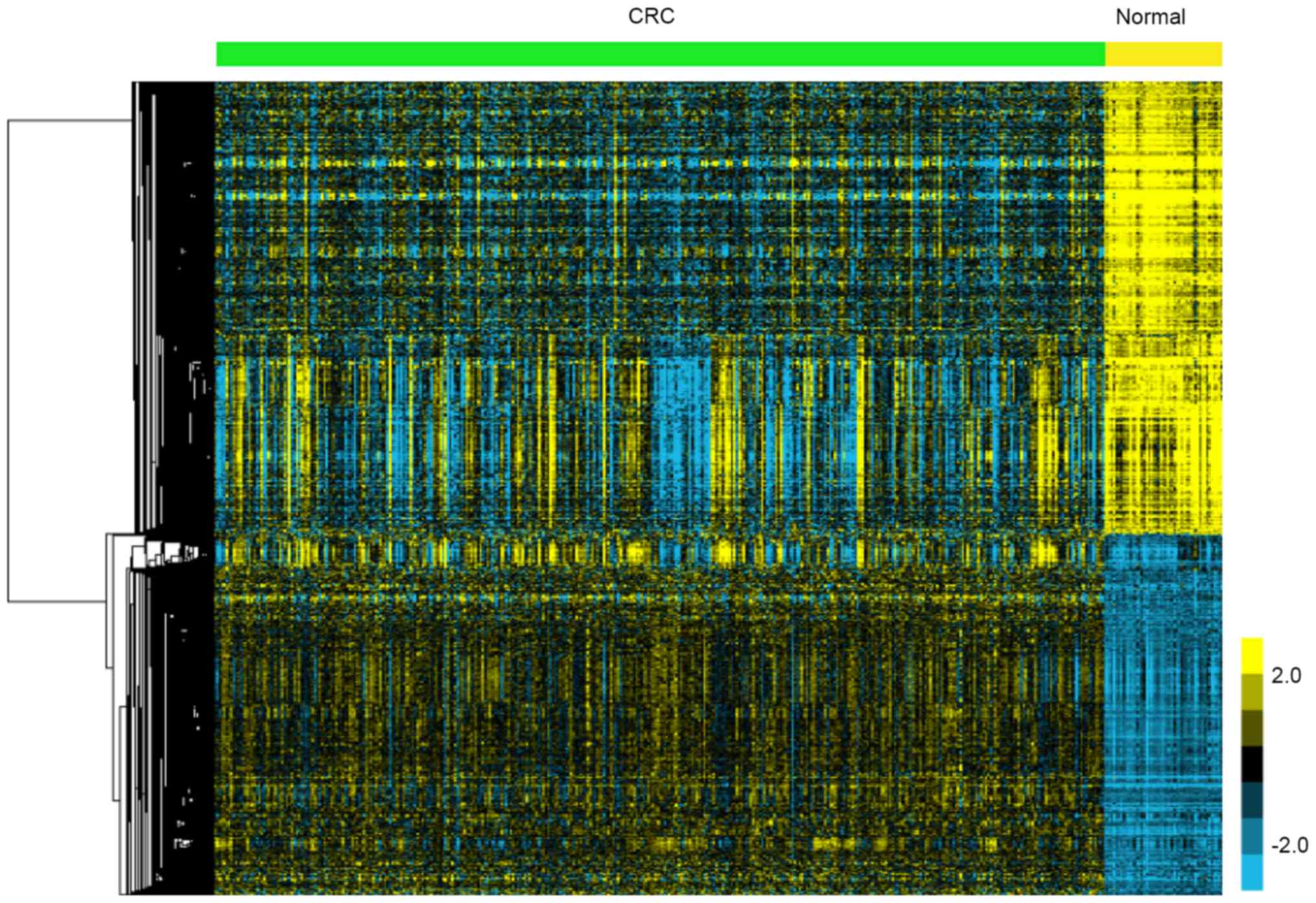
A total of 811 upregulated and 1,024
downregulated genes were identified in CRC
In the present study, TCGA data analysis showed
1,835 genes were identified to be DEGs, including 811 upregulated
and 1,024 downregulated genes in CRC compared to adjacent non-tumor
tissues. The heatmap of DEGs is presented in Fig. 1.
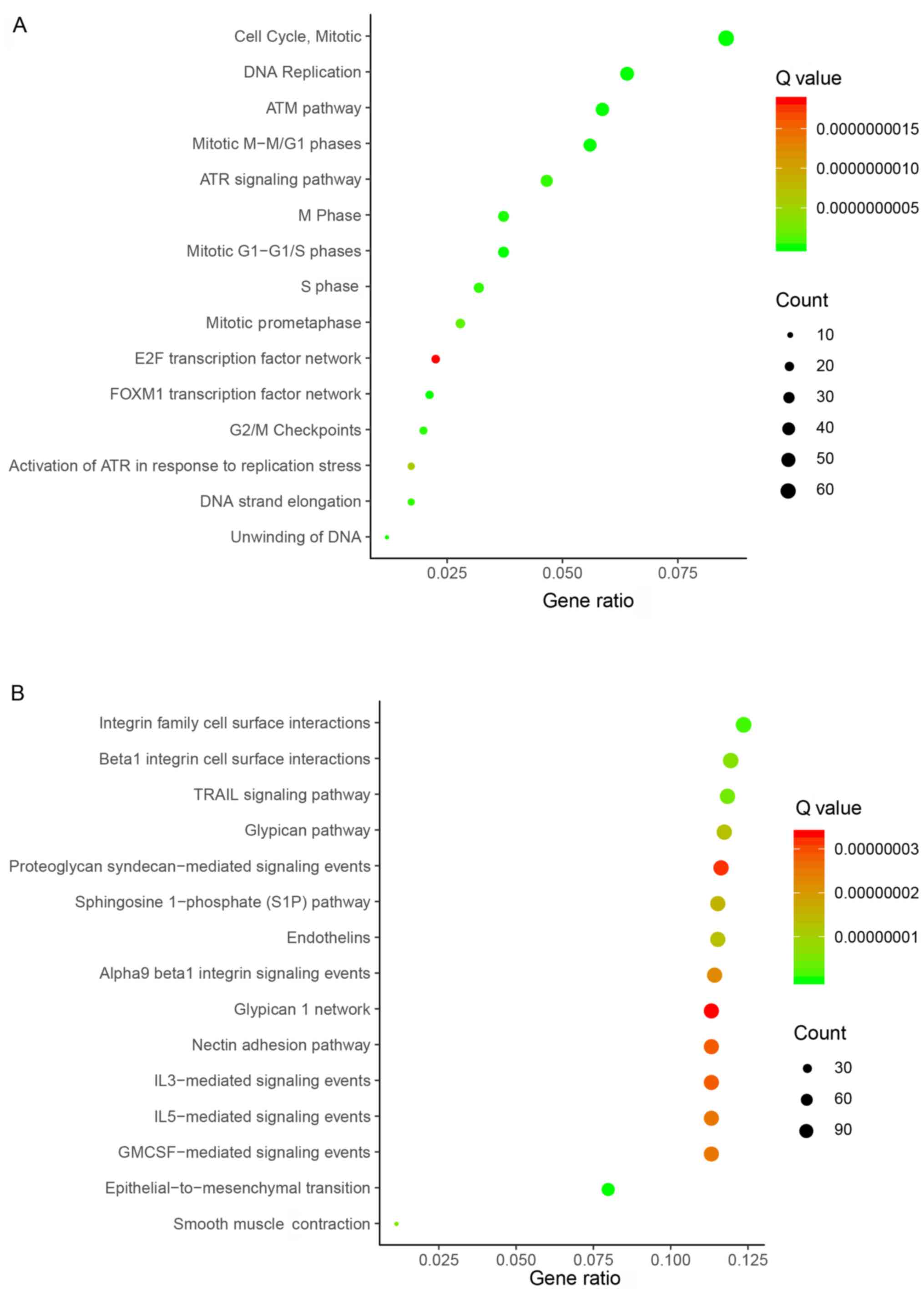
Enrichment analyses of upregulated and
downregulated genes in CRC
To determine the function of the identified DEGs in
CRC, gene enrichment analysis was performed using DAVID. Enrichment
analyses of upregulated and downregulated genes were performed
separately. By subjecting the upregulated genes to enrichment
analysis, numerous enriched gene sets were observed. Based on
bioinformatics analysis, the top 15 biological processes related to
upregulated genes included ‘Cell Cycle, Mitotic’, ‘DNA
Replication’, ‘ATM pathway’, ‘Mitotic M-M/G1 phases’, ‘ATR
signaling pathway’, ‘M Phase’, ‘Mitotic G1-G1/S phases’, ‘S Phase’,
‘Mitotic Prometaphase’, ‘E2F transcription factor network’ ‘FOXM1
transcription factor network’, ‘G2/M Checkpoints’, ‘Activation of
ATR in response to replication stress’, ‘DNA strand elongation’ and
‘Unwinding of DNA’ (Fig. 2A).
By subjecting the downregulated genes to enrichment
analysis, numerous enriched gene sets were observed. Based on
bioinformatics analysis, the top 15 biological processes related to
down-regulated genes included ‘integrin family cell surface
interactions’, ‘β1 integrin cell surface interactions’, ‘TRAIL
signaling pathway’, ‘glypican pathway’, ‘proteoglycan
syndecan-mediated signaling events’, ‘Sphingosine 1-phosphate (S1P)
pathway’, ‘endothelins’, ‘α9 β1 integrin signaling events’,
‘glypican 1 network’, ‘nectin adhesion pathway’, ‘IL3-mediated
signaling events’, ‘IL5-mediated signaling events’, ‘GMCSF-mediated
signaling events’, ‘epithelial-to-mesenchymal transition’ and
‘smooth muscle Contraction’ (Fig.
2B).
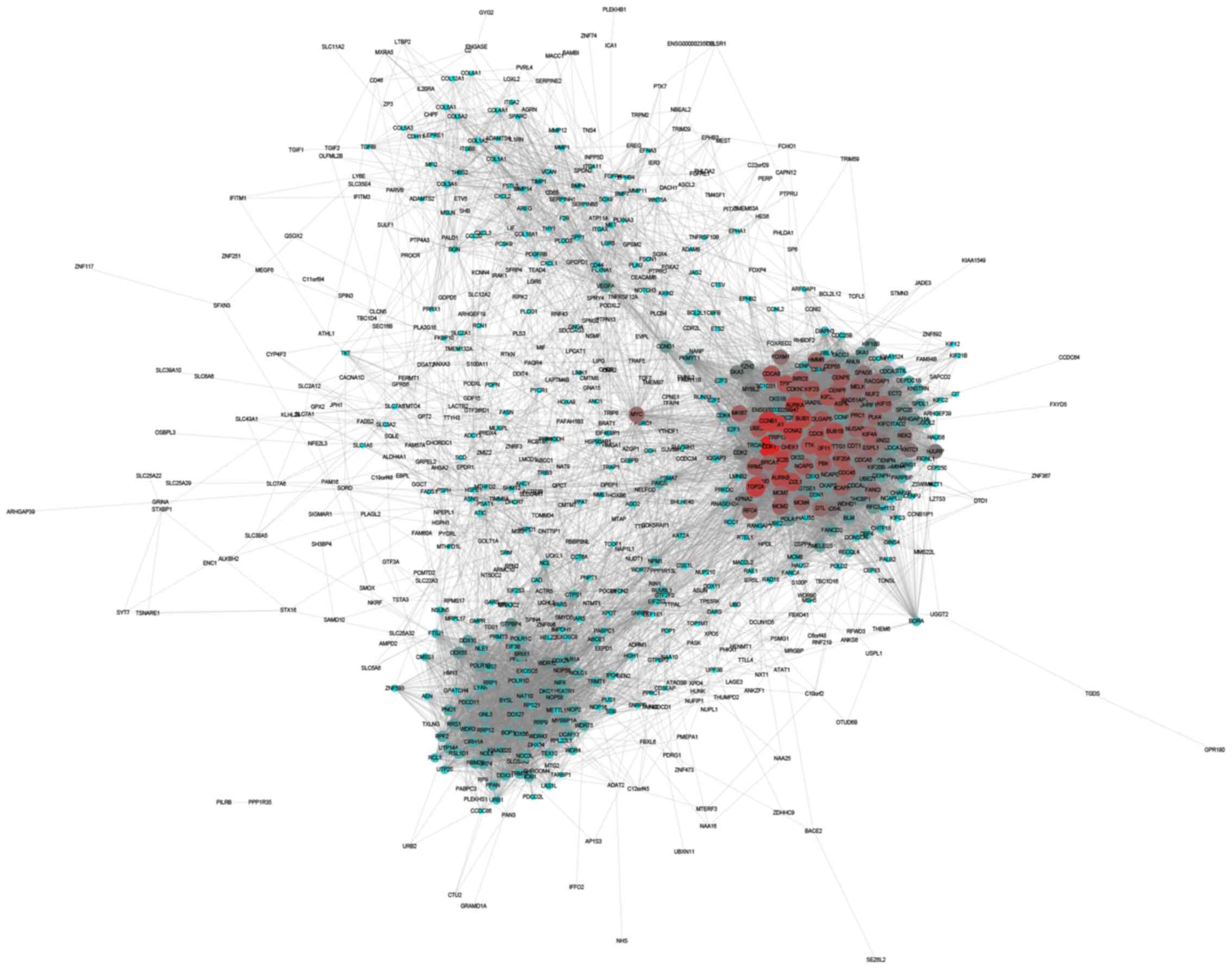
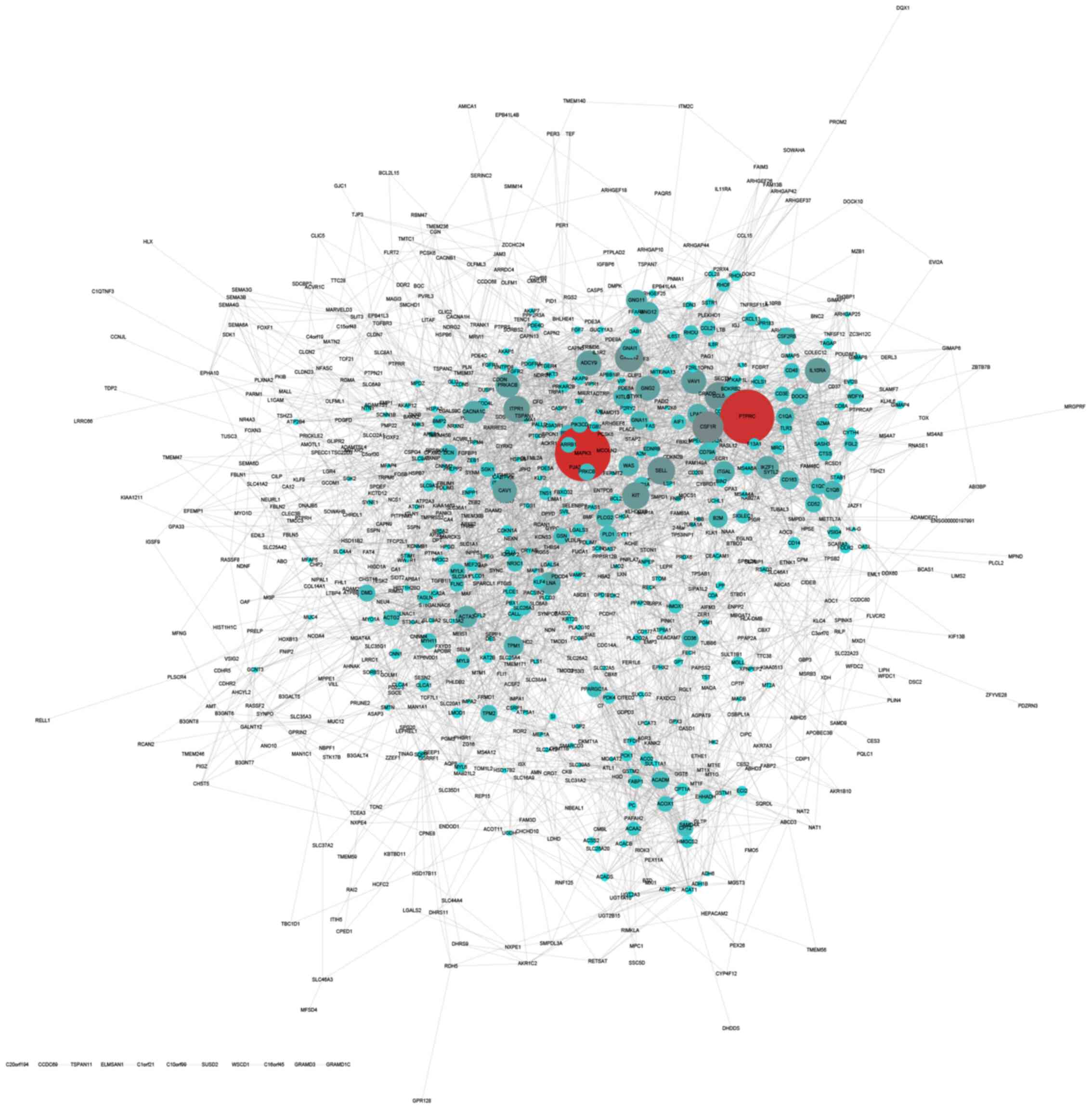
Construction of upregulated and
downregulated PPI networks in CRC
The STRING online database was used to analyze the
interactions among the DEGs. The results were extracted and
visualized using Cytoscape software. After excluding the isolated
nodes, the final upregulated PPI network was composed of 649 nodes
and 11,747 edges (Fig. 3). The
downregulated PPI network was composed of 771 nodes and 4,232 edges
(Fig. 4).
Identification of hub upregulated and
downregulated genes in CRC
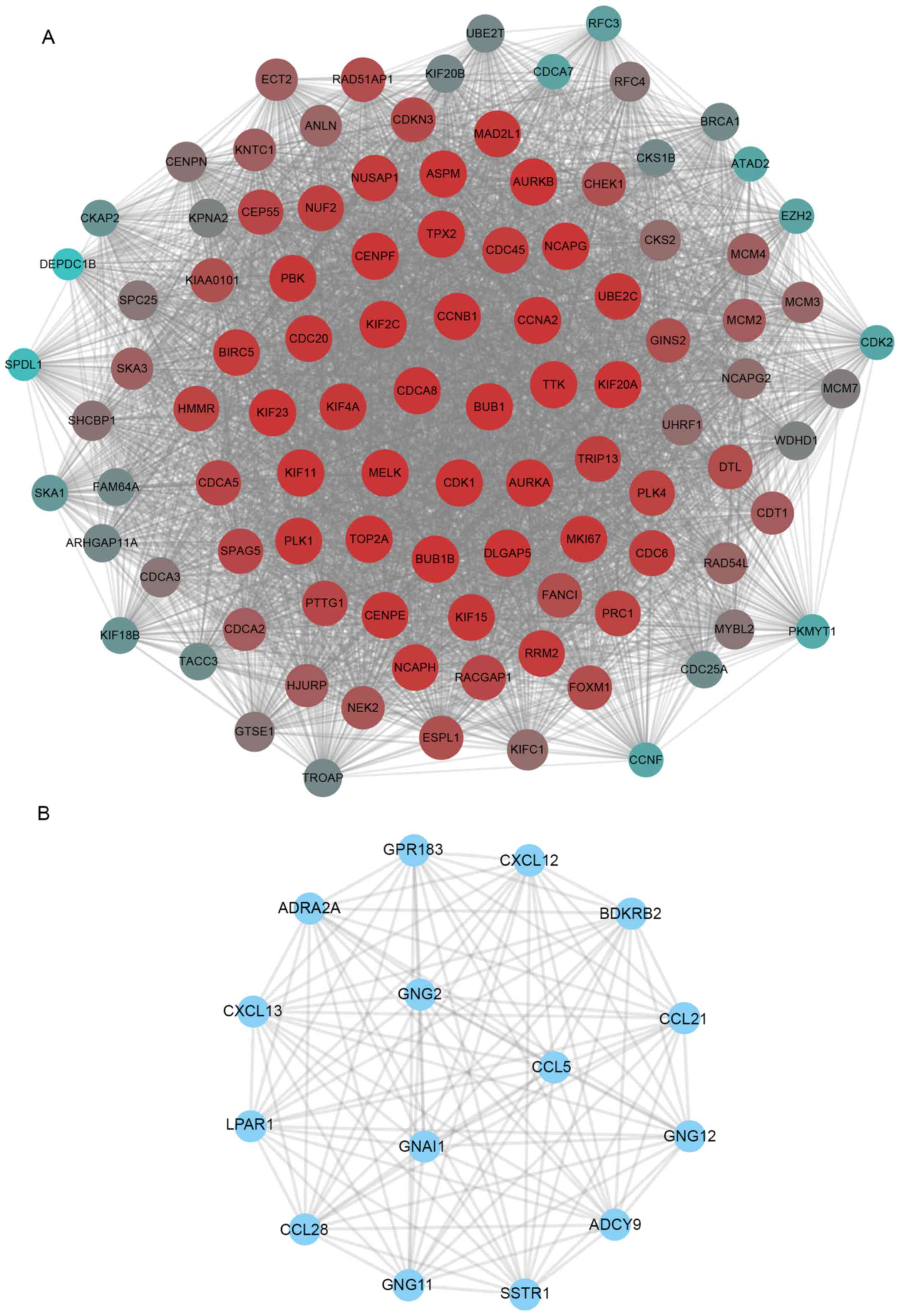
A significant densely connected module was
identified using the MCODE plug-in. The hub upregulated network
included 101 nodes (Fig. 5A).
Notably, we found multiple ubiquitin-conjugating enzyme E family
members served a key role in this network, including UBE2T and
UBE2C (Fig. 5A). The bioinformatics
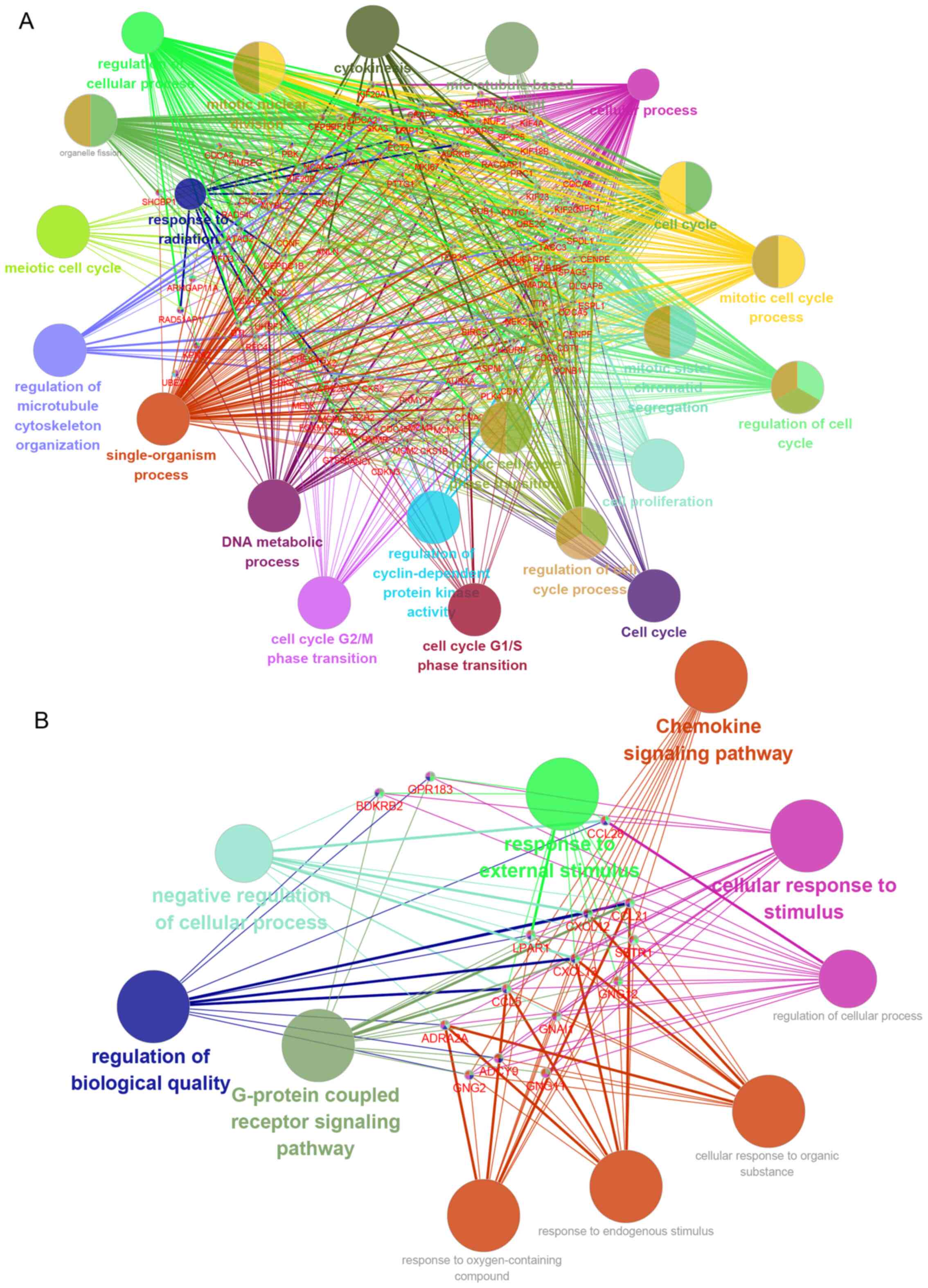
analysis demonstrated that this hub network was associated with
‘DNA metabolic process’, ‘cell cycle G2/M phase
transition’, ‘cellular process’, ‘cell cycle’, ‘regulation of
cyclin-dependent protein kinase activity’, ‘cell cycle
G1/S phase transition’, ‘mitotic cell cycle phase
transition’, ‘regulation of cell cycle’, ‘mitotic sister chromatid
segregation’, ‘cell proliferation’, ‘regulation of cell cycle
process’, ‘cell cycle’, ‘mitotic cell cycle process’,
‘microtubule-based movement’, ‘cytokinesis’, ‘response to
radiation’, ‘single-organism process’, ‘mitotic nuclear division’,
‘regulation of cellular process’, ‘meiotic cell cycle’ and
‘regulation of microtubule cytoskeleton organization’ (Fig. 6A).
The hub downregulated network included 15 nodes
(Fig. 5B). The bioinformatics
analysis demonstrated that this hub network was associated with
‘Chemokine signaling pathway’, ‘response to external stimulus’,
‘G-protein coupled receptor signaling pathway’, ‘cellular response
to stimulus’, ‘regulation of biological quality’ and ‘negative
regulation of cellular processes’ (Fig.
6B).
Higher UBE2T expression in CRC samples
is associated with shorter survival time
The present study focused on UBE2T, which had been
reported to be a key role in the DNA damage repair pathway via
FANCD2 monoubiquitination (22,23). To
the best of our knowledge, the molecular functions of UBE2T in CRC
remain unclear. In aforementioned bioinformatics analysis, it was
reported that UBE2T is a key regulator in hub network 1 (Fig. 5A) and involved in regulating cell
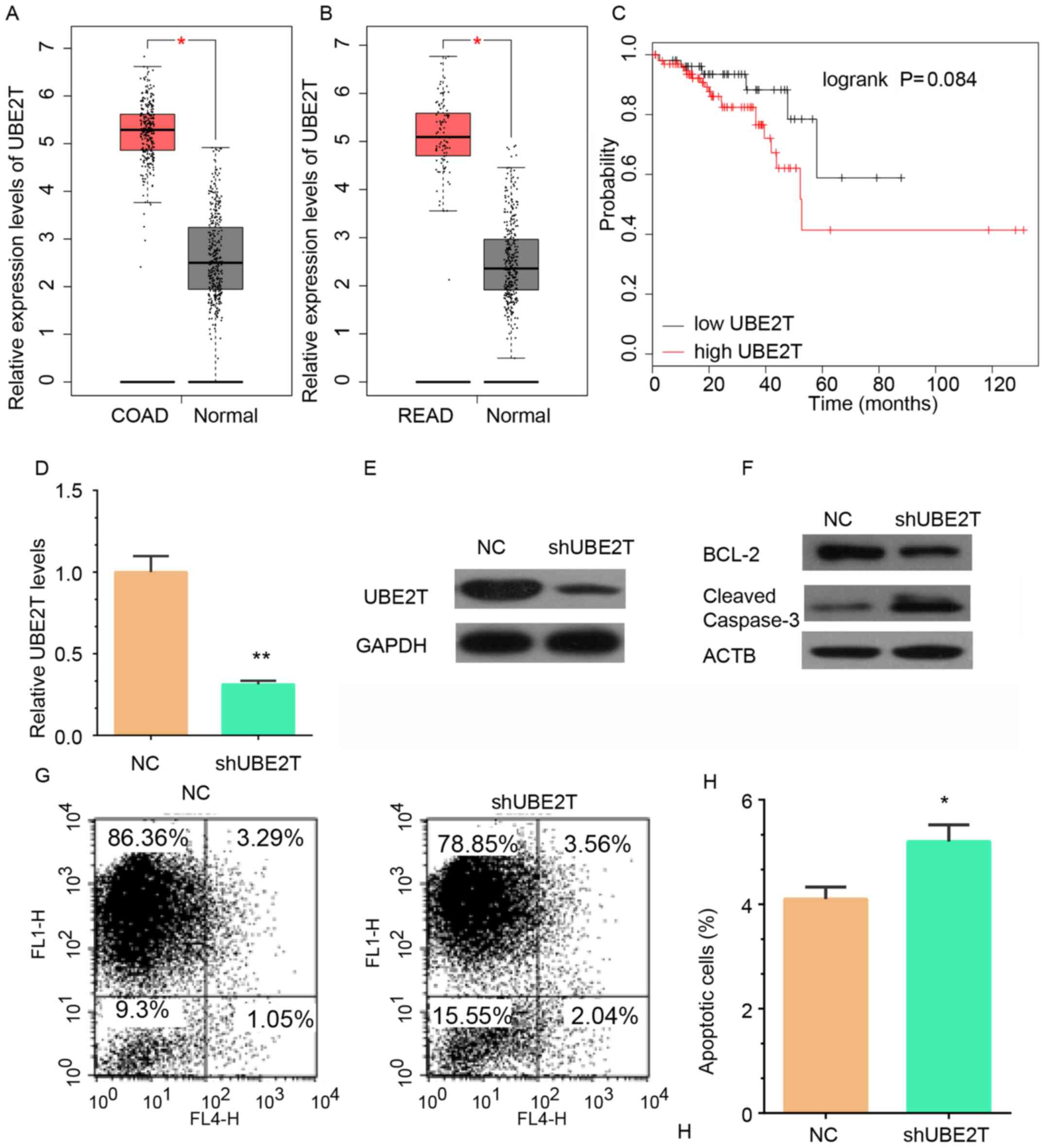
proliferation, cell cycle and DNA metabolic process (Fig. 6A). TCGA analysis revealed that UBE2T
RNA expression was upregulated in colon adenocarcinoma (COAD)
(Fig. 7A) and rectum adenocarcinoma
(READ) (Fig. 7B) samples compared
with adjacent non-tumor tissues. Kaplan-Meier analysis revealed
that high UBE2T expression was associated with shorter overall
survival time in CRC, despite the difference is not significant
enough (P>0.05; Fig. 7C).
Knockdown of UBE2T inhibits CRC cell
proliferation
To investigate the molecular functions of UBE2T in
CRC, lentivirus-mediated knockdown of UBE2T in RKO cells was
performed. RT-qPCR and western blotting demonstrated that both the
RNA and protein expression levels of UBE2T were reduced in RKO
cells infected with shUBE2T lentivirus compared with cells infected
with control lentivirus (Fig. 7D and
E). Furthermore, western blot analysis of Bcl-2 and cleaved
caspase 3 and an Annexin V/FACS kit were used to detect cell
apoptosis after knockdown of UBE2T in RKO cells. UBE2T knockdown
induced a reduction in Bcl-2 expression and an increase in cleaved
caspase 3 protein expression in RKO cells (Fig. 7F). The percentage of apoptotic RKO
cells was significantly increased in the UBE2T knockdown group
compared with the control group (Fig. 7G
and H). These results demonstrated that UBE2T was associated
with the regulation of apoptosis in CRC.
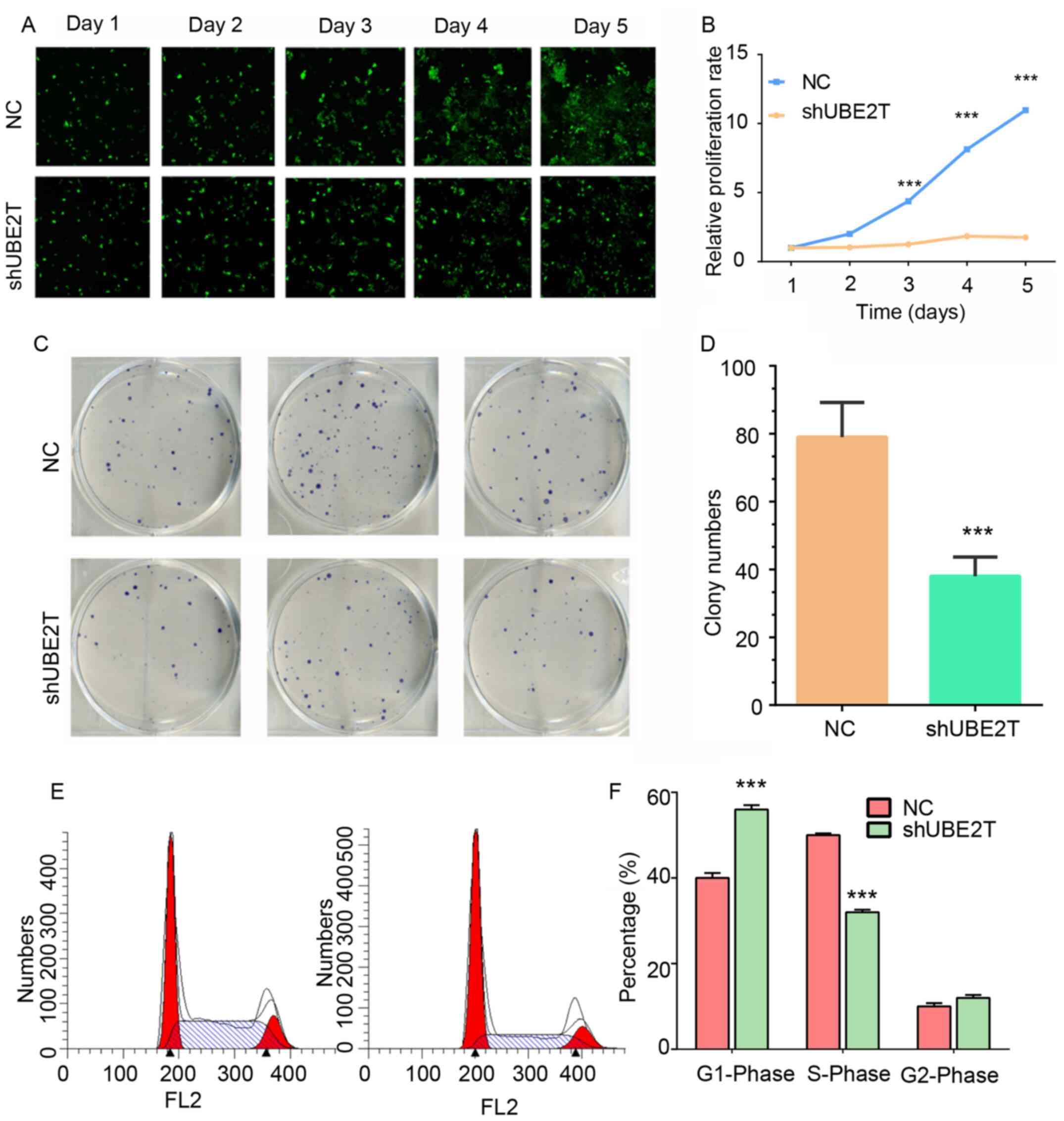
To detect the effects of UBE2T knockdown on
proliferation in RKO cells, a Celigo® assay was used to
monitor cell proliferation for 5 days (Fig. 8A and B). Knockdown of UBE2T exerted a
significant impairment on cell proliferation from day 3 compared
with the normal group (Fig. 8A and
B).
Knockdown of UBE2T induces cell cycle
arrest
A colony formation assay was performed to validate
the role of UBE2T in the regulation of CRC cell proliferation. The
results demonstrated that UBE2T knockdown in RKO cells induced a
decrease in colony formation compared with the control group
(Fig. 8C and D). The number of
colonies in the UBE2T knockdown group was reduced by ~50% compared
with the control group (Fig. 8C and
D).
The abnormal regulation of the cell cycle and
apoptosis contributes to proliferation in cancer cells (24). To investigate the roles of UBE2T in
the cell cycle and apoptosis, a series of flow cytometry assays was
performed. Using a PI/FACS kit, it was revealed that the percentage
of cells in the S phase was significantly reduced in UBE2T
knockdown RKO cells compared with the control group. However, the
percentage of cells in the G1 phase was significantly
increased in UBE2T knockdown RKO cells compared with the control
group, suggesting that the UBE2T gene was associated with cell
cycle regulation (Fig. 8E and
F).
Discussion
Identification of key regulators that contribute to
cancer progression is critical for cancer diagnosis and treatment.
In the present study, 1,835 genes were identified to be DEGs,
including 811 upregulated and 1,024 downregulated genes.
Bioinformatics analysis revealed that these DEGs were associated
with the regulation of cell proliferation of CRC by modulating
multiple pathways, including mitotic nuclear division, response to
radiation, Cell cycle, mitotic cell cycle process, mitotic sister
chromatid segregation, cell proliferation and DNA metabolic
process.
To identify hub genes in CRC, PPI networks were
constructed and two hub networks were revealed. Among them, the
upregulated hub network included 101 nodes and the downregulated
hub network included 15 nodes. Most of these genes have been
reported to serve a crucial role in human CRC (25–28). For
example, high serine/threonine-protein kinase PLK4 expression
promotes tumor progression and induces epithelial-mesenchymal
transition by regulating the Wnt/β-catenin signaling pathway in CRC
(25). Serine/threonine-protein
kinase PLK1 has tumor-suppressive potential in adenomatous
polyposis coli-truncated colon cancer cells (26). Cell division cycle-associated protein
3 mediates p21-dependent proliferation by regulating E2F
transcription factor 1 expression in CRC (27). Mitotic checkpoint kinase monopolar
spindle 1/TTK protein kinase is associated with the prognosis of
patients with colon cancer and regulates tumor cell proliferation
and differentiation via the protein kinase C α/ERK1/2 and PI3K/Akt
signaling pathways (29). PDZ
binding kinase/T-lymphokine-activated killer cell-originated
protein kinase interacts with the DNA-binding domain of tumor
suppressor p53 and modulates the expression of transcriptional
targets, including p21 (30).
Notably, the present study identified that multiple
members of ubiquitin-conjugating enzyme E family served a key role
in CRC, which have been reported to modulate the progression of
human cancer, such as UBE2T (11)
and UBE2C (31). In previous
reports, ubiquitin-conjugating enzyme E family members have been
identified to regulate cancer development (32,33). For
instance, UBE2C expression is higher in late-stage tumors compared
with in early-stage tumors (32).
Ubiquitin-conjugating enzyme UBE2O has been reported to regulate
the cellular clock function by promoting the degradation of the
transcription factor aryl hydrocarbon receptor nuclear
translocator-like protein 1 (33).
UBE2T is a member of the ubiquitinconjugating enzyme family. The
ubiquitin-proteasome signaling pathway serves a key role in cell
proliferation and DNA damage repair (28,34,35).
UBE2T expression has been reported to be upregulated in multiple
types of tumors, such as prostate cancer and lung cancer (6,8,9,11,12,36,37).
However, to the best of our knowledge, the association between
UBE2T and CRC has not been investigated. Furthermore, to the best
of our knowledge, the present study was the first to demonstrate
that UBE2T expression was upregulated in COAD and READ samples
compared with normal tissues. Kaplan-Meier analysis revealed that
higher levels of UBE2T were associated with worse prognosis
compared with low UBE2T expression levels in CRC. Additionally, the
present study revealed that knockdown of UBE2T inhibited CRC cell
proliferation. Flow cytometry assays revealed that UBE2T knockdown
induced cell cycle arrest and apoptosis in vitro.
In summary, the present study screened DEGs in CRC.
Further validation demonstrated that UBE2T expression was
upregulated in CRC and knockdown of UBE2T inhibited the
proliferation of CRC cells by inducing cell cycle arrest and
apoptosis. These results indicated that UBE2T may be a novel
potential biomarker for CRC.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author upon reasonable
request.
Authors' contributions
ML and YZ designed the study and drafted the initial
manuscript. ML performed the experiments. YZ analyzed the data. ML
and YZ confirmed the authenticity of all the raw data. All authors
read and approved the final version of the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Rawla P, Sunkara T and Barsouk A:
Epidemiology of colorectal cancer: Incidence, mortality, survival,
and risk factors. Prz Gastroenterol. 14:89–103. 2019.PubMed/NCBI
|
|
2
|
Siegel RL, Miller KD, Fedewa SA, Ahnen DJ,
Meester RGS, Barzi A and Jemal A: Colorectal cancer statistics,
2017. CA Cancer J Clin. 67:177–193. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Morreale FE, Bortoluzzi A, Chaugule VK,
Arkinson C, Walden H and Ciulli A: Allosteric targeting of the
fanconi anemia ubiquitin-conjugating enzyme Ube2T by fragment
screening. J Med Chem. 60:4093–4098. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Alpi A, Langevin F, Mosedale G, Machida
YJ, Dutta A and Patel KJ: UBE2T, the Fanconi anemia core complex,
and FANCD2 are recruited independently to chromatin: A basis for
the regulation of FANCD2 monoubiquitination. Mol Cell Biol.
27:8421–8430. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Liu LL, Zhu JM, Yu XN, Zhu HR, Shi X,
Bilegsaikhan E, Guo HY, Wu J and Shen XZ: UBE2T promotes
proliferation via G2/M checkpoint in hepatocellular carcinoma.
Cancer Manag Res. 11:8359–8370. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Alpi AF, Pace PE, Babu MM and Patel KJ:
Mechanistic insight into site-restricted monoubiquitination of
FANCD2 by Ube2t, FANCL, and FANCI. Mol Cell. 32:767–777. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hao J, Xu A, Xie X, Hao J, Tian T, Gao S,
Xiao X and He D: Elevated expression of UBE2T in lung cancer tumors
and cell lines. Tumour Biol. 29:195–203. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hao P, Kang B, Li Y, Hao W and Ma F: UBE2T
promotes proliferation and regulates PI3K/Akt signaling in renal
cell carcinoma. Mol Med Rep. 20:1212–1220. 2019.PubMed/NCBI
|
|
10
|
Liu LP, Yang M, Peng QZ, Li MY, Zhang YS,
Guo YH, Chen Y and Bao SY: UBE2T promotes hepatocellular carcinoma
cell growth via ubiquitination of p53. Biochem Biophys Res Commun.
493:20–27. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Luo C, Yao Y, Yu Z, Zhou H, Guo L, Zhang
J, Cao H, Zhang G, Li Y and Jiao Z: UBE2T knockdown inhibits
gastric cancer progression. Oncotarget. 8:32639–32654. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wen M, Kwon Y, Wang Y, Mao JH and Wei G:
Elevated expression of UBE2T exhibits oncogenic properties in human
prostate cancer. Oncotarget. 6:25226–25239. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Huang da W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Huang DW, Sherman BT, Tan Q, Kir J, Liu D,
Bryant D, Guo Y, Stephens R, Baseler MW, Lane HC and Lempicki RA:
DAVID bioinformatics resources: Expanded annotation database and
novel algorithms to better extract biology from large gene lists.
Nucleic Acids Res. 35:W169–W175. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Szklarczyk D, Franceschini A, Wyder S,
Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos
A, Tsafou KP, et al: STRING v10: Protein-protein interaction
networks, integrated over the tree of life. Nucleic Acids Res.
43:D447–D452. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Bader GD and Hogue CW: An automated method
for finding molecular complexes in large protein interaction
networks. BMC Bioinformatics. 4:22003. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Shi J, Zhong X, Song Y, Wu Z, Gao P, Zhao
J, Sun J, Wang J, Liu J and Wang Z: Long non-coding RNA RUNX1-IT1
plays a tumour-suppressive role in colorectal cancer by inhibiting
cell proliferation and migration. Cell Biochem Funct. 37:11–20.
2019. View
Article : Google Scholar : PubMed/NCBI
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chuang KC, Chen FW, Tsai MH and Shieh JJ:
EGR-1 plays a protective role in AMPK inhibitor compound C-induced
apoptosis through ROS-induced ERK activation in skin cancer cells.
Oncol Lett. 21:3042021. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Cui F, Hu J, Ning S, Tan J and Tang H:
Overexpression of MCM10 promotes cell proliferation and predicts
poor prognosis in prostate cancer. Prostate. 78:1299–1310. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kelsall IR, Langenick J, MacKay C, Patel
KJ and Alpi AF: The Fanconi anaemia components UBE2T and FANCM are
functionally linked to nucleotide excision repair. PLoS One.
7:e369702012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Rickman KA, Lach FP, Abhyankar A, Donovan
FX, Sanborn EM, Kennedy JA, Sougnez C, Gabriel SB, Elemento O,
Chandrasekharappa SC, et al: Deficiency of UBE2T, the E2 ubiquitin
ligase necessary for FANCD2 and FANCI ubiquitination, causes FA-T
subtype of fanconi anemia. Cell Rep. 12:35–41. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Seoane J: Cancer: Division hierarchy leads
to cell heterogeneity. Nature. 549:164–166. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Liao Z, Zhang H, Fan P, Huang Q, Dong K,
Qi Y, Song J, Chen L, Liang H, Chen X, et al: High PLK4 expression
promotes tumor progression and induces epithelial-mesenchymal
transition by regulating the Wnt/β-catenin signaling pathway in
colorectal cancer. Int J Oncol. 54:479–490. 2019.PubMed/NCBI
|
|
26
|
Raab M, Sanhaji M, Matthess Y, Hörlin A,
Lorenz I, Dötsch C, Habbe N, Waidmann O, Kurunci-Csacsko E,
Firestein R, et al: PLK1 has tumor-suppressive potential in
APC-truncated colon cancer cells. Nat Commun. 9:11062018.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Qian W, Zhang Z, Peng W, Li J, Gu Q, Ji D,
Wang Q, Zhang Y, Ji B, Wang S, et al: CDCA3 mediates p21-dependent
proliferation by regulating E2F1 expression in colorectal cancer.
Int J Oncol. 53:2021–2033. 2018.PubMed/NCBI
|
|
28
|
Nakayama KI and Nakayama K: Ubiquitin
ligases: Cell-cycle control and cancer. Nat Rev Cancer. 6:369–381.
2006. View
Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhang L, Jiang B, Zhu N, Tao M, Jun Y,
Chen X, Wang Q and Luo C: Mitotic checkpoint kinase Mps1/TTK
predicts prognosis of colon cancer patients and regulates tumor
proliferation and differentiation via PKCα/ERK1/2 and PI3K/Akt
pathway. Med Oncol. 37:52019. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Hu F, Gartenhaus RB, Eichberg D, Liu Z,
Fang HB and Rapoport AP: PBK/TOPK interacts with the DBD domain of
tumor suppressor p53 and modulates expression of transcriptional
targets including p21. Oncogene. 29:5464–5474. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhang HQ, Zhao G, Ke B, Ma G, Liu GL,
Liang H, Liu LR and Hao XS: Overexpression of UBE2C correlates with
poor prognosis in gastric cancer patients. Eur Rev Med Pharmacol
Sci. 22:1665–1671. 2018.PubMed/NCBI
|
|
32
|
Dastsooz H, Cereda M, Donna D and Oliviero
S: A comprehensive bioinformatics analysis of UBE2C in cancers. Int
J Mol Sci. 20:22282019. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Chen S, Yang J, Zhang Y, Duan C, Liu Q,
Huang Z, Xu Y, Zhou L and Xu G: Ubiquitin-conjugating enzyme UBE2O
regulates cellular clock function by promoting the degradation of
the transcription factor BMAL1. J Biol Chem. 293:11296–11309. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Voutsadakis IA: Ubiquitin- and
ubiquitin-like proteins-conjugating enzymes (E2s) in breast cancer.
Mol Biol Rep. 40:2019–2034. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Hoeller D, Hecker CM and Dikic I:
Ubiquitin and ubiquitin-like proteins in cancer pathogenesis. Nat
Rev Cancer. 6:776–788. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhang W, Zhang Y, Yang Z, Liu X, Yang P,
Wang J, Hu K, He X, Zhang X and Jing H: High expression of UBE2T
predicts poor prognosis and survival in multiple myeloma. Cancer
Gene Ther. 26:347–355. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Gong YQ, Peng D, Ning XH, Yang XY, Li XS,
Zhou LQ and Guo YL: UBE2T silencing suppresses proliferation and
induces cell cycle arrest and apoptosis in bladder cancer cells.
Oncol Lett. 12:4485–4492. 2016. View Article : Google Scholar : PubMed/NCBI
|