Introduction
T-cell lymphoma accounts for 10–15% of non-Hodgkin's
lymphomas (1). In 2008, the World
Health Organization classified T-cell lymphoma into different
pathological subtypes: T-cell and natural killer (NK) cell
lymphoma/leukemia, which originate from lymph nodes, extranodal
tissues or skin (2,3). The prognosis of mature or peripheral
T-cell lymphoma is worse compared with that of aggressive B-cell
lymphoma (4,5). Therefore, the key to improving the
prognosis of T-cell lymphoma is to determine the biological
characteristics of T-cell lymphoma cells.
As a core subunit of the nucleosome-remodeling
factor (NURF) complex, bromodomain PHD finger transcription factor
(BPTF) serves crucial roles in chromatin remodeling (6). BPTF is critical for epigenetic
regulation of DNA accessibility and gene expression (7). Recently, its function in tumor
progression has attracted increased attention (8). BPTF has recently been found to
influence the course of cancer, particularly by directly activating
oncogenic signaling or through synergistic interactions with other
key protein factors (9,10). To date, there is no relevant research
report of BPTF in T-cell lymphoma, and thus the present study aimed
to explore its regulatory mechanism and biological function in
T-cell lymphoma.
Abnormal activation of the MAPK signaling pathway
has an important role in cell malignant transformation and
evolution (11). Multiple reports
have demonstrated that MAPKs are significantly associated with the
occurrence and development of breast cancer, ovarian cancer,
esophageal cancer, colon cancer, gastric cancer, liver cancer and
other tumors (12,13). The present study demonstrated that
BPTF was highly expressed in cell lines and tissues of T-cell
lymphoma. In addition, it was observed that BPTF and Raf1 were
coexpressed in T-cell lymphoma cells. Silencing BPTF inhibited the
activation of the MAPK pathway. Raf1 was also demonstrated to be
highly expressed in cell lines and tissues of T-cell lymphoma.
Silencing BPTF or Raf1 induced apoptosis in T-cell lymphoma cells.
In summary, the current results suggested that BPTF promoted the
proliferation of T-cell lymphoma by activating the MAPK pathway.
BPTF may serve as a molecular target for the treatment of T-cell
lymphoma.
Materials and methods
Patients and tissue specimens
In the present study, 30 human cancerous lymph node
tissues and matched adjacent nontumor tissues were obtained from
patients who underwent lymph node resection between November 2016
and December 2019 in the Department of Pathology, the First
Affiliated Hospital of Xiamen University (Xiamen, China). The
overall experimental scheme was approved by the Ethics Committee of
the First Affiliated Hospital of Xiamen University (approval no.
KY-2018-014). All patients had signed informed consent. The
clinical features of the patients, including age, sex, lymph node
metastasis status, lactate dehydrogenase (LDH) levels and clinical
stage, were collected from their medical records and listed in
Table I.
 | Table I.Clinicopathological features of 30
patients with T-cell lymphoma. |
Table I.
Clinicopathological features of 30
patients with T-cell lymphoma.
| Clinicopathological
feature | Samples (n=30) | P-value |
|---|
| Age (years) |
|
|
| ≤60 | 21 | 0.014 |
|
>60 | 9 |
|
| Sex |
|
|
|
Female | 12 | 0.278 |
| Male | 18 |
|
| Nodes |
|
|
|
Intranodal | 23 | 0.021 |
|
Extranodal | 7 |
|
| Serum LDH levels |
|
|
|
Normal | 6 | 0.035 |
| Above
normal | 24 |
|
| Clinical stages |
|
|
| I–II | 23 | 0.041 |
|
III–IV | 7 |
|
Cell lines and culture
The human T cell line (H9) and human T-cell lymphoma
cell line (Hut-102) were purchased from the Cell Resource Center,
Shanghai Institute of Life Sciences, Chinese Academy of Sciences
(Shanghai, China). They were cultured in 1640 medium (cat. no.
22400121; Thermo Fisher Scientific, Inc.) containing 10% fetal
bovine serum (cat. no. 10099141C; Thermo Fisher Scientific, Inc.)
and penicillin-streptomycin (cat. no. C0222; Beyotime Institute of
Biotechnology) at 37°C and 5% CO2.
Mission short hairpin (sh) RNA series (cat. no.
CSTVRS; Sigma-Aldrich; Merck KGaA) vectors were used for RNA
silencing. shRNA probes TRCN0000016819 and TRCN0000001066 were used
to silence BPTF and Raf1, respectively. A nontargeting shRNA (cat.
no. SHC312V; Sigma-Aldrich; Merck KGaA) was used as the negative
control (NC). PCMV3 (cat. no. NM_005228.3; SinoBiological, Inc.)
was used as the overexpression vector to construct PCMV3-BPTF
recombinant plasmid; blank vector was used as the control. The
constructs were transfected into Hut-102 cells using Lipofectamine
3000 transfection reagent (cat. no. L3000001; Thermo Fisher
Scientific, Inc.). Hut-102 cell lines stably transfected with NC,
shBPTF or shRaf1 were constructed.
Tumor xenograft model
Four-week-old female BALB/cA nude mice (n=30) were
purchased from the Shanghai Experimental Animal Center, Chinese
Academy of Sciences. The study protocol was approved by the
Experimental Animal Ethics Committee of the First Affiliated
Hospital of Xiamen University (Xiamen, China; approval no.
2019-231). During the experiment, animal handling and care were
carried out according to the National Institutes of Health Guide
for the Care and Use of Laboratory Animals (NIH Publications no.
8023, revised 1978). Hut-102 cells stably transfected with
nontargeting shRNA or shBPTF were inoculated on the backs of nude
mice (3×106 cells per mouse) after light anesthesia
using 37.5 mg/kg pelltobarbitalum natricum. The total experimental
period was three weeks. The behavior and health of the nude mice
were observed every day and the experiment was terminated in time
for abnormal individuals. The maximum diameter of the tumor was
0.71 cm. The tumors were removed using resection, photographed
using a camera (model E-PL9; Olympus Corporation) and weighed after
all the mice were sacrificed using intraperitoneal injection of 200
mg/kg pelltobarbitalum natricum. Before euthanasia, carprofen (cat.
no. V1074; InvivoChem Co., Ltd.; 5 mg/kg) was injected
subcutaneously to relieve the pain of mice (14,15). The
tumor volume was calculated using the following formula: V =
(length × width2)/2.
MTT assay
Adherent cells (Hut-102 cell lines stably expressing
NC, shBPTF or shRaf1) were cultured in 96-well plates for 24 h at a
density of 5,000 cells per well. The original medium was discarded,
and 100 µl serum-free DMEM and 10 µl (5 mg/ml) MTT (cat. no. 88417;
Sigma-Aldrich; Merck KGaA) were added to each well. After 4 h, the
reaction medium was discarded, and the formazan crystals were fully
dissolved in 100 µl DMSO. The absorbance was measured at 570 nm
using a microplate reader (Multiskan FC; Thermo Fisher Scientific,
Inc.). Each experiment was repeated three times.
Clone formation assay
Adherent cells (Hut-102 cell lines stably expressing
NC, shBPTF or shRaf1) were cultured in 6-well plates for ~2 weeks
at a density of 200 cells per well. When the clone number of a
single group reached >70, the culture medium was discarded, and
the cells were fixed using methanol for 30 min. The fixed cells
were stained with 0.5% crystal violet and photographed using a
camera (model E-PL9; Olympus Corporation).
Annexin V/propidium iodide (PI)
assay
Cells were digested with trypsin-EDTA solution (cat.
no. T4049; Sigma-Aldrich; Merck KGaA), which was removed by
centrifugation at 168 × g at room temperature for 10 min. Then, the
cells were washed three times with sterile PBS and resuspended in
500 µl Annexin V/PI binding solution (cat. no. APOAF-20TST;
Sigma-Aldrich; Merck KGaA). The cell suspension was stained with 10
µl PI and 5 µl Annexin V-FITC (cat. no. APOAF-20TST; Sigma-Aldrich;
Merck KGaA at room temperature for 30 min. The cells were detected
by flow cytometry (model NOVOCyte 2060R; ACEA Biosciences, Inc.)
using the F1 channel (Annexin V-FITC) and F2 channel (PI).
NovoExpress software v.1.2.4.1602 (ACEA Biosciences, Inc.) was used
to analyze the results of flow cytometry.
Cell cycle distribution assays
Cells were digested with trypsin-EDTA solution (cat.
no. T4049; Sigma-Aldrich; Merck KGaA), which was removed by
centrifugation at 161 × g at room temperature for 10 min. Then, the
cells were washed three times with sterile PBS and fixed by adding
70% ethanol at −20°C overnight. The cell suspension was washed
twice with PBS and stained with 400 µl PI (cat. no. P4864;
Sigma-Aldrich; Merck KGaA) in the dark at 4°C for 30 min. Cell
cycle distribution was detected by flow cytometry using the F2
channel. NovoExpress software v.1.2.4.1602 (ACEA Biosciences, Inc.)
was used to analyze the results of flow cytometry.
Western blot analysis
Total protein from H9 cells, Hut-102 cells (shNC,
shBPTF, shRaf1, blank vector or overexpressed BPTF) or tissues
(normal, T-cell lymphoma or xenograft tumors) was extracted by RIPA
lysis buffer (cat. no. R0010; Beijing Solarbio Science &
Technology Co., Ltd.) on ice for 30 min. Bicinchoninic acid (BCA)
method was used to detect the protein concentration. Proteins (15
µg) of different sizes were separated by SDS-PAGE (10% separating
gel and 5% concentrating gel). The protein in the PAGE gel was
transferred to a polyvinylidene difluoride (PVDF) membrane and
blocked in 5% skim milk at room temperature for 3 h. The PVDF
membrane was incubated with the corresponding primary antibodies at
room temperature for 2 h. After rinsing with Tris-buffered saline
containing 0.05% Tween-20 (TBST) three times (10 min each), the
PVDF membrane was incubated with secondary antibodies at room
temperature for 1 h. The ECL Plus western blotting substrate (cat.
no. 32132; Thermo Fisher Scientific, Inc.) was added to the PVDF
membrane and detected by a multifunctional gel imaging system
(model Gel Doc XR+; Bio-Rad Laboratories, Inc.). The relative
amount of protein was quantitatively analyzed by densitometry using
SageDetect software v.2.1.8.160722 (Beijing Sage Creation Science
Co., Ltd.). Antibodies against the following proteins were used:
BPTF (cat. no. ab72036; 1:2,000; Abcam), Raf1 (cat. no. ab137435;
1:1,000; Abcam), phosphorylated (p-) Raf1 S569 (cat. no. ab173539;
1:1,000; Abcam), MEK1 (cat. no. ab32091; 1:1,000; Abcam),
p-MEK1-S298 (cat. no. ab96379; 1:1,000; Abcam), Erk1/2 (cat. no.
WL01864; 1:500; Wanleibio Co., Ltd.), p-Erk1/2-Thr202/Tyr204 (cat.
no. WLP1512; 1:500; Wanleibio Co., Ltd.), and β-actin (cat. no.
WL01372; 1:3,000; Wanleibio Co., Ltd.). The secondary antibody was
HRP-conjugated goat anti-rabbit immunoglobulin G (1:5,000; cat. no.
A-11006; Thermo Fisher Scientific, Inc.).
RNA isolation and reverse
transcription-quantitative PCR (RT-qPCR)
Total RNA was isolated from the cells and tissues
using a MicroElute Total RNA kit (cat. no. R6831-01; Omega Bio-Tek,
Inc.). The RNA was reverse transcribed into cDNA using the
PrimeScript RT reagent kit (cat. no. RR037A; Takara Biotechnology
Co., Ltd.). The reverse transcription reaction conditions were 37°C
for 15 min and 85°C for 5 sec. Reverse Transcription of RNA was
performed using the PrimeScript RT reagent kit (cat. no. RR037A;
TAKARA Inc.) on PCR instrument (model TC-E-48D; Hangzhou Bioer
Technology Co. Ltd.). RT-qCR was performed using the SYBR Green
qPCR Mix (cat. no. D7260; Beyotime Institute of Biotechnology) on
an Applied Biosystems 7500 Real-Time PCR system (Thermo Fisher
Scientific, Inc.). The qPCR reaction conditions were: 1 cycle of
95°C for 10 sec, followed by 40 cycles of 94°C for 5 sec and 60°C
for 34 sec, and finally 1 cycle of 95°C for 15 sec, 60°C for 1 min
and 95°C for 15 sec. The following primer sequences were used for
qPCR: BPTF, forward, 5′-CCCAGGTGGTGATGAAGCAT-3′ and reverse,
5′-CCCAGGTGGTGATGAAGCAT-3′; Raf1, forward,
5′-CGCTTAGATTGGAATACTGA-3′ and reverse, 5′-AAAGGTGAAGGCGTGAG-3′;
and GAPDH, forward, 5′-ATGACATCAAGAAGGTGGTGAAGCAGG-3′ and reverse,
5′-GCGTCAAAGGTGGAGGAGTGGGT-3′. GAPDH was used as an internal
reference gene. Relative fold changes in mRNA expression were
calculated using the formula 2−ΔΔCq (16).
Immunohistochemistry
The transplanted tumor tissues in nude mice and
clinical tumor samples were fixed with 4% paraformaldehyde at room
temperature for 24 h. The samples were embedded in paraffin, and
sectioned (5-µm thickness). After dewaxing and hydration, the
samples were incubated with 3% hydrogen peroxide at room
temperature for 10 min. The samples were treated with 0.01 mol/l
sodium citrate buffer solution (pH 6.0) at 95°C for 10 min for
antigen retrieval. The samples were incubated in goat serum (cat.
no. C0265; Beyotime Institute of Biotechnology) at room temperature
for 20 min. Then, the samples were incubated with the corresponding
primary antibodies at 4°C overnight. After rinsing with PBS three
times (5 min each), the samples were incubated with secondary
antibodies at 37°C for 1 h. The samples were colored according to
the instructions of the 3,3′ diaminobenzidine color developing kit
(cat. no. P0202; Institute of Biotechnology) and visualized using a
light microscope (model CX41; Olympus Corporation).
TUNEL assay
The transplanted tumor tissues in nude mice were
fixed using 4% paraformaldehyde for 24 h at room temperature. The
fixed tumor tissues were embedded in paraffin and sectioned to 5-µm
thickness. Paraffin tissue was dewaxed with xylene and gradient
ethanol. The samples were treated with 20 µg/ml protease K without
DNase at 37°C for 30 min. The samples were washed twice with PBS
for 10 min each time. TUNEL solution (50 µl) was added to the
sample and incubated at 37°C for 60 min. The samples were washed
twice with PBS for 10 min each time. DAPI (5 µg/ml) (cat. no.
C1002; Beyotime Institute of Biotechnology) was used for nuclear
staining at room temperature for 10 min. The samples were washed
twice with PBS for 10 min each time. After drying, the slides were
sealed with a sealing solution containing an anti-fluorescence
quenching agent. The samples were observed under a fluorescence
microscope (model CKX53; Olympus Corporation) in 10 fields of view
with a ×200 magnification.
Statistical analysis
All experimental data were analyzed by SPSS 21.0
software (IBM Corp.) and expressed as the means ± standard
deviation (SD). Experiments were repeated at least three times.
Unpaired Student's t-test was used for statistical analysis between
two independent samples, while paired Student's t-test was used for
statistical analysis between two paired samples. Bonferroni's
correction was used for one-way ANOVA among multiple groups.
Chi-square test was used to analyze the distribution of clinical
variables (age, sex, nodes, serum LDH levels, clinical stages).
P<0.05 was considered to indicate a statistically significant
difference.
Results
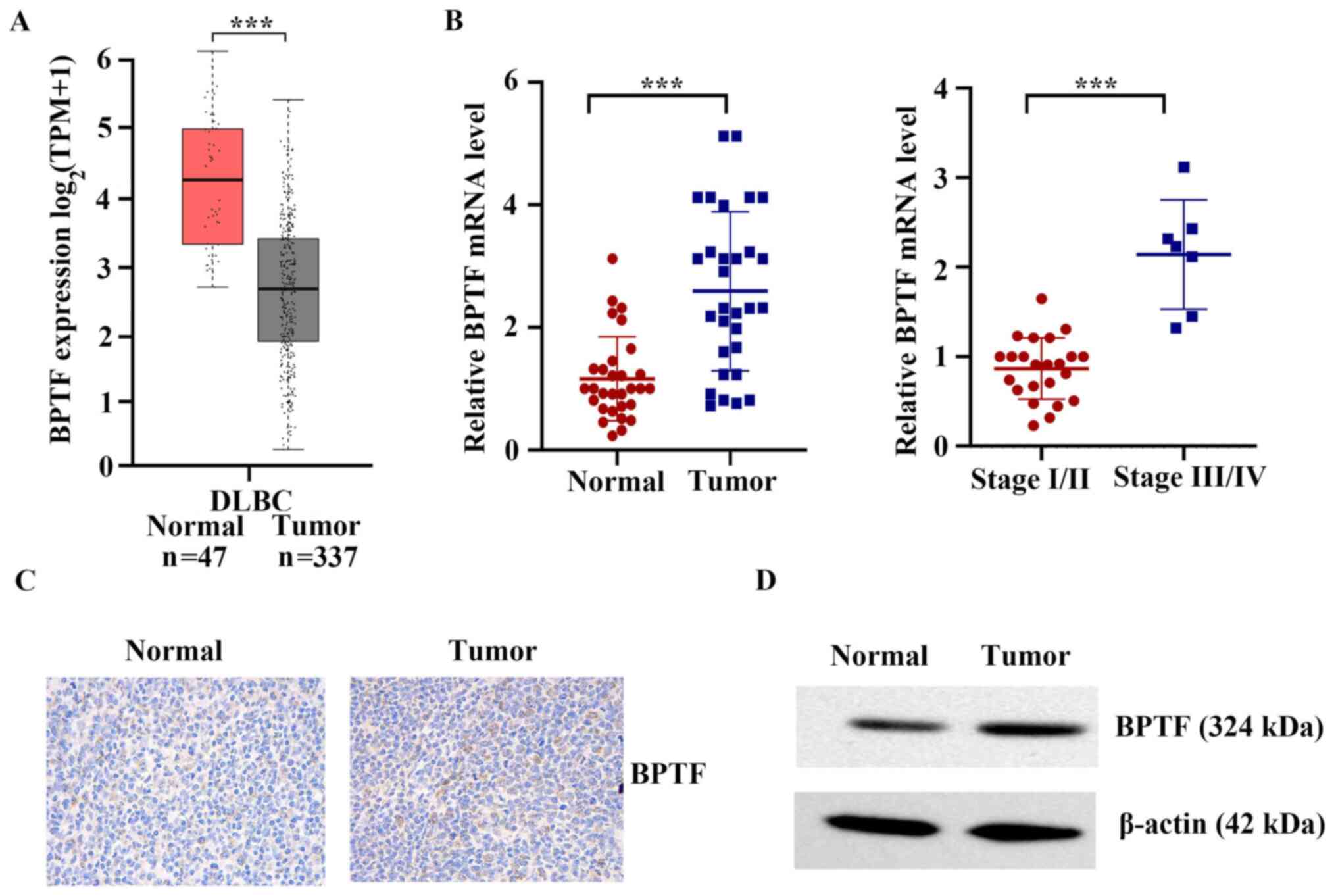
BPTF is upregulated in clinical T-cell
lymphoma tissues
The Cancer Genome Atlas (TCGA) database (https://portal.gdc.cancer.gov/) was used to
analyze the mRNA differences between lymphoma tissues and adjacent
normal tissues. As shown in Fig. 1A,
the mRNA expression levels of BPTF in lymphoid neoplasm diffuse
large B-cell lymphoma (DLBC) were significantly higher compared
with those in normal tissues (P<0.001). The present study
further detected the mRNA expression levels of BPTF in 30 human
cancerous lymph node tissues and matched adjacent nontumor tissues
from patients who were diagnosed with T-cell lymphoma. The results
demonstrated that the mRNA expression levels of BPTF in cancerous
lymph node tissue were significantly higher compared with those in
normal tissues (P<0.001; Fig.
1B). Compared with stage I/II, BPTF mRNA was significantly
higher expressed in stages III/IV T-cell lymphoma (P<0.001;
Fig. 1B). The results of
immunohistochemistry and western blot analyses also indicated that
the protein expression of BPTF in cancerous lymph node tissue was
significantly higher compared with that in normal tissues (Fig. 1C and D). Altogether, the present
results indicated that BPTF upregulation may have a critical role
in the development and progression of T-cell lymphoma.
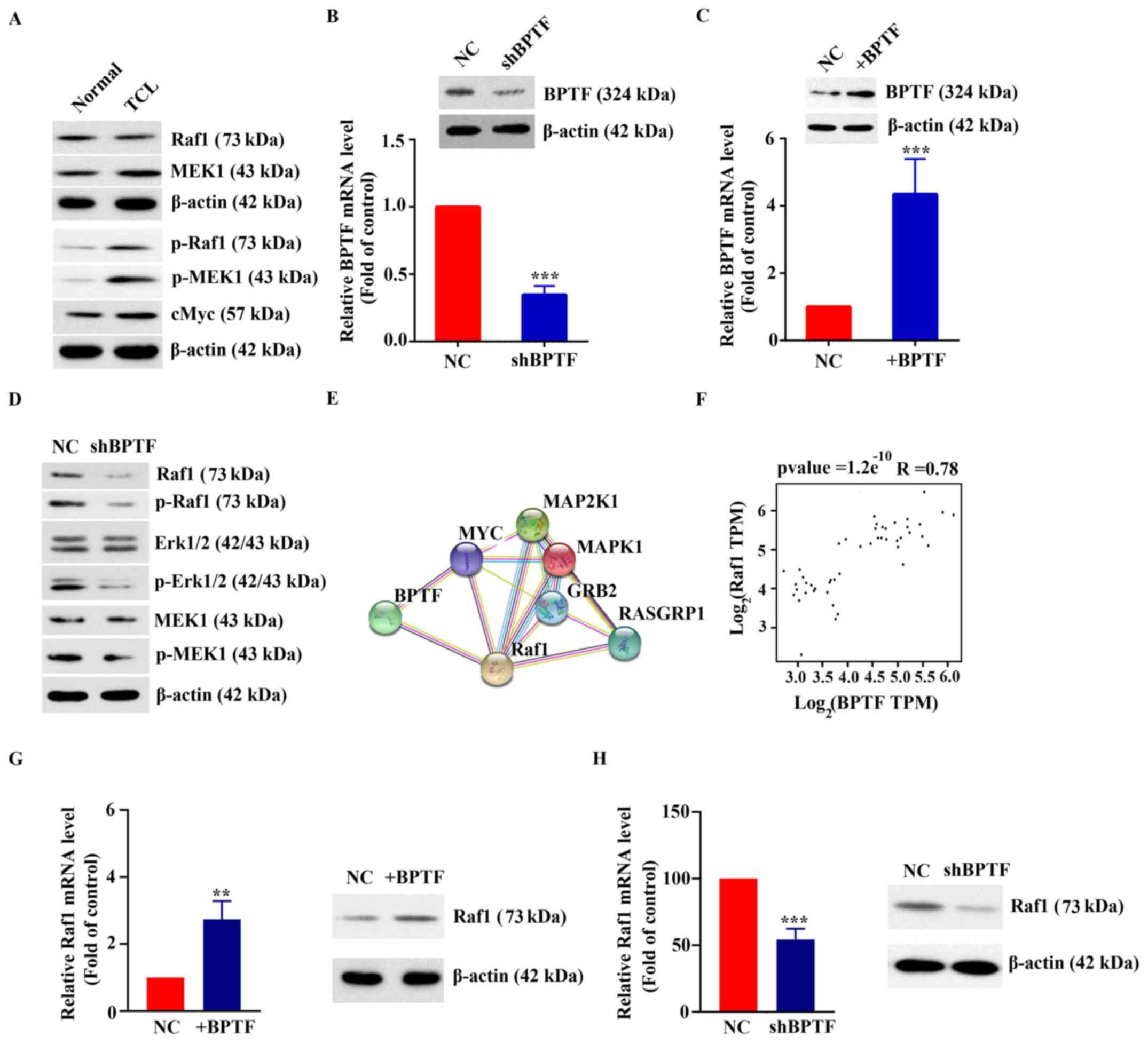
Activation of the MAPK pathway by
coexpression of BPTF and Raf1
Western blot analysis results demonstrated that the
expression and phosphorylation levels of MAPK pathway-related
proteins in T-cell lymphoma tissues were significantly higher
compared with those in normal tissues (Fig. 2A). To further explore the function of
BPTF in T-cell lymphoma, shRNA silencing and overexpression
experiments were performed. As shown in Fig. 2B and C, RT-qPCR results demonstrated
that the expression of BPTF in Hut-102 cells transfected with
shBPTF was 34.67±6.66% of that in the control group (transfected
with NC shRNA) and the expression of BPTF in Hut-102 cells
overexpressing BPTF was 4.35±1.04-fold of that in the control group
(transfected with blank vector). The results of western blot
analysis and RT-qPCR were consistent (Fig. 2C and C). Western blot results showed
that MAPK pathway-related proteins and their phosphorylation levels
were downregulated in BPTF-silenced Hut-102 cells (Fig. 2D). The potential interaction between
BPTF and MAPK pathway-related proteins was analyzed using the
STRING website (https://string-db.org/cgi/input?sessionId=b55z0svNSjXZ&input_page_active_form=multiple_identifiers)
(17). The results showed that BPTF
interacted with MYC and Raf1 (Fig.
2E). Notably, BPTF and Raf1 were coexpressed. The correlation
analysis of BPTF and Raf1 from TCGA database showed that there was
a positive correlation between them (R=0.78, P<0.001; Fig. 2F). RT-qPCR results showed that mRNA
expression levels of Raf1 were increased to 2.74±0.55-fold of the
control (P<0.01) when Hut-102 cells overexpressed BPTF (Fig. 2G). In addition, the mRNA expression
levels of Raf1 were 54.26±8.14% of the control (P<0.001) when
BPTF was silenced in Hut-102 cells (Fig.
2H). Western blot results were consistent with RT-qPCR results
(Fig. 2G and H). The present
findings indicated that high expression of BPTF may activate the
MAPK pathway, and that BPTF and Raf1 may be coexpressed in T-cell
lymphoma.
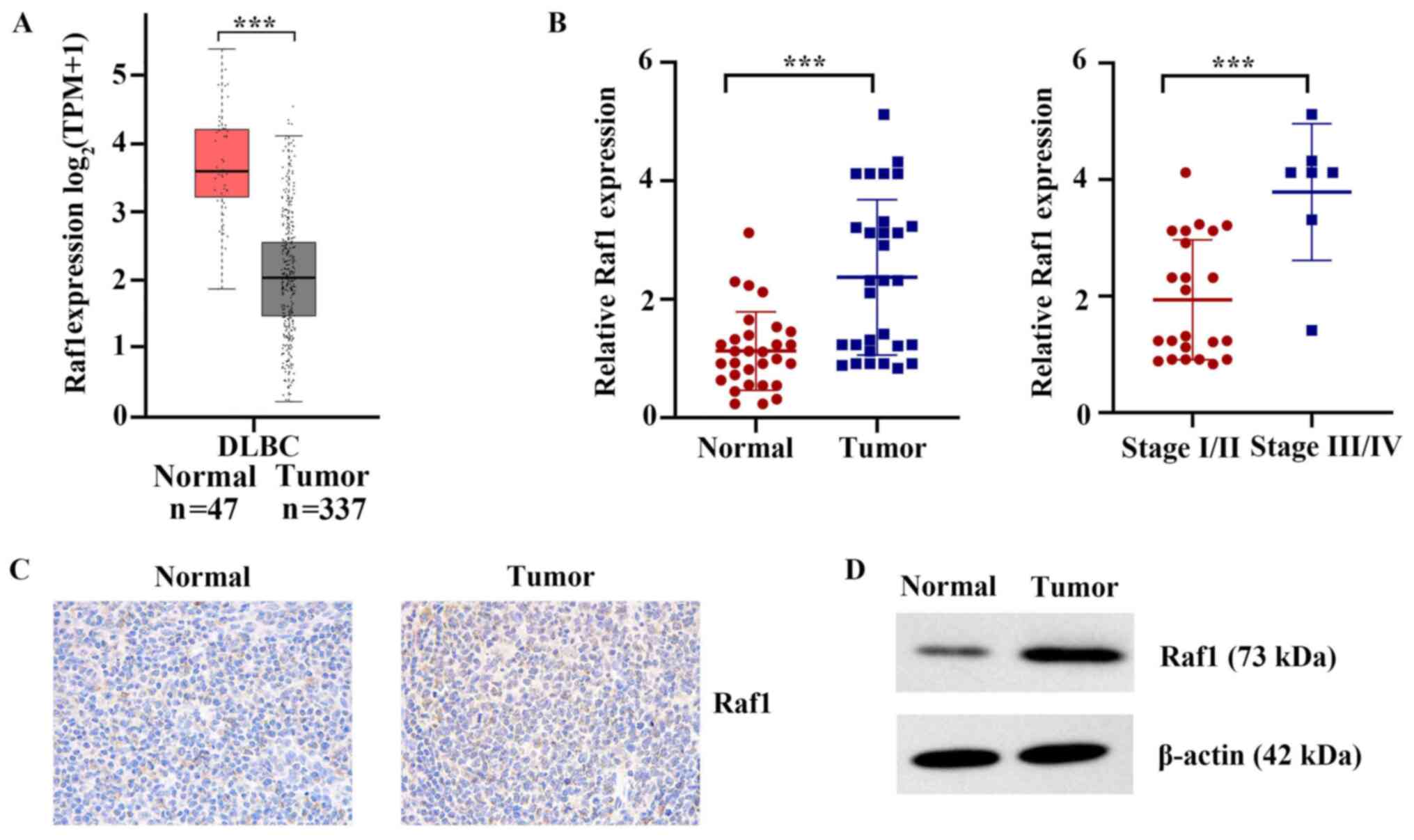
Raf1 is upregulated in clinical T-cell
lymphoma tissues
TCGA database was used to analyze the mRNA
differences between lymphoma tissues and adjacent normal tissues.
As shown in Fig. 3A, the mRNA
expression levels of Raf1 in lymphoid neoplasm DLBC were
significantly higher compared with those in normal tissues
(P<0.001). The mRNA expression of Raf1 was further detected in
the 30 human cancerous lymph node tissues and matched adjacent
nontumor tissues, collected for the presents study. The results
showed that the mRNA expression levels of Raf1 in cancerous lymph
node tissue were significantly higher compared with those in normal
tissues (P<0.001; Fig. 3B).
Compared with stage I/II, Raf1 mRNA was more highly expressed in
stages III/IV T-cell lymphatic carcinoma (P<0.001; Fig. 3B). The results of
immunohistochemistry and western blot also indicated that the
protein expression of Raf1 in cancerous lymph node tissue was
markedly higher compared with that in normal tissues (Fig. 3C and D). Altogether, the present
results indicated that Raf1 upregulation may have a critical role
in the development and progression of T-cell lymphoma.
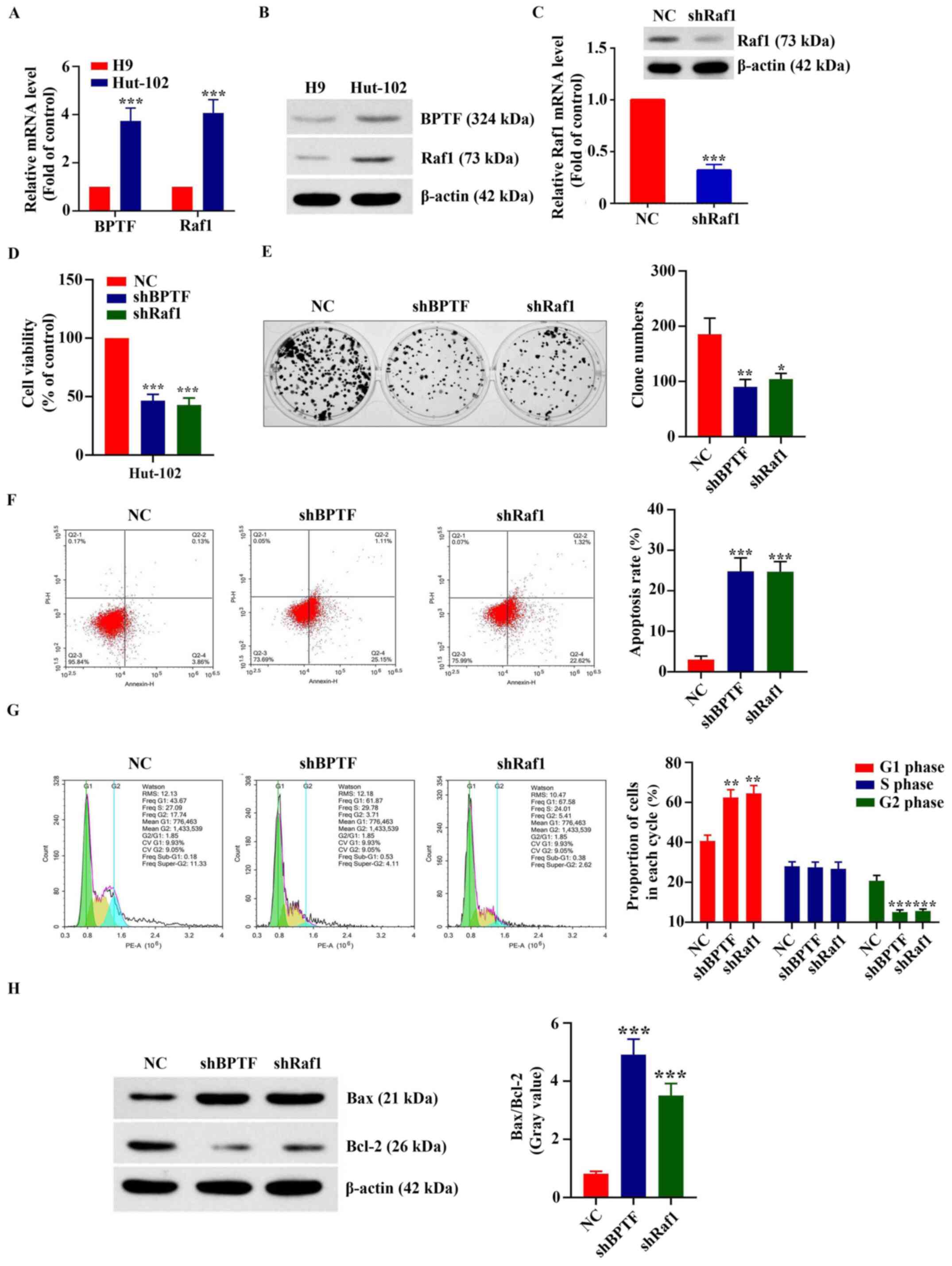
Effect of BPTF and Raf1 on the
proliferation of T-cell lymphoma cells
The human T-cell line H9 and human T-cell lymphoma
cell line Hut-102 were used as cell models. As shown in Fig. 4A, the mRNA expression levels of BPTF
and Raf1 in Hut-102 cells were 3.74±0.54 (P<0.001) and 4.11±0.55
(P<0.001) times higher compared with those in H9 cells,
respectively. Western blot results were consistent with the RT-qPCR
results (Fig. 4B). Fig. 4C demonstrates the successful
silencing of Raf1 in Hut-102 cells by shRNA: RT-qPCR results showed
that the mRNA expression of Raf1 in Hut-102 cells following Raf1
silencing was 32.11±5.57% of that in the control group (transfected
with NC) and the results of western blot analysis were consistent
with that of RT-qPCR. The results of the cell viability assay
showed that the numbers of viable Hut-102 cells were significantly
decreased following BPTF or Raf1 silencing (46.56±5.4 and 42.78±6%
of the NC, respectively; P<0.001; Fig. 4D). The clone formation experiment
showed that the number of clones in the Hut-102 cells transfected
with the NC shRNA was higher than that of Hut-102 cells transfected
with shBPTF or shRaf-1 (2.06±0.04-fold of the shBPTF, P<0.01;
1.78±0.11-fold of the shRaf1, P<0.05; Fig. 4E). Annexin V/PI staining experiments
showed that the apoptosis rate of Hut-102 cells transfected with
the NC shRNA (NC) was significantly lower than that of Hut-102
cells transfected with shBPTF or shRaf-1 (12.12±2.95% with shBPTF,
P<0.001; 12.39±4.22% with shRaf1, P<0.001; Fig. 4F). Cell cycle phase distribution
experiments showed that Hut-102 cells following BPTF or Raf-1
silencing remained in G1 phase compared with those in the NC group
(Fig. 4G). Bax and Bcl-2 protein
expression was detected by western blotting, and the gray value was
used for relative quantification. The results revealed that the
Bax/Bcl-2 signal ratio in Hut-102 cells following BPTF or Raf-1
silencing was significantly higher than that in Hut-102 cells
transfected with the NC shRNA (6.01±0.3-fold of the NC, P<0.001;
4.29±0.23-fold of the NC, P<0.001; Fig. 4H). These results indicated that high
expression of BPTF or Raf1 may promote the proliferation of T-cell
lymphoma.
Effect of BPTF on tumor growth in
vivo
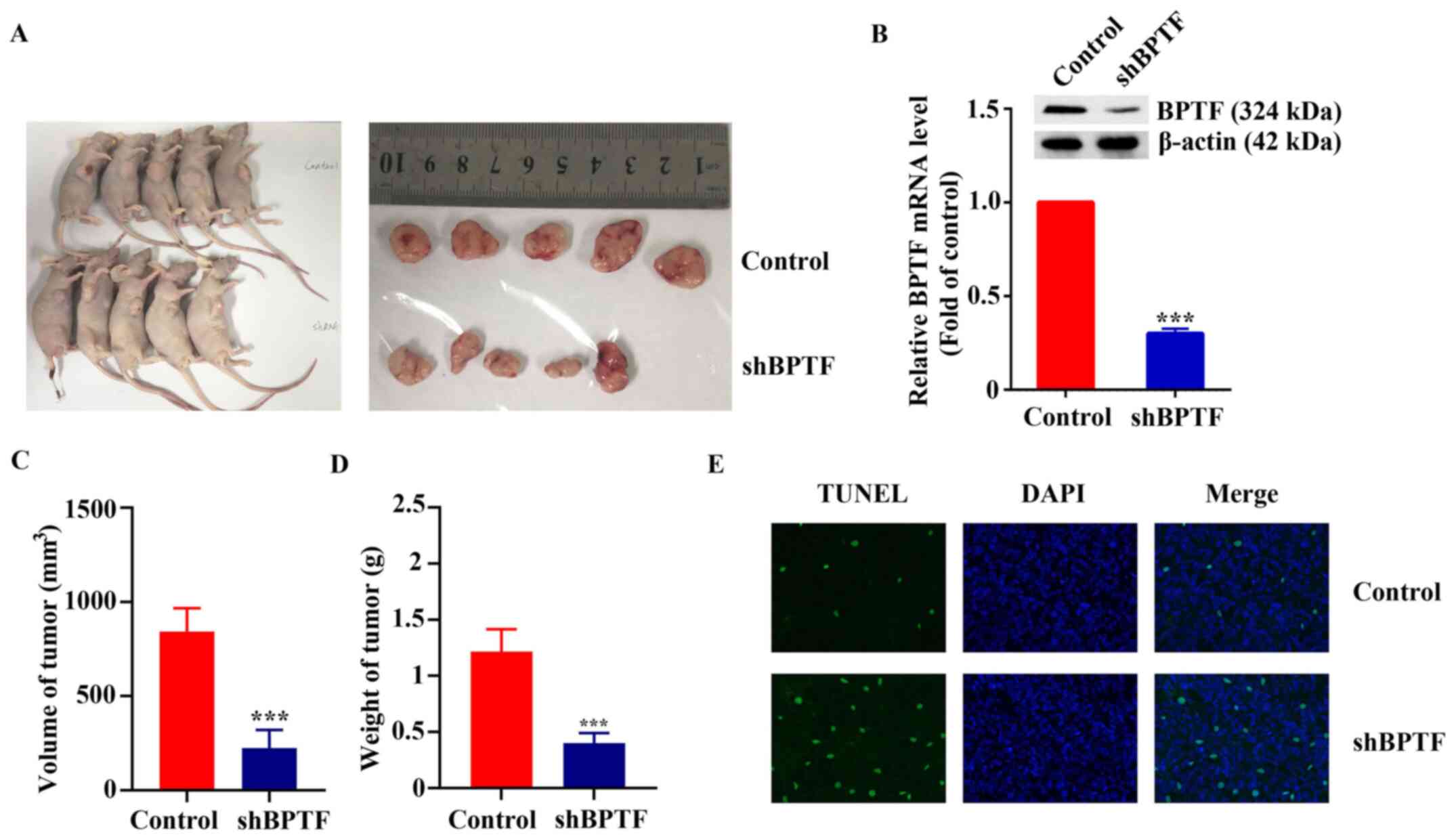
Finally, a xenograft model was used to investigate
the effect of BPTF on tumor growth in vivo. As shown in
Fig. 5A, tumors in the shBPTF group
grew smaller compared with those in the control group. RT-qPCR
results confirmed that the expression of BPTF in the shBPTF
cell-derived tumors was 28.22±2.65% of that in the control tumors
and the result of western blot was consistent with that of RT-qPCR
(Fig. 5B). The average volume of
tumors in the shBPTF group was 26.65±10.63% that of the control
group (P<0.001; Fig. 5C). The
weight of tumors in the shBPTF group was 34.19±11.49% that of the
control group (P<0.001; Fig. 5D).
The results of the TUNEL assay showed that the tumors in the shBPTF
group had a marked increase in apoptosis compared with the control
group (Fig. 5E).
Discussion
Lymphomas mainly originate from lymph nodes and
other lymphoid tissues. In recent years, the incidence rate of
lymphoma has increased rapidly, ranking fifth in the world, which
seriously threatens human health (18,19).
There are many reports about the signaling pathways involved in the
occurrence and development of lymphoma (20,21).
Miller et al (22) reported
that the MAPK signaling pathway influenced apoptosis in malignant
lymphoid cells treated with glucocorticoids. Inhibition of MAPKs
restored the drug sensitivity of a glucocorticoid-resistant clone
in CEM-C1-15 cells. Sun et al (23) reported in a review that multiple
signaling pathways (B cell receptor, NF-κB, PI3K/AKT/mTOR, and
JAK/STAT signaling pathways) were involved in the development of
lymphoma. This suggests that these molecular markers may be used as
targets for diagnosis and treatment. The present study demonstrated
that the MAPK pathway was abnormally activated in T-cell lymphoma
tissues compared with normal tissues. Furthermore, the present
results revealed that the coexpression of BPTF and Raf1 was
involved in the abnormal activation of the MAPK pathway in T-cell
lymphoma.
As the core subunit of the NURF complex, BPTF has an
important role in chromatin remodeling (24). In recent years, increasing attention
has been given to its role in tumor development. Zhao et al
(7) reported that BPTF promoted
hepatocellular carcinoma (HCC) proliferation by targeting human
telomerase reverse transcriptase and suggested that BPTF could be a
potential molecular target for the treatment of HCC. Dai et
al (8) reported that BPTF
promoted lung cancer growth via cooperation with p50 NF-κB and
regulation of cyclooxygenase-2 (COX-2) expression. That study
suggested that the BPTF/p50/COX-2 axis could be a potential
therapeutic target for lung cancer. The present study found that
BPTF and Raf1 were abnormally overexpressed in T-cell lymphoma
cells and tissues. The viability of human T-cell lymphoma cells
(Hut-102) was decreased significantly after silencing BPTF or Raf1.
Several cells showed early apoptosis accompanied by activation of
the apoptosis factor Bax. Additionally, the cell cycle was blocked
in G1 phase when BPTF or Raf1 were silenced in Hut-102 cells.
Therefore, it can be speculated that the coexpression of BPTF and
Raf1 is abnormally elevated, promoting the survival of T-cell
lymphoma cells. The present study further confirmed the
carcinogenic effect of BPTF in nude mice. Results from in
vivo xenografting experiments revealed that tumors derived from
BPTF-silenced cells grew more slowly than those derived from
control cells. The tumors in the shBPTF group had a marked increase
in apoptosis compared with the control group, as demonstrated by a
TUNEL assay. Altogether, the present results indicated that BPTF
and Raf1 upregulation may have a critical role in the development
and progression of T-cell lymphoma. There is a coexpression
relationship between BPTF and Raf1.
Oncogene signal transduction pathways and oncogene
changes have an important role in the development of lymphoma. In
the era of precision medicine, it is necessary and valuable to
recognize the activation of these carcinogenic pathways and
biomarkers (25,26). The present study identified
abnormalities in the MAPK pathway in T cell lymphoma. It was
further found that BPTF could activate the MAPK pathway by
coexpression with Raf1 and promote the proliferation of T-cell
lymphoma cells. These findings enrich the understanding of the
pathogenesis of T-cell lymphoma and may provide a novel target and
strategies for the treatment of T-cell lymphoma.
Acknowledgements
The authors would like to thank The Cancer Genome
Atlas (TCGA) repository for providing data.
Funding
Support for this study was provided by the National Natural
Science Foundation of China (grant no. 81800196).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
DB was responsible for the conception and writing of
the overall research. YZ and FS were responsible for cell and
animal experiments. DG and WS were responsible for the statistical
analysis. HZ and HL were responsible for bioinformatics analysis,
molecular biology experiments and data proofreading. DB and HZ
confirmed the authenticity of all the raw data. All authors read
and approved the final manuscript.
Ethics approval and consent to
participate
Experimental research involving patients was
approved by the Ethics Committee of the First Affiliated Hospital
of Xiamen University (Xiamen, China; approval no. KY-2018-014). All
patients had signed informed consent. The animal protocol was
approved by the Experimental Animal Ethics Committee of the First
Affiliated Hospital of Xiamen University (Xiamen, China; approval
no. 2019-231). The authors state that they have obtained
appropriate institutional review board approval or have followed
the principles outlined in the Declaration of Helsinki for all
human or animal experimental investigations.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Evens AM, Querfeld C and Rosen ST: T-cell
non-Hogdkin's lymphoma. Cancer Treat Res. 131:161–220. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
The world health organization
classification of malignant lymphomas in Japan, . Incidence of
recently recognized entities. Lymphoma study group of Japanese
pathologists. Pathol Int. 50:696–702. 2000. View Article : Google Scholar
|
|
3
|
Mugnaini EN and Ghosh N: Lymphoma. Prim
Care. 43:661–675. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Nair R and Neelapu SS: The promise of CAR
T-cell therapy in aggressive B-cell lymphoma. Best Pract Res Clin
Haematol. 31:293–298. 2018. View Article : Google Scholar
|
|
5
|
Grimm KE and O'Malley DP: Aggressive B
cell lymphomas in the 2017 revised WHO classification of tumors of
hematopoietic and lymphoid tissues. Ann Diagn Pathol. 38:6–10.
2019. View Article : Google Scholar
|
|
6
|
Alkhatib SG and Landry JW: The nucleosome
remodeling factor. FEBS Lett. 585:3197–3207. 2011. View Article : Google Scholar
|
|
7
|
Zhao X, Zheng F, Li Y, Hao J, Tang Z, Tian
C, Yang Q, Zhu T, Diao C, Zhang C, et al: BPTF promotes
hepatocellular carcinoma growth by modulating hTERT signaling and
cancer stem cell traits. Redox Biol. 20:427–441. 2019. View Article : Google Scholar
|
|
8
|
Dai M, Hu S, Liu CF, Jiang L, Yu W, Li ZL,
Guo W, Tang R, Dong CY, Wu TH and Deng WG: BPTF cooperates with p50
NF-κB to promote COX-2 expression and tumor cell growth in lung
cancer. Am J Transl Res. 11:7398–7409. 2019.PubMed/NCBI
|
|
9
|
Dar AA, Majid S, Bezrookove V, Phan B,
Ursu S, Nosrati M, De Semir D, Sagebiel RW, Miller JR III, Debs R,
et al: BPTF transduces MITF-driven prosurvival signals in melanoma
cells. Proc Natl Acad Sci USA. 113:6254–6258. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Richart L, Real FX and Sanchez-Arevalo
Lobo VJ: c-MYC partners with BPTF in human cancer. Mol Cell Oncol.
3:e11523462016. View Article : Google Scholar
|
|
11
|
Peluso I, Yarla NS, Ambra R, Pastore G and
Perry G: MAPK signalling pathway in cancers: Olive products as
cancer preventive and therapeutic agents. Semin Cancer Biol.
56:185–195. 2019. View Article : Google Scholar
|
|
12
|
Zou X and Blank M: Targeting p38 MAP
kinase signaling in cancer through post-translational
modifications. Cancer Lett. 384:19–26. 2017. View Article : Google Scholar
|
|
13
|
Rezatabar S, Karimian A, Rameshknia V,
Parsian H, Majidinia M, Kopi TA, Bishayee A, Sadeghinia A, Yousefi
M, Monirialamdari M and Yousefi B: RAS/MAPK signaling functions in
oxidative stress, DNA damage response and cancer progression. J
Cell Physiol. Feb 27–2019.(Epub ahead of print). View Article : Google Scholar
|
|
14
|
Davis JA: Mouse and rat anesthesia and
analgesia. Curr Protoc Neurosci. Jan 1–2008.(Epub ahead of print).
View Article : Google Scholar
|
|
15
|
Adams S and Pacharinsak C: Mouse
anesthesia and analgesia. Curr Protoc Mouse Biol. 5:51–63. 2015.
View Article : Google Scholar
|
|
16
|
Schmittgen TD and Livak KJ: Analyzing
real-time PCR data by the comparative C(T) method. Nat Protoc.
3:1101–1108. 2008. View Article : Google Scholar
|
|
17
|
Szklarczyk D, Gable AL, Lyon D, Junge A,
Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork
P, et al: STRING v11: Protein-protein association networks with
increased coverage, supporting functional discovery in genome-wide
experimental datasets. Nucleic Acids Res. 47:D607–D613. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Jiang M, Bennani NN and Feldman AL:
Lymphoma classification update: T-cell lymphomas, Hodgkin
lymphomas, and histiocytic/dendritic cell neoplasms. Expert Rev
Hematol. 10:239–249. 2017. View Article : Google Scholar
|
|
19
|
Jaffe ES: Diagnosis and classification of
lymphoma: Impact of technical advances. Semin Hematol. 56:30–36.
2019. View Article : Google Scholar
|
|
20
|
Yu L, Li L, Medeiros LJ and Young KH:
NF-κB signaling pathway and its potential as a target for therapy
in lymphoid neoplasms. Blood Rev. 31:77–92. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yang W, Wang Y, Yu Z, Li Z, An G, Liu W,
Lv R, Ma L, Yi S and Qiu L: SOX11 regulates the pro-apoptosis
signal pathway and predicts a favorable prognosis of mantle cell
lymphoma. Int J Hematol. 106:212–220. 2017. View Article : Google Scholar
|
|
22
|
Miller AL, Geng C, Golovko G, Sharma M,
Schwartz JR, Yan J, Sowers L, Widger WR, Fofanov Y, Vedeckis WV and
Thompson EB: Epigenetic alteration by DNA-demethylating treatment
restores apoptotic response to glucocorticoids in
dexamethasone-resistant human malignant lymphoid cells. Cancer Cell
Int. 14:352014. View Article : Google Scholar
|
|
23
|
Sun RF, Yu QQ and Young KH: Critically
dysregulated signaling pathways and clinical utility of the pathway
biomarkers in lymphoid malignancies. Chronic Dis Transl Med.
4:29–44. 2018.PubMed/NCBI
|
|
24
|
Gong YC, Liu DC, Li XP and Dai SP: BPTF
biomarker correlates with poor survival in human NSCLC. Eur Rev Med
Pharmacol Sci. 21:102–107. 2017.
|
|
25
|
Nakagawa H and Fujita M: Whole genome
sequencing analysis for cancer genomics and precision medicine.
Cancer Sci. 109:513–522. 2018. View Article : Google Scholar
|
|
26
|
Pauli C, Hopkins BD, Prandi D, Shaw R,
Fedrizzi T, Sboner A, Sailer V, Augello M, Puca L, Rosati R, et al:
Personalized in vitro and in vivo cancer models to guide precision
medicine. Cancer Discov. 7:462–477. 2017. View Article : Google Scholar : PubMed/NCBI
|