Introduction
Lung cancer is the most common malignancy with the
highest mortality rate worldwide (1). There are two main histopathological
types of lung cancer: Non-small cell lung cancer (NSCLC) and small
cell lung cancer (2). Lung
adenocarcinoma (LUAD) is one of the most frequently occurring types
of NSCLC and accounts for approximately half of all lung cancers
(3). Despite advancements in early
diagnosis and targeted therapies, the prognosis of patients with
LUAD remains unsatisfactory, and the 5-year survival rate is
<25% (4). New treatment
strategies are required to improve the clinical outcomes of
patients with LUAD, particularly those diagnosed with unresectable,
locally advanced or metastatic LUAD. Therefore, it is urgently
necessary to explore the molecular mechanisms of LUAD to further
understand this disease and discover novel biomarkers for its
diagnosis, prognosis and treatment.
Cyclins are proteins that bind to cyclin-dependent
kinases (CDKs) and thereby regulate the cell division cycle
(5,6); the HUGO Gene Nomenclature Committee
lists 31 members in the cyclin gene group (7). Cyclin B1 (CCNB1) is considered as a
mitotic cyclin, which plays a key role in the regulation of CDK1 by
complexing with it to promote the transition of the cell cycle from
the G2 phase to mitosis (8). In
the metaphase and late stages of mitosis, degradation of CCNB1
occurs through the ubiquitin proteasome pathway, leading to
chromosome depolymerization and nucleolar and nuclear membrane
regeneration (9). Previous studies
have demonstrated that CCNB1 is abnormally expressed in a variety
of tumors and is associated with poor prognosis (10–12).
A study by Gu et al (13)
evaluated the upregulation of CCNB1 in liver cancer tissues
compared with normal liver tissues, and found that a high
expression level of CCNB1 was closely associated with poor
prognosis in patients with hepatocellular carcinoma (HCC).
Furthermore, the study demonstrated that the knockdown of CCNB1
significantly inhibited the proliferation, migration and invasion
of HCC cells. Another study reported that CCNB1 was upregulated in
colorectal cancer tissues and negatively associated with lymph node
metastasis, distant metastasis and TNM stage, and the survival rate
of patients with higher CCNB1 expression was significantly higher
than that of patients with lower CCNB1 expression. In addition,
cell-based experiments in the study revealed that the inhibition of
CCNB1 expression increased the migration and invasion of colorectal
cancer cells (14). Therefore,
CCNB1 may be a prognostic biomarker. In addition, cancers are
associated with increased inflammatory burden. Numerous studies
have shown an association between inflammatory markers and
malignant conditions (15). Also,
a recent study has demonstrated that CCNB1 is involved in
atherosclerosis-induced inflammation in blood vessels (16). Therefore, the expression of CCNB1
in cancer is worthy of evaluation.
A study revealed that in lung cancer, the CCNB1
expression level is upregulated and higher levels of CCNB1 indicate
poorer survival outcomes (17).
Mechanistically, the degradation of CCNB1 by anaphase promoting
complex subunit 11 via ubiquitin-60S ribosomal protein L49
ubiquitylation is critical in the cell cycle progression and
proliferation of NSCLC cell lines (18). In another study, CCNB1
overexpression was shown to promote the progression of LUAD cells,
and it was suggested that microRNA-139-5p negatively regulates
CCNB1 in LUAD, thereby suppressing cell proliferation, migration,
invasion and the cell cycle (19).
However, the clinical characteristics of CCNB1 in
lung cancer, particularly LUAD, remain unclear, and its potential
mechanism requires further exploration. Therefore, in the present
study, the expression of CCNB1 in LUAD was analyzed and its
association with the clinicopathological features and prognosis of
patients with LUAD was explored. Furthermore, the molecular
mechanism of CCNB1 and its use in the prognosis of LUAD were
preliminarily investigated.
Materials and methods
Data mining
The RNA-sequencing (RNA-seq) data and clinical data
of 535 LUAD samples and 59 normal samples were downloaded from The
Cancer Genome Atlas (TCGA; http://portal.gdc.cancer.gov/). After the exclusion of
those samples without completely specific TNM stage and intact
survival data, 334 LUAD samples were finally used in the present
study. The clinicopathological data were downloaded for reanalysis,
including the age at the initial diagnosis, sex, clinical stage,
TNM stage, survival status and overall survival time. The RNA-seq
data of LUAD were also download from the Gene Expression Omnibus
(GEO) database (www.ncbi.nlm.nih.gov/geo) for analysis of the
transcription level of CCNB1 (GSE116959; 11 healthy lung and 57
LUAD samples; http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE116959)
(20). All samples were divided
into low and high expression groups according to the median value
of CCNB1 expression.
Kyoto encyclopedia of genes and
genomes (KEGG) and gene ontology (GO) enrichment analyses
Pathway enrichment analysis of differential genes
(DEGs) was performed using the KEGG (https://www.kegg.jp/) and GO databases (http://geneontology.org/docs/ontology-documentation/).
KEGG combines numerous database resources from high-throughput
experimental technologies at the molecular level, while the GO
database is widely used in bioinformatics to provide information on
cellular components (CC), molecular functions (MF) and biological
processes (BP). The Org.Hs.eg.db R package (version 3.6.0;
http://bioconductor.org/packages/org.Hs.eg.db/) was
used to convert the symbols of DEGs into Entrez IDs. Subsequently,
KEGG analysis was performed using the enrichKEGG function of the
clusterProfiler R package (version 3.6.0; http://bioconductor.org/packages/clusterProfiler/). GO
analysis was performed using the enrichGO function in
clusterProfiler. The results of the KEGG and GO enrichment analyses
were visualized using the ggplot2 R package (version 3.3.5;
http://ggplot2.tidyverse.org).
Weighted gene co-expression network
analysis (WGCNA)
WGCNA involves the construction of a weighted gene
expression network that represents the associations between
different genes and can be used to identify highly coordinated gene
sets. In the present study, the expression data of the DEGs were
used to construct a gene co-expression network using the WGCNA R
package (version 3.6.0; http://horvath.genetics.ucla.edu/html/CoexpressionNetwork/Rpackages/WGCNA/).
The DEGs for the WGCNA were screened using edgeR (version 3.6.0;
http://bioconductor.org/packages/edgeR/) (defined as
fold change ≥1 and P≤0.05). The WGCNA included identification of
the gene expression similarity matrix, adjacency matrix and
co-expression network. A scale-free plot was used to evaluate
whether the network exhibited a scale-free topology. The power
value of the soft threshold of the adjacency matrix was set as 5 to
meet the scale-free topology criterion. The hierarchical clustering
analysis was based on the average linkage generated by a dynamic
analysis using the tree-cut method for branch cutting (cut height,
0.995; minimum cluster size, 30).
Patient inclusion criteria and tissue
sample collection
LUAD tissue samples were collected from 94 patients
undergoing surgical resection in the Department of Thoracic Surgery
of the Affiliated Hospital of Zunyi Medical University (Zunyi,
China) between January 2010 and August 2015. The patients included
54 females and 40 males. The oldest was 76 years old and the
youngest was 21 years old, and the media age was 57 years. Due to
the collection of normal lung tissue being challenging, and
paracancerous tissue being different from normal tissue and
potentially having different biological properties, an independent
normal lung tissue series was used as a control for the
institutional LUAD tissues (21).
This comprised 30 normal lung tissue sections, which were purchased
from Shanghai Xinchao Biological Technology Co., Ltd. The following
criteria were met in all cases: i) Histologically diagnosed LUAD;
ii) complete clinical data; and iii) no other malignant tumor was
present and the patients did not accept tumor-related treatment
before the initial diagnosis, such as radiotherapy, chemotherapy
and immunotherapy. Tissue samples were fixed in 4% paraformaldehyde
and then embedded in paraffin for postoperative
immunohistochemistry. The pathological stage was determined
according to the Union for International Cancer Control and
American Joint Committee on Cancer staging criteria (eighth
edition). The follow-up was initiated on the day of surgery and
terminated in January 2019 or at death. Overall survival (OS) was
defined as the interval from the end of surgery to the date of the
last follow-up or death. This study obtained written consent from
all patients and was approved by the Research Ethics Committee of
the Affiliated Hospital of Zunyi Medical University [no.
(2021)1-098].
Immunohistochemistry (IHC)
The LUAD tumor tissues were fixed in 4%
paraformaldehyde for 24 h at room temperature, dehydrated with
graded ethanol and cleared with xylene. After embedding in
paraffin, the tumor tissues were sectioned into 4-µm slices. The
paraffin sections were dewaxed with xylene and hydrated with
gradient ethanol using standard procedures. After treatment with
citrate buffer (pH 6.0) for antigen retrieval at 95°C for 12 min,
the slices were incubated with 3% hydrogen peroxide for 10 min at
room temperature to block endogenous peroxidase activity and 5%
goat serum (Beijing Solarbio Science & Technology Co., Ltd.)
for 30 min at 25°C to block non-specific binding sites. The
sections were then incubated with the primary antibody anti-cyclin
B1 (cat. no. TA374365; OriGene Technologies, Inc.) at a dilution of
1:300 overnight at 4°C. After warming for 1 h at room temperature,
the sections were washed three times in PBS and then incubated with
the undiluted secondary antibody goat anti-rabbit IgG-HRP (PV-9000;
OriGene Technologies, Inc.) at 37°C for 20 min. The primary
antibody was replaced with PBS to serve as the negative control.
Finally, the sections were stained with DAB and imaged under a
light microscope (DM3000; Leica Microsystems GmbH).
The IHC results were independently assessed by two
experienced pathologists from Zunyi Medical University who were
blinded to the clinical data of the patients. Five random fields
from each section were observed under an optical microscope at ×200
magnification. The expression of CCNB1 was scored according to the
percentage of positive tumor cells and the staining intensity. The
percentage of positive cells was scored according to the following
criteria: 0 (0%), 1 (1–25%), 2 (26–50%), 3 (51–75%) and 4
(76–100%). The staining intensity was scored as follows: 0 (no
staining), 1 (light yellow), 2 (brownish) and 3 (tan). The staining
intensity score and the percentage of positive staining were summed
to obtain the final score, with a total score >2 defined as
positive expression and ≤2 defined as negative expression.
Cell culture and transfection
The PC9, A549, H1299 and H827 LUAD cell lines were
purchased from the Cell Bank of Type Culture Collection of the
Chinese Academy of Sciences, and stored in the Cancer Research
Laboratory of the Affiliated Hospital of Zunyi Medical University.
The cells were cultured in RPMI-1640 (HyClone; Cytiva) supplemented
with 10% fetal bovine serum (Shanghai XP Biomed Ltd.) and 100X
Penicillin-Streptomycin Solution (Sangon Biotech Co., Ltd.) at 37°C
with 5% CO2. Small interfering RNAs (siRNAs) purchased
from Sangon Biotech Co., Ltd. were used to knock down CCNB1. The
sequences were as follows: CCNB1-PLVT7 forward,
CTTGAGTTGGAGTACTATATT and reverse, AATATAGTACTCCAACTCAAG;
CCNB1-PLVT8 forward, GGTTGTTGCAGGAGACCATGT and reverse,
ACATGGTCTCCTGCAACAACC; CCNB1-PLVT9 forward, GATCGGTTCATGCAGAATAAT
and reverse, ATTATTCTGCATGAACCGATC; negative-PLVT forward,
TTCTCCGAACGTGTCACGT and reverse, ACGTGACACGTTCGGAGAA. Cells were
transfected with siRNAs targeting CCNB1 or non-sense control siRNA
using Lipofectamine 2000 (Thermo Fisher Scientific, Inc.) according
to the manufacturer's instructions. Briefly, the LUAD cells were
seeded at a density of 1.5×105 in a 6-well plate.
Lipofectamine 2000 (Thermo Fisher Scientific, Inc.) was used to
transfect the siRNAs into H1299 cell lines to select the most
efficient one for subsequent use (CCNB1-PLVT7, CCNB1-PLVT8,
CCNB1-PLVT9). Following the standard protocol, siR-NC or siR-CCNB1
(100 pmol/well; Shanghai GeneChem Co., Ltd.) was transfected into
H1299 cell lines. After 6 h of culture at 37°C, the medium was
replaced with DMEM containing 10% FBS. After cultivation for 72 h
at 37°C, the cells were collected for further assays.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was extracted from LUAD cells using RNAiso
Plus reagent (Takara Bio, Inc.) according to the manufacturer's
protocol, and cDNAs were reverse transcribed using a
PrimeScript™ RT reagent Kit (Perfect Real Time) (Takara
Bio, Inc.) at 37°C for 15 min. qPCR was performed with an ABI Prism
7500 Real-Time PCR system (Applied Biosystems; Thermo Fisher
Scientific, Inc.) and a ChamQ™ Universal SYBR qPCR
Master Mix Kit (Vazyme Biotech Co., Ltd.) was used to quantify the
expression of CCNB1 and GAPDH. qPCR was initiated at 95°C for 3
min, followed by 40 cycles at 95°C for 20 sec and 60°C for 30 sec.
GAPDH expression was used as the internal control, and the relative
quantification of gene expression was calculated using the
2−ΔΔCq method (22).
The primers were designed and synthesized by Sangon Biotech Co.,
Ltd. and their sequences were as follows: CCNB1 forward,
5′-GGAGAGCATCTAAGATTGGAGAGGTTG-3′ and reverse,
5′-GCTTCGATGTGGCATACTTGTTCTTG-3′; and β-actin forward,
5′-CCTGGCACCCAGCACAAT-3′ and reverse, 5′-GGGCCGGACTCGTCATAC-3′.
RNA extraction and RNA-seq
Total RNA was extracted from the LUAD cells using
RNAiso Plus reagent (Takara Bio, Inc.) according to the
manufacturer's protocol. Samples with RNA an optical density ratio
260 and 280 nm of >1.8 were subjected to subsequent analyses.
Libraries were constructed using the TruSeq Stranded mRNA LT Sample
Prep Kit (Illumina®; cat. no. RS-122-2101.) according to
the manufacturer's instructions. The loading concentration of 30
ng/µl was measured by library quantification using Thermo Fisher
Qubit Flex (Thermo Fisher Scientific, Inc.), and then the library
was sequenced with a NovaSeq 6000 S4 Rgt Kit (20028312) on an
Illumina sequencing platform (NovaSeq 6000; Illumina, Inc.), and
150 bp paired-end reads were generated. Base calling was performed
with RTA v2.7.6 (Illumina, Inc.), and the fastq files were
generated by bcl2fastq v2.15.0 (Illumina, Inc.). Removal of
low-quality bases and adapters from paired-end reads was processed
by fastp v0.22.0 (https://github.com/OpenGene/fastp). Alignment of the
trimmed RNA-seq reads to the ensembl human genome assembly
(GRCh38.p13; http://www.ensembl.org/Homo_sapiens/Info/Index)
employed HISAT2 v2.2 (http://daehwankimlab.github.io/hisat2/). 1 with the
options of ‘-k 3 -p 20 --pen-noncansplice 1000000’. The counts of
the reads mapped to individual genes were calculated by
featureCounts v2.0.3 (http://subread.sourceforge.net/featureCounts.html).
DEGs were identified by the DESeq2 (version 3.15; http://bioconductor.org/packages/DESeq2/), a cut-off
of padj <0.05 and |log2(fold change)|>1.5 was applied.
Statistical analysis
The datasets were mainly analyzed using R package
software (version 3.6.0; http://www.r-project.org/) and integrated using Perl
(version 5.30.0.1; http://strawberryperl.com/). Tools for the analysis
and interpretation of high-throughput genomic data were obtained
from Bioconductor (version 3.15; http://bioconductor.org/). For normally distributed
continuous variables, significant differences were detected using
unpaired t-tests when two groups were compared and one-way ANOVA
followed by Tukey's or Dunnett's post hoc tests when multiple
groups were compared. For categorical variables, including the
associations between CCNB1 and clinicopathological variables,
analyses were performed using Pearson's χ2 and Fisher's
exact tests. The survival analysis was performed using Kaplan-Meier
curves, and the significance of differences in survival was
examined using the log-rank test. The Cox proportional hazards
regression model was used for univariate and multivariate analyses.
The hazard ratio (HR) and 95% confidence interval (CI) were
calculated to estimate the hazard risk of variables. P<0.05 was
considered to indicate a statistically significant result.
Results
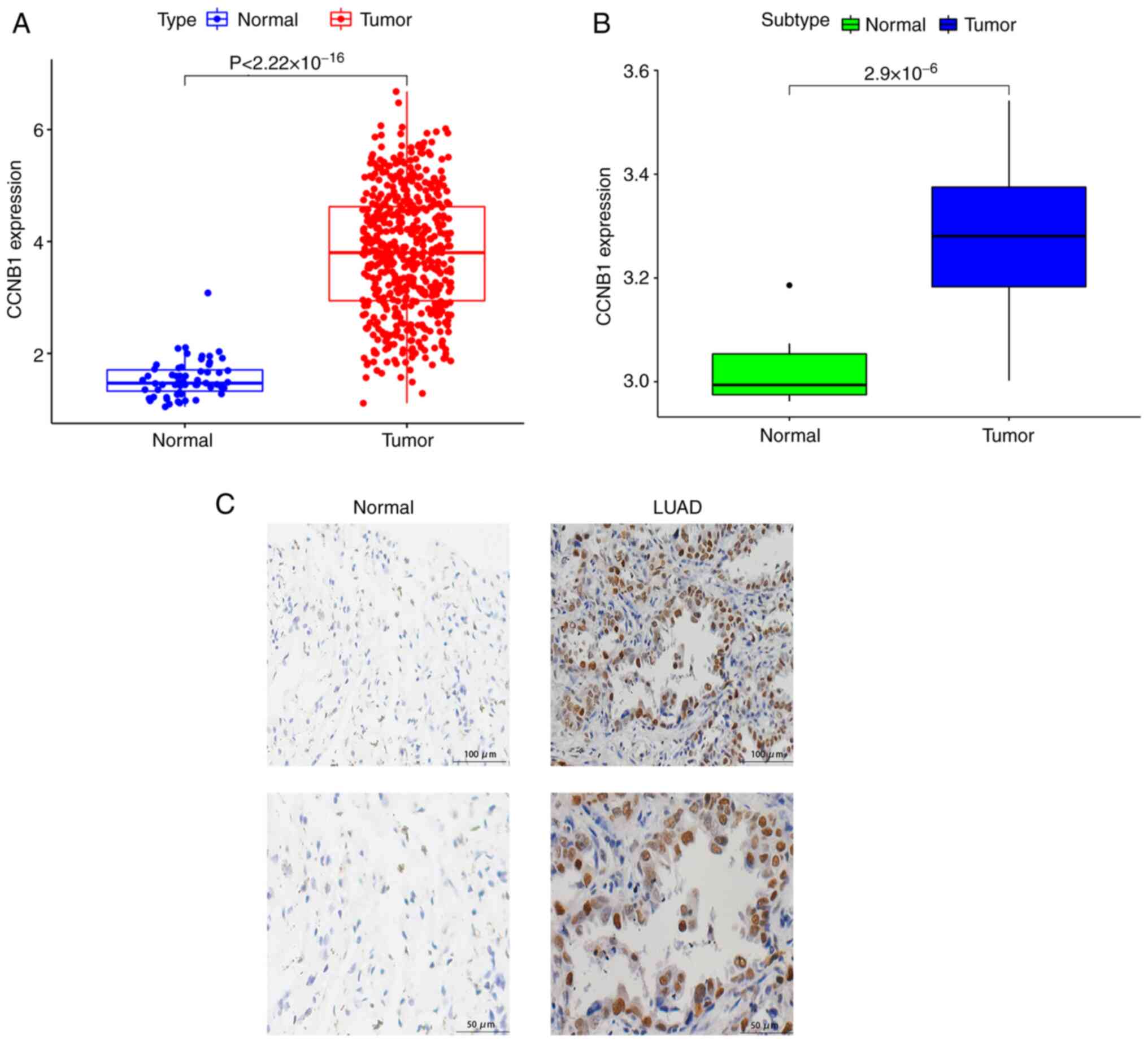
Expression of CCNB1 is higher in LUAD
tissues than in normal lung tissues
To investigate the expression of CCNB1 in LUAD, data
from the TCGA and GEO databases were analyzed. The mRNA expression
of CCNB1 was significantly increased in LUAD tissues compared with
normal lung tissues in both datasets (P<0.05; Fig. 1A and B). In addition, LUAD tissues
were collected from 94 patients undergoing surgical resection and
30 normal lung tissues were acquired for comparison. To verify that
the expression of CCNB1 was higher in LUAD tissues than in normal
lung tissues, the expression levels of CCNB1 in LUAD and normal
tissues were detected using IHC. The IHC staining showed that CCNB1
was localized in the nucleus and cytoplasm (Fig. 1C). CCNB1 staining was negative in
all 30 normal lung tissues, and among the 94 LUAD tissues, the
positive expression of CCNB1 was detected in 27.66% (26/94) of
patients. The frequency of positive expression of CCNB1 in LUAD was
significantly higher than that in normal tissues (P<0.05;
Table I). These results suggest
that CCNB1 is significantly upregulated in LUAD tissues compared
with normal lung tissues.
 | Table I.Expression of cyclin B1 in primary
lung adenocarcinoma and normal lung tissues. |
Table I.
Expression of cyclin B1 in primary
lung adenocarcinoma and normal lung tissues.
|
|
| Expression (n) |
|
|
|---|
|
|
|
|
|
|
|---|
| Group | N | Negative | Positive | χ2 | P-value |
|---|
| Cancer | 94 | 68 | 26 | 10.499 | 0.001 |
| Normal | 30 | 30 | 0 |
|
|
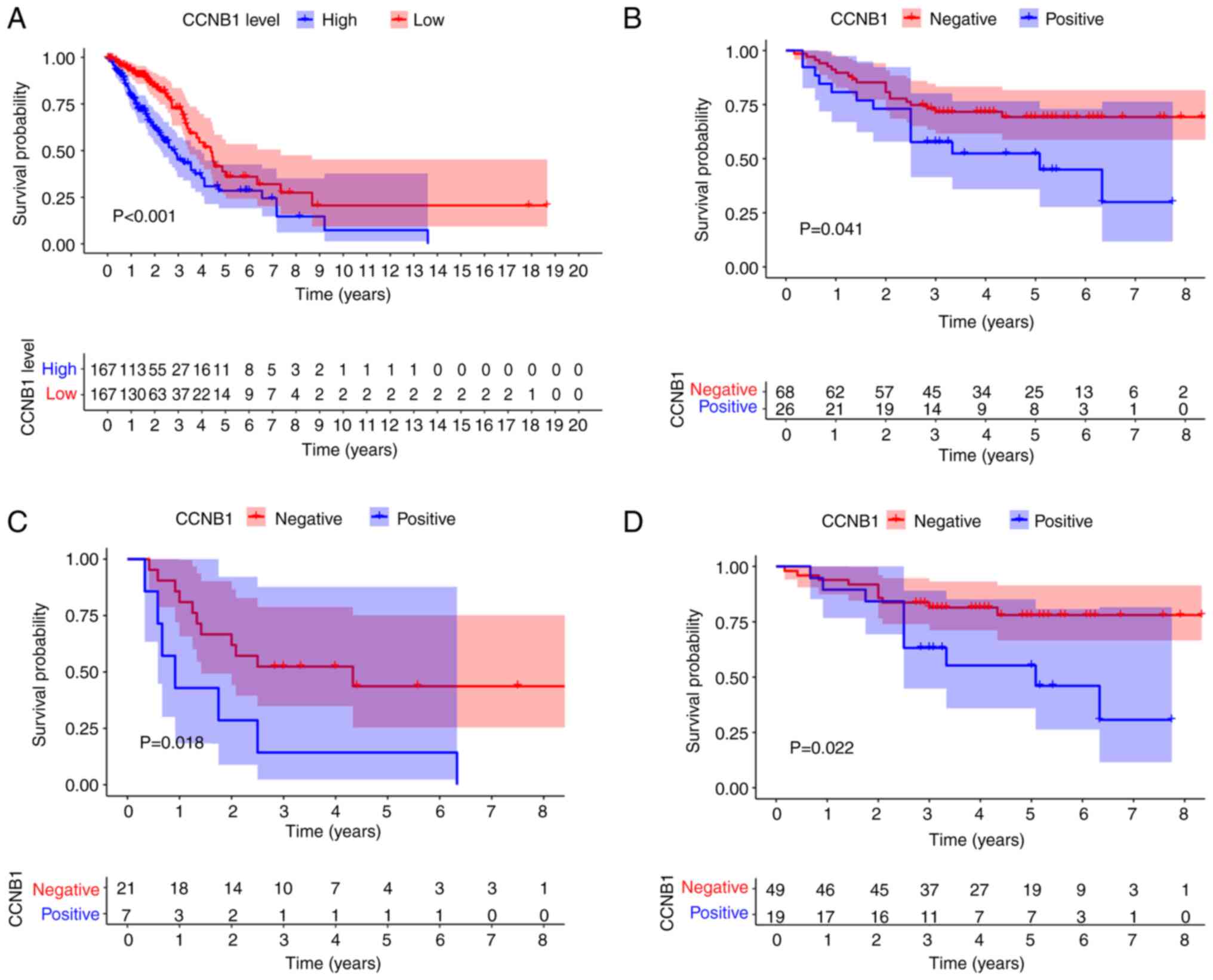
Relationship between CCNB1 expression
and the prognosis of patients with LUAD
The prognostic value of CCNB1 was assessed using
TCGA-LUAD data. Kaplan-Meier survival curves were plotted to
evaluate the relationship between CCNB1 and the prognosis of
patients with LUAD. As shown in Fig.
2A, the OS significantly differed between the CCNB1-high and
CCNB1-low patients (P<0.05). Furthermore, this conclusion was
validated by the primary patient data. Patients with positive CCNB1
expression had a worse prognosis than patients with negative CCNB1
expression (P<0.05; Fig. 2B).
To further analyze the prognostic value of CCNB1 expression in
subgroups of patients, stratification by age, sex, smoking status,
tumor size, lymph node status, pleural invasion status and clinical
stage was performed. Kaplan-Meier analysis revealed that patients
with CCNB1-positive results had a significantly shorter OS than
patients with negative CCNB1 expression in the T3 + T4 and N0 + N1
subgroups (P<0.05; Fig. 2C and
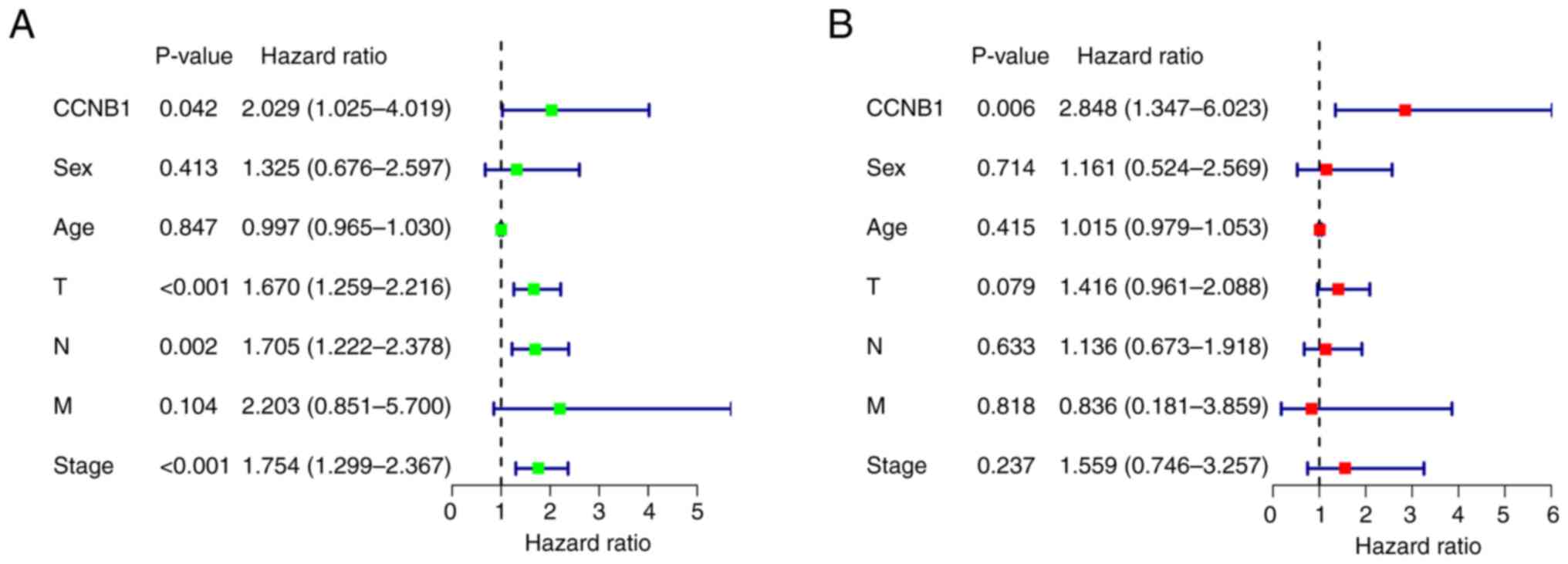
D, respectively). Univariate analysis suggested that patient
survival was influenced by T stage, N state, stage and CCNB1
expression (Fig. 3A). Furthermore,
multivariate analysis indicated that CCNB1 expression is an
independent prognostic factor in patients with LUAD (Fig. 3B).
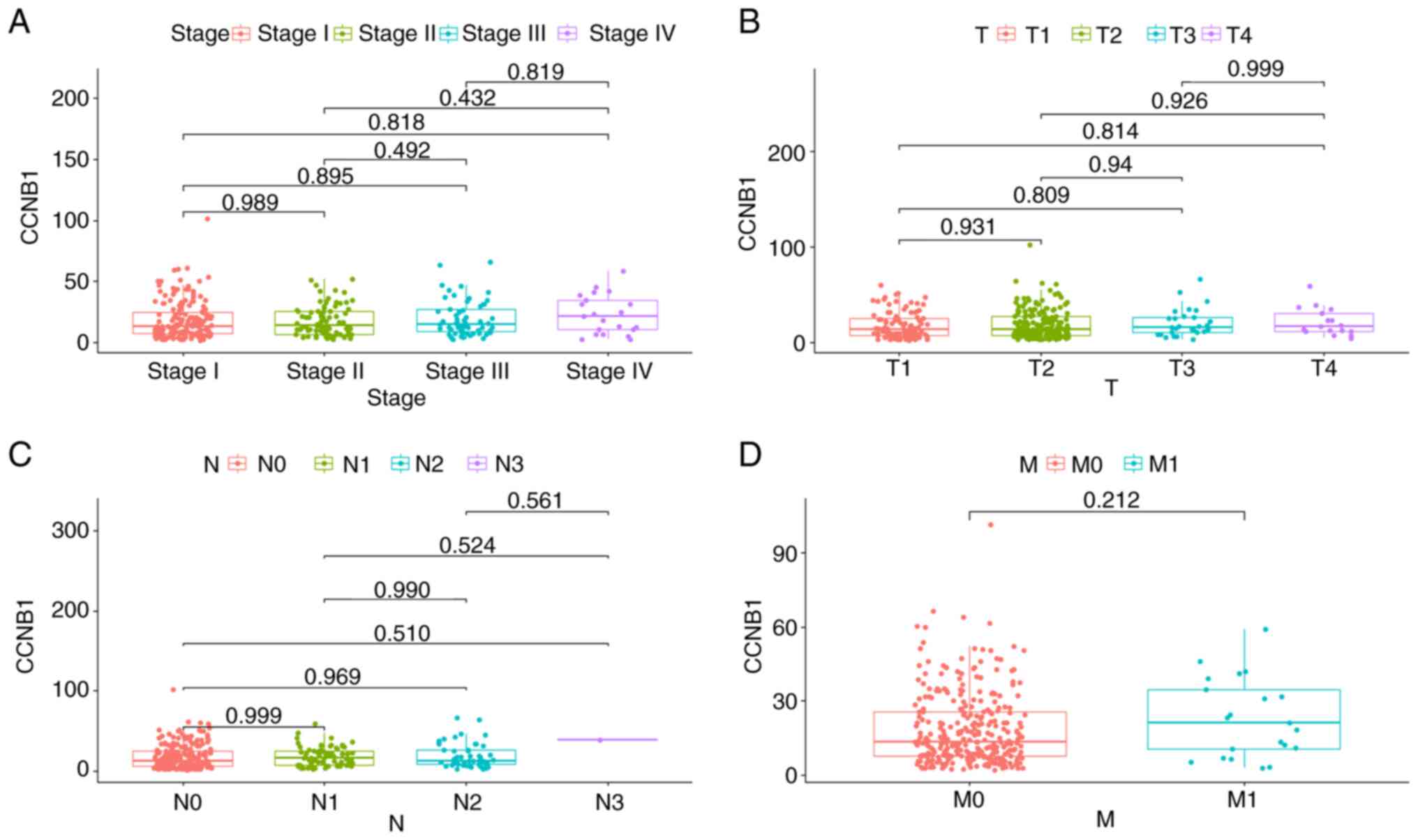
Association of CCNB1 expression with
the clinicopathological parameters of patients with LUAD
The relationship between CCNB1 expression levels and
the clinicopathological parameters of 334 patients in the TCGA-LUAD
dataset were investigated. As shown in Fig. 4A, no significant difference in
CCNB1 expression levels between stages I, II, III and IV was
detected. Following this, the expression of CCNB1 in patients with
different TNM stages was also compared, and no differences were
identified (Fig. 4B-D). These
results were consistent with the primary data collected from 94
patients. The associations between the CCNB1 expression levels and
clinicopathological features of the patients are summarized in
Table II. The results revealed no
significant association between CCNB1 expression and clinical
parameters, including age, sex, smoking, differentiation, bronchial
margin, tumor size, lymph node metastasis, distant metastasis, T
stage, pathological stage, visceral pleural invasion and tumor
type.
 | Table II.Association of cyclin B1 expression
with clinicopathological features in patients with lung
adenocarcinoma. |
Table II.
Association of cyclin B1 expression
with clinicopathological features in patients with lung
adenocarcinoma.
|
|
| CCNB1 expression
(n) |
|
|---|
|
|
|
|
|
|---|
| Feature | N | Negative | Positive | P-value |
|---|
| Age |
|
|
| 0.521 |
|
≤55 | 42 | 29 | 13 |
|
|
>55 | 52 | 39 | 13 |
|
| Sex |
|
|
| 0.367 |
|
Female | 54 | 41 | 13 |
|
|
Male | 40 | 27 | 13 |
|
| Smoking |
|
|
| 0.478 |
|
Yes | 38 | 29 | 9 |
|
| No | 56 | 39 | 17 |
|
| Tumor size
(cm) |
|
|
| 0.775 |
|
≤3.5 | 60 | 44 | 16 |
|
| >3.5
cm | 34 | 24 | 10 |
|
|
Differentiation |
|
|
| 0.112 |
|
Low/moderate | 49 | 32 | 17 |
|
|
High | 45 | 36 | 9 |
|
| T stage |
|
|
| 0.707 |
| T1 +
T2 | 66 | 47 | 19 |
|
| T3 +
T4 | 28 | 21 | 7 |
|
| Lymph node
metastasis |
|
|
| 0.484 |
|
Yes | 31 | 21 | 10 |
|
| No | 63 | 47 | 16 |
|
| Distant
metastasis |
|
|
| 0.704a |
|
Yes | 9 | 6 | 3 |
|
| No | 85 | 62 | 23 |
|
| Pathological
stage |
|
|
| 0.912 |
| I +
II | 57 | 41 | 16 |
|
| III +
IV | 37 | 27 | 10 |
|
| Visceral pleural
invasion |
|
|
| 0.961 |
| No | 51 | 37 | 14 |
|
|
Yes | 43 | 31 | 12 |
|
| Bronchial
margin |
|
|
| 0.732a |
|
Positive | 12 | 8 | 4 |
|
|
Negative | 82 | 60 | 22 |
|
| Tumor type |
|
|
| 1.000a |
|
Central | 11 | 8 | 3 |
|
|
Peripheral | 83 | 60 | 23 |
|
RNA sequencing of H1299 cells with
CCNB1 knockdown
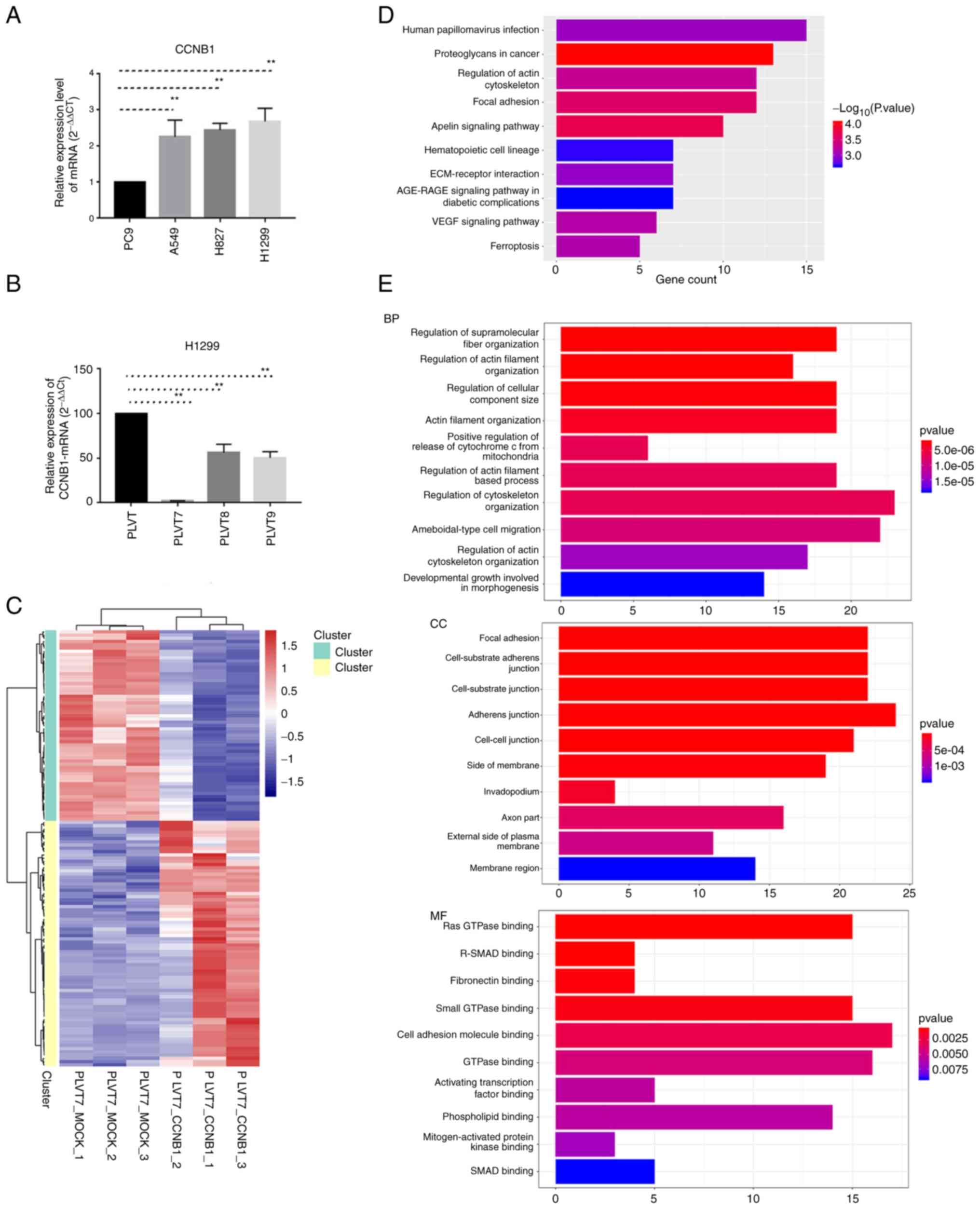
To detect the expression level of CCNB1 in
vitro, four LUAD cell lines, namely A549, H827, H1299 and PC9,
were analyzed. The RT-qPCR results suggested that CCNB1 expression
in H1299 cells was higher than that in the other three cell lines
(Fig. 5A); therefore, H1299 cells
were selected for subsequent experiments. To explore the specific
mechanism of CCNB1 in LUAD, CCNB1 expression was knocked down using
siRNAs in H1299 cells. Among the three CCNB1 siRNAs, CCNB1-PLVT7
was the most effective in knocking down CCNB1 at the mRNA level
(Fig. 5B). Therefore, RNA-seq was
performed on H1299 cells transfected with CCNB1-PLVT7 as the
experimental group and transfected with PLVT7-mock as the control
group, using three repeat samples for each group. The RNAs that
underwent changes in expression in H1299 cells with CCNB1 knockdown
were analyzed, and 135 DEGs in total were identified. By setting
log2(fold change)>1.5 as the upregulated threshold
and <-1.5 as the downregulated threshold, 76 upregulated genes
and 59 downregulated genes were detected (Fig. 5C). To explore the underlying
biological functions of CCNB1 downregulation in LUAD, GO and KEGG
enrichment analyses were performed on the 135 genes. The results of
KEGG pathway analysis indicated that DEGs were mainly enriched in
‘human papillomavirus infection’, ‘proteoglycans in cancer’,
‘regulation of actin cytoskeleton’, ‘focal adhesion’ and the
‘apelin signaling pathway’ (Fig.
5D). In addition, the results of GO enrichment analysis showed
that the BP of the DEGs included ‘regulation of supramolecular
fiber organization’, ‘regulation of actin filament organization’,
‘regulation of cellular component size’ and ‘regulation of
cytoskeleton organization’ (Fig.
5E). The main CC biological processes included ‘focal
adhesion’, ‘cell-substrate adherens junction’ and ‘cell-cell
junction’ (Fig. 5E). The main MFs
of the DEGs were ‘Ras GTPase binding’, ‘R-SMAD binding’,
‘fibronectin binding’ and ‘small GTPase binding’ (Fig. 5E).
Identification of co-expressed genes
based on WGCNA and RNA-seq
CCNB1 has been shown to interact with other genes to
promote the occurrence and development of tumors (23). Therefore, to further study the
interaction between the DEGs identified in the present study,
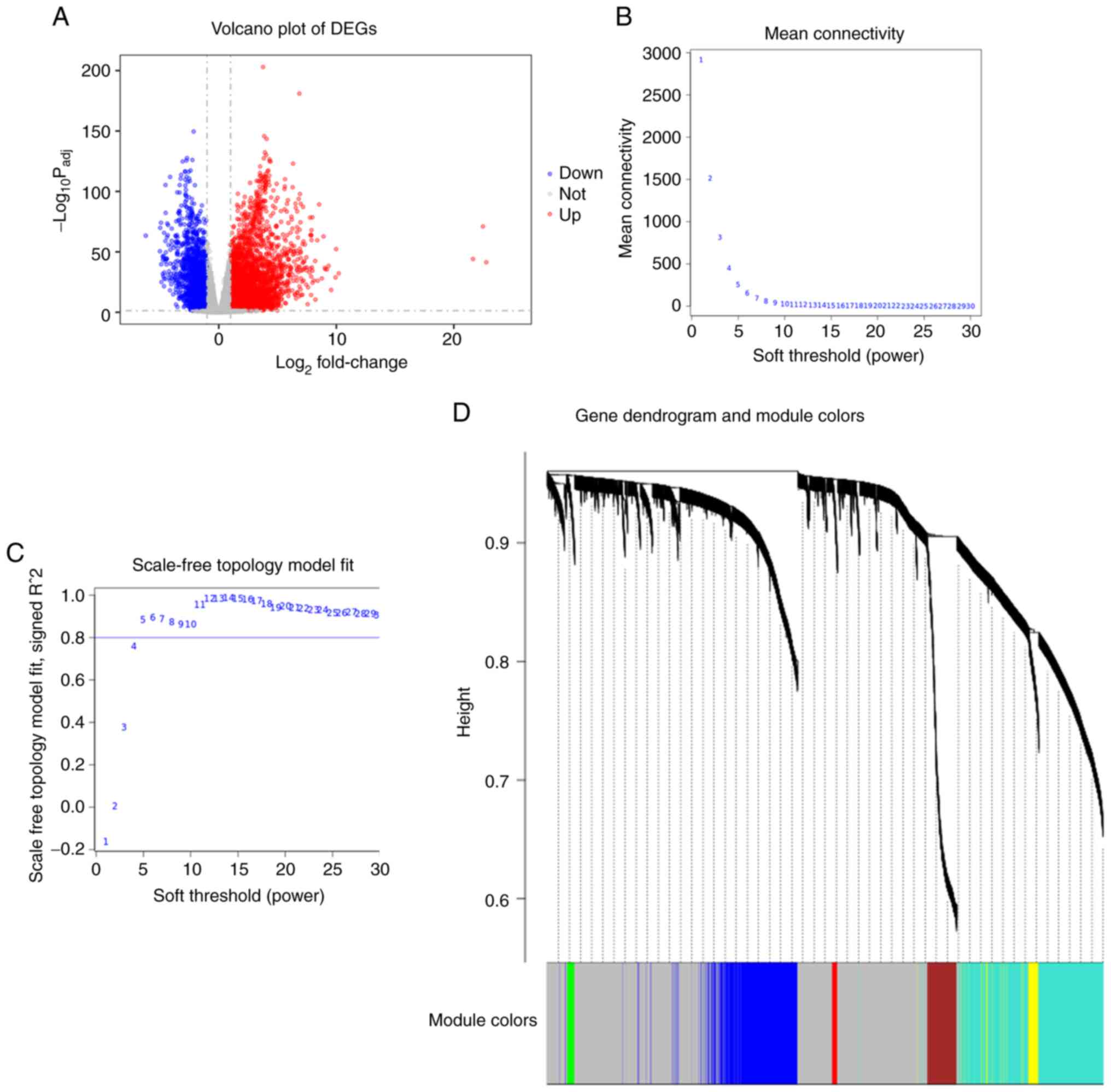
cluster analysis was performed. A total of 5,756 DEGs were screened
from the TCGA-LUAD dataset and are represented as a volcano plot
(Fig. 6A). Co-expression analysis
was carried out to construct a co-expression network. A power of
β=5 was selected as the soft-thresholding parameter to ensure a
scale-free network (Fig. 6B and
C). A total of 7 modules were identified via average lineage
hierarchical clustering (Fig. 6D).
The 850 genes contained in the blue module had the highest
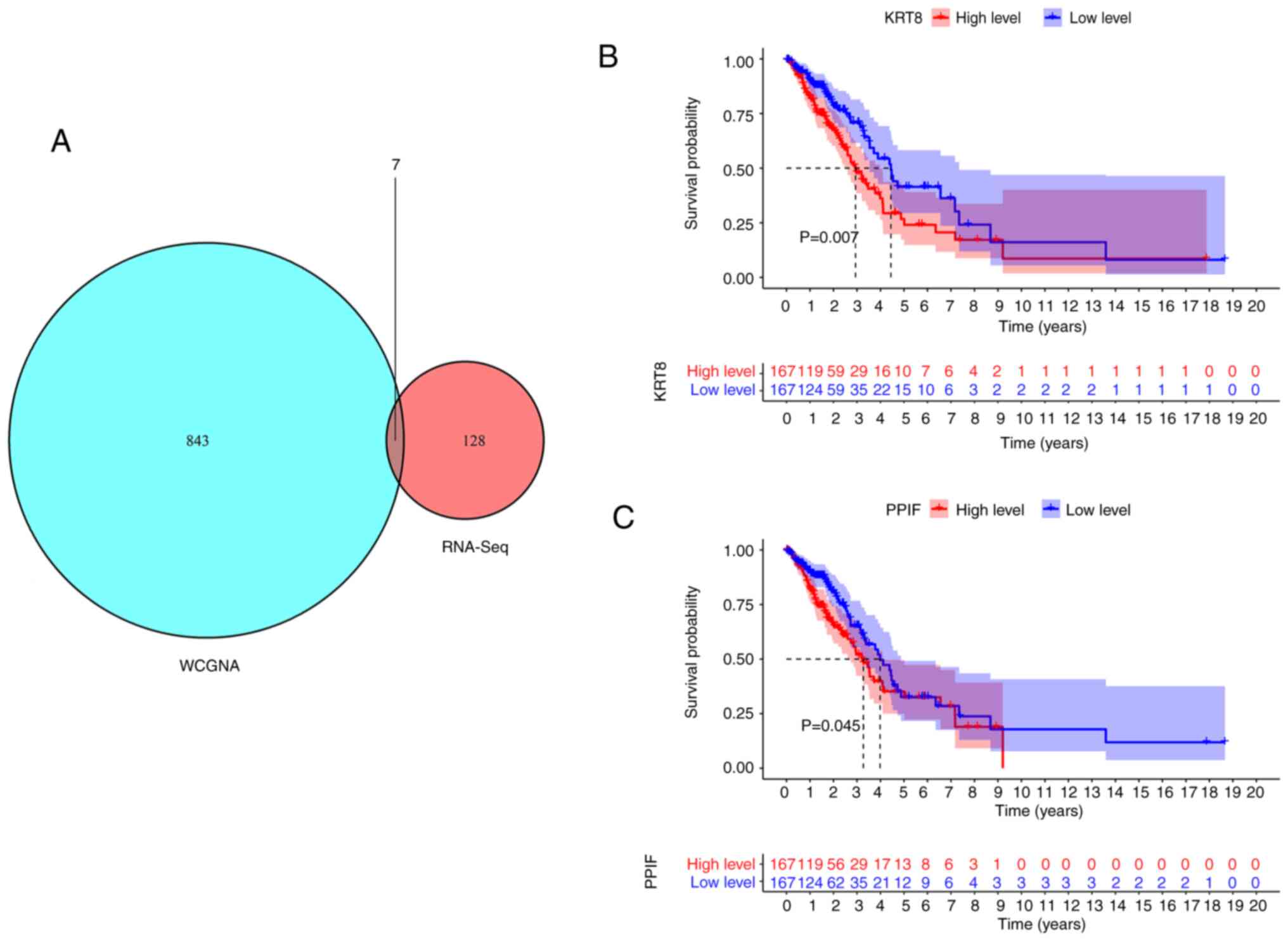
association with CCNB1 expression. Comparison of the RNA-seq and
WGCNA data led to the identification of six common genes: Complexin
1 (CPLX1), peptidylprolyl isomerase F (PPIF), serine-arginine
protein kinase 2 (SRPK2), keratin 8 (KRT8), solute carrier family
member 20 member 1 (SLC20A1) and chromobox 2 (CBX2) (Fig. 7A). Kaplan-Meier survival curves
were plotted for the 334 patients in the TCGA-LUAD dataset to
evaluate the relationship between these genes and the prognosis of
patients with LUAD. As shown in Fig.
7B and C, high KRT8 or PPIF expression levels were unfavorable
to the OS of patients with LUAD (P<0.05), while the other four
genes had no effect on prognosis (data not shown). These results
indicate that CCNB1, as an independent prognostic factor of LUAD,
may interact with CPLX1, PPIF, SRPK2, KRT8, SLC20A1 and CBX2 to
influence the outcome of patients with LUAD.
Discussion
In the present study, clinical samples and
bioinformatics methods were used to show that CCNB1 is highly
expressed in LUAD tissues. Kaplan-Meier survival curves and
multivariate Cox regression analysis confirmed that CCNB1 is an
independent prognostic factor for patients with LUAD. Higher CCNB1
expression predicted worse overall survival, indicating that CCNB1
is an oncogene. However, CCNB1 expression was not found to be
associated with any clinicopathological parameters. Integration of
the results of RNA-seq and WGCNA analyses to identify intersecting
genes indicated that CCNB1 may cooperate with CPLX1, PPIF, SRPK2,
KRT8, SLC20A1 and CBX2 to affect the prognosis of patients with
LUAD. In addition, GO and KEGG pathway analyses showed that a
reduction in CCNB1 expression induces changes in different
pathways.
Although the exact mechanism of CCNB1 upregulation
is unclear, CCNB1 is known to be essential for the survival and
proliferation of tumor cells; upregulated CCNB1 binds to its
partner CDKs and promotes cancer cell growth (24). High levels of CCNB1 are associated
with the immortalization of tumor cells and chromosomal
instability, which contribute to tumor cell invasion and the
prognosis of patients with cancer (25). Conversely, the decreased expression
of CCNB1 causes tumor cell death (26). Downregulation of the expression of
CCNB1 has been shown to activate the p53 signaling pathway and
thereby inhibit HCC cell growth (27). Although the role of CCNB1 has been
reported in several types of cancer (28–32),
its involvement in LUAD has not been elucidated. In the present
study it was found that CCNB1 expression was significantly
upregulated in LUAD tissues. These results suggest that CCNB1 may
be involved in promoting the transformation of normal tissues into
cancerous tissues and could be a cancer promoter. Kaplan-Meier
survival analyses showed that patients with high CCNB1 expression
had worse OS than those with low CCNB1 expression, which is
consistent with previous reports of CCNB1 in hypopharyngeal
squamous cell carcinoma (11),
liver cancer (12,33) and oesophageal cancer (34). However, Chae et al (35) did not detect any association of the
expression of CCNB1 with the prognosis of patients with breast
cancer. These inconsistent findings could potentially be explained
by the different expression patterns of CCNB1 in different types of
tumors.
Furthermore, the associations between CCNB1
expression and clinicopathological parameters were analyzed in the
present study using TCGA data and the immunohistochemical results
of 94 patients with LUAD. However, as there were no positive
findings, CCNB1 appears to be a relatively independent expression
factor. Similar findings have been reported in previous studies on
breast cancer (35), pediatric
embryonic tumors (11) and
pancreatic cancer (36). However,
some studies have identified associations between CCNB1 and
clinical factors in LUAD. For example Wang et al (18) found that CCNB1 expression level was
clinically associated with sex, smoking, T stage and N stage in
institutional and TCGA NSCLC cohorts. Furthermore, Bao et al
(19) determined the expression of
CCNB1 mRNA in patient tissues using RT-qPCR, which demonstrated
that CCNB1 expression was high in LUAD tissues and associated with
advanced tumor stages and shorter overall survival. However, in the
present study, immunohistochemistry was used to detect CCNB1
protein expression, and the results may differ according to the
experimental methods used. It is necessary to further expand the
sample size and continue to explore the effect of CCNB1 on the
clinicopathological factors of LUAD in future studies.
The mechanism of CCNB1 in LUAD was further explored
in the present study by knocking down the expression of CCNB1 in
H1299 cells and performing RNA-seq to detect the changes in gene
expression at the transcriptional level. The GO and KEGG analysis
results showed that the knockdown of CCNB1 caused changes in
pathways associated with cytoskeleton-related proteins, the
formation of focal adhesions, Ras GTPase binding and small GTPase
binding. The increased expression of focal adhesion-associated
proteins affects cell junction functions and suggests a change in
the GTPase pathway. GTPase is a molecular switch for cell-signal
transduction, and it serves an important role in dynamic changes of
the cytoskeleton. Cell movement regulates malignant cell
transformation, proliferation and tumor angiogenesis, invasion and
metastasis (37,38). Another focal adhesion molecule,
namely focal adhesion kinase (FAK), also plays a key role in
numerous signal transduction pathways associated with tumor
proliferation, apoptosis, metastasis, invasion and angiogenesis,
and so is a potential antitumor target (39). FAK is overexpressed in a variety of
cancers and is closely associated with the occurrence and
development of tumors (40). As a
functional protein in the cytoplasm, it usually acts in a
kinase-dependent manner (41). By
studying the expression and distribution of key proteins in the
integrin-FAK-Rho GTPase signaling pathway, Shen et al
(42) elucidated their
relationship with the molecular mechanism of endothelial cell
adhesion and migration; cell migration and FAK phosphorylation
levels are closely associated with the regulation of Rho GTPase
expression. Therefore, we hypothesize that the high expression of
CCNB1 regulates the formation of focal adhesions in LUAD, affects
FAK phosphorylation and then activates the GTPase pathway, thereby
affecting the invasion and migration ability of LUAD cells and
ultimately affecting the prognosis of patients.
The WGCNA results obtained in the present study
showed that 850 genes, including CCNB1, were co-expressed in LUAD.
After identifying the intersecting RNA-seq and WGCNA results, it
was found that the expression of CPLX1, PPIF, SRPK2, KRT8, SLC20A1
and CBX2 was closely associated with that of CCNB1. Notably,
Kaplan-Meier analyses revealed that high expression of KRT8 and
PPIF was associated with poor prognosis. Previous studies have
shown the significant upregulation of KRT8 expression in various
types of human cancer (43–45)
and its predominant expression in epithelial cells. The aberrant
expression of KRT8 in multiple types of tumors has been shown to be
associated with cell migration (46), cell adhesion (47) and drug resistance (48). Other studies have reported that
PPIF is involved in mitochondrial permeability transition-regulated
necrosis and necroptosis (49,50),
and strongly upregulated in endometrial cancer tissues with an
expression profile closely associated with promoter hypomethylation
(51). These data suggest that
CPLX1, PPIF, SRPK2, KRT8, SLC20A1 and CBX2, particularly KRT8 and
PPIF, are key genes involved in the biological effects of CCNB.
However, the relationship between KRT8, PPIF and CCNB1 has not been
reported in other related studies and is worthy of further
exploration.
Interestingly, consistent results were obtained
using TCGA data and clinical samples, both of which indicate that
the increased expression of CCNB1 is a marker of poor prognosis for
LUAD. Moreover, the findings suggest that CCNB1 may affect the
expression of CPLX1, PPIF, SRPK2, KRT8, SLC20A1 and CBX2 genes,
leading to a poor prognosis in patients with LUAD. This study
provides a comprehensive and reliable theoretical basis and data
source for subsequent studies of CCNB1 in LUAD. However, the study
has certain limitations. Firstly, the sample size was small and a
larger sample size should be analyzed to further confirm the
expression and prognostic value of CCNB1. Secondly, the mechanism
merits further study, but no cell experiments were conducted to
verify the potential mechanism. Further intensive in vitro
and in vivo investigations should help to clarify the
underlying mechanism of CCNB1 in the pathogenesis and development
of LUAD. It is hoped that CCNB1 can be applied to the clinical
practice of patients with LUAD to guide their prognosis and
facilitate individualized treatment.
In conclusion, the present study identified that
CCNB1 was highly expressed in patients with LUAD and associated
with a poor prognosis. Patients whose IHC results were positive for
CCNB1 expression had a significantly shorter OS than patients with
whose results were negative. CCNB1 may affect the expression of the
CPLX1, PPIF, SRPK2, KRT8, SLC20A1 and CBX2 genes and be
functionally regulated by different pathways. CCNB1 has the
potential to become a novel prognostic target for LUAD and may
assist physicians in finding new diagnostic and therapeutic methods
for patients with LUAD.
Acknowledgements
Not applicable.
Funding
This study was supported by the National Natural Science
Foundation of China (grant nos. 81860469 and 82160574) and the
Science and Technology Support Program of Guizhou [grant no.
qiankehejichu (2020) 1Z063, qiankehezhicheng (2021 normal
073)].
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request. The raw sequencing datasets generated during the current
study are available in the GEO repository (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE207450).
Authors' contributions
YL, YXL, QL and CC conceived the project and
participated in study design and interpretation of the results. YL
wrote the manuscript. YXS, FC and YDD participated in study design
and helped to revise the manuscript. YL and QYW contributed to
sample collection and acquisition of patients' clinical and
survival data. YXL, NJ and HD conducted experiments. YL and QL
confirm the authenticity of all the raw data. All authors have read
and approved the final manuscript.
Ethics approval and consent to
participate
All experiments using human tissue were approved by
the Ethics Committee of Zunyi Medical University [no. (2021)1-098].
Written informed consent was obtained from all patients for the use
of their tissues in the study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2020. CA Cancer J Clin. 70:7–30. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chen Z, Fillmore CM, Hammerman PS, Kim CF
and Wong KK: Non-small-cell lung cancers: A heterogeneous set of
diseases. Nat Rev Cancer. 14:535–546. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Xia L, Zhu Y, Zhang C, Deng S, Deng Y,
Yang Z, Mei J and Liu L: Decreased expression of EFCC1 and its
prognostic value in lung adenocarcinoma. Ann Transl Med. 7:6722019.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Macheleidt IF, Dalvi PS, Lim SY, Meemboor
S, Meder L, Käsgen O, Müller M, Kleemann K, Wang L, Nurnberg P, et
al: Preclinical studies reveal that LSD1 inhibition results in
tumor growth arrest in lung adenocarcinoma independently of driver
mutations. Mol Oncol. 12:1965–1979. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lim W, Bae H, Bazer FW and Song G: Ephrin
A1 promotes proliferation of bovine endometrial cells with abundant
expression of proliferating cell nuclear antigen and cyclin D1
changing the cell population at each stage of the cell cycle. J
Cell Physiol. 234:4864–4873. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hydbring P, Malumbres M and Sicinski P:
Non-canonical functions of cell cycle cyclins and cyclin-dependent
kinases. Nat Rev Mol Cell Biol. 17:280–292. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Braschi B, Denny P, Gray K, Jones T, Seal
R, Tweedie S, Yates B and Bruford E: Genenames.org: The HGNC and
VGNC resources in 2019. Nucleic Acids Res. 47(D1): D786–D792. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Vishnoi N and Yao J: Single-cell,
single-mRNA analysis of Ccnb1 promoter regulation. Sci Rep.
7:20652017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Liu D, Xu W, Ding X, Yang Y, Su B and Fei
K: Polymorphisms of CCNB1 associated with the clinical outcomes of
platinum-based chemotherapy in Chinese NSCLC patients. J Cancer.
8:3785–3794. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wei LJ, Li JA, Bai DM and Song Y:
miR-223-RhoB signaling pathway regulates the proliferation and
apoptosis of colon adenocarcinoma. Chem Biol Interact. 289:9–14.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Li W, Dong Q, Li L, Zhang Z, Cai X and Pan
X: Prognostic significance of claudin-1 and cyclin B1 protein
expression in patients with hypopharyngeal squamous cell carcinoma.
Oncol Lett. 11:2995–3002. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chai N, Xie HH, Yin JP, Sa KD, Guo Y, Wang
M, Liu J, Zhang XF, Zhang X, Yin H, et al: FOXM1 promotes
proliferation in human hepatocellular carcinoma cells by
transcriptional activation of CCNB1. Biochem Biophys Res Commun.
500:924–929. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Gu J, Liu X, Li J and He Y: MicroRNA-144
inhibits cell proliferation, migration and invasion in human
hepatocellular carcinoma by targeting CCNB1. Cancer Cell Int.
19:152019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Fang Y, Liang X, Jiang W, Li J, Xu J and
Cai X: Cyclin b1 suppresses colorectal cancer invasion and
metastasis by regulating e-cadherin. PLoS One. 10:e01268752015.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Sit M, Aktas G, Ozer B, Kocak MZ, Erkus E,
Erkol H, Yaman S and Savli H: Mean platelet volume: An overlooked
herald of malignant thyroid nodules. Acta Clin Croat. 58:417–420.
2019.PubMed/NCBI
|
|
16
|
Wang Y, Ruan Y and Wu S: ET-1 regulates
the human umbilical vein endothelial cell cycle by adjusting the
ERβ/FOXN1 signaling pathway. Ann Transl Med. 8:14992020. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Brcic L, Heidinger M, Sever AZ, Zacharias
M, Jakopovic M, Fediuk M, Maier A, Quehenberger F, Seiwerth S and
Popper H: Prognostic value of cyclin A2 and B1 expression in lung
carcinoids. Pathology. 51:481–486. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wang F, Chen X, Yu X and Lin Q:
Degradation of CCNB1 mediated by APC11 through UBA52 ubiquitination
promotes cell cycle progression and proliferation of non-small cell
lung cancer cells. Am J Transl Res. 11:7166–7185. 2019.PubMed/NCBI
|
|
19
|
Bao B, Yu X and Zheng W: MiR-139-5p
targeting CCNB1 modulates proliferation, migration, invasion and
cell cycle in lung adenocarcinoma. Mol Biotechnol. 64:852–860.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Leon LM, Gautier M, Allan R, Ilié M,
Nottet N, Pons N, Paquet A, Lebrigand K, Truchi M, Fassy J, et al:
Correction: The nuclear hypoxia-regulated NLUCAT1 long non-coding
RNA contributes to an aggressive phenotype in lung adenocarcinoma
through regulation of oxidative stress. Oncogene. 40:26212021.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Aran D, Camarda R, Odegaard J, Paik H,
Oskotsky B, Krings G, Goga A, Sirota M and Butte AJ: Comprehensive
analysis of normal adjacent to tumor transcriptomes. Nat Commun.
8:10772017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Arora S, Singh P, Rahmani AH, Almatroodi
SA, Dohare R and Syed MA: Unravelling the role of miR-20b-5p,
CCNB1, HMGA2 and E2F7 in development and progression of non-small
cell lung cancer (NSCLC). Biology (Basel). 9:2012020.PubMed/NCBI
|
|
24
|
Fang L, Du WW, Awan FM, Dong J and Yang
BB: The circular RNA circ-Ccnb1 dissociates Ccnb1/Cdk1 complex
suppressing cell invasion and tumorigenesis. Cancer Lett.
459:216–226. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhan Q, Antinore MJ, Wang XW, Carrier F,
Smith ML, Harris CC and Fornace AJ Jr: Association with Cdc2 and
inhibition of Cdc2/Cyclin B1 kinase activity by the p53-regulated
protein Gadd45. Oncogene. 18:2892–2900. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Vairapandi M, Balliet AG, Hoffman B and
Liebermann DA: GADD45b and GADD45g are cdc2/cyclinB1 kinase
inhibitors with a role in S and G2/M cell cycle checkpoints induced
by genotoxic stress. J Cell Physiol. 192:327–338. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Li M, Shang H, Wang T, Yang SQ and Li L:
Huanglian decoction suppresses the growth of hepatocellular
carcinoma cells by reducing CCNB1 expression. World J
Gastroenterol. 27:939–958. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ding K, Li W, Zou Z, Zou X and Wang C:
CCNB1 is a prognostic biomarker for ER+ breast cancer. Med
Hypotheses. 83:359–364. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Chen EB, Qin X, Peng K, Li Q, Tang C, Wei
YC, Yu S, Gan L and Liu TS: HnRNPR-CCNB1/CENPF axis contributes to
gastric cancer proliferation and metastasis. Aging (Albany NY).
11:7473–7491. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Moon SJ, Kim JH, Kong SH and Shin CS:
Protein expression of cyclin B1, transferrin receptor, and
fibronectin is correlated with the prognosis of adrenal cortical
carcinoma. Endocrinol Metab (Seoul). 35:132–141. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Xing Z and Wang X, Liu J, Zhang M, Feng K
and Wang X: Expression and prognostic value of CDK1, CCNA2, and
CCNB1 gene clusters in human breast cancer. J Int Med Res.
49:3000605209806472021.PubMed/NCBI
|
|
32
|
Radhakrishnan A, Nanjappa V, Raja R, Sathe
G, Chavan S, Nirujogi RS, Patil AH, Solanki H, Renuse S,
Sahasrabuddhe NA, et al: Dysregulation of splicing proteins in head
and neck squamous cell carcinoma. Cancer Biol Ther. 17:219–229.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhuang L, Yang Z and Meng Z: Upregulation
of BUB1B, CCNB1, CDC7, CDC20, and MCM3 in tumor tissues predicted
worse overall survival and disease-free survival in hepatocellular
carcinoma patients. Biomed Res Int. 2018:78973462018. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Fu S, Jin L, Gong T, Pan S, Zheng S, Zhang
X, Yang T, Sun Y, Wang Y, Guo J, et al: Effect of sinomenine
hydrochloride on radiosensitivity of esophageal squamous cell
carcinoma cells. Oncol Rep. 39:1601–1608. 2018.PubMed/NCBI
|
|
35
|
Chae SW, Sohn JH, Kim DH, Choi YJ, Park
YL, Kim K, Cho YH, Pyo JS and Kim JH: Overexpressions of cyclin B1,
cdc2, p16 and p53 in human breast cancer: The clinicopathologic
correlations and prognostic implications. Yonsei Med J. 52:445–453.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhou L, Li J, Zhao YP, Cui QC, Zhou WX,
Guo JC, You L, Wu WM and Zhang TP: The prognostic value of cyclin
B1 in pancreatic cancer. Med Oncol. 31:1072014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Woldu SL, Hutchinson RC, Krabbe LM, Sanli
O and Margulis V: The Rho GTPase signalling pathway in urothelial
carcinoma. Nat Rev Urol. 15:83–91. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Lawson CD and Ridley AJ: Rho GTPase
signaling complexes in cell migration and invasion. J Cell Biol.
217:447–457. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhou B, Wang GZ, Wen ZS, Zhou YC, Huang
YC, Chen Y and Zhou GB: Somatic mutations and splicing variants of
focal adhesion kinase in non-small cell lung cancer. J Natl Cancer
Inst. 110:195–204. 2018. View Article : Google Scholar
|
|
40
|
Walker S, Foster F, Wood A, Owens T,
Brennan K, Streuli CH and Gilmore AP: Oncogenic activation of FAK
drives apoptosis suppression in a 3D-culture model of breast cancer
initiation. Oncotarget. 7:70336–70352. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Brami-Cherrier K, Gervasi N, Arsenieva D,
Walkiewicz K, Boutterin MC, Ortega A, Leonard PG, Seantier B, Gasmi
L, Bouceba T, et al: FAK dimerization controls its kinase-dependent
functions at focal adhesions. EMBO J. 33:356–370. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Shen Y, Ma Y, Gao M, Lai Y, Wang G, Yu Q,
Cui FZ and Liu X: Integrins-FAK-Rho GTPases pathway in endothelial
cells sense and response to surface wettability of plasma
nanocoatings. ACS Appl Mater Interfaces. 5:5112–5121. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Selamat SA, Chung BS, Girard L, Zhang W,
Zhang Y, Campan M, Siegmund KD, Koss MN, Hagen JA, Lam WL, et al:
Genome-scale analysis of DNA methylation in lung adenocarcinoma and
integration with mRNA expression. Genome Res. 22:1197–1211. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Sanchez-Carbayo M, Socci ND, Lozano J,
Saint F and Cordon-Cardo C: Defining molecular profiles of poor
outcome in patients with invasive bladder cancer using
oligonucleotide microarrays. J Clin Oncol. 24:778–789. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Curtis C, Shah SP, Chin SF, Turashvili G,
Rueda OM, Dunning MJ, Speed D, Lynch AG, Samarajiwa S, Yuan Y, et
al: The genomic and transcriptomic architecture of 2,000 breast
tumours reveals novel subgroups. Nature. 486:346–352. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Fortier AM, Asselin E and Cadrin M:
Keratin 8 and 18 loss in epithelial cancer cells increases
collective cell migration and cisplatin sensitivity through
claudin1 up-regulation. J Biol Chem. 288:11555–11571. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Galarneau L, Loranger A, Gilbert S and
Marceau N: Keratins modulate hepatic cell adhesion, size and G1/S
transition. Exp Cell Res. 313:179–194. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Wang Y, He QY, Tsao SW, Cheung YH, Wong A
and Chiu JF: Cytokeratin 8 silencing in human nasopharyngeal
carcinoma cells leads to cisplatin sensitization. Cancer Lett.
265:188–196. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Mulay SR, Honarpisheh MM, Foresto-Neto O,
Shi C, Desai J, Zhao ZB, Marschner JA, Popper B, Buhl EM, Boor P,
et al: Mitochondria permeability transition versus necroptosis in
oxalate-induced AKI. J Am Soc Nephrol. 30:1857–1869. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Nakagawa T, Shimizu S, Watanabe T,
Yamaguchi O, Otsu K, Yamagata H, Inohara H, Kubo T and Tsujimoto Y:
Cyclophilin D-dependent mitochondrial permeability transition
regulates some necrotic but not apoptotic cell death. Nature.
434:652–658. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Yang L, Cui Y, Sun X and Wang Y:
Overexpression of TICRR and PPIF confer poor prognosis in
endometrial cancer identified by gene co-expression network
analysis. Aging (Albany NY). 13:4564–4589. 2021. View Article : Google Scholar : PubMed/NCBI
|