Introduction
Pancreatic ductal adenocarcinoma (PDAC) is the
fourth leading cause of tumor-related death worldwide, with a
survival time of only ~6 months and a 5-year survival rate of
<8% (1,2). The poor outcome prospects are
partially attributable to its insidious onset. More than 80% of
patients have metastatic or locally advanced PDAC at first
diagnosis (2,3). In addition, PDAC is not sensitive to
radiotherapy or chemotherapy (4–9).
Yin Yang-1 (YY1) is a widely expressed transcription
factor that belongs to the GLI-Kruppel class of zinc finger
proteins. As its name suggests, YY1 can positively or negatively
control genes depending on the DNA-binding sites or cell types
(6–9). YY1 is highly expressed in PDAC where
it acts as a tumor suppressor (10). According to our previous findings,
YY1 can inhibit the proliferation, migration, and invasion of PDAC
by regulating the expression of different downstream molecules
(10,11).
Regulator of G-protein signaling 22 (RGS22) is a
newly identified protein that belongs to the RGS family, which
negatively regulate heterotrimeric G-protein signaling (12–15).
RGS22 is specifically expressed in the testis and in cancers of
epithelial origin (16).
Furthermore, overexpression of RGS22 can inhibit invasion and
migration in EC9706 esophageal squamous cell carcinoma cells
(17). In PDAC, RGS22 acts as a
tumor suppressor by repressing the migration of PDAC cells via
coupling to GNA12/13 to inhibit stress fiber formation (18). However, the expression pattern and
regulation of RGS22 in PDAC, and its influence on tumor cell
proliferation still requires investigation. Our previous
ChIP-Sequencing (ChIP-Seq) results suggested that YY1 could
directly bind to the promoter region of RGS22 (10) (Table
SI), indicating that YY1 may regulate the transcription of
RGS22. Therefore, the aim of the present study was to investigate
the expression and regulation of RGS22 in PDAC tissues, and the
potential role of YY1 in the regulation of RGS22 and its inhibitory
effects on PDAC.
Materials and methods
Patients and pancreatic tissues
A total of 52 pairs of tumorous and adjacent
nontumorous human pancreatic tissue samples (between February 2015
and August 2016) were collected from patients who underwent
pancreaticoduodenectomy for PDAC at the first Affiliated Hospital
of Nanjing Medical University. The patients were aged between 44
and 91 years, with a median age of 52 years, and included 36 male
and 16 female patients. Each tissue sample was divided into two
parts after surgical resection. One part of the tissue was fixed in
5% formalin and embedded in paraffin after 24 h for
immunohistochemistry (IHC). The other part of the tissue was stored
in liquid nitrogen for subsequent RNA extraction. Pathological
analysis of all the tissue samples was performed by two experienced
pathologists, who confirmed that the patients had PDAC based on the
pathological features of the samples. Patients were followed up
regularly until November 1st, 2018. None of the patients had
received radiation therapy or chemotherapy before cancer resection.
TNM staging was performed based on the 8th edition of the American
Joint Committee on Cancer guidelines (19). Patients were given written notice
before the surgery was performed, and the study was approved by the
Ethics Committees of the first Affiliated Hospital of Nanjing
Medical University. Informed consent was obtained from the
participants or their legal guardians. In addition, TCGA and GTEX
databases (http://gepia.cancer-pku.cn/index.html) were used to
analyze the mRNA expression of RGS22 in pancreatic cancer/adjacent
normal tissues.
Cell lines and culture
PDAC cell lines (PANC-1, CFPAC-1, BXPC-3, and
MiaPaCa-2) and a normal human pancreatic ductal cell line
(hTERT-HPNE, also known as HPNE) were obtained from Shanghai Cell
Bank. Cells were cultured in DMEM (Thermo Fisher Scientific, Inc.)
supplemented with 10% FBS (Wisent Inc.) and 1%
penicillin/streptomycin (Thermo Fisher Scientific, Inc.). Cell
lines underwent routine testing for mycoplasma contamination every
3 months. The genetic identity of the cell lines was confirmed by
short tandem repeat profiling.
Preparation of stable cell lines
PANC-1 cells stably overexpressing YY1 were
constructed as described previously (10). YY1-knockdown lentiviruses
(lentiviral plasmid: pLKD-CMV-G&PR-U6-YY1 shRNA) were
constructed by Sunbio Medical Biotechnology Co., Ltd. as described
previously (10).
RGS22-overexpression (lentiviral plasmid:
pSLenti-EF1a-mCherry-P2A-Puro-CMV-RGS22-3Flag) and RGS22-knockdown
lentiviruses (lentiviral plasmid: pLKD-CMV-G&PR-U6-RGS22 shRNA)
were constructed by Heyuan Biotechnology Co., Ltd. The sequences of
YY1-shRNA, RGS22-shRNA, and scramble shRNA are shown in Table SII. When PANC-1 and CFPAC-1 cells
reached ~40% confluence they were infected with
RGS22-overexpression, RGS22-knockdown, YY1-knockdown, or the
respective control lentiviruses. The multiplicity of infection
(MOI) used to infect PANC-1 and CFPAC-1 cells was 10. Polybrene
(final concentration, 5 µg/ml) was added to the cells before
infection with lentiviruses. After 12–20 h of infection, the medium
was replaced with fresh medium. After 72 h of infection, puromycin
(final concentration, 5 µg/ml; MilliporeSigma) was added and the
medium was replaced every 2–3 days. Stable cell lines
(PANC-1-Vector, PANC-1-RGS22, CFPAC-1-Scramble shRNA,
CFPAC-1-RGS22-shRNA and CFPAC-1-YY1-shRNA) were selected by
culturing for 14 days in media containing 5 µg/ml puromycin. The
puromycin concentration used for maintenance was 5 µg/ml. RGS22 and
YY1 expression was confirmed by reverse transcription-quantitative
PCR (RT-qPCR) and western blotting.
Transient transfection of siRNAs
Three different sequences of RGS22-knockdown siRNAs
were obtained: RS1 sense, 5′-GCUUGCAACCUCUCACAAATT-3′ and
antisense, 5′-UUUGUGAGAGGUUGCAAGCTT-3′; RS2 sense,
5′-GCUUAUCACAACCUCCUAATT-3′ and antisense,
5′-UUAGGAGGUUGUGAUAAGCTT-3′; RS3 sense, 5′-GCACCAAGAUUCUGUGUUATT-3′
and antisense, 5′-UAACACAGAAUCUUGGUGCTT-3′); and scrambled siRNA
negative control sense 5′-UUCUCCGAACGUGUCACGUTT-3′ and antisense,
5′-ACGUGACACGUUCGGAGAATT-3′. siRNAs were synthesized by Shanghai
GenePharma Co., Ltd. These RGS22-knockdown siRNAs (5 µg/ml) and 5
µg/ml Lipofectamine® 3000 (Thermo Fisher Scientific,
Inc.) were transfected into wild-type PANC-1 cells for 72 h at 37°C
according to the manufacturer's instructions. At 72 h after
transfection, the expression of RGS22 was determined by western
blotting to verify transfection efficiency. RS3 was chosen to
construct RGS22-knockdown lentiviruses, as it exhibited the maximum
efficiency out of the three sequences assessed (Fig. S1). CCK-8, colony formation, EdU,
wound healing assays and Transwell assays were performed 72 h after
RS3 transfection.
RNA isolation and RT-qPCR
TRIzol® reagent (Thermo Fisher
Scientific, Inc.) was used to extract total RNA from tissue samples
and cells according to the manufacturer's instructions. The RNA was
reverse transcribed to cDNA using an iScript cDNA Synthesis Kit
(Bio-Rad Laboratories, Inc.). RT-qPCR was performed in a Step One
Plus Real-time PCR System (Thermo Fisher Scientific, Inc.) using
the SYBR green reagent (Thermo Fisher Scientific, Inc.). Each
quantitative round of PCR was performed in triplicate and repeated
independently three times. The β-actin gene was used for
normalization. The following primers constructed by Nanjing
Realgene Biotechnology Co., Ltd. were used: RGS22 forward,
5′-AACTGGAGTTTGAACATTTCCG-3′ and reverse, RGS22,
5′-GCCTTCCTTTGATTTCGATCTC-3′; YY1 forward,
5′-ACGGCTTCGAGGATCAGATTC-3′ and reverse,
5′-TGACCAGCGTTTGTTCAATGT-3′; and β-actin forward,
5′-CTCCATCCTGGCCTCGCTGT-3′ and reverse,
5′-GCTGTCACCTTCACCGTTCC-3′.
Western blotting
Proteins were extracted from PDAC cells using RIPA
lysis buffer (Abcam). Protein concentrations were measured using a
DC protein assay kit (Bio-Rad Laboratories, Inc.). Equal amounts of
proteins (10 µg/lane) were loaded on a 10% SDS-gel, resolved using
SDS-PAGE, and transferred to a PVDF membrane. Nonspecific protein
interactions were blocked by incubation in 5% nonfat dry milk in
TBS with 0.1% Tween 20 (TBST) buffer at room temperature for 1 h
and then washed with TBST. Membranes were then incubated at 4°C
overnight with primary antibodies in fresh blocking buffer. Anti-β
actin (cat. no. ab8226; dilution, 1:1,000), anti-YY1 (cat. no.
ab109228; dilution, 1:1,000), and anti-RGS22 (cat. no. ab131048 and
ab248357; dilution, 1:1,000) antibodies were purchased from Abcam.
The blots were then washed and incubated with HRP-conjugated
secondary antibodies (cat. no. A0208; dilution, 1:1,000; Beyotime
Institute of Biotechnology) for 1 h at room temperature. Each
western blotting experiment was repeated three times
independently.
Tissue microarrays and IHC
Tissue microarrays containing 52 pairs of PDAC
samples, and their corresponding non-tumorous tissues were
constructed, and IHC was performed by Wuhan Servicebio Technology
Co., Ltd. The expression levels of RGS22 were assessed based on the
staining intensity and the percentage of positively stained cells.
Expression levels were evaluated according to the staining
intensity (0, absent; 1, weak; 2, moderate; and 3, strong staining)
and proportion of positive cells (0, <10%; 1, 10 to <50%; and
2, ≥50% of cells). The IHC score was calculated using the following
formula: IHC score=positive staining score × staining intensity
score.
Cell counting kit-8 (CCK-8) assay
A CCK-8 assay (Dojindo Molecular Technologies, Inc.)
was used to assess cell proliferation. A total of 2,000 cells per
well were cultured in 96-well plates for 5 days. At the same time
of each day, 10 µl CCK-8 reagent was mixed with 90 µl DMEM and
added to each well. After 2 h of incubation, the absorbance of each
well was assessed at 450 nm using a spectrophotometer.
Colony formation assay
PDAC cells were plated at a concentration of 500
cells per well in 6-well plates and cultured in supplemented medium
for 20 days. Colonies were stained with a 0.05% crystal violet
solution for 30 min at room temperature. Photos of the colonies
were taken using a Zeiss microscope. The number of colonies
consisting of >50 cells were counted using a light microscope
(magnification, ×100). Each sample consisted of three duplicate
wells, with three repeats.
5-EdU assay
A5-EdU assay was performed using an EdU kit
(Guangzhou RiboBio, Ltd.) according to the manufacturer's
instructions. Each sample consisted of three duplicate wells and
was repeated three times independently.
Wound-healing assay
A wound-healing assay was performed to evaluate cell
migration. Briefly, cells were seeded at a density of
1×103 cells/well into six-well plates. When cells in the
6-well plates formed a confluent monolayer, the monolayer was
scratched with a sterile 200-µl pipette tip. Images were taken
using an inverted fluorescence microscope (magnification, ×100;
Olympus Corporation) immediately and 48 h after scratching. The
relative wound areas were then measured using ImageJ 1.8.0
(National Institutes of Health).
Transwell assays
To assess the migratory and invasive abilities of
the cells, Transwell assays were performed. For the migration
assays, 4×103 cells were suspended in 1 ml serum-free
medium and then added to the upper chamber, and 1 ml supplemented
medium was added to the bottom chamber. After 24 h of culturing,
the cells in the upper chamber were wiped away using a cotton swab,
and the chambers were stained with 1% crystal violet solution for
20 min at room temperature. For the invasion experiments, Matrigel
(BD Bioscience Pharmingen) was first added to the upper chamber and
allowed to solidify, after which the above steps were performed,
except cells were cultured for 48 h. The chambers were imaged using
an inverted fluorescence microscope (magnification, ×100; Olympus
Corporation). All the experiments were repeated three times.
Luciferase activity assay
The results of our previous ChIP-Seq analysis
(10) predicted the key
YY1-binding site of RGS22 (5′-GAAAAACCATTAAAAAGTTT-3′). Luciferase
activity assays were performed based on this result by Heyuan
Biotechnology Co., Ltd. A luciferase reporter construct containing
the human RGS22 promoter (−1,331/151, upstream and downstream of
translation start site, Supplementary Materials and methods) was
prepared using the pGL4.10-basic vector (Promega Corporation).
Heyuan Biotechnology Co., Ltd. synthesized a DNA fragment of the
RGS22 promoter region (including the sites of restriction enzymes).
The DNA fragment was subcloned into the pGL4.10-basic vector to
construct pGL4.10-RGS22-promoter (WT) (WT pRGS22) recombinant
plasmid and then confirmed by sequencing. The mutant construct
containing the RGS22 promoter in which the presumed YY1-binding
site (GAAAAACCATTAAAAAGTTT, nucleotides −554 to −573), was deleted
was also constructed. The construct was named
pGL4.10-RGS22-promoter (MT pRGS22).
Transfections were performed using
Lipofectamine® 2000, according to the manufacturer's
protocol. YY1 overexpression PANC-1 cells or control cells were
plated into 12-well cell culture plates (2×105/well) 1
day before transfection. Each transfection was performed using 1 µg
luciferase reporter construct (WT pRGS22 or MT pRGS22) plus 2.5 ng
Renilla luciferase reporter vector, pRL-SV40 as an internal control
(Promega Corporation). A total of 48 h after transfection, cells
were washed with PBS and lysed using 1× lysis buffer. Firefly and
Renilla luciferase activities were measured using a GloMax-20/20
luminometer (Promega Corporation) using a Dual-Luciferase Reporter
Assay System (Promega Corporation). Firefly luciferase activity was
normalized to the Renilla luciferase activity. Each experiment was
performed in triplicate and independently repeated three times.
Nuclear protein extraction and
electrophoretic mobility shift assay (EMSA)
Nuclear protein extraction was performed using the
NE-PER Nuclear and Cytoplasmic Extraction Reagents (Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol. EMSA
was performed using the DIG Gel Shift Kit (Roche Diagnostics GmbH)
according to the manufacturer's instructions. Probes (wild-type
probe, 5′-GAAAAACCATTAAAAAGTTT-3′; mutant probe,
5′-GAAAACTTCCTAAAAAGTTT-3′) for this experiment were synthesized by
Heyuan Biotechnology Co., Ltd.
Construction of the in vivo model
For the construction of in vivo models of
PDAC, 12 4-week-old female, nude mice (BALB/cA-nu) were purchased
from Beijing Vital River Laboratory Animal Technology Co., Ltd. The
mice were randomly divided into two groups (PANC-1-Vector and
PANC-1-RGS22), and 1×106 PANC-1-Vector or PANC-1-RGS22
cells were injected into the abdominal cavity of each mouse. The
size of the neoplasms was measured every 6 days for 30 days, and
the formula (width2 × length)/2 was used to calculate
the tumor volumes. The mice were anesthetized by intraperitoneal
injection of 5% chloral hydrate (the dose of chloral hydrate was
400 mg/kg body weight). The tumors were excised after the mice were
anesthetized and sacrificed by cervical dislocation. IHC was
performed to determine the Ki-67 levels (1:200; Wuhan Servicebio
Technology Co., Ltd.) of the tumor samples. The animal experiments
were approved by the Ethics Committee of the First Affiliated
Hospital of Nanjing Medical University. All methods were performed
in accordance with the relevant guidelines and regulations.
Statistical analysis
Statistical analysis was performed using SPSS
version 22.0 (IBM Corp.) and GraphPad Prism version 6.0 (GraphPad
Software, Inc.). Quantitative data are presented as the mean ± SD.
The association between RGS22 expression with clinicopathological
features was analyzed using a Pearson's χ2 test. The
Kaplan-Meier test was employed to calculate the survival rates of
the two groups. A Student's t-test was used to analyze differences
in the mean values between two groups. Area calculations were
performed using ImageJ. P<0.05 was considered to indicate a
statistically significant difference.
Results
Expression of RGS22 in PDAC
samples
RT-qPCR was used to examine the mRNA expression
levels of RGS22 in 52 pairs of PDAC tissue samples and adjacent
non-neoplastic tissue samples. RGS22 expression was upregulated in
the PDAC tissues, and its expression was significantly higher than
that in the adjacent non-neoplastic tissues (P<0.0001, Fig. 1A). Similar findings were obtained
with IHC analysis of RGS22 protein expression, and expression
differed significantly between the PDAC and non-neoplastic tissues
(P<0.0001, Fig. 1B and E).
These results are consistent with data from TCGA and GTEx databases
(Fig. 1D) (20).
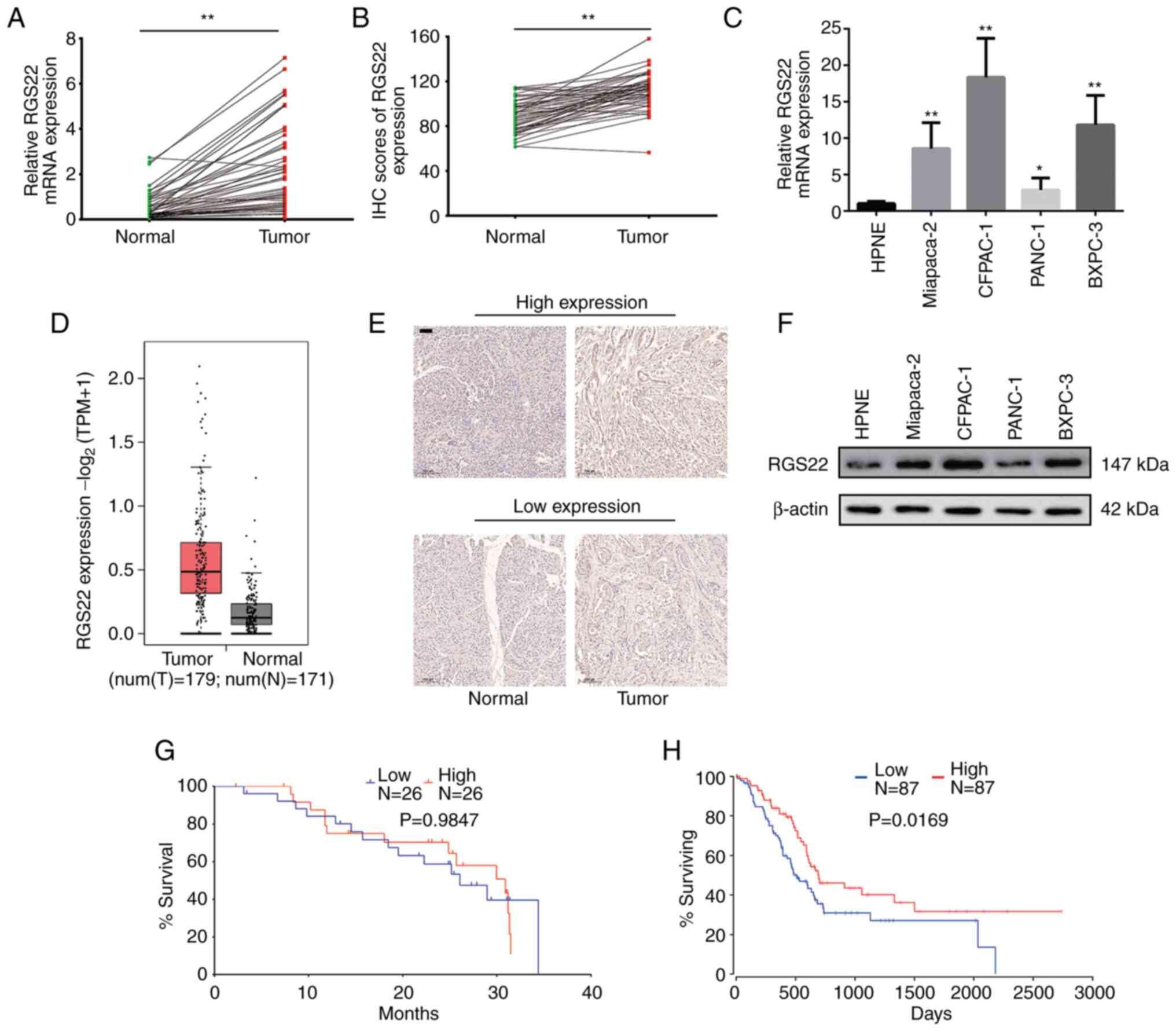 | Figure 1.Expression of RGS22 in PDAC. (A)
Boxplot showing RGS22 RNA expression in 52 pairs of PDAC tissues
and adjacent tissues, as determined by RT-qPCR. (B) A boxplot
showing RGS22 protein expression in 52 pairs of PDAC tissues and
adjacent tissues, as determined by IHC. (C) RT-qPCR analysis of
RGS22 RNA expression in a panel of human PDAC cell lines and the
HPNE cell line. (D) A boxplot showing RGS22 RNA expression in 179
PDAC tissues and 171 normal pancreatic tissues [data from TCGA and
GTEx; unit of measurement: log2(TPM+1)]. (E) RGS22
expression was lower in the PDAC cells than in the adjacent
tissues, as determined by IHC. Scale bar, 50 µm. (F) Western blot
analysis of RGS22 protein expression in a panel of human PDAC cell
lines and the HPNE cell line. (G) Kaplan-Meier curves for OS based
on RGS22 expression in 52 cases of PDAC. (H) Kaplan-Meier curves
for OS according to RGS22 expression in 174 cases of PDAC (data
from TCGA and GTEx). *P≤0.05, **P≤0.01. RGS22, Regulator of
G-protein signaling 22; PDAC, pancreatic ductal adenocarcinoma;
RT-qPCR, reverse transcription-quantitative PCR; IHC,
immunohistochemistry; TCGA, The Cancer Genome Atlas; GTEx,
Genotype-Tissue Expression; TPM, transcripts per million; OS,
overall survival. |
The expression of RGS22 was also assessed using
western blotting and RT-qPCR in four PDAC cell lines (PANC-1,
CFPAC-1, BxPC-3, and MiaPaCa-2) and a normal human pancreatic
ductal cell line (HPNE). Compared with the HPNE cells, the PDAC
cells exhibited higher expression of RGS22 (Fig. 1C and F). PANC-1 (which had
relatively low expression of RGS22) and CFPAC-1 (which had
relatively high expression of RGS2) cells were used for all
subsequent experiments.
Correlation between RGS22 expression
and the prognosis of PDAC
Survival analysis included 52 patients. The cutoff
value for low/high RGS22 expression was determined by the median
expression value based on RT-qPCR data. There was no statistically
significant difference in overall survival between the low RGS22
and high RGS22 groups (Fig. 1G).
However, data from TCGA and GTEx databases showed that patients
with higher RGS22 expression had significantly better overall
survival (Fig. 1H, P=0.0169)
(20). In addition, the
correlation between RGS22 expression and clinical characteristics
of the PDAC patients was analyzed. As shown in Table I, the expression of RGS22 was
associated with blood vessel invasion (P=0.0438); that is, patients
with high RGSS22 expression had a lower degree of blood vessel
invasion.
 | Table I.Association between RGS22 expression
and the clinicopathological features of PDAC. |
Table I.
Association between RGS22 expression
and the clinicopathological features of PDAC.
|
| RGS22
expression |
|
|---|
|
|
|
|
|---|
| Variable | High | Low | P-value |
|---|
| Sex |
|
| 0.0714 |
|
Male | 21 | 15 |
|
|
Female | 5 | 11 |
|
| Age, year |
|
| 0.3965 |
|
≤65 | 14 | 17 |
|
|
>65 | 12 | 9 |
|
| Location |
|
| 0.3584 |
|
Head | 17 | 20 |
|
| Body,
tail | 9 | 6 |
|
|
Differentiation |
|
| 0.4164 |
|
Poor | 2 | 5 |
|
|
Moderate or High | 24 | 21 |
|
|
Tumor-Node-Metastasis stage |
|
| 0.7801 |
|
I–IIA | 11 | 12 |
|
|
IIB-IV | 15 | 14 |
|
| T stage |
|
| 0.1259 |
| T1 or
T2 | 21 | 16 |
|
| T3 or
T4 | 5 | 10 |
|
| N stage |
|
| 0.7814 |
| 0 | 12 | 13 |
|
| 1 or
2 | 14 | 13 |
|
| Blood vessel
invasion |
|
| 0.0438a |
|
Absent | 13 | 6 |
|
|
Present | 13 | 20 |
|
| Serum CA19-9,
kU/l |
|
| 0.2111 |
|
≤39 | 5 | 9 |
|
|
>39 | 21 | 17 |
|
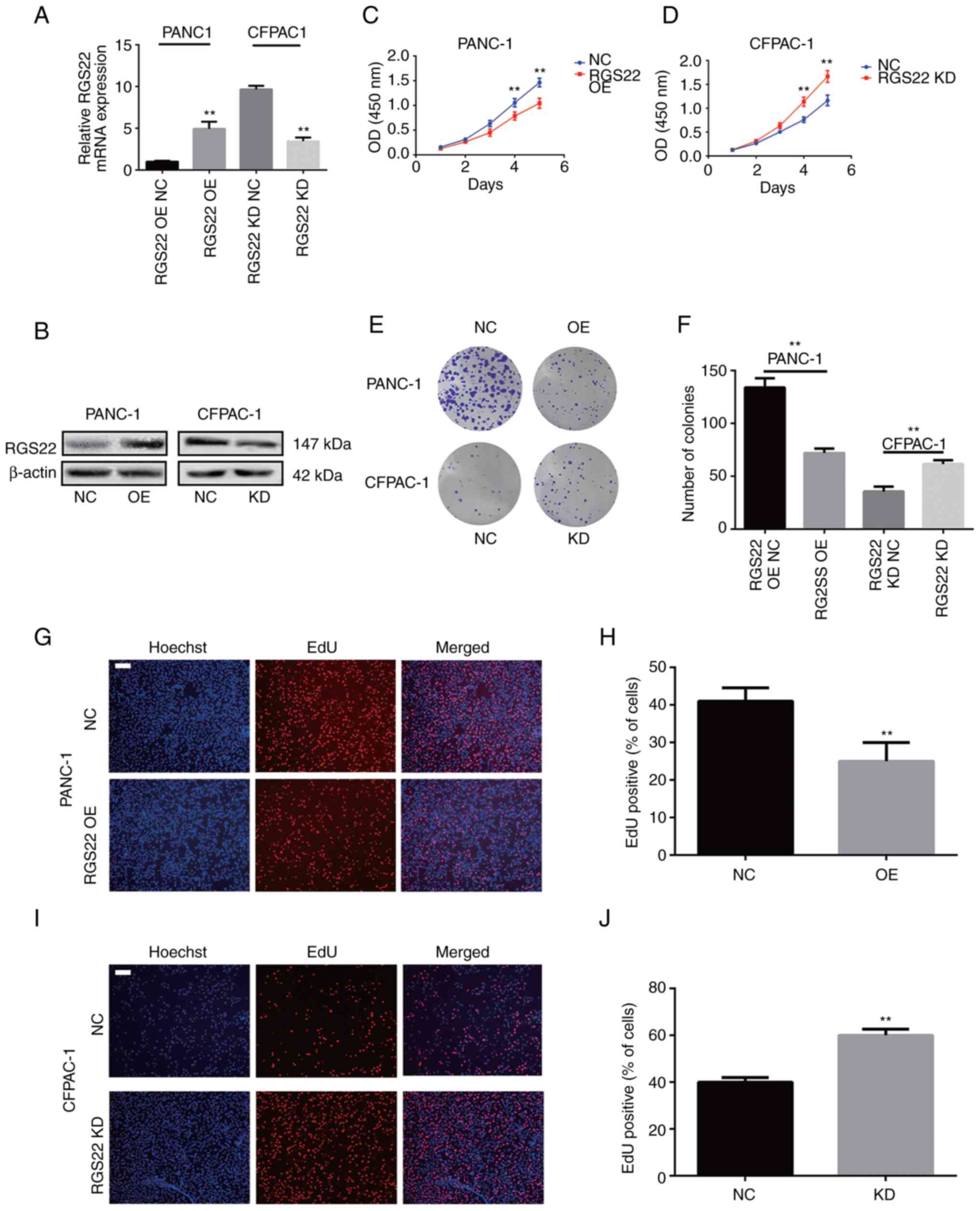
RGS22 suppresses the proliferation,
migration, and invasion of pancreatic cancer cells in vitro
To assess the effect of RGS22 on the development of
PDAC, stable RGS22 overexpression or knockdown cells
(PANC-1-Vector, PANC-1-RGS22, CFPAC-1-Scramble shRNA, and
CFPAC-1-RGS22 shRNA) were constructed using lentiviruses. RT-qPCR
and western blotting confirmed the change in levels of RGS22 in
these cell lines (Fig. 2A and B).
The effects of RGS22 on the proliferation of PDAC cells were
investigated by CCK-8, colony-formation, and EdU assays. RGS22
overexpression resulted in a significant decrease in the
proliferation of PANC-1 cells compared with the control group
(Fig. 2C). Conversely, RGS22
knockdown resulted in a significant increase in the proliferation
of CFPAC-1 cells (Fig. 2D).
Similar results were observed in the colony formation assays. The
number of colonies were significantly lower in RGS22-overexpressing
PANC-1 cells and significantly higher in RGS22-knockdown CFPAC-1
cells compared with their respective controls (Fig. 2E and F); there was a substantially
large difference in colony formation between the two cell lines
(PANC-1 and CFPAC-1) as compared to their growth curves (CCK-8
assay). The difference may be attributed to the duration of each
experiment; in colony formation assays, cells were cultured for 20
days, thus the time scale to allow for measurement of large
differences was larger than in the CCK-8 experiments in which cells
were cultured for only 5 days. The EdU assay (Fig. 2G-J) showed that the proportion of
EdU-positive nuclei was significantly decreased in
RGS22-overexpressing PANC-1 cells and significantly increased in
RGS22-knockdown CFPAC-1 cells. Together, these results demonstrate
that RGS22 suppresses the proliferation of pancreatic cells in
vitro.
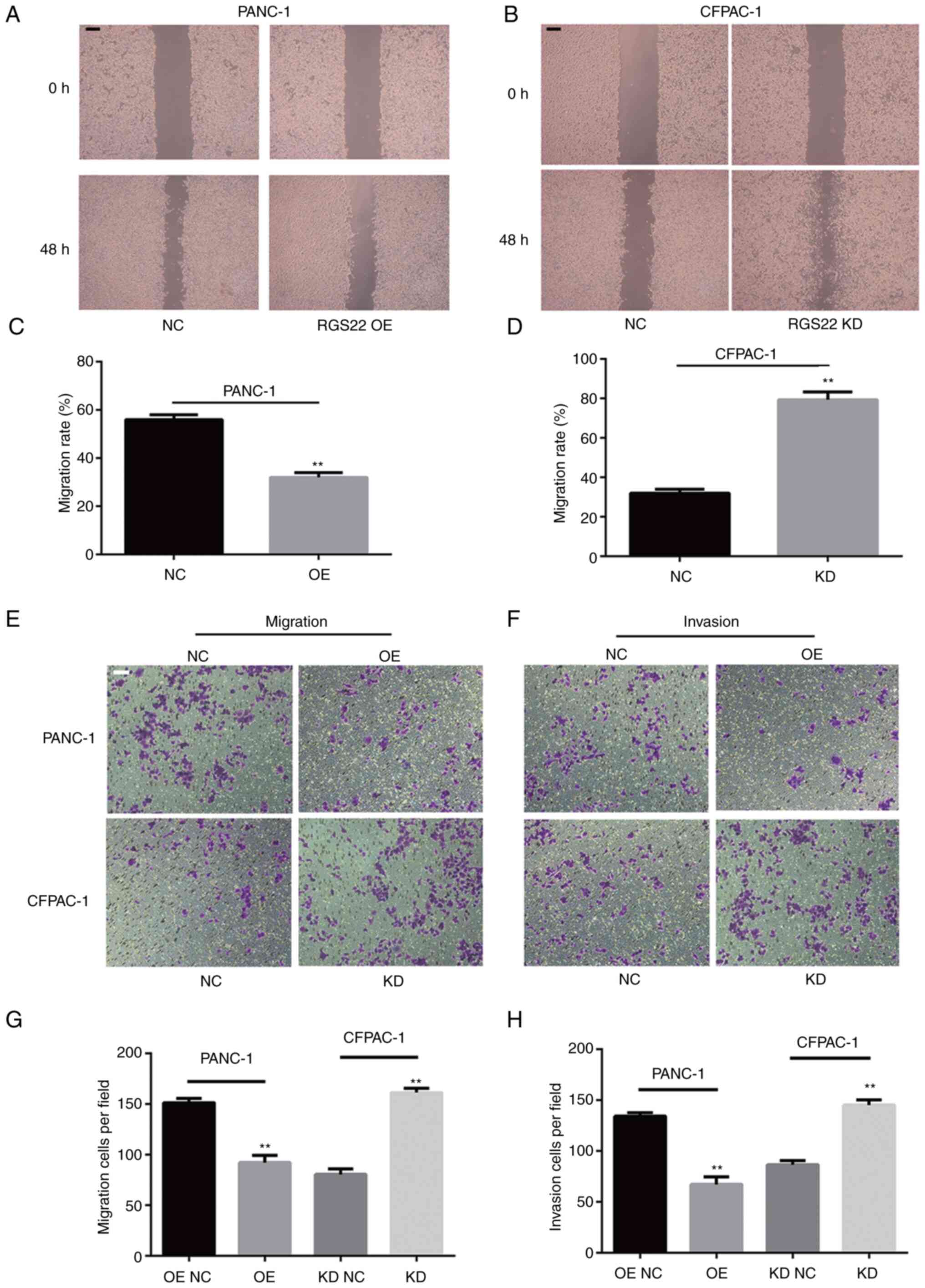
Wound-healing assays and Transwell assays were
performed to investigate the effects of RGS22 on the invasion and
migration of PDAC cells. The results of the wound-healing assays
showed that RGS22 knockdown resulted in an increase in the rate of
wound healing and RGS22 overexpression resulted in a decrease in
the rate of wound healing (Fig.
3A-D). Similar results were obtained with the Transwell assays;
RGS22 overexpression suppressed the migration and invasion of
PANC-1 cells, while RGS22 knockdown promoted the migration and
invasion of CFPAF-1 cells (Fig.
3E-H). These results indicate that RGS22 suppresses the
migration and invasion of pancreatic cancer cells in
vitro.
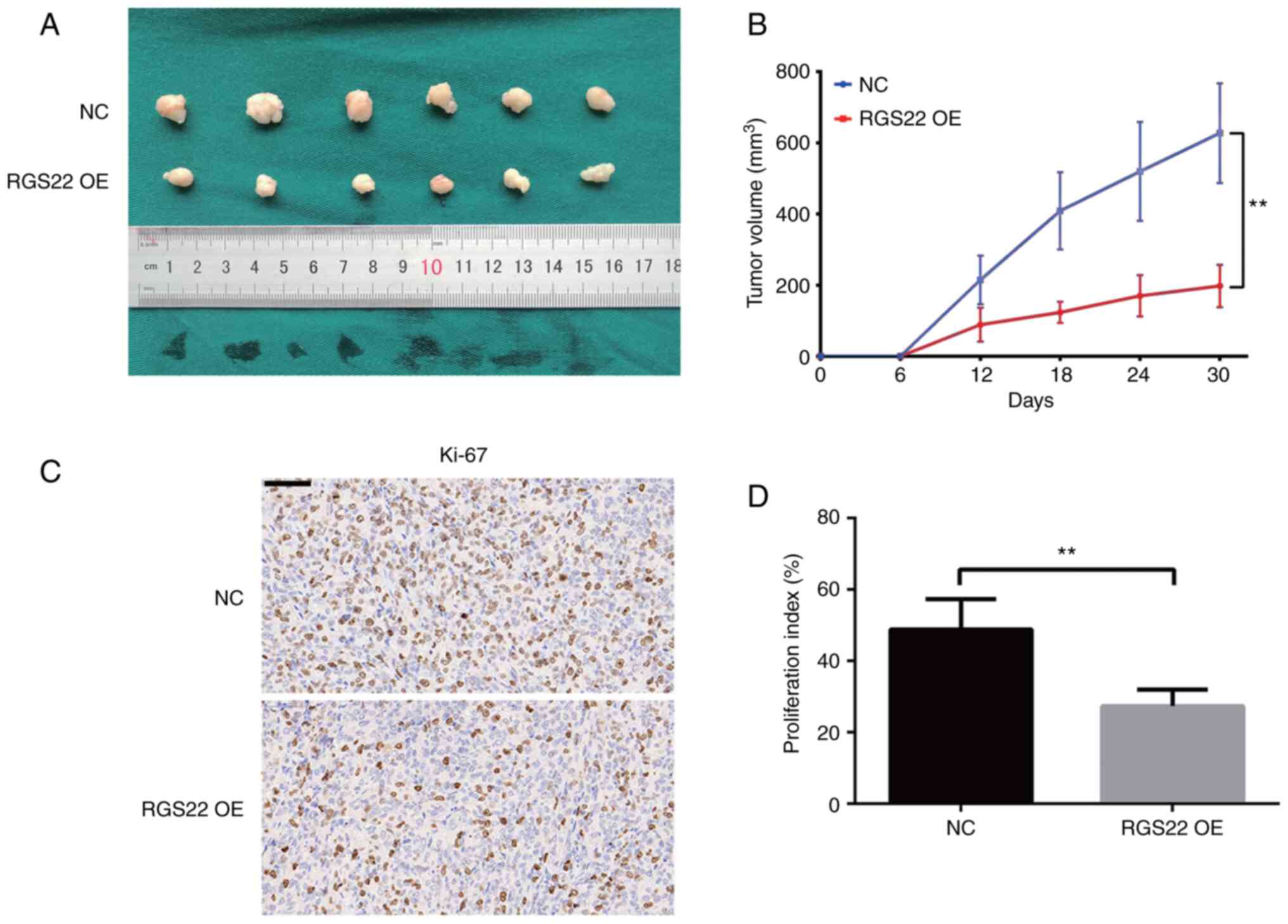
RGS22 suppresses the growth of PDAC in
vivo
To study the in vivo effects of RGS22 on PDAC
cells, stable RGS22-overexpressing PANC-1 cells were subcutaneously
implanted into BALB/cA-nu mice. The size of the xenograft tumors
was measured every 6 days with a pair of calipers for 30 days. The
size of the tumors formed by the RGS22-overexpressing PANC-1 cells
was significantly smaller than that of the control group tumors
(Fig. 4A and B). In addition,
Ki-67 staining showed that RGS22 overexpression decreased the
proliferation index of the xenograft tumors (Fig. 4C and D).
Direct regulation of RGS22 by YY1
The results of the western blotting and RT-qPCR
experiments showed that RGS22 expression was positively correlated
with YY1 expression (Fig. 5A-C).
These results indicate that RGS22 may be regulated by the
transcription factor YY1. To verify this hypothesis, EMSA and
luciferase experiments were performed. Based on the predicted
binding site from the previous analysis, a digoxigenin-labeled
probe was constructed for the EMSA experiment. The probe bound to
YY1 and the YY1 antibody to form a specific super shift (Fig. 5E). These results showed that YY1
directly bound to the promotor region of RGS22.
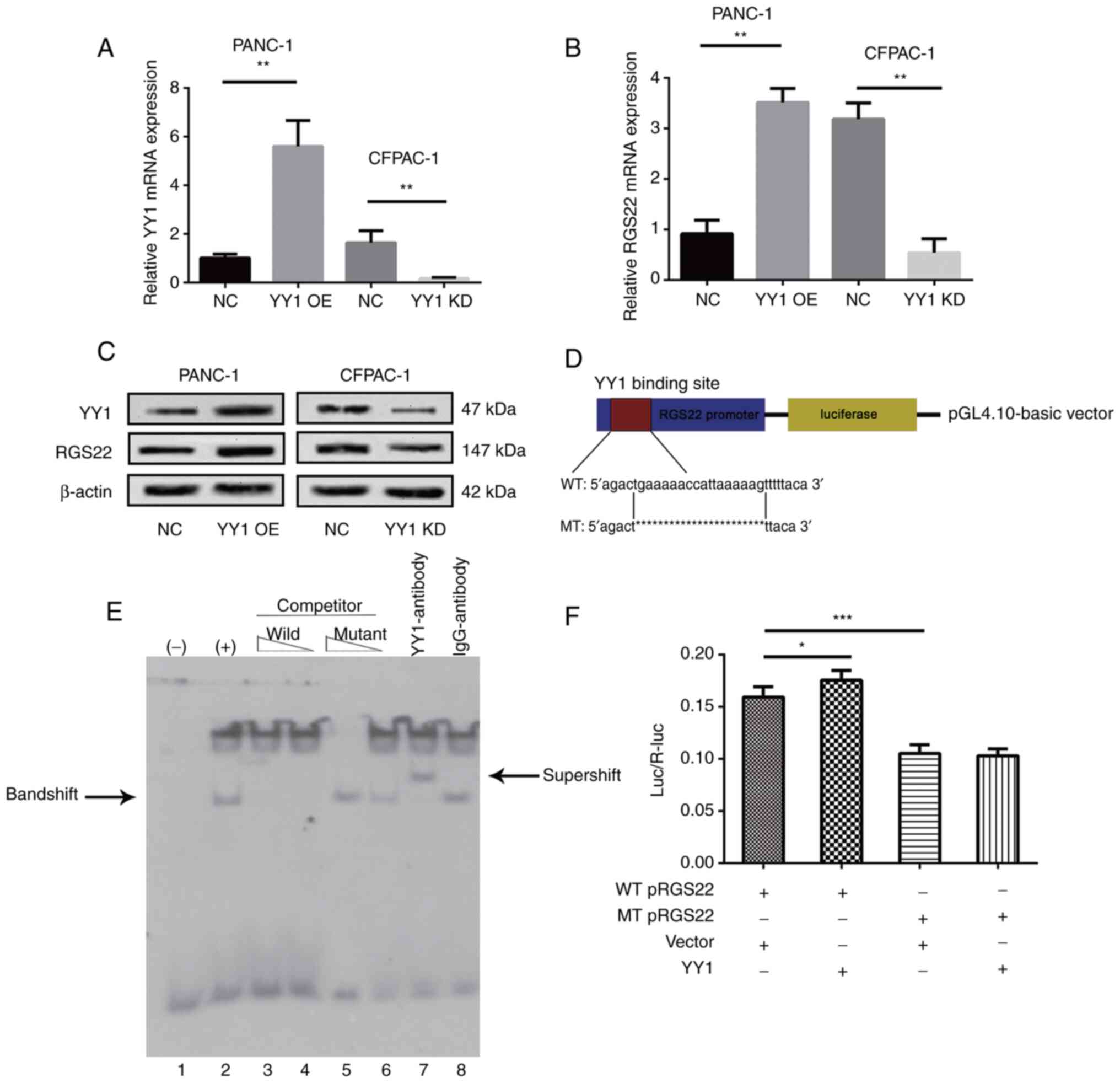 | Figure 5.Regulation of RGS22 expression by the
binding of YY1 directly to the RGS22 promoter. (A-C) RGS22
expression in YY1-overexpressing PANC-1 cells or YY1-knockdown
CFPAC-1 cells was measured by reverse transcription-quantitative
PCR and western blotting. (D) Schematic diagram of the luciferase
reporter construct containing the human RGS22 promoter and the
mutant construct containing the RGS22 promoter in which the
predicted YY1 binding site was mutated. (E) EMSA showing that YY1
binds to the RGS22 promoter. The wild-type probe was incubated
without (lane 1) or with (lane 2) PANC-1-YY1 cell nuclear proteins
in the absence or presence of unlabeled probes (lanes 3–6). Lanes 3
and 4 contain the wild-type probe, and lanes 5 and 6 contain the
mutant probe, each at 50- and 100-fold molar excess. EMSA was
performed using an anti-YY1 antibody (lane 7), and the IgG antibody
was used as a negative control for the YY1 antibody (lane 8). (F)
Luciferase assays demonstrated the luciferase activity of PANC-1
cells transfected with YY1-overexpression or control lentiviruses.
Data are presented as the mean ± SD of three independent
experiments.*P≤0.05, **P≤0.01, ***P≤0.001. EMSA, electrophoretic
mobility shift assay; RGS22, Regulator of G-protein signaling 22;
YY1, Yin Yang-1; OE, overexpression; NC, negative control; KD,
knockdown; WT, wildtype; MT, mutant. |
To further investigate the effect of YY1 on RGS22
expression, luciferase assays were performed. RGS22 reporter gene
plasmids and its mutant plasmids were constructed as shown in
Fig. 5D. The reporter plasmids
were co-transfected with YY1-overexpression plasmids.
Overexpression of YY1 significantly reduced the luciferase values
compared with those observed in the vector cells (Fig. 5F). In addition, when the putative
binding site of YY1 was mutated, the luciferase value decreased;
this demonstrates the specificity of the binding site. These
results show that YY1 binds to a specific region of the RGS22
promotor to positively control the expression of RGS22.
RGS22 as a functional target of
YY1
Previous studies have shown that YY1 acts as a tumor
suppressor in PDAC. To investigate whether the effects of YY1 on
pancreatic cells are mediated by RGS22, RGS22 expression was
knocked down using a specific siRNA construct in PANC-1 cells
stably overexpressing YY1 for the recovery experiments. The results
of the CCK-8, colony formation, and EdU assays showed that
downregulation of RGS22 in YY1-overexpressing cells restored the
inhibition of PANC-1 cell proliferation by YY1 (Fig. 6A-E). Cellular Transwell assays and
wound-healing assays illustrated that downregulation of RGS22
restored the inhibitory effect of YY1 overexpression on the
invasion and migration of PANC-1 cells (Fig. 6F-J). These results indicate that
YY1 inhibits the proliferation, invasion, and migration of PDAC
cells by targeting RGS22.
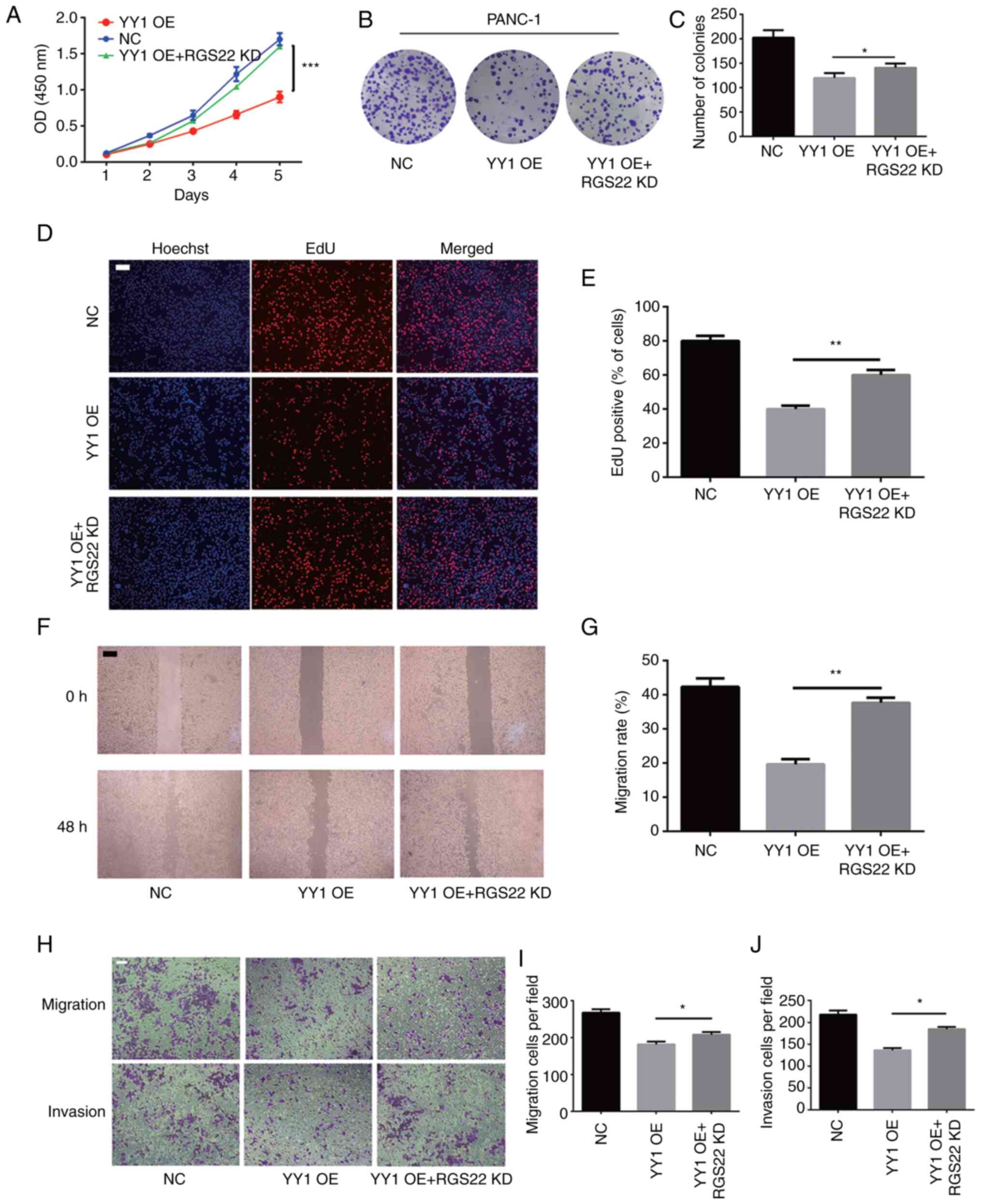 | Figure 6.RGS22 as a functional target of YY1.
(A) Cell Counting Kit-8, (B and C) colony formation, and (D and E)
EdU assays were performed to analyze the proliferation of
PANC-1-YY1 cells transfected with RGS22 siRNA. (F and G) Wound
healing assays were performed to analyze the migration of
PANC-1-YY1 cells transfected with RGS22 siRNA. (H-J) Cell migration
and invasion assays were performed in PANC-1-YY1 cells transfected
with RGS22 siRNA (magnification, ×100; scale bar, 100 µm). Data are
presented as the mean ± SD value of three independent experiments.
*P≤0.05, **P≤0.01, ***P≤0.001. RGS22, Regulator of G-protein
signaling 22; YY1, Yin Yang-1; siRNA, small interfering RNA; OD,
optical density; OE, overexpression; NC, negative control; KD,
knockdown. |
Discussion
The in vitro and in vivo findings of
the present study indicate that RGS22 inhibited tumor
proliferation, invasion, and metastasis of PDAC. These findings
were corroborated by the clinical findings, which showed that RGS22
was overexpressed in PDAC tissues and was correlated with a better
prognosis for PDAC patients. Our previous ChIP-Seq results
indicated that the transcriptional factor YY1 could directly bind
to the promoter region of RGS22, and the present results indicate
that YY1 binds to a specific region of the promotor to positively
regulate RGS22 expression. Thus, RGS22 may play an important role
in the tumor-suppressing effect of YY1 in pancreatic cancer.
High expression of RGS22 in PDACs was shown using
the IHC analysis of the 52 pairs of tissues obtained from PDAC
patients and the results of in vitro western blotting and
RT-qPCR experiments on PDAC cells, which were consistent with data
from TCGA and GTEx (20). A
previous study analyzed the expression of RGS22 in several tumor
types using RT-qPCR and IHC and found that RGS22 is specifically
expressed in the testis and in epithelial cancers (16). This is consistent with the results
of the present study, as pancreatic cancer is a classical tumor of
epithelial origin.
The results of the present study showed that RGSS22
exerted a protective effect in pancreatic cancer. Data from TCGA
and GTEx and the clinical data of the 52 pancreatic patients
demonstrated a correlation between high RGS22 levels and longer
overall survival, as well as with a low degree of blood vessel
invasion. Our in vitro and in vivo studies revealed
that RGS22 suppresses the proliferation, migration, and invasion of
PDAC. RGS22 was first reported in 2008 as a testis-specific gene by
Hu et al (17), and their
study highlighted RGS22 as a cancer/testis antigen. They found that
RGS22 was specifically expressed in the testis and epithelial
cancers and acted as a tumor suppressor, repressing tumor cell
invasion and migration. As far as we know, only one study has
investigated the role of RGS22 in pancreatic cancer and reported
the suppressive effect of RGS22 on cell migration and invasion, but
not proliferation (18).
Therefore, the present findings make an important contribution to
the literature on this topic by demonstrating the in vitro
and in vivo suppressive role of RGS2 in PDAC.
RGS22 is a member of the RGS family; this means that
it can interact with G proteins and negatively regulate the GPCR
signaling pathway (12–14,21,22).
Considering the complexity of this pathway and the limited scope of
the present study, we did not investigate the pathways downstream
of RGS22 in pancreatic cancer. However, we investigated the
involvement of the transcription factor YY1, which is an upstream
regulator of RGS22. The results of EMSA and luciferase experiments
showed that YY1 directly binds to the promotor region of RGS22 and
positively regulates its expression. Moreover, the recovery
experiments proved that RGS22 is a functional target of YY1; this
means that YY1 inhibits the proliferation, invasion, and migration
of pancreatic cells by targeting RGS22.
The expression pattern of RGS22 and its function in
pancreatic cancer seem to be contradictory: That is, it is
overexpressed in cancer cells, but it acts as a tumor suppressor.
It is hypothesized that RGS22 is not a causal factor of PDAC but is
part of a feedback mechanism for the prevention of cancer
progression. While RGS22 cannot prevent pancreatic cancer, it may
be able to delay tumor progression and inhibit tumor metastasis.
Although RGS22 expression is relatively high in pancreatic cancer
cells compared with normal pancreatic cells, the absolute amount of
RGS22 in PDAC is still very low. As data from TCGA and GTEx
databases showed (20), RGS22 gene
expression is only 0.4 TPM, which is 0.005× lower than the RGS22
expression levels in the testis. These findings point to the
possibility of using exogenous RGS22 to prevent tumor progression
in patients with pancreatic cancer.
Although there are no studies regarding the clinical
application of RGS22 to the best of our knowledge, other members of
the RGS protein family have been extensively studied for their
clinical use. For example, it is well known that the GPCR signaling
pathway is involved in a variety of pathophysiological processes,
including tumor growth. GPCRs have long served as extraordinarily
successful drug targets (23).
However, due to the broad spectrum of GPCR action, drugs targeting
GPCRs often lack specificity and tend to interfere with normal
physiological processes. In contrast, the expression and action of
the RGS proteins are tissue-specific; therefore, RGS proteins are
better targets than upstream GPCRs in terms of specificity. To
date, expression analyses, purification techniques, structural
studies, cell line development, and screening methods related to
RGS proteins have been developed, and these provide a solid
foundation for designing drugs that target RGS22 (23). Furthermore, the present findings of
the present study lay the basis for potential future treatments on
RGS22 as a target for management of PDAC.
In conclusion, the present study demonstrated that
RGS22 expression was upregulated in PDAC tissues, and YY1-mediated
RGS22 regulation suppressed the proliferation, migration and
invasion of PDAC cells.
Supplementary Material
Supporting Data
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
This study was supported by funding from that National Natural
Science Foundation of China (grant nos. 81572337 and 81672471), the
Outstanding Young and Middle-aged Talents Support Program of the
First Affiliated Hospital of Nanjing Medical University, the
Innovation Capability Development Project of Jiangsu Province
(grant no. BM2015004), Jiangsu Key Medical Discipline (General
Surgery) (grant no. ZDXKA2016005), and the Priority Academic
Program Development of Jiangsu Higher Education Institutions (grant
no. PAPD, JX10231801).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Author's contributions
SC, WG, and LM drafted the article. KJ and JZ
critically revised the article for important intellectual content.
KJ and JZ conceived and designed the study. SC, WG, LM, QC, YM, KJ
and JZ contributed to acquisition of data, analysis, and
interpretation of data. WG and LM confirm the authenticity of all
the raw data. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
This study was approved by the Ethics Committee of
the First Affiliated Hospital of Nanjing Medical University
(approval no. 2009-SR-031) and all patients provided written
informed consent. All animal experiments were performed in
accordance with animal protocols approved by the Nanjing Medical
University (approval no. 2022-SRFA-016).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics. CA Cancer J Clin. 69:7–34. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Griffin JF, Poruk KE and Wolfgang CL:
Pancreatic cancer surgery: Past, present, and future. Chin J Cancer
Res. 27:332–348. 2015.PubMed/NCBI
|
|
3
|
Dal Molin M, Zhang M, de Wilde RF,
Ottenhof NA, Rezaee N, Wolfgang CL, Blackford A, Vogelstein B,
Kinzler KW, Papadopoulos N, et al: Very long-term survival
following resection for pancreatic cancer is not explained by
commonly mutated genes: Results of whole-exome sequencing analysis.
Clin Cancer Res. 21:1944–1950. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chin V, Nagrial A, Sjoquist K, O'Connor
CA, Chantrill L, Biankin AV, Scholten RJ and Yip D: Chemotherapy
and radiotherapy for advanced pancreatic cancer. Cochrane. Database
Syst Rev. 3:CD0110442018.PubMed/NCBI
|
|
5
|
Grasso C, Jansen G and Giovannetti E: Drug
resistance in pancreatic cancer: Impact of altered energy
metabolism. Crit Rev Oncol Hematol. 114:139–152. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Gordon S, Akopyan G, Garban H and Bonavida
B: Transcription factor YY1: Structure, function, and therapeutic
implications in cancer biology. Oncogene. 25:1125–1142. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Shi Y, Lee JS and Galvin KM: Everything
you have ever wanted to know about Yin Yang 1. Biochim Biophys
Acta. 1332:F49–F66. 1997.PubMed/NCBI
|
|
8
|
Shi Y, Seto E, Chang LS and Shenk T:
Transcriptional repression by YY1, a human GLI-Kruppel-related
protein, and relief of repression by adenovirus E1A protein. Cell.
67:377–388. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Thomas MJ and Seto E: Unlocking the
mechanisms of transcription factor YY1: Are chromatin modifying
enzymes the key? Gene. 236:197–208. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhang JJ, Zhu Y, Xie KL, Peng YP, Tao JQ,
Tang J, Li Z, Xu ZK, Dai CC, Qian ZY, et al: Yin Yang-1 suppresses
invasion and metastasis of pancreatic ductal adenocarcinoma by
downregulating MMP10 in a MUC4/ErbB2/p38/MEF2C-dependent mechanism.
Mol Cancer. 13:1302014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yang C, Zhang JJ, Peng YP, Zhu Y, Yin LD,
Wei JS, Gao WT, Jiang KR and Miao Y: A Yin-Yang 1/miR-30a
regulatory circuit modulates autophagy in pancreatic cancer cells.
J Transl Med. 15:2112017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Koelle MR: A new family of G-protein
regulators-the RGS proteins. Curr Opin Cell Biol. 9:143–147. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Siderovski DP, Hessel A, Chung S, Mak TW
and Tyers M: A new family of regulators of G-protein-coupled
receptors? Curr. Biol. 6:211–212. 1996.PubMed/NCBI
|
|
14
|
Watson N, Linder ME, Druey KM, Kehrl JH
and Blumer KJ: RGS family members: GTPase-activating proteins for
heterotrimeric G-protein alpha-subunits. Nature. 383:172–175. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Hunt TW, Fields TA, Casey PJ and Peralta
EG: RGS10 is a selective activator of G alpha i GTPase activity.
Nature. 383:175–177. 1996. View
Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hu Y, Xing J, Wang L, Huang M, Guo X, Chen
L, Lin M, Zhou Y, Liu Z, Zhou Z and Sha J: RGS22, a novel
cancer/testis antigen, inhibits epithelial cell invasion and
metastasis. Clin Exp Metastasis. 28:541–549. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hu Y, Xing J, Chen L, Guo X, Du Y, Zhao C,
Zhu Y, Lin M, Zhou Z and Sha J: RGS22, a novel testis-specific
regulator of G-protein signaling involved in human and mouse
spermiogenesis along with GNA12/13 subunits. Biol Reprod.
79:1021–1029. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hu Y, Xing J, Chen L, Zheng Y and Zhou Z:
RGS22 inhibits pancreatic adenocarcinoma cell migration through the
G12/13 α subunit/F-actin pathway. Oncol Rep. 34:2507–2514. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kamarajah SK, Burns WR, Frankel TL, Cho CS
and Nathan H: Validation of the American joint commission on cancer
(AJCC) 8th edition staging system for patients with pancreatic
adenocarcinoma: A surveillance, epidemiology and end results (SEER)
analysis. Ann Surg Oncol. 24:2023–2030. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Tang Z, Li C, Kang B, Gao G, Li C and
Zhang Z: GEPIA: A web server for cancer and normal gene expression
profiling and interactive analyses. Nucleic Acids Res. 45:W98–W102.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Sriram K and Insel PA: G protein-coupled
receptors as targets for approved drugs: How many targets and how
many drugs? Mol Pharmacol. 93:251–258. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wang D, Xu Y, Feng L, Yin P, Song SS, Wu
F, Yan P and Liang Z: RGS5 decreases the proliferation of human
ovarian carcinomaderived primary endothelial cells through the
MAPK/ERK signaling pathway in hypoxia. Oncol Rep. 41:165–177.
2019.PubMed/NCBI
|
|
23
|
O'Brien JB, Wilkinson JC and Roman DL:
Regulator of G-protein signaling (RGS) proteins as drug targets:
Progress and future potentials. J Biol Chem. 294:18571–18585. 2019.
View Article : Google Scholar : PubMed/NCBI
|