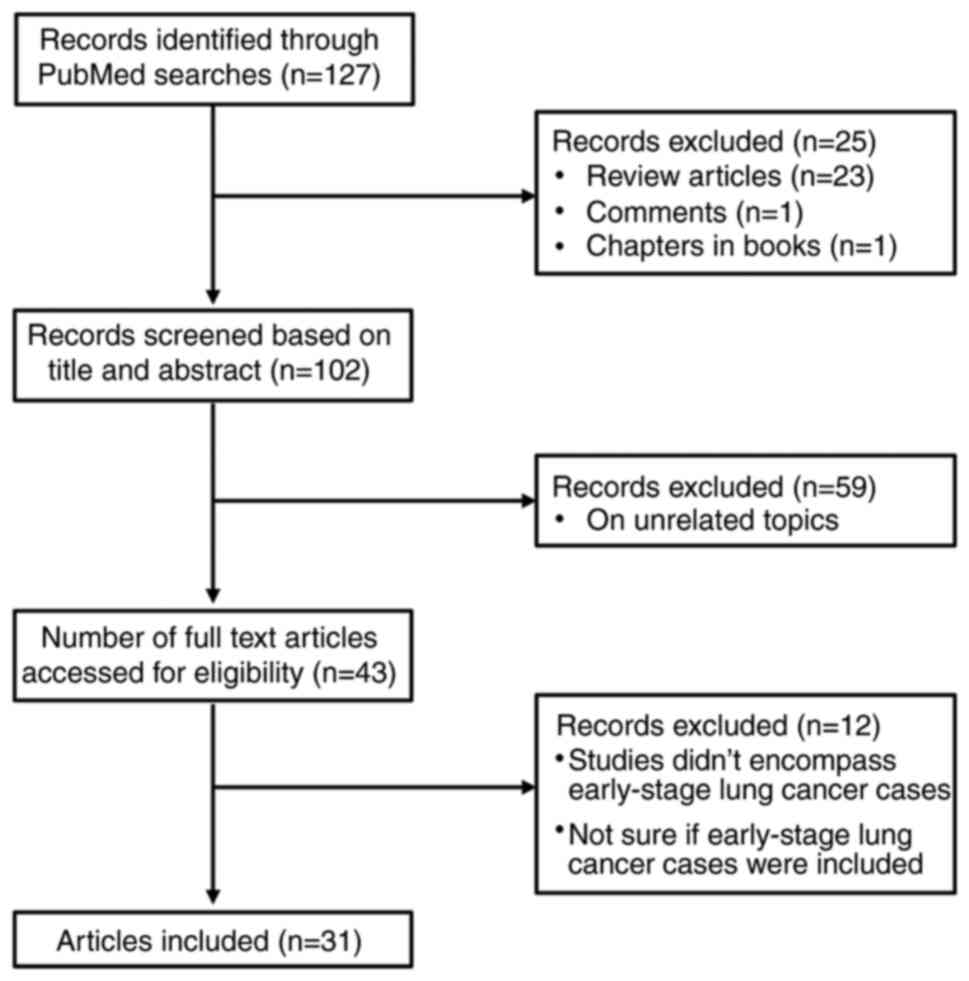
Lung cancer is one of the most common malignancies
and is the leading cause of cancer-associated mortality worldwide
(1). The poor prognosis and high
mortality rate of lung cancer are mainly due to late diagnosis
(2). At present, only ~15% of newly
diagnosed lung cancer cases are diagnosed in the early stages
(stages I–II), which contributes to >60% probability of 5-year
survival when effective treatment is available (3–5).
However, >60% patients with lung cancer are first diagnosed
already in the advanced stages (stage IV) or already with
metastatic tumors, who typically only have 5-year survival rates of
<5% (6).
In addition to the reduction in exposure to tobacco
smoke, screening for the early detection of lung cancer has been
considered to be a major strategy for decreasing the rate of lung
cancer mortality (7). At present,
low-dose CT (LDCT) screening in the high-risk population is the
predominant tool used for detecting lung cancer in the early stages
(2). The results of the US National
Lung Screening Trial (ClinicalTrials.gov number, NCT00047385) found
that compared with chest X-ray examination, LDCT screening was
associated with a 20% reduction in lung cancer-specific mortality
in a high-risk group of participants defined by their smoking
status (8). In addition, other
previous studies have also confirmed the validity of LDCT screening
for the early detection of lung cancer to reduce mortality rate
(9,10). However, potentially healthy
individuals are also at risk of being subjected to expensive and
potentially harmful diagnostic procedures, such as positron
emission tomography, transthoracic/bronchoscopic biopsy or even
surgery, due to the considerably high false-positive rate of LDCT
(nearly 96.4%) (8). Therefore, the
combination of LDCT with additional biomarker-based tests has been
proposed to be a more favorable strategy for improving the
effectiveness of lung cancer screening programs whilst reducing the
cost and harmfulness to otherwise healthy individuals (11). Such tests can either pre-select
individuals from a high-risk population for LDCT examination or
discriminate between benign and malignant pulmonary nodules
detected by LDCT screening (12).
Several different components of blood, including
specific serum/plasma proteins, autoantibodies, microRNA, cell-free
DNA and circulating tumor cells, have all been proposed to be
potential lung cancer biomarkers (13–15).
However, they typically have low specificities and few were found
for wider beneficial application in clinical practice, especially
for the early detection of lung cancer. Instead, monitoring
cancer-related metabolites is an emerging and promising approach
for the detection and diagnosis of a number of malignant tumors,
including colorectal, gastric, gynecological and lung cancer
(16–20).
Metabolomics, also known as metabolic profiling,
uses quantitative and qualitative analyses to determine key
metabolism-associated molecules of different molecular masses
(21,22). It reveals information into specific
states of cancer that are otherwise not apparent (22). Previously, the assessment of
metabolic changes is limited to measuring the levels of individual
hormones and metabolites using imaging modalities and standard
clinical laboratory tests (23). By
contrast, metabolomics involves the measurement of vast numbers of
metabolites systematically, including carbohydrates, nucleotides,
carboxylic acids, amino acids and lipids in blood, urine, or other
body fluids (24). Metabolomics has
emerged to be a potentially powerful approach for identifying
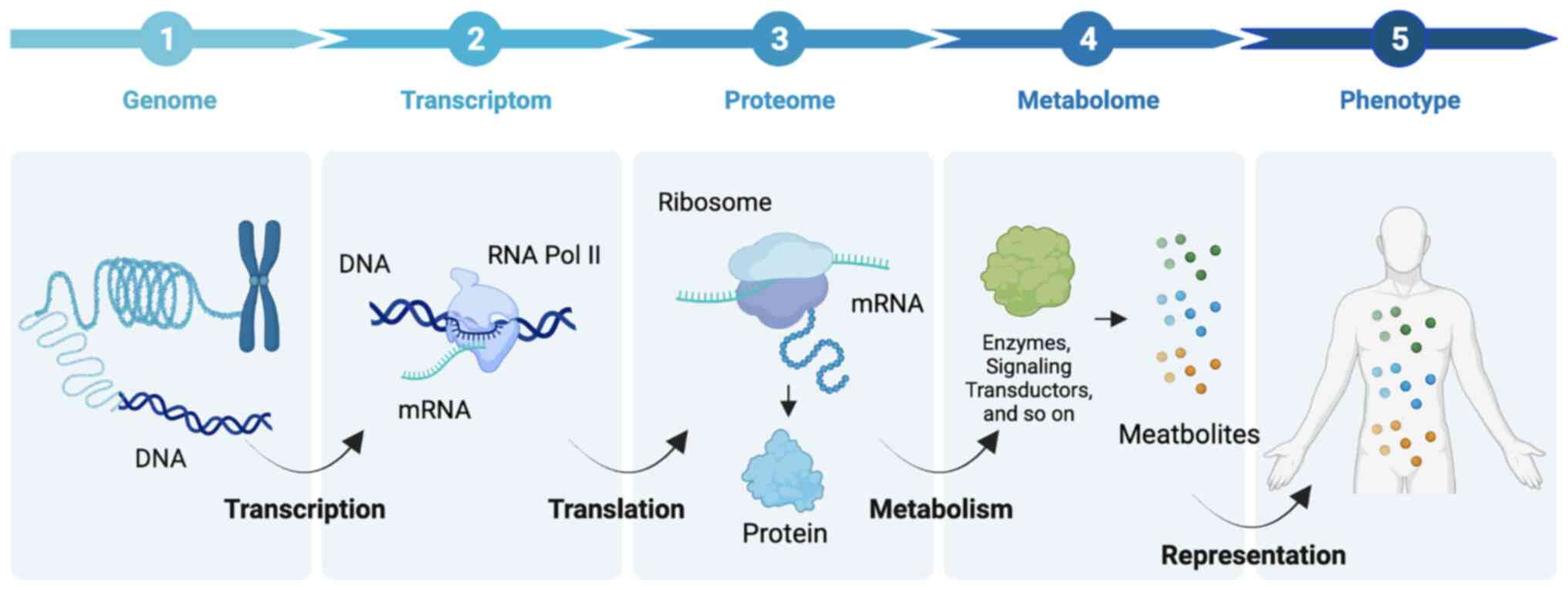
cancer biomarkers and drivers of tumorigenesis (25). In addition, compared with other
‘omes’, such as genome, transcriptome, and the proteome, the
metabolome reflects the real-time status of a particular phenotype
to reveal what exactly has happened in the organisms exactly,
providing bona fide biomarkers for disease surveillance
(Fig. 2).
The main methodologies involved in metabolomics have
been based on nuclear magnetic resonance (NMR) spectroscopy and
mass spectrometry (MS) techniques, coupled with either gas
chromatography (GS) or liquid chromatography (LC), each with their
specific advantages and limitations (Table I). NMR is highly selective and
non-destructive, rendering it recognized to be the gold standard
for elucidating the structure of metabolites. However, the
sensitivity of NMR is relatively low, only being able to detect
metabolites with concentrations >10−5 M (26). By contrast, MS has higher
sensitivity and selectivity (26).
Modern MS provides highly specific chemical information, such as
accurate molecular mass, isotope distribution patterns for element
formulation determination, and characteristic fragment-ion
information directly related to the chemical structure of
metabolites (27). In addition, the
high sensitivity of MS allows for the detection and measurement of
a large number of primary metabolites (the initial end products
created by a live organism as a result of growth) and secondary
metabolites (aid in the performance of various biological tasks
that are not engaged in the growth and maintenance of cellular
activity) at picomolar to femtomolar levels. As one of the major
tools for the collection of ‘omic’ information, MS techniques use
big data for processing and interpretation by machine learning
(28,29). These unique advantages make MS an
essential tool for metabolomics analysis (30). Over the past decade, various
comprehensive reviews have already discussed how NMR and MS work,
and how each can be used for metabolomics (31–33).
Despite their own advantages and disadvantages, several studies
have shown how they can be used to complement each other (31–35).
Indeed, the use of multiple technologies greatly broadens the level
of metabolite coverage and the types of samples that can be studied
(36).
Over the past two decades, a number of metabolomics
studies have been performed based on NMR and/or MS techniques to
generate metabolite profiles that can discriminate patients with
lung cancer from healthy individuals using different types of
biological samples. The present review summarizes the studies that
include early-stage lung cancer cases to evaluate the viability of
using metabolomics for the early detection of lung cancer.
Blood samples, including plasma and serum, are the
most widely studied biological fluids for lung cancer research. It
can be used for characterizing metabolic markers, using both
targeted (measuring a specific set or family of compounds only) and
untargeted (global profiling) approaches (Table II) (37–54).
Previous studies have described alterations in the
levels of amino acids, especially in alanine, glutamine, histidine,
leucine, isoleucine, lysine and serine, in the serum or plasma of
patients with lung cancer (37–42,44).
These alterations may be associated with the increase in amino acid
demand caused by the proliferation of the tumor cells, highlighting
the important role of amino acid metabolic pathways in lung cancer
progression.
Lactic acid is another commonly altered metabolite
in patients with lung cancer (37,39).
Previous studies on lactate metabolism in patients with cancer have
suggested that changes in the level of this metabolite are due to
the increased glucose uptake and lactate production by the tumor in
the absence of oxygen (55,56). In support of this, in various tumor
cells such as lung carcinoma, renal carcinoma and gastric carcinoma
cells, glucose is preferentially catabolized through fermentation
into lactate even when oxygen is not limiting, in a phenomenon
known as the Warburg effect (55,57).
Compared with other biospecimens, urine can be
obtained from larger cohorts non-invasively and is therefore
accepted more easily by the public. Similar to blood samples, urine
also contains useful metabolic information for detecting the
occurrence of lung cancer (59).
The composition of urine is naturally less complex in comparison
with that of plasma or serum, making it more popular for
metabolomics analysis (59). In
addition, once the blood is filtered by the glomerulus, certain
components like metabolites can be concentrated in the urine,
making their detection easier compared with that of other types of
biological fluids. The first study to measure potential urinary
metabolomic cancer biomarkers based on NMR and MS was published in
2006 and the concentration of nucleosides was found to be
significantly increased in patients with breast cancer when
compared with the normal controls (60). Carrola et al (61) first showed the valuable potential of
using NMR-based metabolomics for finding putative biomarkers of
lung cancer in the urine in 2011. Previous studies have since
analyzed urinary metabolites employing either NMR or MS for early
detection of lung cancer (Table
II) (62–65).
The majority of urine metabolomic studies into
patients with lung cancer found alterations in creatine and
creatinine, making creatine and creatinine potentially valuable
biomarkers for early lung cancer detection (61,63).
To further assess whether creatine was elevated in the urine
samples of subjects prior to lung cancer diagnosis, a detailed
prospective study based on LC-MS was previously conducted (64). Urine samples from 178 patients with
lung cancer and 351 volunteers were collected and examined, where
it was revealed that elevated creatine levels were associated with
lung cancer risk in both European- and African-Americans (64). Consistently, creatine and creatinine
have been shown to be upregulated in other biofluids, such as serum
and saliva, in patients with lung cancer (39,66).
In the human body, creatine is synthesized from methionine,
glycine, and arginine, which is then converted into creatinine
(67). Therefore, the increase in
creatine and creatinine levels may be associated with upregulated
amino acid metabolism. Nevertheless, the promising results of
urinary creatine and creatinine in early lung cancer detection
remain to be validated by independent studies based on material
collected real-time during lung cancer screening.
Normal metabolism produces a variety of volatile
organic compounds (VOCs), which can be expelled through respiration
(68). Therefore, exhaled breath
has also been explored as a potential source of cancer biomarkers.
It was first shown that VOCs in the exhaled breath could be used to
differentiate patients with lung cancer from healthy individuals in
1999 (69). Since then,
accumulating evidence has been supporting the utility of VOC
detections in the exhaled breath for the early detection of lung
cancer, most yielding high degrees of sensitivity and specificity
(Table III) (70–78).
Lung cancer causes oxidative stress and induces
oxidase enzymes in tumor tissues, which in turn produce higher
concentrations of specific VOCs, especially carbonyl VOCs in the
exhaled breath. Carbonyl VOCs are produced by biochemical pathways,
such as the respiratory chain and oxidative phosphorylation
pathway, as intermediates, some of which can yield unique
information into specific pathways, such as lipid oxidation induced
by free radicals (79,80).
Consequently, several studies have focused on the
identification of carbonyl VOCs as markers of lung cancer in the
exhaled breath (73,74,81).
Fu et al (73) previously
found that the concentrations of 2-butanone, 2-hydroxyacetaldehyde,
3-hydroxy-2-butanone and 4-hydroxyhexenal (4-HHE) in the exhaled
breath of patients with lung cancer were significantly higher
compared to those in the exhaled breath of healthy controls and
patients with benign pulmonary nodules (BPN). This was found using
Fourier transform-ion cyclotron resonance MS (73). Bousamra et al (81) then showed that the sensitivity and
specificity of breath analysis was associated with the number of
the elevated VOCs. Among patients with lung cancer, three or four
elevated cancer markers (2-butanone, 2-hydroxyacetaldehyde,
3-hydroxy-2-butanone and 4-HHE) produced a specificity of 95% to
discriminate patients with lung cancer from healthy controls.
Furthermore, an enhanced model based on the elevated levels of the
six carbonyl VOCs, including the four markers identified in Fu's
work (73), plus acrolein and
malondialdehyde, was found to effectively discriminate patients
with lung cancer from healthy individuals in addition to patients
with BPN (73). The sensitivity in
each case was ≥96%, with specificity ranging from 64% for BPN to
86% for smokers and 100% for non-smokers and for the three groups
combined 84% (74).
Other potential metabolic biomarkers that can be
used for the early detection of lung cancer were also obtained in
previous studies. Typical examples are isoprene, methanol and
acetone (71,75,76).
Despite these promising advances, the lack of normalization and
standardization has led to significant variations in the VOC
profiles and/or concentrations among the different studies and no
commercial products have been used in clinical practice due to the
lack of uniform sampling standards and sample storage methods.
The metabolome is the most representative of the
molecular phenotype of an organism, where the concentrations of
metabolites directly reflect the current biochemical activity of
the organism. Therefore, metabolomics is considered a suitable
approach for increasing the efficacy of detecting early-stage lung
cancer. However, such applications require a deeper understanding
into how these measurements relate are associated with human
physiology and cancer pathology. It remains to be elucidated which
metabolites can be measured in biofluids which are readily
accessible to accurately reflect cancer status. Although progress
has been made, it remains unclear to what extent the metabolites in
biofluids can reveal about the metabolic activity of the tumor.
Additional metabolomics experiments in fluids associating these
measurements to the physiology of cancer would be a promising
future direction.
In addition, global metabolic alterations in
biofluids do not allow for the differentiation of cancer from other
diseases with systemic metabolic alterations such as
hypercholesteremia and phenylketonuria. The issue of such
confounding effects in metabolomic analysis will be minimized if
the tumor tissues are tested appropriately using NMR and MS.
Therefore, it would be of benefit to identify differentiating
metabolites in tissues through untargeted metabolomics before
testing them in the biofluids through targeted metabolomics.
Several studies have recently highlighted the role of extracellular
vesicles (EVs) and their cargo (protein and RNAs) in lung cancer
diagnosis (82–85), proposing EVs to be another potential
source of cancer biomarkers. Therefore, combined metabolomics
approaches for EVs phenotyping would provide vital insights into
the characteristics of EVs in cancer and potentially identify novel
strategies for the early detection of lung cancer.
One of the challenges with metabolomics is the vast
number and chemical complexity of metabolites that exist, such that
no current metabolomics approach can cover these complexities
comprehensively. This leads to inaccuracy in the early detection of
lung cancer. The present review proposes that currently, the
optimal metabolomics method for research would be combination with
other ‘omics’ approaches to comprehensively elucidate the changes
in metabolites in lung cancer whilst also to improve the accuracy
of lung cancer screening.
Not applicable.
The present study was funded by grants from the National Key
Research and Development Program of China (grant no.
2018YFC1315000) and the National Natural Science Foundation of
China (grant no. 82273722).
Not applicable.
YJX and XSD were involved in conceptualization,
writing, and reviewing. CQ, FW and JL were involved in writing, and
reviewing. YWY, LZ, FWT, WQC and WC were involved in reviewing and
editing. JH and NL were involved in study concept and design, draft
manuscript preparation and analysis and interpretation. All authors
reviewed the paper and approved the final version of the
manuscript. Data authentication is not applicable.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
International Early Lung Cancer Action
Program Investigators, . Henschke CI, Yankelevitz DF, Libby DM,
Pasmantier MW, Smith JP and Miettinen OS: Survival of patients with
stage I lung cancer detected on CT screening. N Engl J Med.
355:1763–1771. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Blandin Knight S, Crosbie PA, Balata H,
Chudziak J, Hussell T and Dive C: Progress and prospects of early
detection in lung cancer. Open Biol. 7:1700702017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Eggert JA, Palavanzadeh M and Blanton A:
Screening and early detection of lung cancer. Semin Oncol Nurs.
33:129–140. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Shlomi D, Abud M, Liran O, Bar J, Gai-Mor
N, Ilouze M, Onn A, Ben-Nun A, Haick H and Peled N: Detection of
lung cancer and EGFR mutation by electronic nose system. J Thorac
Oncol. 12:1544–1551. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Arbour KC and Riely GJ: Systemic therapy
for locally advanced and metastatic non-small cell lung cancer: A
Review. JAMA. 322:764–774. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hoffman PC, Mauer AM and Vokes EE: Lung
cancer. Lancet. 355:479–485. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
National Lung Screening Trial Research
Team, . Aberle DR, Adams AM, Berg CD, Black WC, Clapp JD,
Fagerstrom RM, Gareen IF, Gatsonis C, Marcus PM and Sicks JD:
Reduced lung-cancer mortality with low-dose computed tomographic
screening. N Engl J Med. 365:395–409. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Pastorino U, Silva M, Sestini S, Sabia F,
Boeri M, Cantarutti A, Sverzellati N, Sozzi G, Corrao G and
Marchiano A: Prolonged lung cancer screening reduced 10-year
mortality in the MILD trial: New confirmation of lung cancer
screening efficacy. Ann Oncol. 30:16722019. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
de Koning HJ, van der Aalst CM, de Jong
PA, Scholten ET, Nackaerts K, Heuvelmans MA, Lammers JJ, Weenink C,
Yousaf-Khan U, Horeweg N, et al: Reduced lung-cancer mortality with
volume CT Screening in a randomized trial. N Engl J Med.
382:503–513. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chu GCW, Lazare K and Sullivan F: Serum
and blood based biomarkers for lung cancer screening: A systematic
review. BMC Cancer. 18:1812018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Priola AM, Priola SM, Giaj-Levra M, Basso
E, Veltri A, Fava C and Cardinale L: Clinical implications and
added costs of incidental findings in an early detection study of
lung cancer by using low-dose spiral computed tomography. Clin Lung
Cancer. 14:139–148. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hassanein M, Rahman JS, Chaurand P and
Massion PP: Advances in proteomic strategies toward the early
detection of lung cancer. Proc Am Thorac Soc. 8:183–188. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hassanein M, Callison JC, Callaway-Lane C,
Aldrich MC, Grogan EL and Massion PP: The state of molecular
biomarkers for the early detection of lung cancer. Cancer Prev Res
(Phila). 5:992–1006. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Sozzi G and Boeri M: Potential biomarkers
for lung cancer screening. Transl Lung Cancer Res. 3:139–148.
2014.PubMed/NCBI
|
|
16
|
Spratlin JL, Serkova NJ and Eckhardt SG:
Clinical applications of metabolomics in oncology: A review. Clin
Cancer Res. 15:431–440. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Li X, Kulkarni AS, Liu X, Gao WQ, Huang L,
Hu Z and Qian K: Metal-Organic framework hybrids aid metabolic
profiling for colorectal cancer. Small Methods. 5:e20010012021.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Su H, Li X, Huang L, Cao J, Zhang M,
Vedarethinam V, Di W, Hu Z and Qian K: Plasmonic alloys reveal a
distinct metabolic phenotype of early gastric cancer. Adv Mater.
33:e20079782021. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Pei C, Liu C, Wang Y, Cheng D, Li R, Shu
W, Zhang C, Hu W, Jin A, Yang Y and Wan J: simpleFeOOH@Metal-Organic
framework core-satellite nanocomposites for the serum metabolic
fingerprinting of gynecological cancers. Angew Chem Int Ed Engl.
59:10831–10835. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Duarte IF, Rocha CM and Gil AM: Metabolic
profiling of biofluids: Potential in lung cancer screening and
diagnosis. Expert Rev Mol Diagn. 13:737–748. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
German JB, Hammock BD and Watkins SM:
Metabolomics: Building on a century of biochemistry to guide human
health. Metabolomics. 1:3–9. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Clish CB: Metabolomics: An emerging but
powerful tool for precision medicine. Cold Spring Harb Mol Case
Stud. 1:a0005882015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Schmidt DR, Patel R, Kirsch DG, Lewis CA,
Vander Heiden MG and Locasale JW: Metabolomics in cancer research
and emerging applications in clinical oncology. CA Cancer J Clin.
71:333–358. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Liu X and Locasale JW: Metabolomics: A
primer. Trends Biochem Sci. 42:274–284. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Rinschen MM, Ivanisevic J, Giera M and
Siuzdak G: Identification of bioactive metabolites using activity
metabolomics. Nat Rev Mol Cell Biol. 20:353–367. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Tsedilin AM, Fakhrutdinov AN, Eremin DB,
Zalesskiy SS, Chizhov AO, Kolotyrkina NG and Ananikov VP: How
sensitive and accurate are routine NMR and MS measurements?
Mendeleev Commun. 25:454–456. 2015. View Article : Google Scholar
|
|
27
|
Dervilly-Pinel G, Courant F, Chereau S,
Royer AL, Boyard-Kieken F, Antignac JP, Monteau F and Le Bizec B:
Metabolomics in food analysis: Application to the control of
forbidden substances. Drug Test Anal. 4 (Suppl 1):S59–S69. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Li XJ, Hayward C, Fong PY, Dominguez M,
Hunsucker SW, Lee LW, McLean M, Law S, Butler H, Schirm M, et al: A
blood-based proteomic classifier for the molecular characterization
of pulmonary nodules. Sci Transl Med. 5:207ra1422013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhang J, Rector J, Lin JQ, Young JH, Sans
M, Katta N, Giese N, Yu W, Nagi C, Suliburk J, et al:
Nondestructive tissue analysis for ex vivo and in vivo cancer
diagnosis using a handheld mass spectrometry system. Sci Transl
Med. 9:eaan39682017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Lei Z, Huhman DV and Sumner LW: Mass
spectrometry strategies in metabolomics. J Biol Chem.
286:25435–25442. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wishart DS: Advances in metabolite
identification. Bioanalysis. 3:1769–1782. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhang A, Sun H, Wang P, Han Y and Wang X:
Modern analytical techniques in metabolomics analysis. Analyst.
137:293–300. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Dunn WB, Bailey NJ and Johnson HE:
Measuring the metabolome: Current analytical technologies. Analyst.
130:606–625. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Psychogios N, Hau DD, Peng J, Guo AC,
Mandal R, Bouatra S, Sinelnikov I, Krishnamurthy R, Eisner R,
Gautam B, et al: The human serum metabolome. PLoS One.
6:e169572011. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Bouatra S, Aziat F, Mandal R, Guo AC,
Wilson MR, Knox C, Bjorndahl TC, Krishnamurthy R, Saleem F, Liu P,
et al: The human urine metabolome. PLoS One. 8:e730762013.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wishart DS: Emerging applications of
metabolomics in drug discovery and precision medicine. Nat Rev Drug
Discov. 15:473–484. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Rocha CM, Carrola J, Barros AS, Gil AM,
Goodfellow BJ, Carreira IM, Bernardo J, Gomes A, Sousa V, Carvalho
L and Duarte IF: Metabolic signatures of lung cancer in biofluids:
NMR-based metabonomics of blood plasma. J Proteome Res.
10:4314–4324. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Deja S, Porebska I, Kowal A, Zabek A, Barg
W, Pawelczyk K, Stanimirova I, Daszykowski M, Korzeniewska A,
Jankowska R and Mlynarz P: Metabolomics provide new insights on
lung cancer staging and discrimination from chronic obstructive
pulmonary disease. J Pharm Biomed Anal. 100:369–380. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Puchades-Carrasco L, Jantus-Lewintre E,
Perez-Rambla C, Garcia-Garcia F, Lucas R, Calabuig S, Blasco A,
Dopazo J, Camps C and Pineda-Lucena A: Serum metabolomic profiling
facilitates the non-invasive identification of metabolic biomarkers
associated with the onset and progression of non-small cell lung
cancer. Oncotarget. 7:12904–12916. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Ros-Mazurczyk M, Wojakowska A, Marczak L,
Polanski K, Pietrowska M, Polanska J, Dziadziuszko R, Jassem J,
Rzyman W and Widlak P: Panel of serum metabolites discriminates
cancer patients and healthy participants of lung cancer screening-a
pilot study. Acta Biochim Pol. 64:513–518. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Klupczynska A, Derezinski P, Garrett TJ,
Rubio VY, Dyszkiewicz W, Kasprzyk M and Kokot ZJ: Study of early
stage non-small-cell lung cancer using Orbitrap-based global serum
metabolomics. J Cancer Res Clin Oncol. 143:649–659. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Chen Y, Ma Z, Min L, Li H, Wang B, Zhong J
and Dai L: Biomarker identification and pathway analysis by serum
metabolomics of lung cancer. Biomed Res Int.
2015:1836242015.PubMed/NCBI
|
|
43
|
Ros-Mazurczyk M, Jelonek K, Marczyk M,
Binczyk F, Pietrowska M, Polanska J, Dziadziuszko R, Jassem J,
Rzyman W and Widlak P: Serum lipid profile discriminates patients
with early lung cancer from healthy controls. Lung Cancer.
112:69–74. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Klupczynska A, Derezinski P, Dyszkiewicz
W, Pawlak K, Kasprzyk M and Kokot ZJ: Evaluation of serum amino
acid profiles' utility in non-small cell lung cancer detection in
Polish population. Lung Cancer. 100:71–76. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Louis E, Adriaensens P, Guedens W,
Bigirumurame T, Baeten K, Vanhove K, Vandeurzen K, Darquennes K,
Vansteenkiste J, Dooms C, et al: Detection of lung cancer through
metabolic changes measured in blood plasma. J Thorac Oncol.
11:516–523. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Mazzone PJ, Wang XF, Beukemann M, Zhang Q,
Seeley M, Mohney R, Holt T and Pappan KL: Metabolite profiles of
the serum of patients with non-small cell carcinoma. J Thorac
Oncol. 11:72–78. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Yu Z, Chen H, Ai J, Zhu Y, Li Y, Borgia
JA, Yang JS, Zhang J, Jiang B, Gu W and Deng Y: Global lipidomics
identified plasma lipids as novel biomarkers for early detection of
lung cancer. Oncotarget. 8:107899–107906. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Xiang C, Jin S, Zhang J, Chen M, Xia Y,
Shu Y and Guo R: Cortisol, cortisone, and 4-methoxyphenylacetic
acid as potential plasma biomarkers for early detection of
non-small cell lung cancer. Int J Biol Markers. 33:314–320. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Klupczynska A, Plewa S, Kasprzyk M,
Dyszkiewicz W, Kokot ZJ and Matysiak J: Serum lipidome screening in
patients with stage I non-small cell lung cancer. Clin Exp Med.
19:505–513. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Zhang L, Zheng J, Ahmed R, Huang G, Reid
J, Mandal R, Maksymuik A, Sitar DS, Tappia PS, Ramjiawan B, et al:
A High-Performing plasma metabolite panel for early-stage lung
cancer detection. Cancers (Basel). 12:6222020. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Huang L, Wang L, Hu X, Chen S, Tao Y, Su
H, Yang J, Xu W, Vedarethinam V, Wu S, et al: Machine learning of
serum metabolic patterns encodes early-stage lung adenocarcinoma.
Nat Commun. 11:35562020. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Derveaux E, Thomeer M, Mesotten L,
Reekmans G and Adriaensens P: Detection of lung cancer via blood
plasma and 1H-NMR metabolomics: Validation by a
semi-targeted and quantitative approach using a protein-binding
competitor. Metabolites. 11:5372021. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Qi SA, Wu Q, Chen Z, Zhang W, Zhou Y, Mao
K, Li J, Li Y, Chen J and Huang Y and Huang Y: High-resolution
metabolomic biomarkers for lung cancer diagnosis and prognosis. Sci
Rep. 11:118052021. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Wang G, Qiu M, Xing X, Zhou J, Yao H, Li
M, Yin R, Hou Y, Li Y, Pan S, et al: Lung cancer scRNA-seq and
lipidomics reveal aberrant lipid metabolism for early-stage
diagnosis. Sci Transl Med. 14:eabk27562022. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Warburg O: On the origin of cancer cells.
Science. 123:309–314. 1956. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Mathupala SP, Ko YH and Pedersen PL:
Hexokinase-2 bound to mitochondria: Cancer's stygian link to the
‘Warburg Effect’ and a pivotal target for effective therapy. Semin
Cancer Biol. 19:17–24. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Dang CV: Links between metabolism and
cancer. Genes Dev. 26:877–890. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Rocha CM, Barros AS, Gil AM, Goodfellow
BJ, Humpfer E, Spraul M, Carreira IM, Melo JB, Bernardo J, Gomes A,
et al: Metabolic profiling of human lung cancer tissue by 1H high
resolution magic angle spinning (HRMAS) NMR spectroscopy. J
Proteome Res. 9:319–332. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Maksymiuk AW, Tappia PS, Sitar DS, Akhtar
PS, Khatun N, Parveen R, Ahmed R, Ahmed RB, Cheng B, Huang G, et
al: Use of amantadine as substrate for SSAT-1 activity as a
reliable clinical diagnostic assay for breast and lung cancer.
Future Sci OA. 5:FSO3652018. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Cho SH, Jung BH, Lee SH, Lee WY, Kong G
and Chung BC: Direct determination of nucleosides in the urine of
patients with breast cancer using column-switching liquid
chromatography-tandem mass spectrometry. Biomed Chromatogr.
20:1229–1236. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Carrola J, Rocha CM, Barros AS, Gil AM,
Goodfellow BJ, Carreira IM, Bernardo J, Gomes A, Sousa V, Carvalho
L and Duarte IF: Metabolic signatures of lung cancer in biofluids:
NMR-based metabonomics of urine. J Proteome Res. 10:221–230. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Hanai Y, Shimono K, Matsumura K, Vachani
A, Albelda S, Yamazaki K, Beauchamp GK and Oka H: Urinary volatile
compounds as biomarkers for lung cancer. Biosci Biotechnol Biochem.
76:679–684. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Mathe EA, Patterson AD, Haznadar M, Manna
SK, Krausz KW, Bowman ED, Shields PG, Idle JR, Smith PB, Anami K,
et al: Noninvasive urinary metabolomic profiling identifies
diagnostic and prognostic markers in lung cancer. Cancer Res.
74:3259–3270. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Haznadar M, Cai Q, Krausz KW, Bowman ED,
Margono E, Noro R, Thompson MD, Mathe EA, Munro HM, Steinwandel MD,
et al: Urinary metabolite risk biomarkers of lung cancer: A
prospective cohort study. Cancer Epidemiol Biomarkers Prev.
25:978–986. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Funai K, Honzawa K, Suzuki M, Momiki S,
Asai K, Kasamatsu N, Kawase A, Shinke T, Okada H, Nishizawa S and
Takamoto H: Urinary fluorescent metabolite O-aminohippuric acid is
a useful biomarker for lung cancer detection. Metabolomics.
16:1012020. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Jiang X, Chen X, Chen Z, Yu J, Lou H and
Wu J: High-Throughput salivary metabolite profiling on an ultralow
noise tip-enhanced laser desorption ionization mass spectrometry
platform for noninvasive diagnosis of early lung cancer. J Proteome
Res. 20:4346–4356. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Slack A, Yeoman A and Wendon J: Renal
dysfunction in chronic liver disease. Crit Care. 14:2142010.
View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Farraia MV, Cavaleiro Rufo J, Paciencia I,
Mendes F, Delgado L and Moreira A: The electronic nose technology
in clinical diagnosis: A systematic review. Porto Biomed J.
4:e422019. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Phillips M, Gleeson K, Hughes JM,
Greenberg J, Cataneo RN, Baker L and McVay WP: Volatile organic
compounds in breath as markers of lung cancer: A cross-sectional
study. Lancet. 353:1930–1933. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Phillips M, Cataneo RN, Cummin AR,
Gagliardi AJ, Gleeson K, Greenberg J, Maxfield RA and Rom WN:
Detection of lung cancer with volatile markers in the breath.
Chest. 123:2115–2123. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Poli D, Carbognani P, Corradi M, Goldoni
M, Acampa O, Balbi B, Bianchi L, Rusca M and Mutti A: Exhaled
volatile organic compounds in patients with non-small cell lung
cancer: Cross sectional and nested short-term follow-up study.
Respir Res. 6:712005. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Phillips M, Altorki N, Austin JH, Cameron
RB, Cataneo RN, Greenberg J, Kloss R, Maxfield RA, Munawar MI, Pass
HI, et al: Prediction of lung cancer using volatile biomarkers in
breath. Cancer Biomark. 3:95–109. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Fu XA, Li M, Knipp RJ, Nantz MH and
Bousamra M: Noninvasive detection of lung cancer using exhaled
breath. Cancer Med. 3:174–181. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Li M, Yang D, Brock G, Knipp RJ, Bousamra
M, Nantz MH and Fu XA: Breath carbonyl compounds as biomarkers of
lung cancer. Lung Cancer. 90:92–97. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Sakumura Y, Koyama Y, Tokutake H, Hida T,
Sato K, Itoh T, Akamatsu T and Shin W: Diagnosis by volatile
organic compounds in exhaled breath from lung cancer patients using
support vector machine algorithm. Sensors (Basel). 17:2872017.
View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Rudnicka J, Kowalkowski T and Buszewski B:
Searching for selected VOCs in human breath samples as potential
markers of lung cancer. Lung Cancer. 135:123–129. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Chen X, Muhammad KG, Madeeha C, Fu W, Xu
L, Hu Y, Liu J, Ying K, Chen L and Yurievna GO: Calculated indices
of volatile organic compounds (VOCs) in exhalation for lung cancer
screening and early detection. Lung Cancer. 154:197–205. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Tsou PH, Lin ZL, Pan YC, Yang HC, Chang
CJ, Liang SK, Wen YF, Chang CH, Chang LY, Yu KL, et al: Exploring
volatile organic compounds in breath for high-accuracy prediction
of lung cancer. Cancers (Basel). 13:14312021. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Hakim M, Broza YY, Barash O, Peled N,
Phillips M, Amann A and Haick H: Volatile organic compounds of lung
cancer and possible biochemical pathways. Chem Rev. 112:5949–5966.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Fan TW, Lane AN, Higashi RM, Farag MA, Gao
H, Bousamra M and Miller DM: Altered regulation of metabolic
pathways in human lung cancer discerned by (13)C stable
isotope-resolved metabolomics (SIRM). Mol Cancer. 8:412009.
View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Bousamra M II, Schumer E, Li M, Knipp RJ,
Nantz MH, van Berkel V and Fu XA: Quantitative analysis of exhaled
carbonyl compounds distinguishes benign from malignant pulmonary
disease. J Thorac Cardiovasc Surg. 148:1074–1080; discussion
1080-1. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Paramanantham A, Asfiya R, Das S, McCully
G and Srivastava A: Extracellular Vesicle (EVs) associated
non-coding RNAs in lung cancer and therapeutics. Int J Mol Sci.
23:136372022. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Hua Y, Dai C, He Q, Cai X and Li M:
Autoantibody panel on small extracellular vesicles for the early
detection of lung cancer. Clin Immunol. 245:1091752022. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Cammarata G, de Miguel-Perez D, Russo A,
Peleg A, Dolo V, Rolfo C and Taverna S: Emerging noncoding RNAs
contained in extracellular vesicles: Rising stars as biomarkers in
lung cancer liquid biopsy. Ther Adv Med Oncol.
14:175883592211312292022. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Pedraz-Valdunciel C, Giannoukakos S,
Gimenez-Capitan A, Fortunato D, Filipska M, Bertran-Alamillo J,
Bracht JWP, Drozdowskyj A, Valarezo J, Zarovni N, et al: Multiplex
Analysis of CircRNAs from plasma extracellular vesicle-enriched
samples for the detection of early-stage non-small cell lung
cancer. Pharmaceutics. 14:20342022. View Article : Google Scholar : PubMed/NCBI
|