Introduction
Acute myeloid leukemia (AML) is a blood cancer
characterized by the abnormal growth and accumulation of cells in
the hematopoietic system (1). It is
the most common type of AML in adults, with ~20,380 new cases and
11,310 deaths in 2023 (2). Despite
extensive research on prognostic biomarkers, the prognosis of AML
remains highly variable, with a <50% 5-year overall survival
(OS) rate and only a 20% survival rate for elderly patients 2 years
post-diagnosis (3). Currently,
cytogenetic and molecular abnormalities at diagnosis are considered
the most important prognostic factors, predicting complete
remission rates, disease-free survival, relapse risk and OS.
Anoikis, a form of programmed cell death, occurs
when cell-cell or cell-extracellular matrix attachments are
disrupted, contributing to tissue homeostasis maintenance by
eliminating misplaced or dislodged cells (4). Cancer cells often evade anoikis
through several mechanisms, resulting in enhanced invasiveness and
metastatic potential (5).
Anoikis-related genes (ARGs) are crucial in driving the overall
progression and metastatic cascade across several cancers, such as
gastric carcinoma (6), lung cancer
(7), breast carcinoma (8) and endometrial carcinoma (9).
Patients with AML and elevated lectin
galactoside-binding soluble 1 (LGALS1) mRNA levels exhibit reduced
disease-free survival (10).
Additionally, hepatocyte growth factor (HGF) affects leukemic cell
proliferation and migration (11),
whilst integrin subunit α 4 (ITGA4) mediates anti-apoptotic
signals, conferring chemoresistance (12). Both in vivo and in
vitro studies have demonstrated the critical role of Ras
homolog gene family member C (RhoC) in promoting metastasis by
protecting metastatic cells from apoptosis, influencing cell
motility and modulating chemokine secretion (13). Despite its substantial impact on
tumorigenesis and metastasis, the role of anoikis in AML remains
unclear.
The present study aimed to identify hub genes
associated with anoikis in patients with AML and enhance the
predictive power of highly influential genes through several
analyses, including univariate and multivariate Cox regressions,
differential expression analysis and Least Absolute Shrinkage and
Selection Operator (LASSO) regression. Using these identified ARGs,
the present study performed prognostic assessments, functional
enrichment analysis and principal component analysis (PCA) in
patients with AML. Ultimately, a risk signature was developed to
assess the predictive value of ARGs in AML, aiming to provide a
novel prognostic tool for patients with this pathology.
Materials and methods
Data acquisition of patients with
AML
The survival data and RNA-sequencing (RNA-seq) data
of 151 patients with AML were obtained from The Cancer Genome Atlas
(TCGA) using the publicly available University of California, Santa
Cruz Xena database (https://xenabrowser.net/datapages/). A training set
comprising 132 patients with AML with comprehensive clinical
information and survival data was used for subsequent analysis. The
GSE71014 dataset, containing RNA-seq and survival data from 104
patients with AML, was sourced from the Gene Expression Omnibus
(GEO) database (https://www.ncbi.nlm.nih.gov/geo/) and served as the
validation set. In a previous study, 434 ARGs were identified
(14).
Identification of anoikis-related
subtypes in the training set
The R package ‘ConsensusClusterPlus’ version 1.54.0
(15) was used to identify subtypes
associated with anoikis, based on the expression of the 434 ARGs.
The clustering results were validated using PCA. The OS among
different subtypes was further assessed using the ‘Survival’
package version 3.2–3.
Simultaneously, the ‘limma’ package version 3.52.4
(16) was used for differential
expression analysis to identify differentially expressed ARGs
(DE-ARGs) between the two identified subtypes. The screening
criteria were set as P<0.05 and
|log2FoldChange|>0.5. The ‘clusterProfiler’ version
4.4.4 (15) was used to perform
Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes
(KEGG) enrichment analyses on these DE-ARGs. The results were
visualized using bubble plots with the R package ‘ggplot2’ version
3.3.2 (The R Foundation) (17).
Construction and validation of the
prognostic risk model
A univariate Cox analysis of DE-ARGs was performed
in the training set to identify the prognosis-related genes
(P<0.05). The most predictive prognostic genes were identified
through using LASSO (18) and
multivariate Cox analyses. Subsequently, patients in the training
set were stratified into two groups based on the median risk score.
The differences in OS between the two groups were visualized using
Kaplan-Meier (KM) curves using the ‘survminer’ package 3.6.0
(19). To assess the prognostic
capability of the model, receiver operating characteristic (ROC)
curves were developed using the ‘survival ROC’ package 1.42.0
(20). Finally, the prognostic
model was validated using the external validation dataset
GSE71014.
Analysis of independent prognostic
factors
Univariate and multifactorial Cox analyses were
performed to determine the association between clinicopathological
characteristics and risk scores, and identify independent
predictive factors for AML. The ‘rms’ package version 6.0–1
(21) was used to develop a
nomogram predicting survival probability based on independent
prognostic criteria. Calibration curve and decision curve analyses
were used to validate the suitability of the nomogram for clinical
decision-making.
Biological differences between the two
groups
The limma package version 3.52.4 (16) was used to identify differentially
expressed genes (DEGs), with criteria set at
|log2FC|>0.5 and adjusted P-value (P.adjust)
<0.05. Subsequently, a functional enrichment analysis on these
DEGs was performed using the R package ‘clusterProfiler’ version
4.4.4 (The R Foundation).
Distribution of clinicopathological
features for risk score determination
In the training set, clinical information was
extracted, such as cytogenetic risk categories mentioned in the
published literature (intermediate/normal, favorable, unknown or
poor) (22), sex (female or male),
prior treatment and diagnosis (No or Yes) and age (>60 or ≤60
years) of patients with AML. The phenotype data of the TCGA AML
dataset were downloaded from the Xena database (https://gdc-hub.s3.us-east-1.amazonaws.com/download/TCGA-LAML.GDC_phenotype.tsv.gz)
to extract clinical features including cytogenetics risk category,
sex, ‘prior_treatment.diagnoses’ and age. The cytogenetics risk
category was categorized into intermediate/normal, favorable, poor
and unknown groups according to the cytogenetics risk category
column in the downloaded phenotype file. Differences in risk scores
between subgroups with different clinical characteristics were
subsequently compared using Wilcoxon rank-sum tests (comparison
between two groups) and the Kruskal-Wallis test (comparison between
multiple groups) to determine significant differences between
clinical conditions (P<0.05. Dunn's test was used as the post
hoc test. The ‘ComplexHeatmap’ version 1.14.0 (23) was used to visualize the results.
Reverse transcription
(RT)-quantitative (q)PCR) analyses
Bone marrow samples were collected from 20
individuals diagnosed with AML, including newly diagnosed patients
and those who had relapsed. The control group consisted of healthy
donors matched for age and sex with the patients. Allogeneic
hematopoietic stem cell donors were recruited from patients
scheduled for hematopoietic stem cell transplantation at Guizhou
Medical University (Guiyang, China) between February 2022 and
October 2023. Ethical approval for the present study was obtained
from The Ethics Committee of the Affiliated Hospital of Guizhou
Medical University (approval no. 2023-744). The donors were all
family members or friends of patients who were then hospitalized.
Prior to the donation, all patients underwent bone marrow
aspiration to assess the normality of bone marrow morphology
according to hospital requirements. The present study was performed
in accordance with the principles of the Declaration of Helsinki,
and all patients provided written informed consent prior to
enrollment. Table SI presents
information on the patients with AML.
RNA extraction was performed using an Ultra Pure RNA
Extraction kit (cat. no. CW0581; Jiangsu CoWin Biotech Co, Ltd.),
followed by RT at 55°C for 5 min to generate cDNA using a Reverse
Transcription cDNA kit (cat. no. K1622; Thermo Fisher Scientific,
Inc.). qPCR was performed with SYBR Green Master Mix (cat. no.
NVZ-Q221-01; Vazyme Biotech Co., Ltd.), using an Applied Biosystems
7500 Real-Time Cycler (QuantStudio 1; Applied Biosystems; Thermo
Fisher Scientific, Inc.). PCR conditions included an initial
denaturation step at 95°C for 10 min, followed by 40 cycles of
denaturation at 95°C for 15 sec, and annealing/extension at 60°C
for 1 min, with a standard melting curve analysis performed
afterward. All samples were analyzed in triplicate, and gene
expression levels were quantified using the comparative threshold
cycle method (2−ΔΔCq) with GAPDH serving as the
reference gene for normalization (24). Primer pairs and corresponding
sequences used in the present study are detailed in Table SII.
Prediction of chemotherapy drug
The oncoPredict tool version 0.2 (25) was used to predict chemotherapy
agents for AML using data from the Genomics of Drug Sensitivity in
Cancer (GDSC) database (https://www.cancerrxgene.org/). The half-maximal
inhibitory concentration (IC50) values were calculated
for each patient with AML in the two groups. To compare differences
in drug sensitivity between the two groups, the Wilcoxon rank-sum
test was used.
Statistical analysis
R software (version 4.2.2; The R Foundation) was
used for all analyses. The Wilcoxon rank-sum test was used to
compare data between the two groups. The Kruskal-Wallis test was
used for multiple comparisons, followed by Dunn's post hoc test.
P<0.05 was considered to indicate a statistically significant
difference.
Results
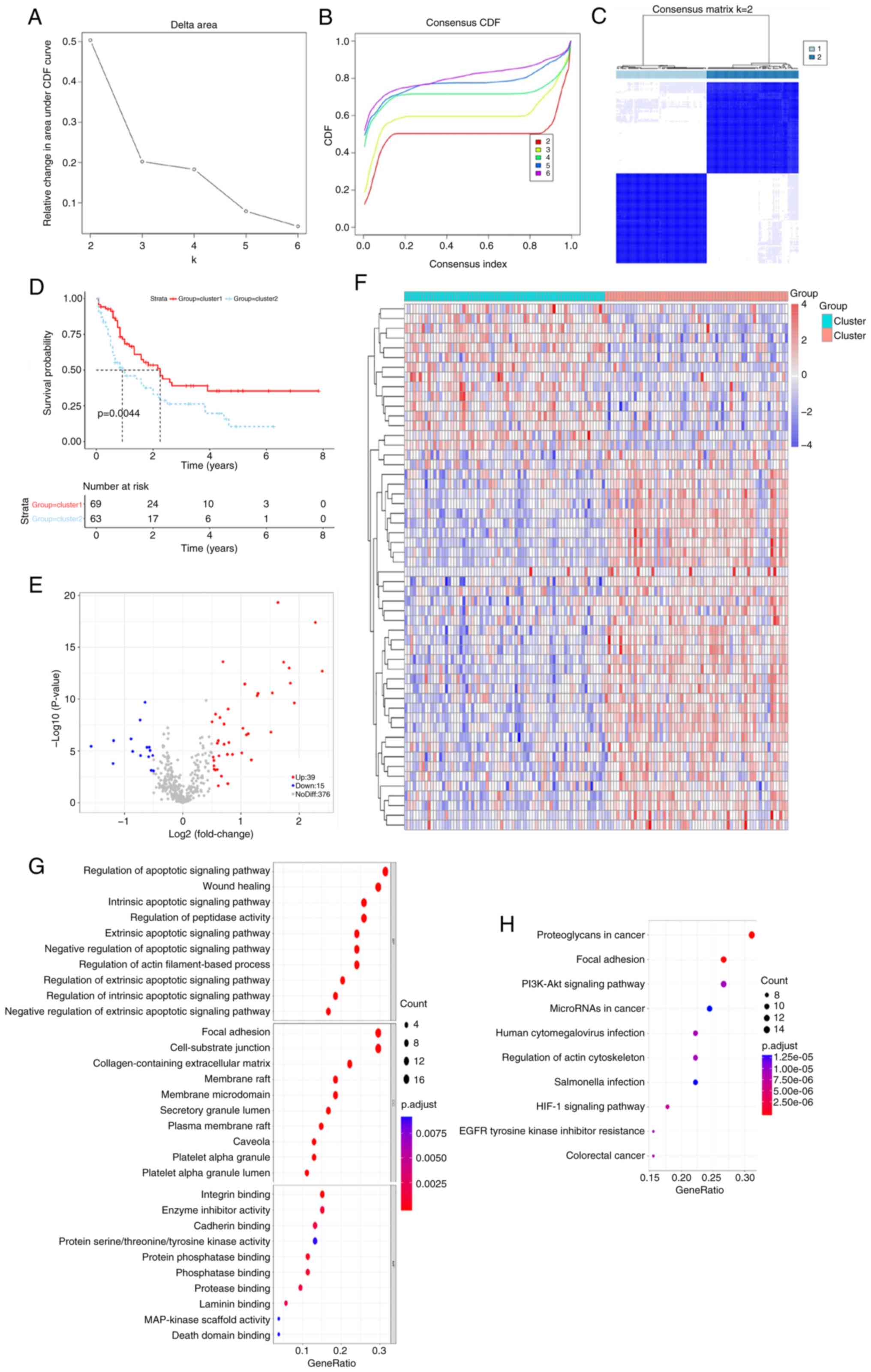
Identification of anoikis-related
subtypes
Using the expression profiles of ARGs, 132 AML
samples were classified through the consensus clustering analysis
method. The consistency distribution for k values ranging from 2–6
was displayed in an empirical cumulative distribution function
plot. The consensus matrix heatmap revealed that k=2 was optimal
for classification, dividing AML samples into cluster 1 and cluster
2 (Figs. 1A-C and S1; Table
SIII). Patients in cluster 2 exhibited inferior prognoses
compared with those in cluster 1 (Fig.
1D), indicating that AML prognosis is influenced by ARG
expression levels and supporting the subsequent screening of
survival-related ARGs. A difference in the expression of 54 ARGs
was observed between the two subtypes (Fig. 1E and F). Using a significance
threshold of P.adjust <0.05, 1,067 GO terms and 104 KEGG
pathways were associated with these DEGs (Fig. 1G and H). GO analysis revealed that
these DEGs were involved in the regulation of the intrinsic
apoptotic signaling pathway, regulation of peptidase activity,
apoptotic signaling pathway, focal adhesion and cell-substrate
junction. Additionally, DEGs were significantly associated with
microRNA (miR) in cancer, phosphatidylinositol 3-kinase
(PI3K)/protein kinase B (Akt) signaling pathway and proteoglycans
in cancer.
Development of an effective prognostic
risk model associated with anoikis in AML
Using DE-ARGs, 21 genes with P<0.05 were
identified in the training set (Fig.
2A). Subsequently, LASSO regression analysis was performed to
exclude false positive genes (Fig. 2B
and C). Finally, four prognostic ARGs were determined using
multifactorial Cox analysis: LGALS1, ITGA4, HGF and RHOC (Fig. 2D).
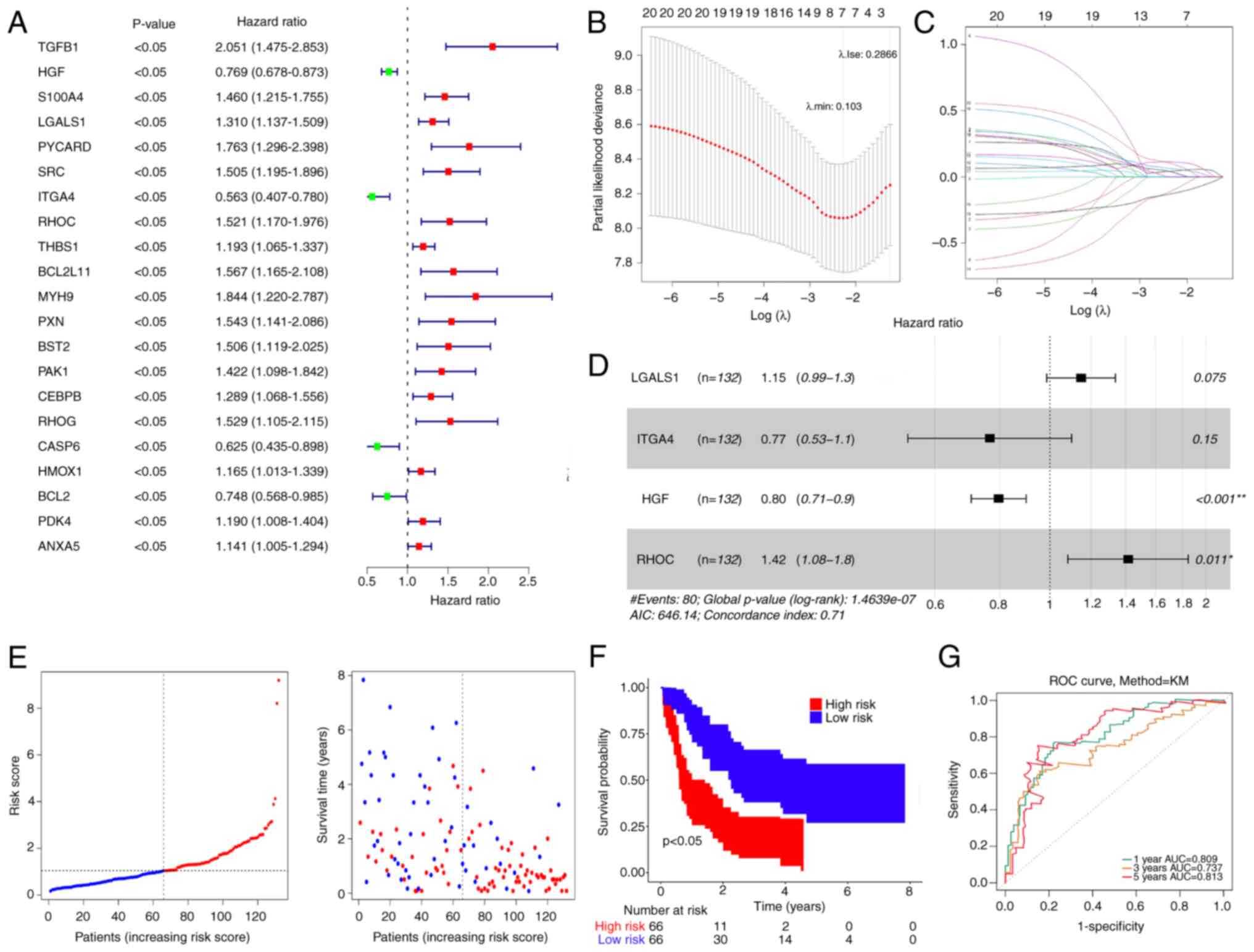 | Figure 2.Prognostic risk model development and
validation. (A) Univariate Cox analysis of DE-ARGs. (B) Abscissa
represents log (λ) and the ordinate denotes the error of
cross-validation. (C) Each curve represents the change trajectory
for each independent variable coefficient. (D) Multifactorial Cox
analysis of DE-ARGs that passed least absolute shrinkage and
selection operator regression analysis. (E) Distribution of
patients into high- and low-risk groups in the training set. (F) KM
survival analysis of patients in the high- and low-risk groups. (G)
ROC curves of patients at 1, 3 and 5 years. DE-ARGs, differentially
expressed anoikis-related genes, ROC, receiver operating
characteristic, KM, Kaplan-Meier, AUC, area under the curve;
LGALS1, lectin galactoside-binding soluble 1; ITGA4, integrin
subunit α4; HGF, hepatocyte growth factor; RHOC, Ras homolog gene
family member C. |
The risk score was calculated as follows: Risk score
= 0.13836534 × LGALS1-0.26749323 × ITGA4-0.227481177 × HGF +
0.3471619 × RHOC.
Patients were stratified into two groups based on a
median risk of 1.042299 (Fig. 2E).
Patients with low-risk scores demonstrated significantly higher OS
rates compared with those with high-risk scores (Fig. 2F). The validity of the risk
signature was further assessed by computing the ROC curve for OS.
The area under the curve (AUC) values were >0.70 at 1, 3 and 5
years, indicating enhanced efficacy of the prognostic risk model
(Fig. 2G).
The model was validated using the GSE71014 dataset,
and AML samples were stratified based on the median risk score. As
the risk score increased, the OS of patients with AML gradually
decreased, accompanied by a steady rise in mortality rates
(Fig. S2A and B). KM curves in the
GSE71014 dataset revealed that patients with AML in the low-risk
group had significantly longer OS rates compared with those in the
high-risk group (P<0.05; Fig.
S2C). Furthermore, the AUC values for 1-, 3- and 5-year
survival rates based on this model were 0.809, 0.737 and 0.813,
respectively, indicating enhanced efficacy of the prognostic risk
model (Fig. S2D).
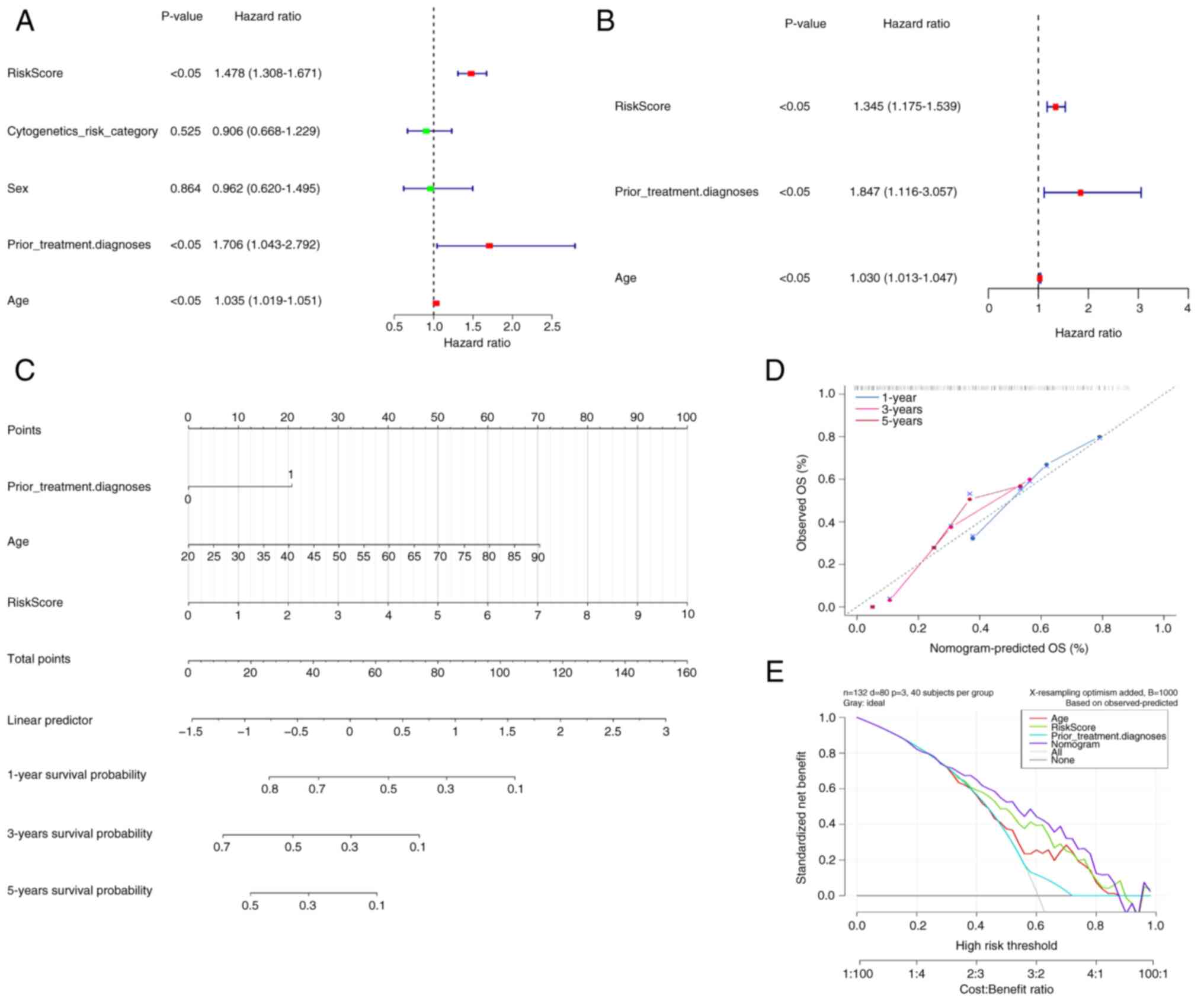
Construction of a nomogram model with
accurate prediction
The clinicopathological variables and risk scores
from 132 patients were combined to perform univariate and
multivariate Cox regression analyses (Fig. 3A and B). Risk scores, prior
treatment, diagnosis and age were demonstrated to be independent
prognostic factors for patients with AML. Based on these factors, a
nomogram model was constructed (Fig.
3C), indicating a marked decrease in survival rate with an
increasing overall score. The calibration curve yielded a c-index
of 0.942 for this nomogram model, demonstrating its high predictive
accuracy and reliability (Fig. 3D).
Therefore, the nomogram emerged as the optimal model (Fig. 3E).
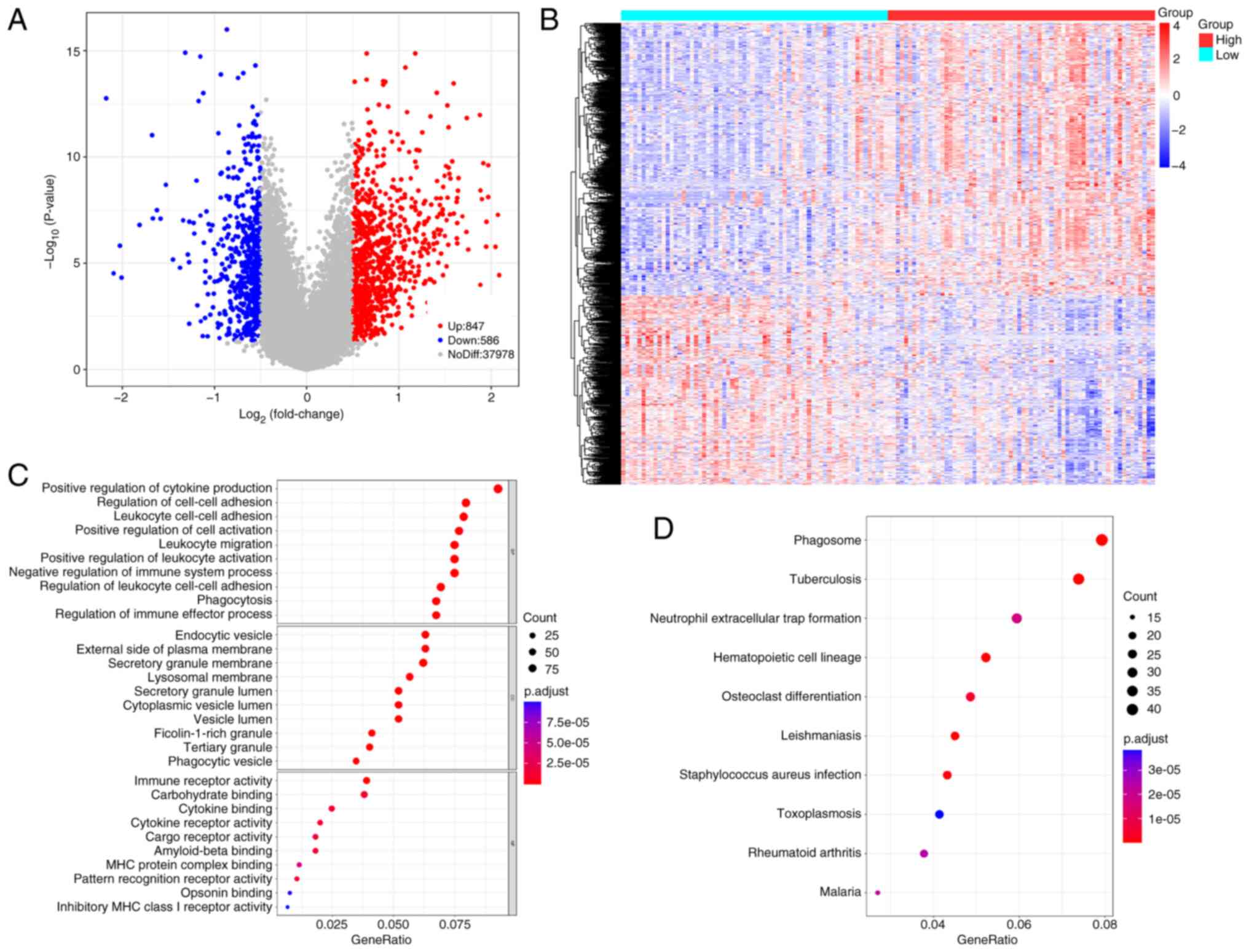
Identification of DEGs and their
functional enrichment analysis
A total of 1,433 DEGs between the two groups were
identified (Table SIV). Volcano
and heatmap representations of these DEGs are presented in Fig. 4A and B. Screening based on P.adjust
values <0.05 yielded 1,218 GO terms and 59 KEGG pathways
(Fig. 4C and D). These DEGs were
significantly associated with regulation of cell-cell adhesion,
positive regulation of cytokine production, endocytic vesicles and
leukocyte cell-cell adhesion. DEGs were significantly involved in
phagosome formation, neutrophil extracellular trap formation,
hematopoietic cell lineage and osteoclast differentiation.
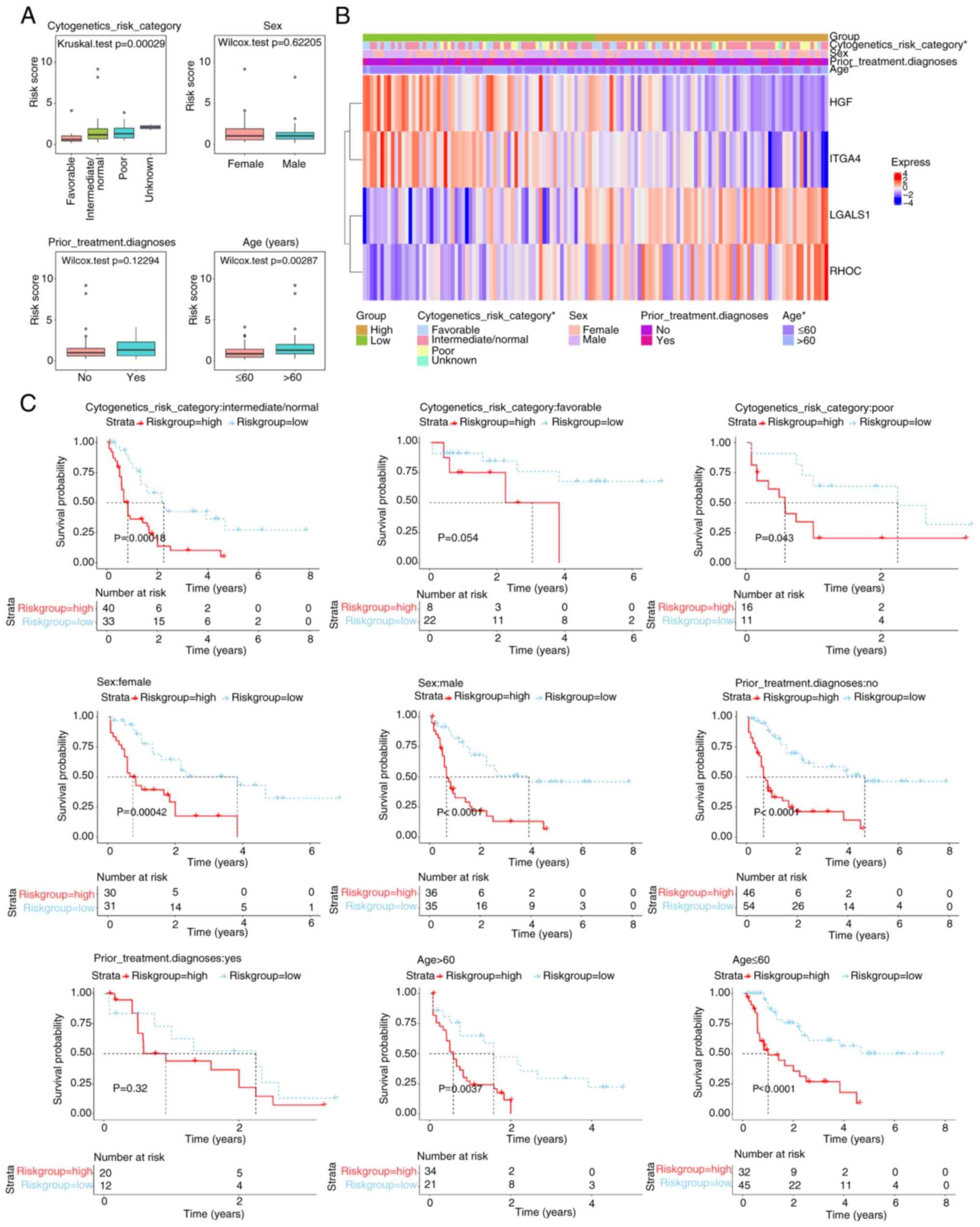
Association between risk scores, age
and cytogenetics risk category
Significant differences in risk scores (P<0.05)
were demonstrated across cytogenetics risk categories (favorable
vs. intermediate/normal, favorable vs. poor and favorable vs.
unknown) and age groups (Fig. 5A;
Table SV). A total of four genes
were notably associated with different clinical characteristics
(Fig. 5B). Survival analysis
stratified by clinical data revealed no significant differences in
the cytogenetics risk category-favorable and prior treatment
diagnosis-Yes subgroups, whilst significant differences were
demonstrated in the remaining subgroups (Fig. 5C).
LGALS1, ITGA4, HGF and RHOC can be
used as prognostic genes of AML
Multivariate Cox regression analysis revealed that
LGALS1, RHOC, ITGA4 and HGF were notably associated with a
favorable prognostic impact on patients with AML. As diagnostic
biomarkers, the AUC of LGALS1, RHOC, ITGA4 and HGF were >0.6,
indicating their high predictive accuracy in AML diagnosis
(Fig. 6A).
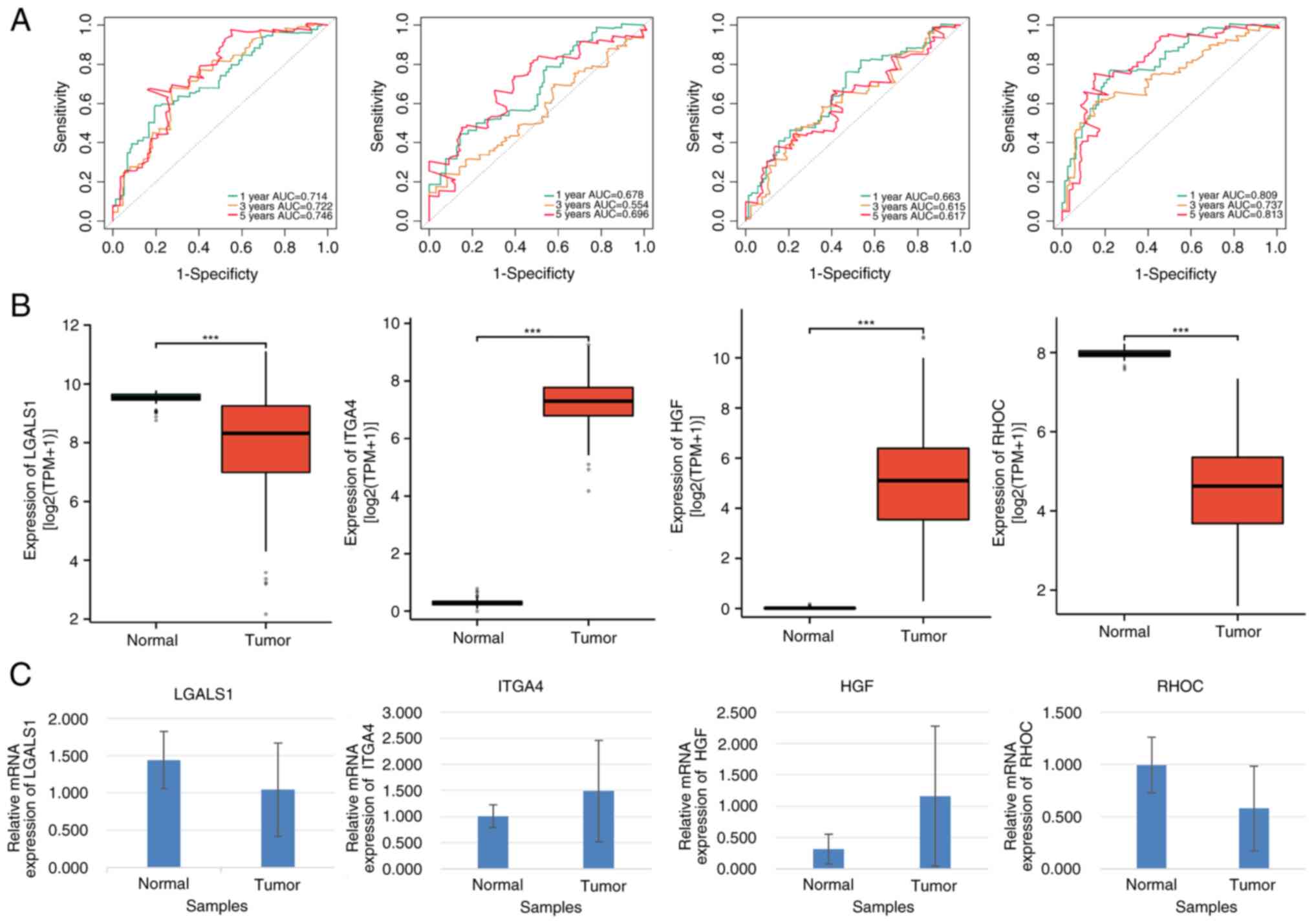 | Figure 6.Prognostic and clinical value
analyses of LGALS1, ITGA4, HGF and RHOC in patients with AML. (A)
Receiver operating characteristic curves of LGALS1, ITGA4, HGF and
RHOC. (B) Gene expression levels of LGALS1, ITGA4, HGF and RHOC in
AML and normal control samples. (C) Reverse
transcription-quantitative PCR analysis of LGALS1, ITGA4, HGF and
RHOC expression (P<0.05). ***P≤0.001. AML, acute myeloid
leukemia, AUC, area under the curve. |
Furthermore, LGALS1 and HGF expression levels were
significantly lower, whilst ITGA4 and RHOC expression levels were
significantly higher in patients with AML compared with the
corresponding controls (all P<0.05; Fig. 6B). These results were further
confirmed by RT-qPCR analysis (Fig.
6C). Additionally, mRNA expression levels of LGALS1 and RHOC
were significantly higher, whilst those of ITGA4 and HGF were
significantly lower in patients with AML compared with their
healthy counterparts (all P<0.05; Table SVI).
Sensitivity of AML-related drugs
varies between high- and low-risk groups
The IC50 values were calculated for each
patient with AML in the two groups. In total, 138 drugs
demonstrated significant IC50 values (Table SVII). Box plots in Fig. 7 demonstrate the IC50
values for the top 10 significantly different treatment-sensitive
drugs. These results indicate substantial disparity between the
high- and low-risk groups, with the former exhibiting considerably
higher IC50 values.
Discussion
AML is an aggressive form of cancer characterized by
the rapid proliferation of immature myeloid leukemia cells
(26). Whilst it primarily affects
the bone marrow, malignant cells may also be found in the
peripheral blood or other tissues (27,28).
Despite advancements in therapeutic and diagnostic techniques,
early diagnosis and treatment of AML remain challenging. Therefore,
identifying new and highly accurate prognostic indicators for AML
is an urgent and unmet need.
Anoikis, a form of programmed cell death, is crucial
for tissue homeostasis and development by preventing the attachment
or growth of dysplastic cells (29). Its dysregulation has been linked to
cancer progression, promotion of tumor invasion and migration, and
the development of drug resistance (30–32).
However, limited research exists on the impact of ARGs on invasive
mobility and drug resistance in AML, as well as their role in
predicting AML prognosis.
The present study used the TCGA database and
existing literature (14) to
acquire relevant data and identify genes associated with the
anoikis. Through consistent cluster, differential gene expression
and functional enrichment analyses, four biomarker genes were
identified (LGALS1, ITGA4, HGF and RHOC). Subsequently, a risk
model was constructed using single-factor Cox, LASSO and stepwise
multi-factor Cox regression analysis. The risk model was evaluated
using the TCGA training set, stratifying it into high- and low-risk
groups based on the median quantitative risk calculated from the
four biomarkers. Moreover, the present study validated the
effectiveness of the risk model through KM survival curves, ROC
curves, risk curves and PCA. External validation using the GSE71014
dataset further confirmed the efficacy of the risk model.
Prognostic analysis identified risk score, prior treatment,
diagnosis and age as significant independent prognostic factors.
Significant differences were also demonstrated in risk scores among
cytogenetics risk categories and age groups. Finally, by evaluating
TCGA training set with the GDSC database, 50 drugs with significant
differences in efficacy between the high- and low-risk groups were
identified.
The anoikis-related model (ARS model) proposed in
the present study demonstrated a significant association with
survival outcomes in AML cases. The ARS model comprises four ARGs:
LGALS1, ITGA4, HGF and RHOC.
LGALS1, a member of the galectin family, is a
protein with a strong affinity for β galactosides, regulating
several tumor suppressors and promoters (33). LGALS1 is highly expressed in AML
cells and is associated with a poor prognosis in affected patients.
Furthermore, it promotes the survival and proliferation of AML
cells by regulating the expression of apoptosis- and cell
cycle-related proteins (10).
ITGA4 is a protein-coding gene belonging to the
integrin α chain family. ITGA4 serves as a key molecule that allows
AML cells to bind to bone marrow stromal elements and facilitates
cellular migration. Methyltransferase-like 3 has been reported to
increase the stability of ITGA4 mRNA transcripts through
N6-methyladenosine modification, leading to its upregulation on the
cell surface and promoting AML cell homing and engraftment
(34).
The HGF gene, located on the long arm of chromosome
7 (7q2111), encodes a precursor protein consisting of 728 amino
acids. Under normal conditions, the HGF/mesenchymal-epithelial
transition factor (c-MET) signaling pathway serves a critical role
in mediating interactions between epithelial and mesenchymal cells,
which is essential for tissue repair, inflammation control and
immune regulation (35). HGF
upregulation emerged as a prominent compensatory mechanism,
contributing to resistance against MET inhibition in AML (36). A 29-fold higher expression of HGF
was reported in bone marrow samples during refractory disease
compared with remission. Additionally, HGF induces upregulation of
matrix metalloproteinase (MMP)2 and MMP9 expression, facilitates
cell cycle progression, suppresses apoptosis and enhances cell
proliferation through activation of the PI3K/AKT and
mitogen-activated protein kinase (MAPK)/extracellular
signal-regulated kinase (ERK) signaling pathways (37). Collectively, these investigations
highlight the pivotal role of the HGF/c-MET signaling pathway in
AML.
RhoC, a member of the Rho family of small GTPases,
regulates several cellular processes (38). Recent research assessed the
involvement of the Rho subfamily in cellular migration (39). This subfamily includes highly
homologous RhoA, RhoB and RhoC, which regulate actin cytoskeleton
dynamics. Overexpression of miR-372 has been reported to lead to
downregulation of RhoC expression via its 3′ untranslated region
(3′ UTR), thereby suppressing the proliferation, migration and
invasion abilities of endometrial adenocarcinoma cells (40). Additionally, miR-10b has been
reported to inhibit homeobox D10 in colorectal cancer, metastatic
breast cancer and malignant glioma cells, upregulating RhoC
expression (41,42). In ovarian cancer, miR-519d binds
directly to the 3′ UTR of RhoC mRNA, suppressing its expression, as
reported in a nude mouse xenotransplantation model (43). miR-493 directly regulates RhoC,
leading to a marked decrease in its mRNA and protein expression
levels, effectively suppressing the growth, invasion and metastasis
of gastric cancer cells (44).
Moreover, upregulation of RhoC expression has been reported to be
notably associated with an unfavorable prognosis. Therefore, a
strong negative association exists between RhoC expression and
cancer prognosis, and these signature genes are closely associated
with tumors.
Using the Gene set enrichment analysis algorithm,
the present study identified several tumor signaling pathways
activated in the high-risk group compared with that in the low-risk
group. These included the PI3K/AKT and hypoxia-inducible factor 1
signaling pathways, which have been previously associated with AML
growth and development. Furthermore, programmed death-1 ligand 1
was reported to facilitate AML progression through the PI3K/AKT
signaling pathway (32). These
results underscore the importance of exploring the ARS model
importance in AML.
The present study analyzed the TCGA and GSE71014
cohorts, revealing that LGALS1, ITGA4, HGF and RHOC were
significantly associated with AML prognosis. Meanwhile, ITGA4 and
HGF were positively associated with AML, whilst LGALS1 and RHOC
demonstrated a negative association with it, consistent with
previous studies (10,11,34).
Future research should involve larger sample sizes and
cellular-level experiments to clarify the specific roles of each
prognostic gene in AML.
Conventional induction chemotherapy has
traditionally been the frontline therapy for AML. However, therapy
resistance remains a challenge, necessitating the development of
new chemotherapeutic drugs. Dasatinib was reported to induce
c-KIT-positive AML cell death via caspase-dependent apoptosis
(45), BI 2536 was reported to
induce mitotic arrest and apoptosis in AML cells (46), and taxol was reported to suppress
microtubule dynamics, inducing mitotic arrest, triggering caspase-3
cleavage and inducing apoptosis in human AML HL-60 cells (47). Furthermore, lapatinib was reported
to effectively suppress the proliferation of AML cell lines in a
dose- and time-dependent manner, inducing either autophagic or
apoptotic cell death (48).
PD0325901 also effectively blocks MEK/ERK signaling, with strong
inhibitory and apoptotic effects, especially in AML (49), Jw-7-52-1 was effective in treating
AML (50), and erlotinib was
reported to target Fms-related tyrosine kinase 3 and Lyn,
overcoming intratumoral heterogeneity in AML (51). Moreover, the interruption of the
canonical NF-κB pathway may enhance the lethality of belinostat
when combined with bortezomib in AML cells (52). Roscovitine, combined with all-trans
retinoic acid, was reported to induce nuclear enrichment of
proteins promoting differentiation and cell cycle arrest in
t(15;17)-negative HL-60 human myeloblastic leukemia cells (53). The dual insulin-like growth factor 1
receptor/insulin receptor, inhibitor BMS-536924 was also reported
to reduce autophosphorylation of its target receptors through the
PI3K/AKT and MAPK pathways and inhibit proliferation and colony
formation in AML cell lines and clinical AML samples (54).
Although the proposed ARS model in the present study
demonstrates promising predictive power for AML prognosis, the
present study has certain limitations. First, all clinical AML
cohorts analyzed were sourced solely from TCGA website,
necessitating validation of the ARS model using external cohorts.
Second, the expression patterns of the ARS genes need to be
confirmed in clinical specimens using molecular biology techniques,
and further research is required to elucidate the underlying
mechanisms of ARS genes through experimental analyses.
In summary, the present study developed a novel gene
signature related to anoikis in AML. The inclusion of ARS genes
significantly enhances the prediction of AML survival outcomes and
effectively stratifies the risk among patients with AML. The
present study offers a fresh perspective on therapeutic strategies
for individuals with AML.
Supplementary Material
Supporting Data
Supporting Data
Supporting Data
Supporting Data
Supporting Data
Supporting Data
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The TCGA datasets used in the present study can be
accessed at https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga,
whilst the GEO datasets are available at https://www.ncbi.nlm.nih.gov/geo/ as dataset GSE71014.
The data generated in the present study are included in the
supplementary figures and/or tables of this article.
Authors' contributions
YDC, YH and JSW conceived the study, wrote the
manuscript and revised it. YDC, WCL and MYH performed the
experiments and contributed to data analysis. WCL and XYY collected
clinical sample data. JSW also revised the manuscript. All authors
have read and approved the final manuscript, agreed to be
accountable for all aspects of the work and contributed to data
analysis as well as drafting or revising the article. YDC and YH
confirm the authenticity of all the raw data.
Ethics approval and consent to
participate
Prior to performing the present study, ethical
approval was obtained from The Ethics Committee of the Affiliated
Hospital of Guizhou Medical University (Guiyang, China; approval
no. 2023-744). The participants were informed about the purpose of
the research, the assurance of anonymity and the storage procedures
for their collected data. Informed consent was obtained from all
participants before commencing with the study and all participants
consented to the disclosure of their medical data.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Izadirad M, Jafari L, James AR, Unfried
JP, Wu ZX and Chen ZS: Long noncoding RNAs have pivotal roles in
chemoresistance of acute myeloid leukemia. Drug Discov Today.
26:1735–1743. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
SEER. Cancer Stat Facts, . Leukemia-Acute
Myeloid Leukemia (AML). 2023.Available from:. https://seer.cancer.gov/statfacts/html/amyl.html
|
|
3
|
Riva L, Luzi L and Pelicci PG: Genomics of
acute myeloid leukemia: The next generation. Front Oncol. 2:402012.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Han HJ, Sung JY, Kim SH, Yun UJ, Kim H,
Jang EJ, Yoo HE, Hong EK, Goh SH, Moon A, et al: Fibronectin
regulates anoikis resistance via cell aggregate formation. Cancer
Lett. 508:59–72. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Adeshakin FO, Adeshakin AO, Afolabi LO,
Yan D, Zhang G and Wan X: Mechanisms for modulating anoikis
resistance in cancer and the relevance of metabolic reprogramming.
Front Oncol. 11:6265772021. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ye G, Yang Q, Lei X, Zhu X, Li F, He J,
Chen H, Ling R, Zhang H, Lin T, et al: Nuclear MYH9-induced CTNNB1
transcription, targeted by staurosporin, promotes gastric cancer
cell anoikis resistance and metastasis. Theranostics. 10:7545–7560.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Jin L, Chun J, Pan C, Kumar A, Zhang G, Ha
Y, Li D, Alesi GN, Kang Y, Zhou L, et al: The PLAG1-GDH1 axis
promotes anoikis resistance and tumor metastasis through
CamKK2-AMPK signaling in LKB1-Deficient lung cancer. Mol Cell.
69:87–99.e7. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Buchheit CL, Angarola BL, Steiner A,
Weigel KJ and Schafer ZT: Anoikis evasion in inflammatory breast
cancer cells is mediated by Bim-EL sequestration. Cell Death
Differ. 22:1275–1286. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Chen J, Gao F and Liu N: L1CAM promotes
epithelial to mesenchymal transition and formation of cancer
initiating cells in human endometrial cancer. Exp Ther Med.
15:2792–2797. 2018.PubMed/NCBI
|
|
10
|
Ruvolo PP, Ma H, Ruvolo VR, Zhang X, Post
SM and Andreeff M: LGALS1 acts as a pro-survival molecule in AML.
Biochim Biophys Acta Mol Cell Res. 1867:1187852020. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Nie D, Ma P, Chen Y, Zhao H, Liu L, Xin D,
Cao W, Wang F, Meng X, Liu L, et al: MiR-204 suppresses the
progression of acute myeloid leukemia through HGF/c-Met pathway.
Hematology. 26:931–939. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhang J, Qi L, Wang T, An J, Zhou B, Fang
Y, Liu Y, Shan M, Hong D, Wu D, et al: FEV maintains homing and
expansion by activating ITGA4 transcription in primary and relapsed
AML. Front Oncol. 12:8903462022. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lou Y, Jiang Y, Liang Z, Liu B, Li T and
Zhang D: Role of RhoC in cancer cell migration. Cancer Cell Int.
21:5272021. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Chen S, Gu J, Zhang Q, Hu Y and Ge Y:
Development of biomarker signatures associated with anoikis to
predict prognosis in endometrial carcinoma patients. J Oncol.
2021:33752972021. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yu G, Wang LG, Han Y and He QY:
clusterProfiler: An R package for comparing biological themes among
gene clusters. OMICS. 16:284–287. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW,
Shi W and Smyth GK: Limma powers differential expression analyses
for RNA-sequencing and microarray studies. Nucleic Acids Res.
43:e472015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wu X, Sui Z, Zhang H, Wang Y and Yu Z:
Integrated analysis of lncRNA-mediated ceRNA network in lung
adenocarcinoma. Front Oncol. 10:5547592020. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Friedman J, Hastie T and Tibshirani R:
Regularization paths for generalized linear models via coordinate
descent. J Stat Softw. 33:1–22. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Liu TT, Li R, Huo C, Li JP, Yao J, Ji XL
and Qu YQ: Identification of CDK2-related immune forecast model and
ceRNA in lung adenocarcinoma, a Pan-cancer analysis. Front Cell Dev
Biol. 9:6820022021. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Heagerty PJ, Lumley T and Pepe MS:
Time-dependent ROC curves for censored survival data and a
diagnostic marker. Biometrics. 56:337–344. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Xu J, Yang T, Wu F, Chen T, Wang A and Hou
S: A nomogram for predicting prognosis of patients with cervical
cerclage. Heliyon. 9:e211472023. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
De Kouchkovsky I and Abdul-Hay M: ‘Acute
myeloid leukemia: A comprehensive review and 2016 update’. Blood
Cancer J. 6:e4412016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Gu Z, Eils R and Schlesner M: Complex
heatmaps reveal patterns and correlations in multidimensional
genomic data. Bioinformatics. 32:2847–2849. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Maeser D, Gruener RF and Huang RS:
oncoPredict: An R package for predicting in vivo or cancer patient
drug response and biomarkers from cell line screening data. Brief
Bioinform. 22:bbab2602021. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Döhner H, Estey E, Grimwade D, Amadori S,
Appelbaum FR, Büchner T, Dombret H, Ebert BL, Fenaux P, Larson RA,
et al: Diagnosis and management of AML in adults: 2017 ELN
recommendations from an international expert panel. Blood.
129:424–447. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Almond LM, Charalampakis M, Ford SJ,
Gourevitch D and Desai A: Myeloid SArcoma: Presentation, diagnosis,
and treatment. Clin Lymphoma Myeloma Leuk. 17:263–267. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Bakir B, Chiarella AM, Pitarresi JR and
Rustgi AK: EMT, MET, plasticity, and tumor metastasis. Trends Cell
Biol. 30:764–776. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Jin L, Chun J, Pan C, Alesi GN, Li D,
Magliocca KR, Kang Y, Chen ZG, Shin DM, Khuri FR, et al:
Phosphorylation-mediated activation of LDHA promotes cancer cell
invasion and tumour metastasis. Oncogene. 36:3797–3806. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Jiang K, Yao G, Hu L, Yan Y, Liu J, Shi J,
Chang Y, Zhang Y, Liang D, Shen D, et al: MOB2 suppresses GBM cell
migration and invasion via regulation of FAK/Akt and cAMP/PKA
signaling. Cell Death Dis. 11:2302020. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kim H, Choi P, Kim T, Kim Y, Song BG, Park
YT, Choi SJ, Yoon CH, Lim WC, Ko H and Ham J: Ginsenosides Rk1 and
Rg5 inhibit transforming growth factor-β1-induced
epithelial-mesenchymal transition and suppress migration, invasion,
anoikis resistance, and development of stem-like features in lung
cancer. J Ginseng Res. 45:134–148. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wang F, Yang L, Xiao M, Zhang Z, Shen J,
Anuchapreeda S, Tima S, Chiampanichayakul S and Xiao Z: PD-L1
regulates cell proliferation and apoptosis in acute myeloid
leukemia by activating PI3K-AKT signaling pathway. Sci Rep.
12:114442022. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Kamili NA, Arthur CM, Gerner-Smidt C,
Tafesse E, Blenda A, Dias-Baruffi M and Stowell SR: Key regulators
of galectin-glycan interactions. Proteomics. 16:3111–3125. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Li M, Ye J, Xia Y, Li M, Li G, Hu X, Su X,
Wang D, Zhao X, Lu F, et al: METTL3 mediates chemoresistance by
enhancing AML homing and engraftment via ITGA4. Leukemia.
36:2586–2595. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wang SH, Wu XC, Zhang MD, Weng MZ, Zhou D
and Quan ZW: Upregulation of H19 indicates a poor prognosis in
gallbladder carcinoma and promotes epithelial-mesenchymal
transition. Am J Cancer Res. 6:15–26. 2016.PubMed/NCBI
|
|
36
|
Chen EC, Gandler H, Tošić I, Fell GG,
Fiore A, Pozdnyakova O, DeAngelo DJ, Galinsky I, Luskin MR,
Wadleigh M, et al: Targeting MET and FGFR in relapsed or refractory
acute myeloid leukemia: Preclinical and clinical findings, and
signal transduction correlates. Clin Cancer Res. 29:878–887. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Guo JR, Li W, Wu Y, Wu LQ, Li X, Guo YF,
Zheng XH, Lian XL, Huang HF and Chen YZ: Hepatocyte growth factor
promotes proliferation, invasion, and metastasis of myeloid
leukemia cells through PI3K-AKT and MAPK/ERK signaling pathway. Am
J Transl Res. 8:3630–3644. 2016.PubMed/NCBI
|
|
38
|
Lawson CD and Ridley AJ: Rho GTPase
signaling complexes in cell migration and invasion. J Cell Biol.
217:447–457. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Stanley A, Thompson K, Hynes A, Brakebusch
C and Quondamatteo F: NADPH oxidase complex-derived reactive oxygen
species, the actin cytoskeleton, and Rho GTPases in cell migration.
Antioxid Redox Signal. 20:2026–2042. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Liu BL, Sun KX, Zong ZH, Chen S and Zhao
Y: MicroRNA-372 inhibits endometrial carcinoma development by
targeting the expression of the Ras homolog gene family member C
(RhoC). Oncotarget. 7:6649–6664. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Knirsh R, Ben-Dror I, Modai S, Shomron N
and Vardimon L: MicroRNA 10b promotes abnormal expression of the
proto-oncogene c-Jun in metastatic breast cancer cells. Oncotarget.
7:59932–59944. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Wang YF, Li Z, Zhao XH, Zuo XM, Zhang Y,
Xiao YH, Li J and Peng ZH: MicroRNA-10b is upregulated and has an
invasive role in colorectal cancer through enhanced Rhoc
expression. Oncol Rep. 33:1275–1283. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Sang XB, Zong ZH, Wang LL, Wu DD, Chen S,
Liu BL and Zhao Y: E2F-1 targets miR-519d to regulate the
expression of the ras homolog gene family member C. Oncotarget.
8:14777–14793. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Zhou W, Zhang C, Jiang H, Zhang Z, Xie L
and He X: MiR-493 suppresses the proliferation and invasion of
gastric cancer cells by targeting RhoC. Iran J Basic Med Sci.
18:1027–1033. 2015.PubMed/NCBI
|
|
45
|
Heo SK, Noh EK, Kim JY, Jeong YK, Jo JC,
Choi Y, Koh S, Baek JH, Min YJ and Kim H: Targeting c-KIT (CD117)
by dasatinib and radotinib promotes acute myeloid leukemia cell
death. Sci Rep. 7:152782017. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Müller-Tidow C, Bug G, Lübbert M, Krämer
A, Krauter J, Valent P, Nachbaur D, Berdel WE, Ottmann OG, Fritsch
H, et al: A randomized, open-label, phase I/II trial to investigate
the maximum tolerated dose of the Polo-like kinase inhibitor BI
2536 in elderly patients with refractory/relapsed acute myeloid
leukaemia. Br J Haematol. 163:214–222. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Ibrado AM, Kim CN and Bhalla K: Temporal
relationship of CDK1 activation and mitotic arrest to cytosolic
accumulation of cytochrome C and caspase-3 activity during
Taxol-induced apoptosis of human AML HL-60 cells. Leukemia.
12:1930–1936. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Chen YJ, Fang LW, Su WC, Hsu WY, Yang KC
and Huang HL: Lapatinib induces autophagic cell death and
differentiation in acute myeloblastic leukemia. Onco Targets Ther.
9:4453–4464. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Ricciardi MR, Scerpa MC, Bergamo P,
Ciuffreda L, Petrucci MT, Chiaretti S, Tavolaro S, Mascolo MG,
Abrams SL, Steelman LS, et al: Therapeutic potential of MEK
inhibition in acute myelogenous leukemia: Rationale for ‘vertical’
and ‘lateral’ combination strategies. J Mol Med (Berl).
90:1133–1144. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Wang Z, Liu Y, Mo Y, Zhang H, Dai Z, Zhang
X, Ye W, Cao H, Liu Z and Cheng Q: The CXCL family contributes to
immunosuppressive microenvironment in gliomas and assists in
gliomas chemotherapy. Front Immunol. 12:7317512021. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Cao ZX, Guo CJ, Song X, He JL, Tan L, Yu
S, Zhang RQ, Peng F, Peng C and Li YZ: Erlotinib is effective
against FLT3-ITD mutant AML and helps to overcome intratumoral
heterogeneity via targeting FLT3 and Lyn. FASEB J. 34:10182–10190.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Dai Y, Chen S, Wang L, Pei XY, Kramer LB,
Dent P and Grant S: Bortezomib interacts synergistically with
belinostat in human acute myeloid leukaemia and acute lymphoblastic
leukaemia cells in association with perturbations in NF-κB and Bim.
Br J Haematol. 153:222–235. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Rashid A, Duan X, Gao F, Yang M and Yen A:
Roscovitine enhances all-trans retinoic acid (ATRA)-induced nuclear
enrichment of an ensemble of activated signaling molecules and
augments ATRA-induced myeloid cell differentiation. Oncotarget.
11:1017–1036. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Wahner Hendrickson AE, Haluska P,
Schneider PA, Loegering DA, Peterson KL, Attar R, Smith BD,
Erlichman C, Gottardis M, Karp JE, et al: Expression of insulin
receptor isoform A and insulin-like growth factor-1 receptor in
human acute myelogenous leukemia: Effect of the dual-receptor
inhibitor BMS-536924 in vitro. Cancer Res. 69:7635–7643. 2009.
View Article : Google Scholar : PubMed/NCBI
|