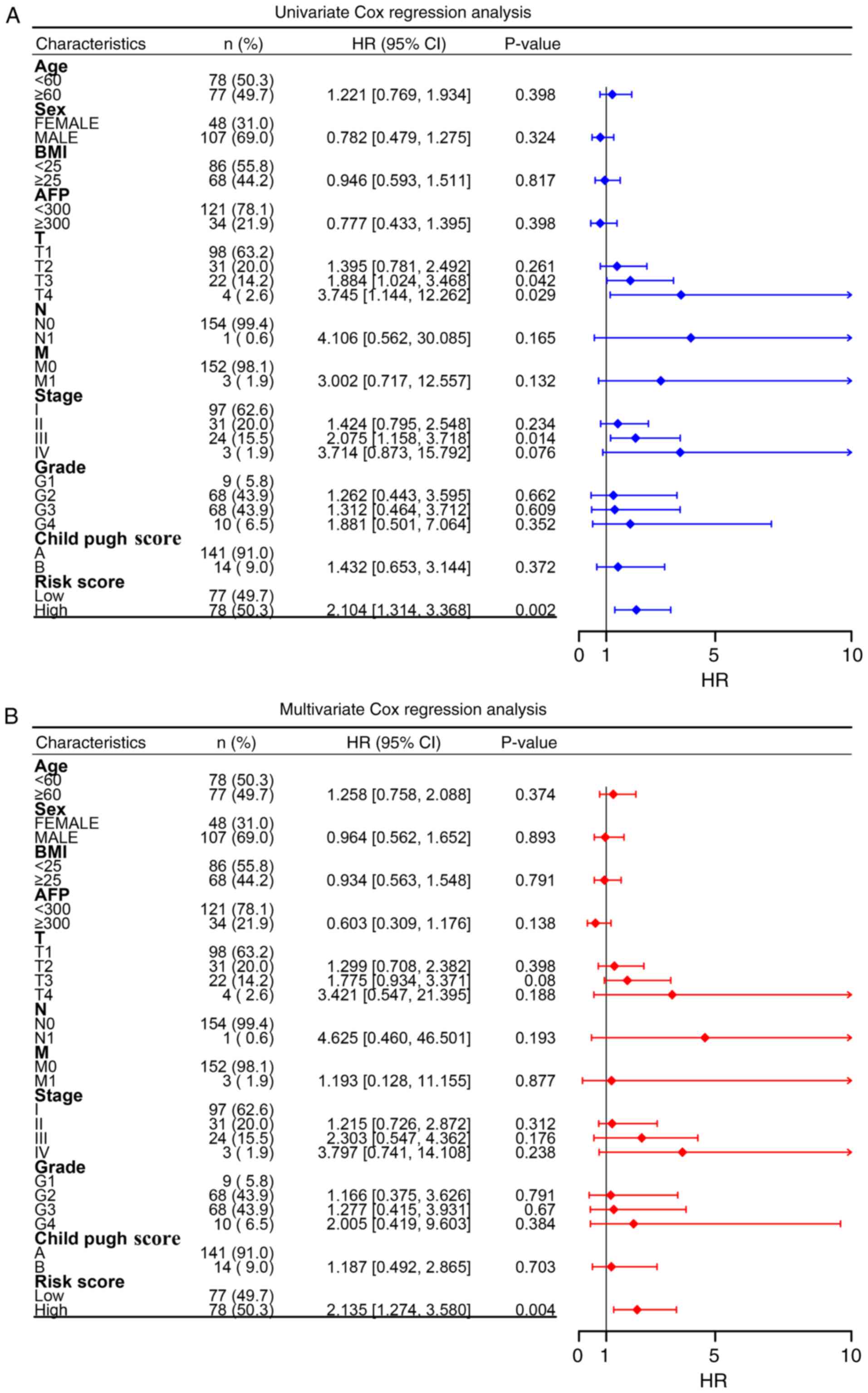
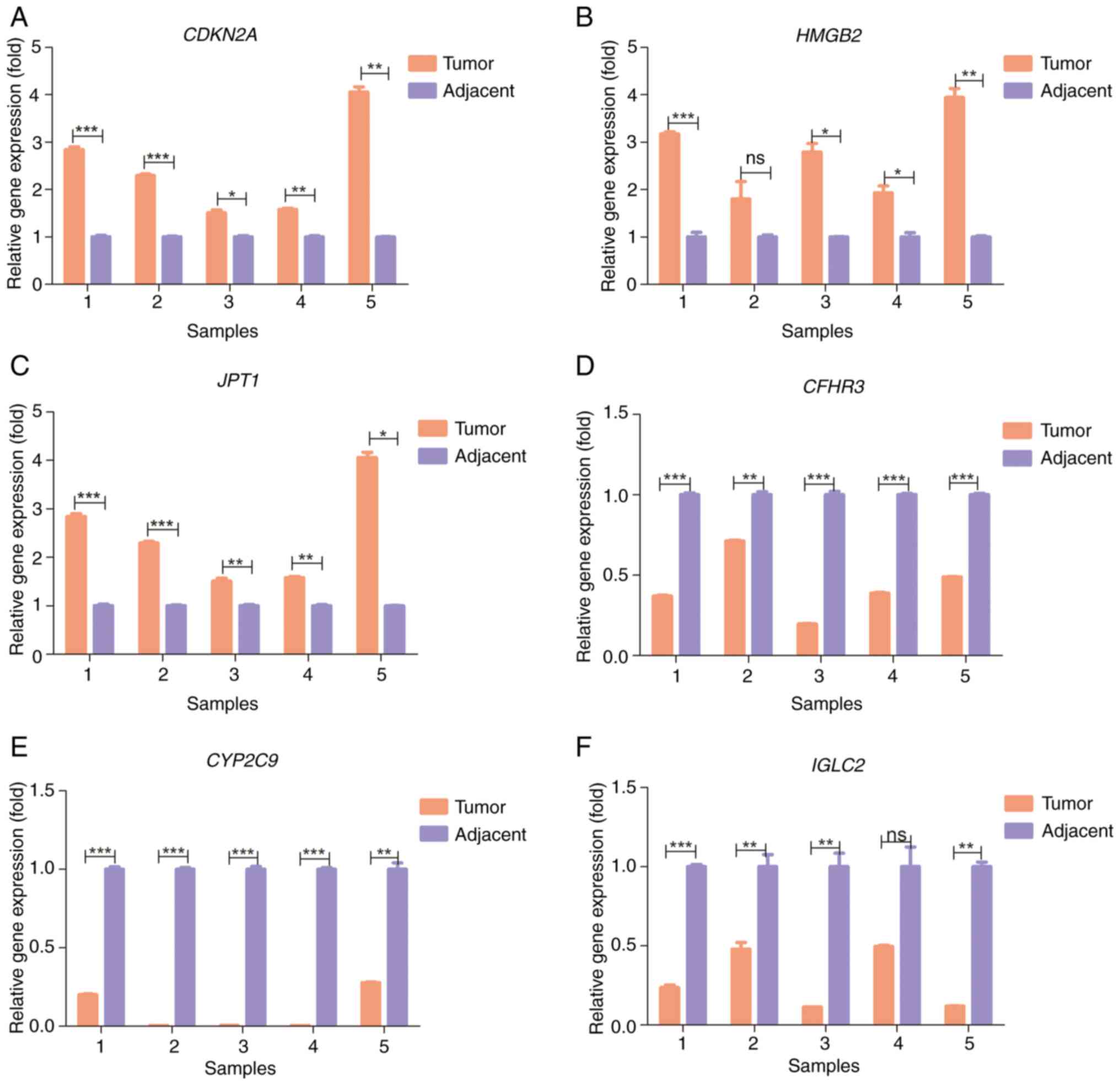
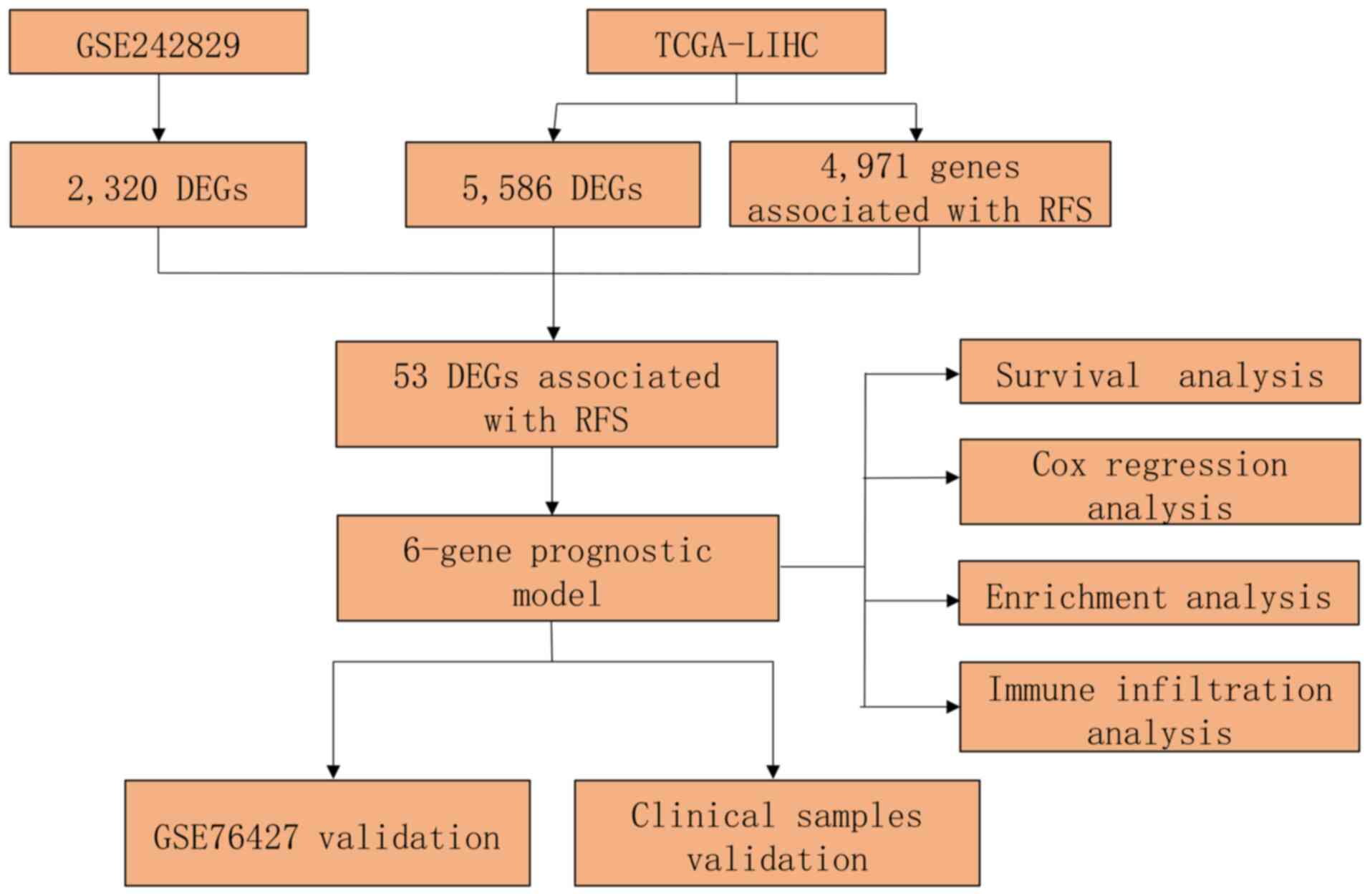
|
1
|
Bray F, Laversanne M, Sung H, Ferlay J,
Siegel RL, Soerjomataram I and Jemal A: Global cancer statistics
2022: GLOBOCAN estimates of incidence and mortality worldwide for
36 cancers in 185 countries. CA Cancer J Clin. 74:229–263.
2024.PubMed/NCBI
|
|
2
|
Arnold M, Abnet CC, Neale RE, Vignat J,
Giovannucci EL, McGlynn KA and Bray F: Global burden of 5 major
types of gastrointestinal cancer. Gastroenterology.
159:335–349.e15. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Daher S, Massarwa M, Benson AA and Khoury
T: Current and future treatment of hepatocellular carcinoma: An
updated comprehensive review. J Clin Transl Hepatol. 6:69–78. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Villanueva A: Hepatocellular carcinoma. N
Engl J Med. 380:1450–1462. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Martínez-Chantar ML, Avila MA and Lu SC:
Hepatocellular carcinoma: Updates in pathogenesis, detection and
treatment. Cancers (Basel). 12:27292020. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Singal AG, Kudo M and Bruix J:
Breakthroughs in hepatocellular carcinoma therapies. Clin
Gastroenterol Hepatol. 21:2135–2149. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Gu JX, Zhang X, Miao RC, Xiang XH, Fu YN,
Zhang JY, Liu C and Qu K: Six-long non-coding RNA signature
predicts recurrence-free survival in hepatocellular carcinoma.
World J Gastroenterol. 25:220–232. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wang W, Wang L, Xie X, Yan Y, Li Y and Lu
Q: A gene-based risk score model for predicting recurrence-free
survival in patients with hepatocellular carcinoma. BMC Cancer.
21:62021. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Guo DZ, Zhang X, Zhang SQ, Zhang SY, Zhang
XY, Yan JY, Dong SY, Zhu K, Yang XR, Fan J, et al: Single-cell
tumor heterogeneity landscape of hepatocellular carcinoma:
Unraveling the pro-metastatic subtype and its interaction loop with
fibroblasts. Mol Cancer. 23:1572024. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Li X, Li R, Miao X, Zhou X, Wu B, Cao J,
Wang C, Li S and Cai J: Integrated single cell analysis reveals an
atlas of tumor associated macrophages in hepatocellular carcinoma.
Inflammation. 47:2077–2093. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sun Y, Wu L, Zhong Y, Zhou K, Hou Y, Wang
Z, Zhang Z, Xie J, Wang C, Chen D, et al: Single-cell landscape of
the ecosystem in early-relapse hepatocellular carcinoma. Cell.
184:404–421.e16. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhou Q, Tao C, Ge Y, Yuan J, Pan F, Lin X
and Wang R: A novel single-cell model reveals
ferroptosis-associated biomarkers for individualized therapy and
prognostic prediction in hepatocellular carcinoma. BMC Biol.
22:1332024. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yu L, Shen N, Shi Y, Shi X, Fu X, Li S,
Zhu B, Yu W and Zhang Y: Characterization of cancer-related
fibroblasts (CAF) in hepatocellular carcinoma and construction of
CAF-based risk signature based on single-cell RNA-seq and bulk
RNA-seq data. Front Immunol. 13:10097892022. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wang T, Dang N, Tang G, Li Z, Li X, Shi B,
Xu Z, Li L, Yang X, Xu C and Ye K: Integrating bulk and single-cell
RNA sequencing reveals cellular heterogeneity and immune
infiltration in hepatocellular carcinoma. Mol Oncol. 16:2195–2213.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Li K, Zhang R, Wen F, Zhao Y, Meng F, Li
Q, Hao A, Yang B, Lu Z, Cui Y and Zhou M: Single-cell dissection of
the multicellular ecosystem and molecular features underlying
microvascular invasion in HCC. Hepatology. 79:1293–1309. 2024.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Grinchuk OV, Yenamandra SP, Iyer R, Singh
M, Lee HK, Lim KH, Chow PK and Kuznetsov VA: Tumor-adjacent tissue
co-expression profile analysis reveals pro-oncogenic ribosomal gene
signature for prognosis of resectable hepatocellular carcinoma. Mol
Oncol. 12:89–113. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Korsunsky I, Millard N, Fan J, Slowikowski
K, Zhang F, Wei K, Baglaenko Y, Brenner M, Loh PR and Raychaudhuri
S: Fast, sensitive and accurate integration of single-cell data
with Harmony. Nat Methods. 16:1289–1296. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lai W, Xie R, Chen C, Lou W, Yang H, Deng
L, Lu Q and Tang X: Integrated analysis of scRNA-seq and bulk
RNA-seq identifies FBXO2 as a candidate biomarker associated with
chemoresistance in HGSOC. Heliyon. 10:e284902024. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Butler A, Hoffman P, Smibert P, Papalexi E
and Satija R: Integrating single-cell transcriptomic data across
different conditions, technologies, and species. Nat Biotechnol.
36:411–420. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW,
Shi W and Smyth GK: limma powers differential expression analyses
for RNA-sequencing and microarray studies. Nucleic Acids Res.
43:e472015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Friedman J, Hastie T and Tibshirani R:
Regularization paths for generalized linear models via coordinate
descent. J Stat Softw. 33:1–22. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yu G, Wang LG, Han Y and He QY:
clusterProfiler: An R package for comparing biological themes among
gene clusters. OMICS. 16:284–287. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Newman AM, Liu CL, Green MR, Gentles AJ,
Feng W, Xu Y, Hoang CD, Diehn M and Alizadeh AA: Robust enumeration
of cell subsets from tissue expression profiles. Nat Methods.
12:453–457. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yoshihara K, Shahmoradgoli M, Martínez E,
Vegesna R, Kim H, Torres-Garcia W, Treviño V, Shen H, Laird PW,
Levine DA, et al: Inferring tumour purity and stromal and immune
cell admixture from expression data. Nat Commun. 4:26122013.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Amin MB, Greene FL, Edge SB, Compton CC,
Gershenwald JE, Brookland RK, Meyer L, Gress DM, Byrd DR and
Winchester DP: The eighth edition AJCC cancer staging manual:
Continuing to build a bridge from a population-based to a more
‘personalized’ approach to cancer staging. CA Cancer J Clin.
67:93–99. 2017.PubMed/NCBI
|
|
26
|
Daniele B, Annunziata M, Barletta E,
Tinessa V and Di Maio M: Cancer of the liver italian program (CLIP)
score for staging hepatocellular carcinoma. Hepatol Res. 37 (Suppl
2):S206–S209. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Moris D, Martinino A, Schiltz S, Allen PJ,
Barbas A, Sudan D, King L, Berg C, Kim C, Bashir M, et al: Advances
in the treatment of hepatocellular carcinoma: An overview of the
current and evolving therapeutic landscape for clinicians. CA
Cancer J Clin. May 20–2025.(Epub ahead of print). PubMed/NCBI
|
|
28
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kong J, Wang T, Shen S, Zhang Z, Yang X
and Wang W: A genomic-clinical nomogram predicting recurrence-free
survival for patients diagnosed with hepatocellular carcinoma.
PeerJ. 7:e79422019. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Luo JP, Wang J and Huang JH: CDKN2A is a
prognostic biomarker and correlated with immune infiltrates in
hepatocellular carcinoma. Biosci Rep. 41:BSR202111032021.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Skerka C, Chen Q, Fremeaux-Bacchi V and
Roumenina LT: Complement factor H related proteins (CFHRs). Mol
Immunol. 56:170–180. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wan Z, Li X, Luo X, Wang B, Zhou X and
Chen A: The miR-590-3p/CFHR3/STAT3 signaling pathway promotes cell
proliferation and metastasis in hepatocellular carcinoma. Aging
(Albany NY). 14:5783–5799. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yu D, Green B, Marrone A, Guo Y, Kadlubar
S, Lin D, Fuscoe J, Pogribny I and Ning B: Suppression of CYP2C9 by
microRNA hsa-miR-128-3p in human liver cells and association with
hepatocellular carcinoma. Sci Rep. 5:85342015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Starkova T, Polyanichko A, Tomilin AN and
Chikhirzhina E: Structure and functions of HMGB2 protein. Int J Mol
Sci. 24:83342023. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Lu K, Zhao T, Yang L, Liu Y, Ruan X, Cui L
and Zhang Y: HMGB2 upregulation promotes the progression of
hepatocellular carcinoma cells through the activation of
ZEB1/vimentin axis. J Gastrointest Oncol. 14:2178–2191. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Li M, Pang X, Xu H and Xiao L: CircSCMH1
accelerates sorafenib resistance in hepatocellular carcinoma by
regulating HN1 expression via miR-485-5p. Mol Biotechnol.
67:304–316. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wang R, Fu Y, Yao M, Cui X, Zhao Y, Lu X,
Li Y, Lin Y and He S: The HN1/HMGB1 axis promotes the proliferation
and metastasis of hepatocellular carcinoma and attenuates the
chemosensitivity to oxaliplatin. FEBS J. 289:6400–6419. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Qu M, Zhang G, Qu H, Vu A, Wu R, Tsukamoto
H, Jia Z, Huang W, Lenz HJ, Rich JN and Kay SA: Circadian regulator
BMAL1::CLOCK promotes cell proliferation in hepatocellular
carcinoma by controlling apoptosis and cell cycle. Proc Natl Acad
Sci USA. 120:e22148291202023. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Wu D, Zhang C, Liao G, Leng K, Dong B, Yu
Y, Tai H, Huang L, Luo F, Zhang B, et al: Targeting
uridine-cytidine kinase 2 induced cell cycle arrest through dual
mechanism and could improve the immune response of hepatocellular
carcinoma. Cell Mol Biol Lett. 27:1052022. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Wang S, Meng L, Xu N, Chen H, Xiao Z, Lu
D, Fan X, Xia L, Chen J, Zheng S, et al: Hepatocellular
carcinoma-specific epigenetic checkpoints bidirectionally regulate
the antitumor immunity of CD4 + T cells. Cell Mol Immunol.
21:1296–1308. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Zhang Z, Ma L, Goswami S, Ma J, Zheng B,
Duan M, Liu L, Zhang L, Shi J, Dong L, et al: Landscape of
infiltrating B cells and their clinical significance in human
hepatocellular carcinoma. Oncoimmunology. 8:e15713882019.
View Article : Google Scholar : PubMed/NCBI
|