Introduction
Globally, pancreatic ductal adenocarcinoma (PAAD)
accounted for 511,000 new cases and 467,000 deaths in 2022
(according to GLOBOCAN data), ranking as the sixth leading cause of
cancer-related mortality worldwide at present and contributing to
5% of all cancer-associated deaths. This mortality rate places PAAD
among the malignancies with the poorest prognosis (1). PAAD, while exhibiting a relatively low
incidence rate, carries a disproportionately high mortality rate.
Global epidemiological data indicate a rising incidence of PAAD
annually (2). Although less
prevalent than a number of other malignancies, PAAD imparts a worse
prognosis due to challenges in early detection and effective
treatment (3). Established risk
factors for PAAD development include smoking, obesity, high-fat
diets, chronic pancreatitis and genetic predisposition. Smoking
constitutes a primary modifiable risk factor, while genetic and
environmental influences also contribute markedly to disease
susceptibility (4). Current
therapeutic modalities for PAAD are primarily limited to surgical
resection, supplemented by radiotherapy, chemotherapy or targeted
agents such as Erlotinib (5).
However, clinical outcomes often remain suboptimal. Immunotherapy
has emerged as a promising therapeutic paradigm, aiming to harness
the host immune system to eradicate cancer cells (6). Consequently, immune checkpoint
inhibitors and related immunotherapeutics are being evaluated in
subsets of patients with PAAD (7).
However, the inherent immunological ‘cold’ nature of PAAD presents
a major barrier to immunotherapy efficacy. Converting these
immunologically quiescent tumors into ‘hot’, immune-responsive
lesions represents an important requirement for immunotherapy
efficacy (8).
The stimulator of interferon genes (STING) pathway
represents a pivotal innate immune modulator with the potential to
convert immunologically ‘cold’ tumors into ‘hot’ microenvironments
(9). STING activation triggers the
production of type I interferons (IFNs) and other cytokines,
thereby reshaping the tumor microenvironment (TME) and modulating
immune cell activity (10). While
STING pathway activation has been linked to tumor progression and
metastasis in certain contexts, often regulated by epigenetic
mechanisms such as DNA methylation (11), its therapeutic manipulation holds
promise. At present, STING agonists are undergoing clinical trials
to evaluate their efficacy in patients with advanced cancer.
Furthermore, the potential application of STING agonists in
early-stage disease (12),
particularly before the establishment of robust immune evasion
mechanisms within the TME, is an active area of investigation
(13). Within the specific context
of PAAD, preclinical studies suggest that STING activation can
stimulate type I IFN production and promote dendritic cell (DC)
maturation (14). Despite this
promise, successful clinical translation faces notable hurdles,
including: i) An incomplete understanding of PAAD-specific STING
interactome dynamics (15); ii) the
induction of compensatory immunosuppressive mechanisms (e.g.,
myeloid-derived suppressor cell expansion) that counteract agonist
efficacy (16); and notably, iii)
the absence of validated predictive biomarkers to stratify patients
likely to respond to STING activation treatment (17). Moreover, the efficacy of STING
agonists in PAAD may be contingent upon the specific TME
composition, patient genetics and the specific combination partners
used (e.g., chemotherapy or immune checkpoint inhibitors), as
synergistic interactions can significantly enhance their
therapeutic effect (18,19). In conclusion, elucidating the
mechanistic role and therapeutic potential of the STING pathway in
cancer immunotherapy has become a major research focus. Such
investigations are important not only for deepening the present
understanding of PAAD pathogenesis but also for developing novel
and effective therapeutic strategies against recalcitrant PAAD
malignancy.
The present study aimed to systematically map the
core regulatory components within the STING signaling-protein
interactome to construct a prognostic signature for PAAD.
Interactome mapping was pursued through comprehensive
protein-protein interaction network-centrality analysis.
Collectively, by decoding the STING-centered protein network, the
present study seeks to identify novel tractable targets for
potentiating STING agonism in PAAD treatment.
Material and methods
Bioinformatics
Patient data
Samples from a total of 353 patients with pancreatic
cancer were integrated from The Cancer Genome Atlas
(TCGA)-pancreatic adenocarcinoma (n=178) and GSE224564 (n=175)
datasets, and cases with incomplete clinical data were excluded.
The RNA sequencing and clinical data of 178 patients with PAAD were
obtained from TCGA (https://www.cancer.gov/). The PAAD dataset GSE224564
(20) was downloaded from GEO
(www.ncbi.nlm.nih.gov/geo/query/acc.cgi) and included
the RNA sequencing and clinical data of 175 patients with PAAD.
Bioinformatic analysis
Differential gene expression between STING-based
subtypes (C1 vs. C2) was analyzed using DESeq2 (v1.38.3).
Functional enrichment was performed via clusterProfiler (v4.0)
using annotations from the Gene Ontology Consortium (http://geneontology.org) and KEGG Pathway Database
(https://www.genome.jp/kegg/), with
significance defined as a Benjamini-Hochberg adjusted P-value
<0.05. TME scores (immune/stromal) and tumor purity were
calculated using the ESTIMATE algorithm (v1.0.13; http://bioinformatics.mdanderson.org/estimate/).
Immune cell infiltration was deconvoluted with CIBERSORTx
(https://cibersortx.stanford.edu) using
the LM22 signature matrix and 1,000 permutations. Mutation profiles
were processed via Maftools (v2.16.0) with TCGA somatic mutation
data. All statistical thresholds are explicitly stated in figure
legends. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and
Genomes (KEGG) pathway enrichment analyses were performed to
interpret the functional significance of the identified genes. GO
analysis (http://geneontology.org) was utilized
to categorize genes into biological processes, molecular functions
and cellular components. KEGG pathway analysis (https://www.genome.jp/kegg/) was used to identify
significantly enriched signaling and metabolic pathways. The
analyses were performed using the clusterProfiler R package (v4.0),
with a significance threshold of an adjusted P-value <0.05.
Construction of STING pathway-related
gene prognostic signature in PAAD
Curated Cancer Cell Atlas (www.weizmann.ac.il/sites/3CA/) was used to identify
the expression of STING in the TME. STRING (cn.string-db.org) and
Genemiania (genemania.org) were used to identify 26 STING
pathway-related genes (Table SI).
The association between STING pathway-related genes and the
prognosis of patients with PAAD was evaluated via univariate cox
regression. A least absolute shrinkage and selection operator
(LASSO) Cox regression analysis was used for constructing the
signature for STING pathway-related genes. Bioinformatics analysis
of PAAD transcriptomic data from public cohorts (TCGA and GEO)
revealed that the majority of STING-related genes were highly
expressed in PAAD (significance defined as |log2(fold change)|>1
and adjusted P-value <0.05). Based on the expression levels of
these genes, PAAD patients from these cohorts were stratified into
high-risk and low-risk groups. These STING-related genes exerted
differential impacts on tumor mutational burden, immune
infiltration and response to immunotherapy. Prognostic STING
pathway-related genes were then selected via stepwise multivariate
Cox regression using the Akaike information criterion for model
optimization. Utilizing these genes, a prognostic model was
constructed (hazard ratio, 2.639; P<0.001) and externally
validated. Risk Score= ∑k=0ncoep (k)*x(k), where coef(k) and x(k)
represented the regression coefficients. The groups were divided
into high- and low-risk groups using the median risk score (1.476)
as the cut-off. Univariate and multivariate Cox regression were
used to evaluate independent factors, including clinical features
and the risk score in PAAD. The Tumor Immune Dysfunction and
Exclusion (TIDE) computational platform (https://tide.dfci.harvard.edu) was used to predict
response to immunotherapy in high- and low-risk PAAD patient groups
stratified by the STING-signature risk score.
Online database
The GEPIA (http://gepia2.cancer-pku.cn/), Cbioportal (www.cbioportal.org), HPA (www.proteinatlas.org) and TISIDB
(cis.hku.hk/TISIDB/index.php) databases were used to evaluate gene
expression, survival and clinical stage and grade (21) in patients with PAAD.
In vitro assays
Cell culture
293T cells (ATCC® CRL-3216™) and murine
pancreatic ductal adenocarcinoma Panc02 cells (cat. no. CRL-2553)
were cultured in high-glucose DMEM medium (Corning, Inc.)
supplemented with 10% fetal bovine serum (Gibco; Thermo Fisher
Scientific, Inc.) and 1% penicillin/streptomycin (HyClone™;
Cytiva). Cells were maintained at 37°C in a humidified 5%
CO2 incubator (Thermo Fisher Scientific, Inc.) and
routinely subcultured every 2–3 days using 0.25% trypsin-EDTA
(Gibco; Thermo Fisher Scientific, Inc.). Cell density was
maintained at 60–80% confluency.
Lentiviral transduction
Lentiviral vectors expressing short hairpin RNAs
(shRNAs) targeting interferon-induced protein with
tetratricopeptide repeats (IFIT2), or corresponding scrambled
control shRNAs (sh-NC), were constructed using the pLKO.1-puro
backbone (cat. no. 8453; Addgene, Inc.). The shRNA sequences
(Table SII) were designed using
the Broad Institute TRC portal (v2.0) and validated by BLAST
(https://blast.ncbi.nlm.nih.gov/Blast.cgi) to ensure
target specificity. For overexpression, full-length zinc finger
DHHC-type containing 1 (ZDHHC1) complementary DNA (OriGene
Technologies, Inc.) was cloned into the pCDH-CMV-MCS-EF1-Puro
vector (cat. no. CD510B-1; System Biosciences, LLC). In the ZDHHC1
overexpression experiment, cells transfected with the empty vector
(pCDNA3.1) served as the negative control to confirm that any
observed phenotypic changes were attributable to the expression of
the target gene rather than the vector itself. All constructs were
verified by Sanger sequencing. Lentiviral particles were produced
using a second-generation system by co-transfecting 293T cells
[American Type Culture Collection (ATCC)] cultured at 37°C with the
transfer vector (shRNA or overexpression plasmid), packaging
plasmid psPAX2 (cat. no. 12260; Addgene, Inc.) and envelope plasmid
pMD2.G (cat. no. 12259; Addgene, Inc.) at a 4:3:1 ratio (6:4.5:1.5
µg) using Lipofectamine® 3000 (cat. no. L3000015; Thermo
Fisher Scientific, Inc.). Following a 48- to 72-h transfection
period, viral supernatants were harvested, filtered through 0.45-µm
PVDF membranes, and concentrated by ultracentrifugation (25,000 ×
g, at 4°C for 2 h). Panc02 cells were seeded in 6-well plates
(5×104 cells/well) and incubated until 40% confluency.
Cells were transduced with a multiplicity of infection of 15 used
to infect cells with lentivirus in the presence of 8 µg/ml
polybrene (cat. no. H9268; MilliporeSigma). After 24 h, the medium
was replaced with fresh complete DMEM. Transduced cells were
selected using 5 µg/ml puromycin (cat. no. ant-pr-1; InvivoGen) for
72 h. Positively selected cells were then maintained in 2 µg/ml
puromycin-containing medium for all subsequent experiments to
ensure stable expression. Knockdown or overexpression efficacy was
validated 7 days post-selection.
Western blotting
Panc02 cells (ATCC CRL-2553™) were lysed in RIPA
buffer (25 mM Tris-HCl pH 7.6, 150 mM NaCl, 1% NP-40, 1% sodium
deoxycholate and 0.1% SDS) supplemented with protease inhibitor
cocktail (cat. no. 11836170001; Roche Diagnostics, GmbH). Protein
concentration was determined by BCA assay. Samples (40 µg/lane)
were denatured in Laemmli buffer at 100°C for 10 min, separated on
10% gels using SDS-PAGE (GenScript), and transferred to PVDF
membranes (cat. no. IPVH00010; MilliporeSigma). Membranes were
blocked with 5% non-fat skimmed milk (cat. no. 1706404; Bio-Rad
Laboratories, Inc.) in 0.1% TBST for 1 h at 25°C, followed by
overnight incubation at 4°C with the following primary antibodies:
Anti-IFIT2 (1:1,000; cat. no. ab305231; Abcam), anti-ZDHHC1
(1:1,000; cat. no. ab223042; Abcam) and anti-GAPDH (1:1,000; cat.
no. CST 2118; Cell Signaling Technology, Inc.). After TBST washes,
membranes were incubated with HRP-conjugated goat anti-rabbit IgG
(1:100,000; cat. no. 31460; Invitrogen; Thermo Fisher Scientific,
Inc.) for 1 h at 25°C. Signals were developed using SuperSignal™
West Pico PLUS substrate (cat.no. 34577; Thermo Fisher Scientific,
Inc.) and imaged with a ChemiDoc™ MP System (Bio-Rad Laboratories,
Inc.) controlled by Image Lab™ Software v6.1. Band intensity was
quantified by built-in densitometry tools.
Cell Counting Kit-8 (CCK-8 assay)
In a standard 96-well plate, cells were seeded at
5,000 Panc02 cells per well (in 100 µl DMEM). The cells were then
incubated in a 37°C, 5% CO2 cell culture incubator for
24 h. Having pre-calculated the corresponding seeding volume per
well, CCK-8 solution (cat. no. CK04-13; Dojindo Laboratories, Inc.)
was added to each well at 10% of the total volume of the medium,
and the reaction proceeded in the dark for 1 h. The difference
between the OD values of the treatment group and sh-NC or vehicle
was calculated, then divided by the difference between the OD
values of the control cells. The resulting value was multiplied by
100% to obtain the cell survival rate.
TUNEL assay
Cells were seeded onto sterile glass coverslips
placed in 24-well culture plates at a density of 5×104
cells per well and allowed to adhere for 24 h. After adherence,
cells were washed once with phosphate-buffered saline (PBS) and
fixed with 4% paraformaldehyde in PBS at room temperature for 30
min. Fixed cells were washed three times with PBS (5 min per wash).
Cell membranes were permeabilized by incubation with pre-chilled
PBS containing 0.1% Triton X-100 on ice for 2 min, followed by two
washes with PBS. The TUNEL assay was performed using the In Situ
Cell Death Detection Kit, Fluorescein (cat. no. 11684795910; Roche
Diagnostics, GmbH). The TUNEL reaction mixture was prepared
according to the manufacturer's protocol by combining 50 µl Enzyme
Solution (terminal deoxynucleotidyl transferase) with 450 µl Label
Solution (fluorescein-dUTP). This mixture was applied to cover the
samples, which were then incubated at 37°C in a humidified dark
chamber for 60 min. Unbound reagents were removed by washing three
times with PBS (5 min per wash). Nuclei were counterstained with
4′,6-diamidino-2-phenylindole (DAPI) at 1 µg/ml in PBS for 10 min
at room temperature in the dark, followed by two final washes with
PBS. Samples were mounted using ProLong Gold Antifade Mountant
(cat. no. P36930; Thermo Fisher Scientific, Inc.). Apoptotic cells
(green fluorescence) and total nuclei (blue fluorescence) were
visualized using an Olympus BX53 fluorescence microscope (Olympus
Corporation). Quantification was performed by counting
TUNEL-positive cells and total DAPI-stained nuclei in ≥10 randomly
selected fields of view per sample at ×200 magnification.
Statistical analysis
For the statistical analysis component of this
study, R software (version 4.3.2; The R Foundation for Statistical
Computing) was used. This open-source software was downloaded from
the Comprehensive R Archive Network (https://cran.r-project.org/). Continuous variables
(expressed as mean ± standard deviation upon confirmation of
normality by Shapiro-Wilk test) and categorical variables (reported
as frequencies with proportions) were compared through
multivariable regression models adjusted for demographic/clinical
confounders; statistical significance was defined as bidirectional
P<0.05 with Benjamini-Hochberg correction for multiple
comparisons. Survival endpoints were evaluated via stratified
log-rank tests and hazard ratios (95% confidence intervals). Data
from CCK-8, TUNEL and western blots were treated as continuous
variables. Differences between two experimental groups were
assessed exclusively by unpaired two-tailed Student's t-tests.
Differences between multiple experimental groups and a single
control group were assessed using one-way ANOVA followed by
Dunnett's multiple comparisons test. To evaluate the monotonic
relationship between risk score and dendritic cells activated
score, Spearman's rank-order correlation analysis was employed.
This non-parametric method was selected since the data were either
ordinal or continuous but failed to meet the assumption of
normality, as assessed by the Shapiro-Wilk test. P<0.05 was
considered to indicate a statistically significant difference.
Results
STING as an adverse prognostic factor
in PAAD
To elucidate the clinical significance of the STING
pathway in PAAD, its expression profile and prognostic relevance
were systematically analyzed. Analysis of the Curated Cancer Cell
Atlas revealed predominant STING expression within tumor cells,
macrophages, T cells and endothelial cells of the PAAD TME
(Fig. 1A) (The Cancer Genome Atlas
PAAD cohort; http://portal.gdc.cancer.gov/projects/TCGA-PAAD).
Utilizing GEPIA2, significantly elevated STING expression was
identified in PAAD compared with normal tissue (Fig. 1B), which was associated with a poor
patient prognosis (Fig. 1C) and
with progressively with advancing clinical stage (22) (Fig.
1D). Genomic analysis demonstrated that STING alterations in
PAAD primarily involved gene amplification (Fig. 1E). Immunohistochemical validation
via The Human Protein Atlas database (https://www.proteinatlas.org) confirmed stronger STING
protein staining in PAAD tissues compared with that in normal
pancreatic ductal epithelium, with STING predominantly localized
within cancer nests (Fig. 1F)
Collectively, these findings established STING as an adverse
prognostic factor in PAAD.
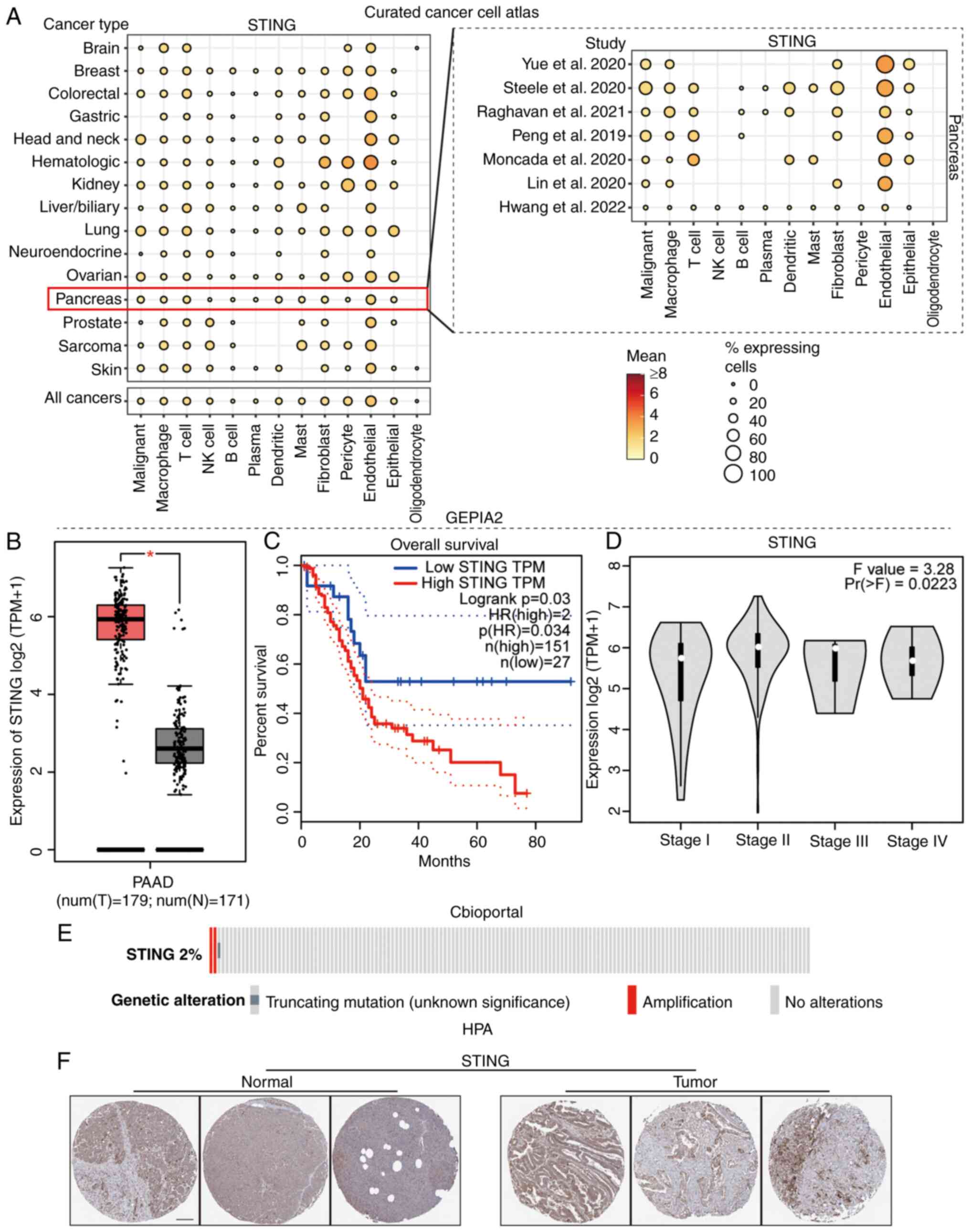 | Figure 1.Role of STING expression in PAAD. (A)
Heatmap comparing STING and immune-related gene expression profiles
in PAAD tumor microenvironment vs. adjacent normal tissue. (B) Box
plot demonstrating significantly elevated STING mRNA expression in
PAAD tumors (T) vs. normal pancreatic tissues (N) (*P<0.05)
(TCGA-PAAD (T) + Genotype-Tissue Expression; normal controls;
http://gtexportal.org). (C) Kaplan-Meier curves
comparing overall survival between PAAD patients with low vs. high
STING expression (log-rank test). (D) Violin plots showing STING
expression distribution across AJCC stages (I–IV) of PAAD. (E)
Oncoprint of STING genomic alterations (mutations, copy number
changes) in PAAD. (F) Human Protein Atlas representative
immunohistochemistry images of STING protein expression (PAAD
tissue; proteinatlas.org). Scale bar, 100 µm. STING, stimulator of
interferon genes; PAAD, pancreatic ductal adenocarcinoma; PAAD,
pancreatic adenocarcinoma; IHC, immunohistochemistry; NK, natural
killer; HR, hazard ratio; T, tumor; N, normal; TPM, transcripts per
million. |
Molecular subtyping of PAAD based on
STING pathway genes
Given the prognostic relevance of STING, its core
signaling network was characterized to define molecular subtypes.
Protein-protein interaction analysis (performed using STRING
database v12.0; cn.string-db.org) identified 26 STING signaling
pathway-associated genes (Pearson r>0.6; all correlations
significant at P<0.001 after Benjamini-Hochberg correction)
(Fig. 2A and B). Unsupervised
clustering based on the expression profiles of these genes
stratified patients with PAAD (TCGA cohort) into two distinct
molecular subtypes: Cluster 1 (C1, STING-high) and Cluster 2 (C2,
STING-low) (Fig. 2C). Genes within
the STING pathway were significantly upregulated in the C1 subtype
(Fig. 2D). Survival analysis
confirmed that patients with the C1 subtype exhibited significantly
worse overall survival (Fig.
2E).
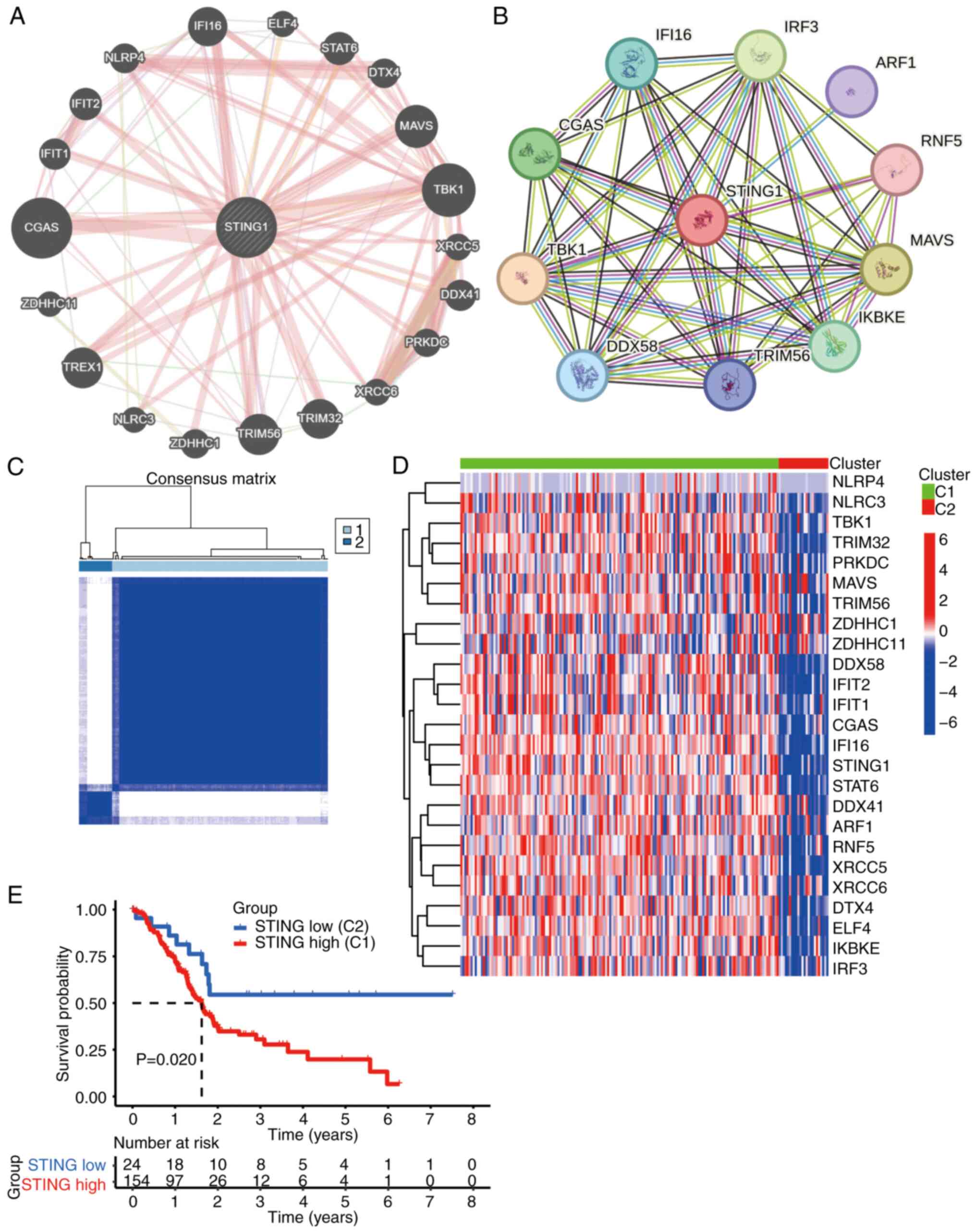 | Figure 2.Molecular characterization of STING
signaling pathway-related genes in PAAD. (A) Protein-protein
interaction (PPI) network of core STING signaling components
identified through STRING database analysis (minimum interaction
score: 0.4). (B) Expanded PPI network showing secondary interactors
of STING pathway genes (minimum interaction score: 0.7). (C)
Consensus matrix heatmap classifying patients with PAAD into two
molecular subtypes (C1 and C2) based on STING-related gene
expression patterns. (D) Heatmap comparing expression profiles of
STING-related genes across PAAD subtypes (C1 vs. C2) with
expression gradient key (red, high expression; blue, low
expression). (E) Kaplan-Meier survival analysis showing
significantly reduced overall survival in the STING-enriched
subtype (C1) compared with the STING-depleted subtype (C2). STING,
stimulator of interferon genes; PAAD, pancreatic ductal
adenocarcinoma; CGAS, cyclic GMP-AMP synthase; TBK1, TANK-binding
kinase 1; MAVS, mitochondrial antiviral-signaling protein; TRIM56,
tripartite motif-containing 56; TREX1, three prime repair
exonuclease 1; TRIM32, tripartite motif-containing 32; IFI16,
interferon γ inducible protein 16; IFIT1, interferon-induced
protein with tetratricopeptide repeats 1; DDX41, DEAD-box helicase
41; NLRP4, NLR family pyrin domain containing 4; DTX4, deltex E3
ubiquitin ligase 4; ZDHHC1, zinc finger DHHC-type containing 1;
PRKDC, protein kinase DNA-activated catalytic subunit; XRCC, X-ray
repair cross complementing; ELF4, E74-like ETS transcription factor
4; NLRC3, NLR family CARD domain containing 3; ARF1, ARF GTPase 1;
DDX56, RNA sensor RIG-I; IKBKE, inhibitor of nuclear factor κB
kinase subunit ε; IRF3, interferon regulatory factor 3; RNF5, ring
finger protein 5. |
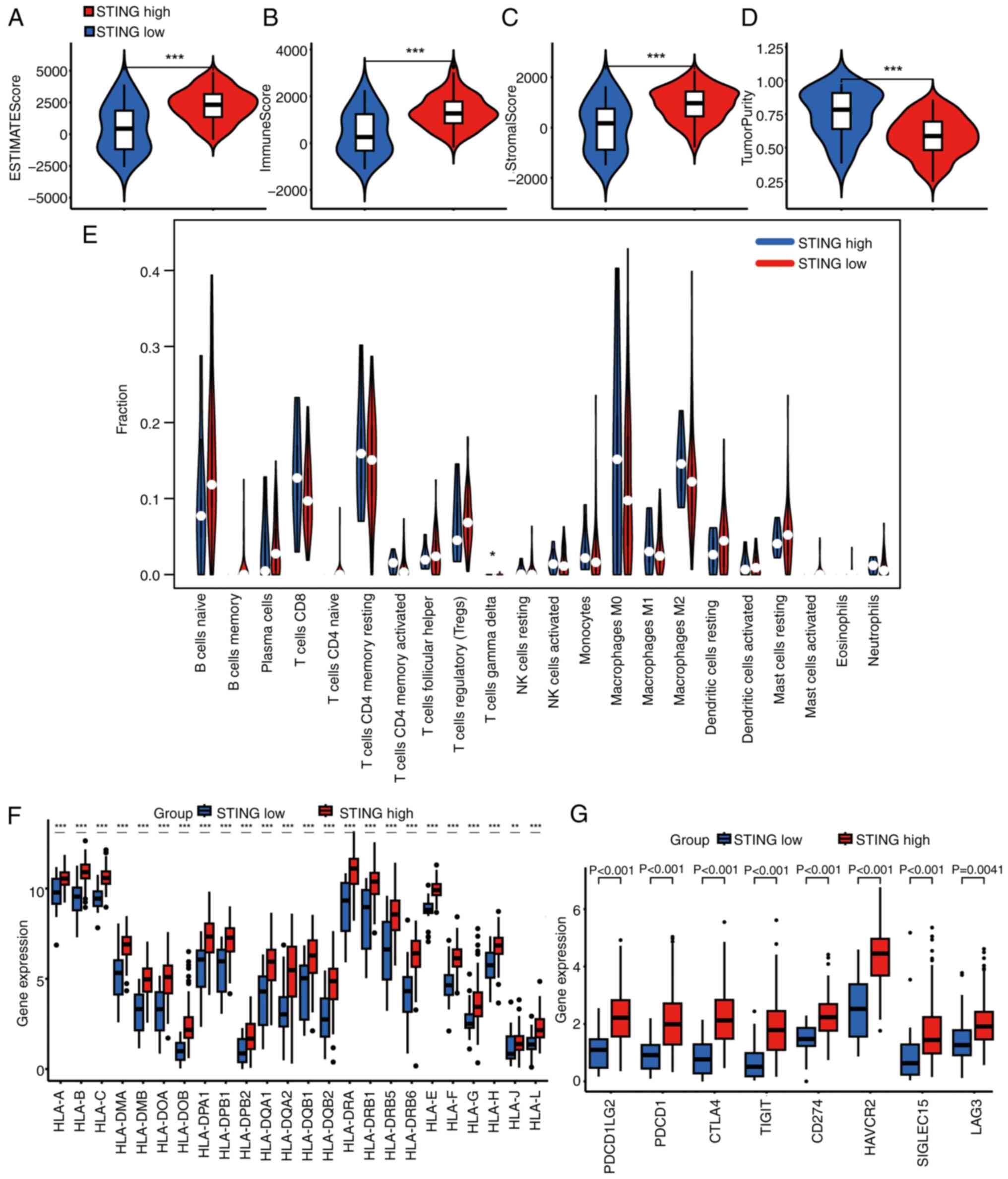
Genomic and immune landscape of
STING-defined subtypes
Differential gene expression analysis between C1 and
C2 subtypes revealed distinct transcriptional profiles (Fig. 3A and B). Gene Ontology (GO)
enrichment analysis of differentially expressed genes highlighted
associations with ‘adaptive immune response’, ‘immunoglobulin
complex’ and ‘production of molecular mediator of immune response’
(Fig. 3C). Kyoto Encyclopedia of
Genes and genomes (KEGG) pathway analysis implicated
‘cytokine-cytokine receptor interaction’, ‘hematopoietic cell
lineage’ and ‘primary immunodeficiency’ (Fig. 3D). Mutation profiling demonstrated
significantly higher tumor mutational burden in the C1 subtype
compared with that in the C2 subtype, characterized by frequent
mutations in key oncogenes (GTPase KRas, TP53-binding protein 1,
mothers against decapentaplegic homolog 4, tumor suppressor ARF and
titin) (Fig. 3E and F). Evaluation
of TME composition using ESTIMATE revealed significantly higher
immune and stromal scores and lower tumor purity in C1 vs. C2
(Fig. 4A-D). CIBERSORTx
deconvolution further indicated elevated infiltration of T cells γ
δ in the C1 subtype (Fig. 4E).
Consistent with enhanced immunogenicity, C1 exhibited significantly
higher expression of human leukocyte antigen family genes and
immune checkpoint molecules relative to C2 (Fig. 4F and G).
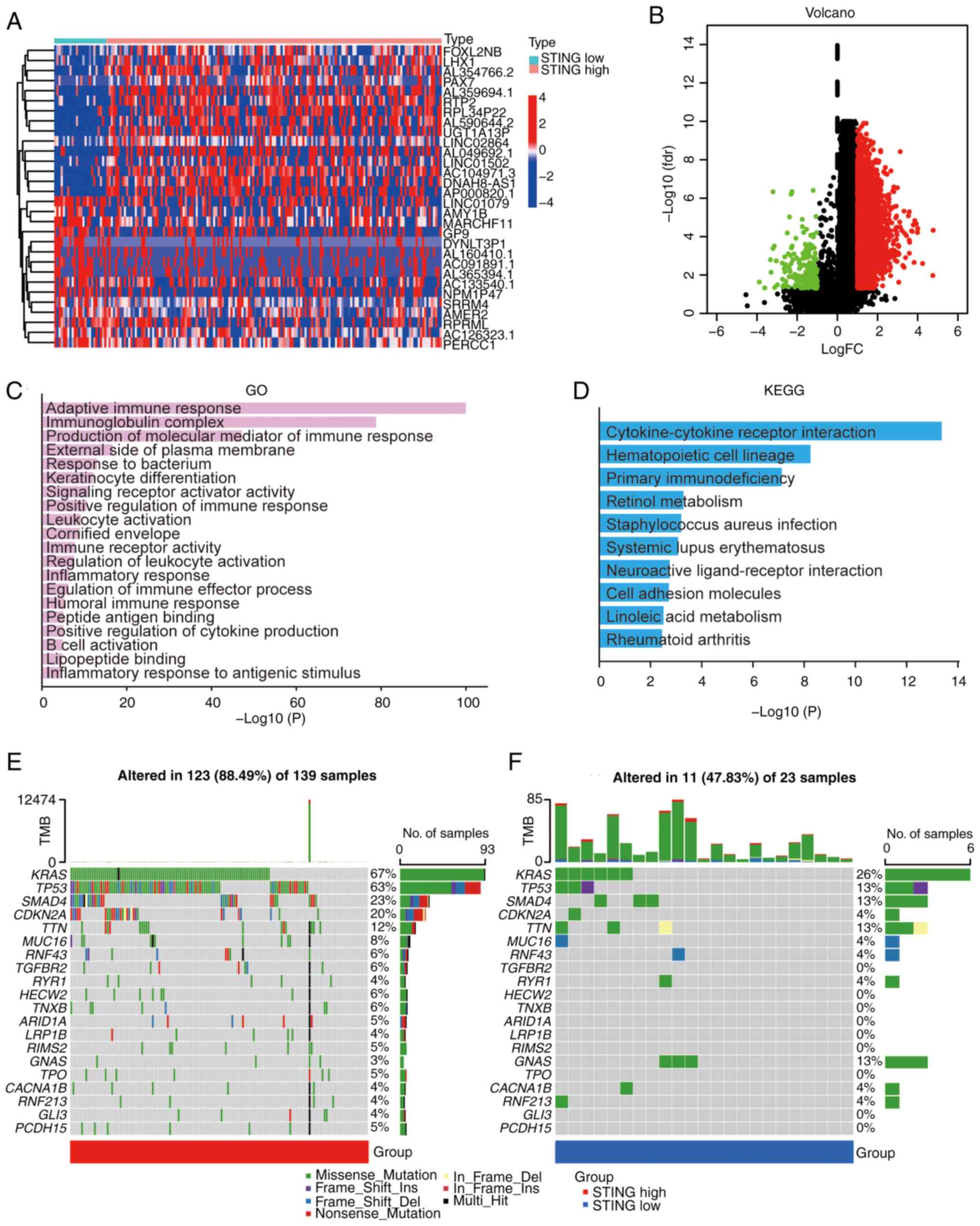 | Figure 3.Gene characteristics of different
PAAD subtypes. (A) Heatmap of upregulated genes in STING1-enriched
subtype (C1) vs. STING1-depleted subtype (C2). (B) Heatmap of
downregulated genes in STING1-enriched subtype (C1) vs.
STING1-depleted subtype (C2). (red, high expression; blue, low
expression). (C) GO enrichment analysis showing top biological
processes in STING1-enriched subtype. (D) KEGG pathway enrichment
analysis showing top signaling pathways in STING1-enriched subtype.
(E) Mutational landscape of STING1-enriched subtype (C1) showing
frequent mutations in KRAS, TP53 and CDKN2A. (F) Mutational
landscape of STING1-depleted subtype (C2) showing distinct mutation
profile with SMAD4 alterations. PAAD, pancreatic ductal
adenocarcinoma; STING, stimulator of interferon genes; FC,
fold-change; fdr, false discovery rate; TMB, tumor mutational
burden; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and
Genomes. |
Development and validation of a STING
pathway prognostic signature
A risk prediction model was constructed to quantify
the prognostic heterogeneity associated with STING pathway
activity. Univariate Cox regression analysis identified 10 genes
among the 26 STING pathway genes that were significantly associated
with PAAD prognosis (Fig. 5A).
LASSO Cox regression refined these into a 6-gene prognostic
signature [deltex E3 ubiquitin ligase 4, interferon γ inducible
protein 16, mitochondrial antiviral-signaling protein (MAVS),
protein kinase DNA-activated catalytic subunit, ZDHHC1 and IFIT2]
(Fig. 5B and C). This signature
effectively stratified patients with PAAD into high- and low-risk
groups. Survival outcomes were significantly different in the TCGA
discovery cohort (Fig. 5D) and the
independent GSE224564 validation cohort (Fig. 5E). Expression patterns of the 6
signature genes were consistent across both TCGA (Fig. 5F) and validation cohort (Fig. 5G) datasets. Notably, multivariate
Cox regression analysis confirmed both the prognostic risk score
and N stage as independent predictors of survival in patients with
PAAD (Fig. 6A and B).
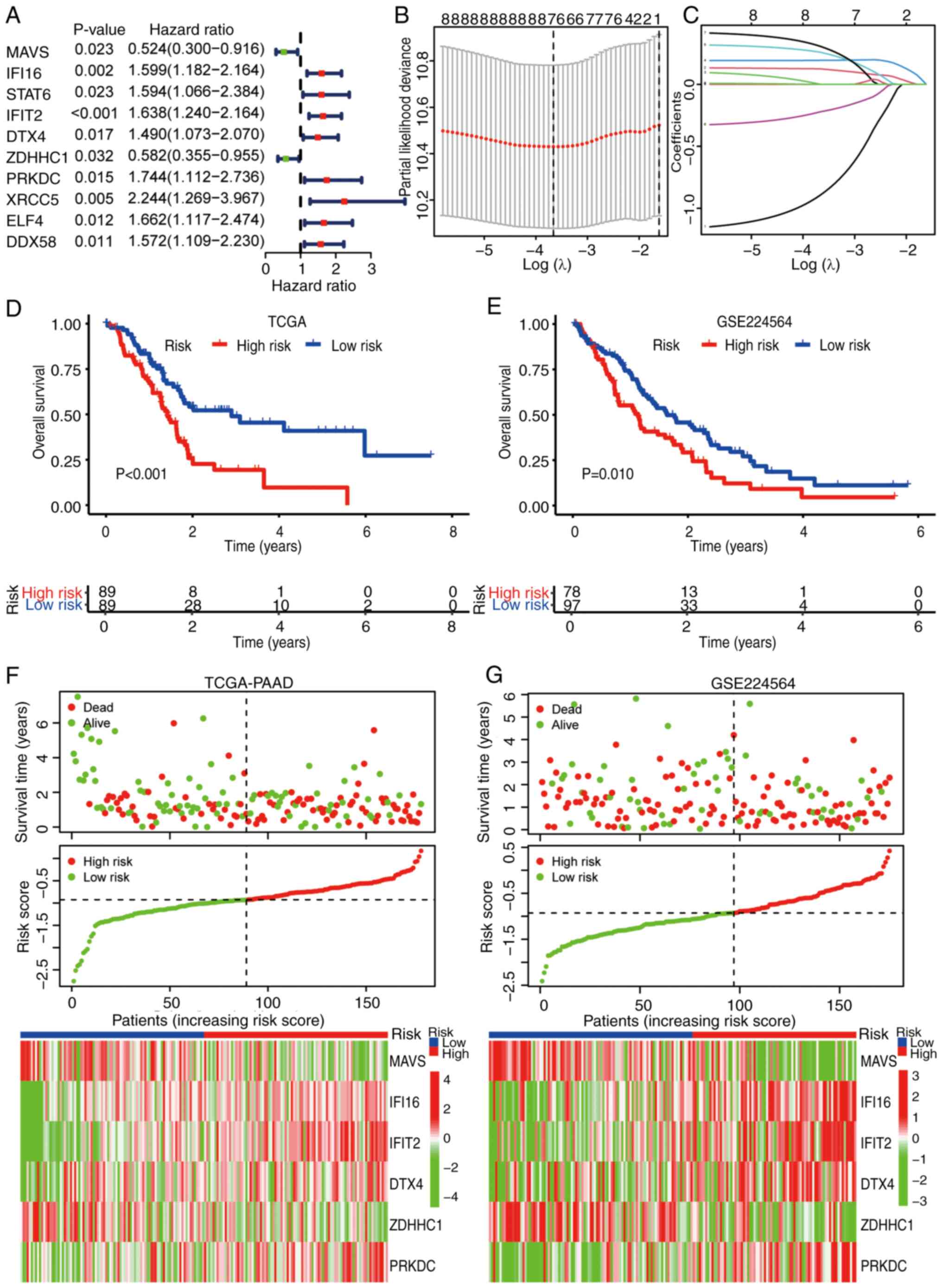 | Figure 5.Development and validation of a
STING1 signaling-related prognostic signature for PAAD. (A)
Univariate Cox regression analysis of STING1 pathway-related genes
in PAAD. Genes with significant prognostic value (P<0.05) are
included. (B) LASSO regression 10-fold cross-validation curve
showing optimal λ selection (minimum criteria). (C) LASSO
coefficient profiles of STING1-related genes across log(λ)
sequence. (D) Kaplan-Meier survival analysis for the prognostic
signature in TCGA-PAAD cohort (high-risk vs. low-risk groups;
cutoff: median risk score). (E) Validation of prognostic signature
in GSE224564 cohort. (F) Risk score distribution plot with
corresponding survival status and expression heatmap for 6
signature genes in TCGA-PAAD. (G) Validation in GSE224564 cohort
showing identical configuration to (F). STING, stimulator of
interferon genes; PAAD, pancreatic ductal adenocarcinoma; LASSO,
least absolute shrinkage and selection operator; TCGA, The Cancer
Genome Atlas; MAVS, mitochondrial antiviral-signaling protein;
IFI16, interferon γ inducible protein 16; IFIT2, interferon-induced
protein with tetratricopeptide repeats 2; DTX4, deltex E3 ubiquitin
ligase 4; ZDHHC1, zinc finger DHHC-type containing 1; PRKDC,
protein kinase DNA-activated catalytic subunit; STAT6, signal
transducer and activator of transcription 6; XRCC5, X-ray repair
cross complementing: ELF4, E74-like ETS transcription factor 4;
DDX58, RNA sensor RIG-I. |
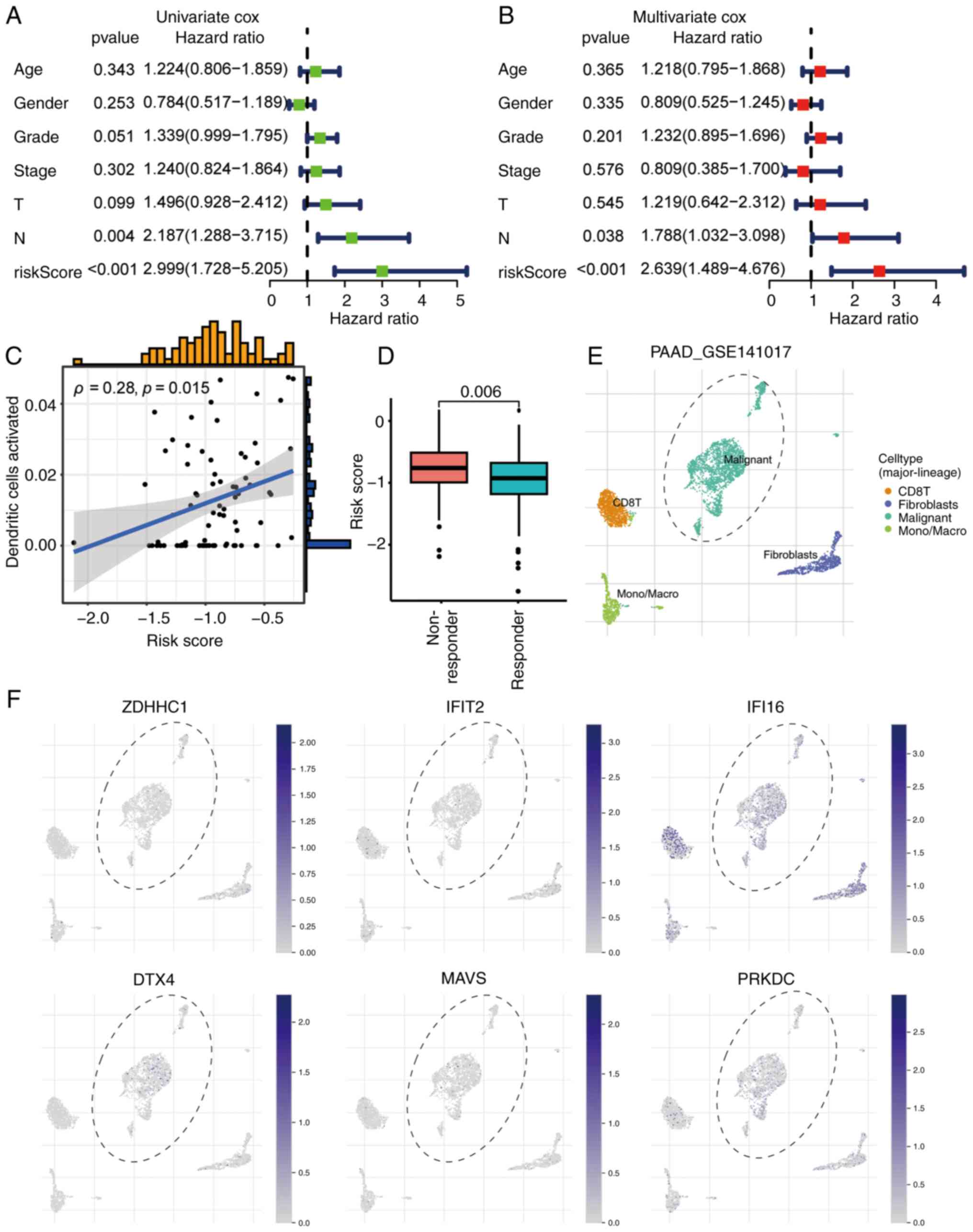 | Figure 6.Independent prognostic value and
immunotherapy relevance of the STING-related risk signature in
PAAD. (A) Univariate Cox regression analysis of clinical parameters
and risk score. (B) Multivariate Cox regression confirming the risk
score as an independent prognostic factor after adjusting for
covariates in (A). (C) Correlation analysis of risk score and
activated dendritic cell abundance. ‘Dendritic cells activated’
represents proportion scores derived from the xCell algorithm (0–1
scale), quantifying relative abundance in the tumor
microenvironment. Correlation was assessed using Spearman's rank
correlation analysis (Spearman's ρ=0.28; P=0.015). (D) Higher risk
scores in immunotherapy non-responders vs. responders (Mann-Whitney
U test). Single-cell RNA sequencing (GSE141017) visualization of
STING1 signature gene expression: (E) t-SNE plot showing cell-type
distribution. (F) Feature plots of signature gene expression across
the PAAD tumor microenvironment. STING, stimulator of interferon
genes; PAAD, pancreatic ductal adenocarcinoma; T, tumor; N, node;
CD8T, T-cell surface glycoprotein CD8; ZDHHC1, zinc finger
DHHC-type containing 1; IFIT2, interferon-induced protein with
tetratricopeptide repeats 2; IFI16, interferon γ inducible protein
16; DTX4, deltex E3 ubiquitin ligase 4; MAVS, mitochondrial
antiviral-signaling protein; PRKDC, protein kinase DNA-activated
catalytic subunit; PAAD, pancreatic adenocarcinoma. |
Association of prognostic risk score
with immune features
The STING pathway-derived prognostic risk score
demonstrated significant biological relevance, showing a positive
correlation with DC activation as determined by Spearman's rank
correlation analysis (Fig. 6C).
Assessment using the TIDE algorithm revealed significantly higher
scores in high-risk vs. low-risk patients, indicating greater
immune evasion potential and reduced likelihood of response to
immune checkpoint blockade (Fig.
6D). Analysis of spatial expression (GSE141017) confirmed that
the 6 signature genes were predominantly expressed within tumor
cells of the PAAD TME (Fig. 6E and
F).
Functional validation of hub genes
IFIT2 and ZDHHC1
Given the established prognostic relevance of STING
in PAAD (characterized by high expression correlating with poor
patient outcomes in the present study), its core signaling network
was characterized to define molecular subtypes. While MAVS
demonstrated strong prognostic associations, its functional role in
PAAD has been extensively explored in prior literature (23). By contrast, ZDHHC1 and IFIT2, which
also showed significant prognostic value (Fig. S1), have received considerably less
attention in PAAD research. These genes were therefore prioritized
for validation, observing distinct expression dynamics: IFIT2
expression significantly increased with advancing AJCC 8th edition
tumor grade/stage (22) up to stage
III/grade 3 before declining in advanced disease, associated with a
poor prognosis, while ZDHHC1 expression decreased progressively
with tumor progression and was associated with favorable outcomes
(Fig. S1A-E). Immunohistochemical
validation using The Human Protein Atlas (https://www.proteinatlas.org) confirmed these protein
patterns in PAAD tissues (Fig. 7A).
Overexpression of ZDHHC1 was confirmed in the Panc-02 cells line
(Fig. 7B). Based on the confirmed
superior knockdown efficiency of sh2, the stable cell line
expressing sh2 was selected for all subsequent functional
experiments (Fig. 7C). Functional
studies in Panc02 cells revealed that overexpression of ZDHHC1
inhibited proliferation (Fig. 7D),
as did knockdown of IFIT2 (Fig.
7E). Overexpression of ZDHHC1 (Fig.
7F) and knockdown of IFIT2 (Fig.
7G) both promoted apoptosis. These results established IFIT2 as
a potential oncogene and ZDHHC1 as a potential tumor suppressor
within the STING pathway in PAAD.
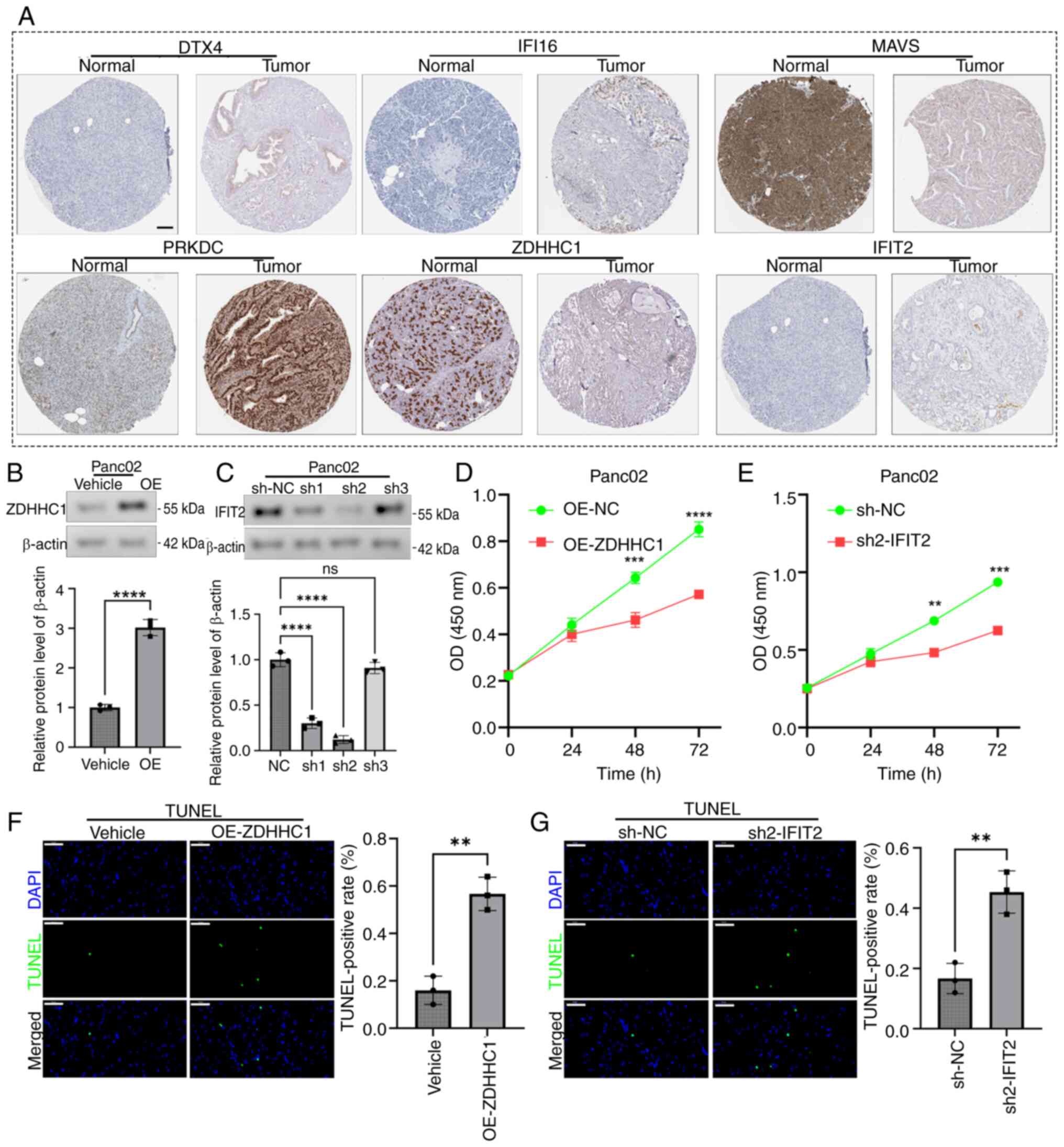 | Figure 7.Functional roles of IFIT2 and ZDHHC1
in PAAD oncobiology. (A) Human Protein Atlas representative
immunohistochemical staining of six prognostic signature genes in
PAAD tissues (proteinatlas.org). Scale bar, 100 µm. (B) Western
blot and semi-quantification confirming successful ZDHHC1
overexpression in Panc02 cells (C) Western blot and
semi-quantification confirming IFIT2 knockout efficiency in Panc02
cells (****P<0.0001). (D) CCK-8 assay: ZDHHC1 overexpression
enhances Panc02 proliferation (***P<0.001 and ****P<0.0001 vs
OE-NC). (E) CCK-8 assay: IFIT2 knockout enhances Panc02
proliferation (**P<0.01 and ***P<0.001 vs. sh-NC). (F) TUNEL
assay (scale bar, 50 µm) demonstrating reduced apoptosis in
ZDHHC1-overexpressing Panc02 cells. (G) TUNEL assay (scale bar 50
µm) showing decreased apoptosis in IFIT2-knockout Panc02 cells
(**P<0.01). IFIT2, interferon-induced protein with
tetratricopeptide repeats 2; ZDHHC1, zinc finger DHHC-type
containing 1; PAAD, pancreatic ductal adenocarcinoma; OE,
overexpression; CCK-8, Cell Counting Kit-8; DTX4, deltex E3
ubiquitin ligase 4; IFI16, interferon γ inducible protein 16; MAVS,
mitochondrial antiviral-signaling protein; PRKDC, protein kinase
DNA-activated catalytic subunit; sh-NC, scrambled negative control
shRNA; ns, not significant. |
Discussion
PAAD presents a notable global health challenge,
ranking 12th in global cancer incidence but 7th in cancer-related
mortality rate according to 2020 data (24), with similar trends observed in China
(25). Bioinformatics has emerged
as a powerful tool for dissecting PAAD pathogenesis (26), enabling the identification of
potential biomarkers, therapeutic targets and immune evasion
mechanisms through the integrated analysis of genomic,
transcriptomic and tumor microenvironmental data (27).
Research into the STING signaling pathway in PAAD
reveals its complex duality. While STING agonists activate the
cyclic GMP-AMP synthase-STING pathway to induce IFN-I, trigger
adaptive immunity and exert cytotoxic effects on tumor cells
(11), clinical translation has
been hampered by intrinsic resistance mechanisms (28). These include STING-induced expansion
of regulatory B cells that suppress natural killer cell activity
(29) and the potential for chronic
pathway activation to promote immunosuppressive signaling
downstream of chromosomal instability (30). Nevertheless, STING agonism
demonstrates significant preclinical promise as it remodels the
PAAD tumor stroma and immune landscape, enhances cytotoxic T cell
infiltration while reducing the frequency of regulatory T cells
(31), activates DCs and reprograms
macrophages towards an immunostimulatory phenotype (32). Furthermore, synergistic strategies,
such as combining STING agonists with interleukin-35 blockade, show
enhanced antitumor efficacy (29).
Future progress may hinge on developing novel agonists (for
example, non-nucleotide compounds) and targeted delivery systems to
improve efficacy and minimize toxicity.
The present study provides novel insights into the
immune-related functions of STING and its associated genes within
the distinct context of PAAD. Notably, high expression of STING
pathway genes was identified to be associated with a poor prognosis
in PAAD. This association contrasts with observations in some other
cancer types (such as gastric cancer, colorectal cancer and
hepatocellular carcinoma) and potentially reflects the unique
immunobiology of the ‘cold’ nature of PAAD tumors. Using this, a
robust prognostic signature centered on STING pathway activity was
developed. Further functional prioritization within this signature
highlighted the notable, yet opposing, roles of IFIT2 and
ZDHHC1.
In the present study, IFIT2 emerged as a potential
oncogenic driver in PAAD. Elevated IFIT2 expression was associated
with advanced tumor grade/stage (22) and worse survival outcomes. This
association aligns with studies linking IFIT2 to chemotherapy
resistance in PAAD and its established role in regulating
inflammatory responses downstream of IFN signaling and pathogen
recognition (33). IFIT2 may
modulate the tumor immune microenvironment (TIME); evidence in
esophageal squamous cell carcinoma shows that
methyltransferase-like protein 3-mediated neuronal membrane
glycoprotein M6-a modification regulates IFIT2, influencing
malignant progression and the TIME (34). While prognostic in oral squamous
cell carcinoma, the specific pro-tumorigenic mechanisms of IFIT2 in
PAAD warrant further investigation. Conversely, in the present
study, ZDHHC1 functioned as a putative tumor suppressor. ZDHHC1 is
frequently epigenetically silenced via promoter methylation across
various cancer types, such as gastric cancer, colorectal cancer and
hepatocellular carcinoma. Restoring ZDHHC1 expression exerts broad
antitumor effects, including the induction of apoptosis and
autophagy, cell cycle arrest, the inhibition of
migration/metastasis, the reversal of epithelial-mesenchymal
transition and stemness (35), and
metabolic reprogramming via suppression of glycolysis and the
pentose phosphate pathway (36).
Mechanistically, ZDHHC1 acts as an S-palmitoyltransferase critical
for palmitoylating p53 at residues C135, C176 and C275, thereby
stabilizing and activating the master tumor suppressor activity of
p53 (37). Loss of ZDHHC1-mediated
p53 palmitoylation represents a novel evasion mechanism in cancer.
Although the role of ZDHHC1 in PAAD was previously undefined, the
present data positions ZDHHC1 as a compelling therapeutic target
meriting further exploration in PAAD.
While providing valuable mechanistic and prognostic
insights into STING signaling in PAAD, the present study has
several limitations: i) The reliance on protein-protein interaction
networks for gene selection may have excluded key epigenomic
regulators or microenvironmental modulators, limiting pathway
comprehensiveness; ii) despite internal-external validation (TCGA +
GSE224564, n=353), cohort size constraints limit statistical power
for detailed subtype-specific analyses and robust assessment of
treatment interactions; iii) while in vitro validation
confirmed IFIT2/ZDHHC1 functionality in Panc02 cells, the present
study lacked in vivo models to evaluate stromal crosstalk,
combinatorial gene perturbation assays to assess IFIT2-ZDHHC1
interplay and spatial resolution of immune effects (for example,
multiplexed T-cell infiltration mapping); iv) clinical translation
requires standardized detection protocols for biopsy specimens and
prospective trials stratifying immunotherapy based on risk scores;
and v) the cellular origin (cancer vs. immune) and temporal
dynamics of STING pathway gene expression during progression remain
unresolved. Future research should prioritize multi-center
signature validation, developing CRISPR-engineered dual-gene models
(IFIT2 knockout + ZDHHC1 overexpression) for in vivo
efficacy testing, employing spatial multi-omics (single cell RNA
sequencing and spatial proteomics) to resolve compartment-specific
expression and conducting STING agonist trials within
signature-defined patient subgroups.
In summary, the present study established and
validated a STING signaling pathway-derived prognostic gene
signature that effectively stratifies patients with PAAD. High-risk
patients identified by this signature exhibited significantly
poorer survival outcomes compared with low-risk patients, and
distinct immune microenvironment profiles. Notably, the
antagonistic roles of two core signature genes were functionally
validated: IFIT2 as a potential oncogene promoting proliferation
and suppressing apoptosis, and ZDHHC1 as a putative tumor
suppressor with the converse effects. The findings of the present
study underscore the notable clinical and biological relevance of
the STING pathway in PAAD. The identified genes, particularly IFIT2
and ZDHHC1, represent promising candidates for further mechanistic
exploration and potential targets for developing novel therapeutic
strategies against this lethal malignancy.
Supplementary Material
Supporting Data
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
WY and JLia contributed to the conceptualization,
reviewing and editing of the manuscript. JLi was responsible for
the curation of the data. YW provided the formal analysis and
supervision. All authors read and approved the final version of the
manuscript. JLi and YW confirm the authenticity of all the raw
data.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
PAAD
|
pancreatic ductal adenocarcinoma
|
|
STING
|
stimulator of interferon genes
|
|
IFIT2
|
interferon-induced protein with
tetratricopeptide repeats 2
|
|
ZDHHC1
|
zinc finger DHHC-type containing 1
|
|
LASSO
|
least absolute shrinkage and selection
operator
|
|
TME
|
tumor microenvironment
|
|
TIDE
|
Tumor Immune Dysfunction and
Exclusion
|
|
CCK-8
|
Cell Counting Kit-8
|
|
TCGA
|
The Cancer Genome Atlas
|
References
|
1
|
Bray F, Laversanne M, Sung H, Ferlay J,
Siegel RL, Soerjomataram I and Jemal A: Global cancer statistics
2022: GLOBOCAN estimates of incidence and mortality worldwide for
36 cancers in 185 countries. CA Cancer J Clin. 74:229–263. 2024.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Blackford AL, Canto MI, Dbouk M, Hruban
RH, Katona BW, Chak A, Brand RE, Syngal S, Farrell J, Kastrinos F,
et al: Pancreatic cancer surveillance and survival of high-risk
individuals. JAMA Oncol. 10:1087–1096. 2024. View Article : Google Scholar
|
|
3
|
Park W, Chawla A and O'Reilly EM:
Pancreatic cancer: A review. JAMA. 326:851–862. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Peduzzi G, Archibugi L, Farinella R, de
Leon Pisani RP, Vodickova L, Vodicka P, Kraja B, Sainz J,
Bars-Cortina D, Daniel N, et al: The exposome and pancreatic
cancer, lifestyle and environmental risk factors for PDAC. Semin
Cancer Biol. 113:100–129. 2025. View Article : Google Scholar
|
|
5
|
Springfeld C, Ferrone CR, Katz MHG, Philip
PA, Hong TS, Hackert T, Büchler MW and Neoptolemos J: Neoadjuvant
therapy for pancreatic cancer. Nat Rev Clin Oncol. 20:318–337.
2023. View Article : Google Scholar
|
|
6
|
Hu ZI and O'Reilly EM: Therapeutic
developments in pancreatic cancer. Nat Rev Gastroenterol Hepatol.
21:7–24. 2024. View Article : Google Scholar
|
|
7
|
Sang W, Zhou Y, Chen H, Yu C, Dai L, Liu
Z, Chen L, Fang Y, Ma P, Wu X, et al: Receptor-interacting protein
kinase 2 is an immunotherapy target in pancreatic cancer. Cancer
Discov. 14:326–347. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ullman NA, Burchard PR, Dunne RF and
Linehan DC: Immunologic strategies in pancreatic cancer: Making
cold tumors hot. J Clin Oncol. 40:2789–2805. 2022. View Article : Google Scholar
|
|
9
|
Zhao K, Huang J, Zhao Y, Wang S, Xu J and
Yin K: Targeting STING in cancer: Challenges and emerging
opportunities. Biochim Biophys Acta Rev Cancer. 1878:1889832023.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhang B, Xu P and Ablasser A: Regulation
of the cGAS-STING pathway. Annu Rev Immunol. 43:667–692. 2025.
View Article : Google Scholar
|
|
11
|
Tani T, Mathsyaraja H, Campisi M, Li ZH,
Haratani K, Fahey CG, Ota K, Mahadevan NR, Shi Y, Saito S, et al:
TREX1 inactivation unleashes cancer cell STING-interferon signaling
and promotes antitumor immunity. Cancer Discov. 14:752–765. 2024.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Luo J, Wang S, Yang Q, Fu Q, Zhu C, Li T,
Yang S, Zhao Y, Guo R, Ben X, et al: γδ T Cell-mediated tumor
immunity is tightly regulated by STING and TGF-β signaling
pathways. Adv Sci (Weinh). 12:e24044322025. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Liu X, Hogg GD, Zuo C, Borcherding NC,
Baer JM, Lander VE, Kang LI, Knolhoff BL, Ahmad F, Osterhout RE, et
al: Context-dependent activation of STING-interferon signaling by
CD11b agonists enhances anti-tumor immunity. Cancer Cell.
41:1073–1090.e12. 2023. View Article : Google Scholar
|
|
14
|
Tian X, Ai J, Tian X and Wei X: cGAS-STING
pathway agonists are promising vaccine adjuvants. Med Res Rev.
44:1768–1799. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chen X, Xu Z, Li T, Thakur A, Wen Y, Zhang
K, Liu Y, Liang Q, Liu W, Qin JJ and Yan Y:
Nanomaterial-encapsulated STING agonists for immune modulation in
cancer therapy. Biomark Res. 12:22024. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chin EN, Sulpizio A and Lairson LL:
Targeting STING to promote antitumor immunity. Trends Cell Biol.
33:189–203. 2023. View Article : Google Scholar
|
|
17
|
Chen X, Meng F, Xu Y, Li T, Chen X and
Wang H: Chemically programmed STING-activating nano-liposomal
vesicles improve anticancer immunity. Nat Commun. 14:45842023.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Dosta P, Cryer AM, Dion MZ, Shiraishi T,
Langston SP, Lok D, Wang J, Harrison S, Hatten T, Ganno ML, et al:
Investigation of the enhanced antitumour potency of STING agonist
after conjugation to polymer nanoparticles. Nat Nanotechnol.
18:1351–1363. 2023. View Article : Google Scholar
|
|
19
|
Wen Z, Sun F, Wang R, Wang W, Zhang H,
Yang F, Wang M, Wang Y and Li B: STING agonists: A range of eminent
mediators in cancer immunotherapy. Cell Signal. 134:1119142025.
View Article : Google Scholar
|
|
20
|
Ohara Y, Tang W, Liu H, Yang S, Dorsey TH,
Cawley H, Moreno P, Chari R, Guest MR, Azizian A, et al:
SERPINB3-MYC axis induces the basal-like/squamous subtype and
enhances disease progression in pancreatic cancer. Cell Rep.
42:1134342023. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Amin MB, Greene FL, Edge SB, Compton CC,
Gershenwald JE, Brookland RK, Meyer L, Gress DM, Byrd DR and
Winchester DP: The eighth edition AJCC cancer staging manual:
Continuing to build a bridge from a population-based to a more
‘personalized’ approach to cancer staging. CA Cancer J Clin.
67:93–99. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lee M, Thomas AS, Lee SY, Cho YJ, Jung HS,
Yun WG, Han Y, Jang JY, Kluger MD and Kwon W: Reconsidering the
absence of extrapancreatic extension in T staging for pancreatic
adenocarcinoma in the AJCC (8th ed) staging manual using the
national cancer database. J Gastrointest Surg. 27:2484–2492. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kerschbaum-Gruber S, Kleinwächter A,
Popova K, Kneringer A, Appel LM, Stasny K, Röhrer A, Dias AB,
Benedum J, Walch L, et al: Cytosolic nucleic acid sensors and
interferon beta-1 activation drive radiation-induced anti-tumour
immune effects in human pancreatic cancer cells. Front Immunol.
15:12869422024. View Article : Google Scholar
|
|
24
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yin L, Wei J, Lu Z, Huang S, Gao H, Chen
J, Guo F, Tu M, Xiao B, Xi C, et al: Prevalence of germline
sequence variations among patients with pancreatic cancer in China.
JAMA Netw Open. 5:e21487212022. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Larson NB, Oberg AL, Adjei AA and Wang L:
A clinician's guide to bioinformatics for next-generation
sequencing. J Thorac Oncol. 18:143–157. 2023. View Article : Google Scholar
|
|
27
|
Petralia F, Ma W, Yaron TM, Caruso FP,
Tignor N, Wang JM, Charytonowicz D, Johnson JL, Huntsman EM, Marino
GB, et al: Pan-cancer proteogenomics characterization of tumor
immunity. Cell. 187:1255–1277.e27. 2024. View Article : Google Scholar
|
|
28
|
Lv M, Chen M, Zhang R, Zhang W, Wang C,
Zhang Y, Wei X, Guan Y, Liu J, Feng K, et al: Manganese is critical
for antitumor immune responses via cGAS-STING and improves the
efficacy of clinical immunotherapy. Cell Res. 30:966–979. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Li S, Mirlekar B, Johnson BM, Brickey WJ,
Wrobel JA, Yang N, Song D, Entwistle S, Tan X, Deng M, et al:
STING-induced regulatory B cells compromise NK function in cancer
immunity. Nature. 610:373–380. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Li J, Hubisz MJ, Earlie EM, Duran MA, Hong
C, Varela AA, Lettera E, Deyell M, Tavora B, Havel JJ, et al:
Non-cell-autonomous cancer progression from chromosomal
instability. Nature. 620:1080–1088. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Jneid B, Bochnakian A, Hoffmann C, Delisle
F, Djacoto E, Sirven P, Denizeau J, Sedlik C, Gerber-Ferder Y,
Fiore F, et al: Selective STING stimulation in dendritic cells
primes antitumor T cell responses. Sci Immunol. 8:eabn66122023.
View Article : Google Scholar
|
|
32
|
Wang J, Li S, Wang M, Wang X, Chen S, Sun
Z, Ren X, Huang G, Sumer BD, Yan N, et al: STING licensing of type
I dendritic cells potentiates antitumor immunity. Sci Immunol.
9:eadj39452024. View Article : Google Scholar
|
|
33
|
Yang Y, Song J, Zhao H, Zhang H and Guo M:
Patients with dermatomyositis shared partially similar
transcriptome signature with COVID-19 infection. Autoimmunity.
56:22209842023. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Xu QC, Tien YC, Shi YH, Chen S, Zhu YQ,
Huang XT, Huang CS, Zhao W and Yin XY: METTL3 promotes intrahepatic
cholangiocarcinoma progression by regulating IFIT2 expression in an
m6A-YTHDF2-dependent manner. Oncogene. 41:1622–1633.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhou B, Liu Y, Ma H, Zhang B, Lu B, Li S,
Liu T, Qi Y, Wang Y, Zhang M, et al: Zdhhc1 deficiency mitigates
foam cell formation and atherosclerosis by inhibiting PI3K-Akt-mTOR
signaling pathway through facilitating the nuclear translocation of
p110α. Biochim Biophys Acta Mol Basis Dis. 1871:1675772025.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Le X, Mu J, Peng W, Tang J, Xiang Q, Tian
S, Feng Y, He S, Qiu Z, Ren G, et al: DNA methylation downregulated
ZDHHC1 suppresses tumor growth by altering cellular metabolism and
inducing oxidative/ER stress-mediated apoptosis and pyroptosis.
Theranostics. 10:9495–9511. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Tang J, Peng W, Feng Y, Le X, Wang K,
Xiang Q, Li L, Wang Y, Xu C, Mu J, et al: Cancer cells escape p53′s
tumor suppression through ablation of ZDHHC1-mediated p53
palmitoylation. Oncogene. 40:5416–5426. 2021. View Article : Google Scholar : PubMed/NCBI
|