Introduction
Esophageal squamous cell carcinoma (ESCC) has one of
the highest mortality rates among patients with solid tumors due to
the fact that it is a highly aggressive malignancy with early
lymphatic and hematogenous dissemination (1,2).
Recent advances in perioperative management, combined with
chemotherapy or chemoradiotherapy, have led to improved survival
rates. However, the prognosis for patients with advanced disease
remains poor and unsatisfactory (3–5).
Discovering suitable biomarkers of malignancy could enhance
monitoring for cancer recurrence, and the development of new
therapeutic approaches is essential for the improvement of survival
rates.
CD47 is a widely expressed transmembrane protein
that has been implicated in multiple cellular processes, including
a function as a ‘do not eat me’ signal inhibiting macrophage
activity by binding the signal regulatory protein α, which is
expressed on phagocytes (6–8). Whereas this function is partly
attributed to ‘self recognition’ in normal physiological
conditions, several human cancers appear to upregulate CD47 to
evade the tumor immunosurveillance system (9–11).
Overexpression of CD47 correlates with poor prognosis in
acute myeloid leukemia (12), acute
lymphoblastic leukemia (13) and
breast cancer (14). However, the
clinicopathological and prognostic significance of CD47
expression in ESCC remains unclear.
It was reported that CD47 expression in
multiple sclerosis is regulated by microRNAs (miRNAs) including
miR-34a, miR-155 and miR-326 (15).
However, the mechanism of CD47 regulation in cancer tissues
has not yet been clarified. miRNAs are small non-coding RNAs of
18–25 nucleotides that partially bind the 3′ untranslated region
(3′-UTR) of their target mRNA, resulting in mRNA degradation and/or
translational repression (16). By
downregulating their target gene expression, miRNAs play an
essential role in several cellular processes, such as
proliferation, differentiation and apoptosis (17,18).
Moreover, miRNAs also function as oncogenes or tumor suppressors
depending on their targets. Therefore, miRNA has garnered attention
as a new diagnostic and therapeutic tool for human malignancies
(19,20).
In a search for CD47 regulatory miRNAs
downregulated in ESCC, we re-analyzed GSE6188, the Gene Expression
Omnibus public microarray database from the National Center for
Biotechnical Information (21).
Five downregulated miRNAs were found in ESCC. Among them, miR-133a
is downregulated in the miRNA expression signatures of several
types of human malignancies relative to normal adjacent tissue:
head and neck squamous cell carcinoma (22–24),
bladder cancer (25,26), colorectal cancer (27), and rhabdomyosarcoma (28,29).
Transfection of miR-133a induced reductions in cell proliferation,
migration and invasion (25). These
data suggested that miR-133a may function as a tumor suppressor.
Furthermore, the TargetScan 5.0 in silico prediction
algorithm (30) also indicated that
miR-133a is a conserved regulatory miRNA of CD47. Based on
these results, we focused on the miR-133a tumor suppressor as a
direct regulator of CD47.
The purpose of this study was to clarify the
clinical significance and regulatory mechanism of CD47 in
ESCC. Firstly, we examined the expression level of CD47 in
ESCC and adjacent non-cancerous tissue. Secondly, we evaluated the
CD47 regulation in ESCC by miR-133a in silico and
in vitro, and investigated the functional analysis of the
miR-133a using a mouse xenograft model.
Materials and methods
Clinical samples and RNA isolation
The pair of primary ESCC and corresponding normal
esophageal epithelia were obtained from 102 ESCC patients (89 males
and 13 females), who had undergone potentially curative surgery at
the Department of General Surgical Science, Gunma University,
between 1990 and 2007, after obtaining their written informed
consent. The patients’ ages ranged from 42 to 83 years, with a mean
of 64.9. The median follow-up period for survivors was 40 months
(range, 1–126 months). The pathological features of the specimens
were classified based on the 6th edition of the TNM classification
of the International Union against Cancer (UICC). The operations
were classified as curative surgeries; there was no evidence of
residual tumors, and the resected margins were microscopically free
of tumors (R0). Normal tissues were obtained far from the center of
the cancer in surgical specimens. All specimens were immediately
frozen in liquid nitrogen and stored at −80°C until RNA extraction.
Total RNA was extracted using the miRNeasy Mini kit (Qiagen)
according to the manufacturer’s instructions. The quantity of
isolated RNA was measured by the ND-1000 spectrophotometer
(NanoDrop Technologies).
Evaluation of CD47 expression in clinical
samples
cDNA for CD47 mRNA quantitative real-time
reverse transcriptase PCR (RT-PCR) was synthesized from 1 μg total
RNA with the Omniscript Reverse Transcriptase kit (Qiagen) in a
reaction volume of 20 μl (60 min at 37°C and 5 min at 93°C before
being put on ice). The cDNA samples were stored at −30°C until
needed for analyses. The sequences of the CD47 primers were
as follows: sense primer, 5′-GGCAATGACGAAGGA GGTTA-3′; antisense
primer, 5′-ATCCGGTGGTATGGAT GAGA-3′. The following β-actin (the
internal control) primers were used: sense primer,
5′-CTCCTCCTGAGCGCAAGT ACTC-3′; antisense primer,
5′-TCCTGCTTGCTGATCCA CATC-3′. Real-time monitoring of PCR reactions
was performed using the Light Cycler System and SYBR-Green I Master
(Roche) according to the manufacturer’s instructions. Quantitative
real-time PCR was performed with the following cycling conditions;
initial denaturation at 95°C for 10 min, followed by 40 cycles of
95°C for 10 sec, annealing at 60°C for 10 sec, and extension at
72°C for 10 sec. After amplification, the relative expression level
of CD47 was obtained by dividing the amount of CD47
mRNA by the amount of β-actin mRNA in each sample.
Re-analysis for GSE6188
The Gene Expression Omnibus public microarray
database GSE6188 from the National Center for Biotechnical
Information (21) was re-analyzed
by Subio Platform Inc., Tokyo, Japan. Significantly different
(≥2-fold change) probes between cancerous tissues (153 samples) and
normal tissues (104 samples) were extracted. The clinical status of
the original tissue samples was determined in over half of the 48
donors with 2 cancerous tissues and 2 adjacent normal tissues.
Validation of miR-133a expression in
clinical samples
cDNA for miR-133a quantitative real-time RT-PCR was
synthesized from 10 ng of total RNA using the TaqMan microRNA
Reverse Transcription Kit and specific stem-loop reverse
transcription primers (Applied Biosystems) according to the
manufacturer’s protocol. The 15 μl reactions were incubated in a
96-well plate using the following temperature profile: 16°C for 30
min followed by 40°C for 30 min and 85°C for 5 min. PCR was
performed in a LightCycler™ 480 System (Roche). The 20 μl of PCR
mix including the LightCycler 480 Probes Master kit (Roche) was
incubated in a 96-well optical plate at 95°C for 10 min and then
followed by 45 cycles of 95°C for 10 sec and 60°C for 30 sec. The
expression levels of miR-133a were normalized to that of the small
nuclear RNA RNU6B and analyzed using the 2−ΔΔCt
method.
ESCC cell line
The human ESCC cell line TE-8 was kindly provided by
Dr T. Nishihira (Institute of Development, Aging and Cancer, Tohoku
University School of Medicine, Sendai, Japan). TE-8 cells were
maintained in Roswell Park Memorial Institute (RPMI-1640 medium)
(Wako Pure Chemical Industries) supplemented with 10% fetal bovine
serum and antibiotics (100 U/ml penicillin and 100 μg/ml
streptomycin) and cultured in a humidified 5% CO2
incubator at 37°C.
Transfection of miR-133a precursor
(Pre-miR-133a)
Pre-miR miRNA Precursor Molecule mimicking miR-133a
(Pre-miR-133a; Applied Biosystems) or non-specific control miRNA
(Pre-miR Negative Control #1; Applied Biosystems) was transfected
at 30 nmol/l into TE-8 cells using Lipofectamine RNAiMAX
(Invitrogen) according to the manufacturer’s instructions.
Plasmid construction
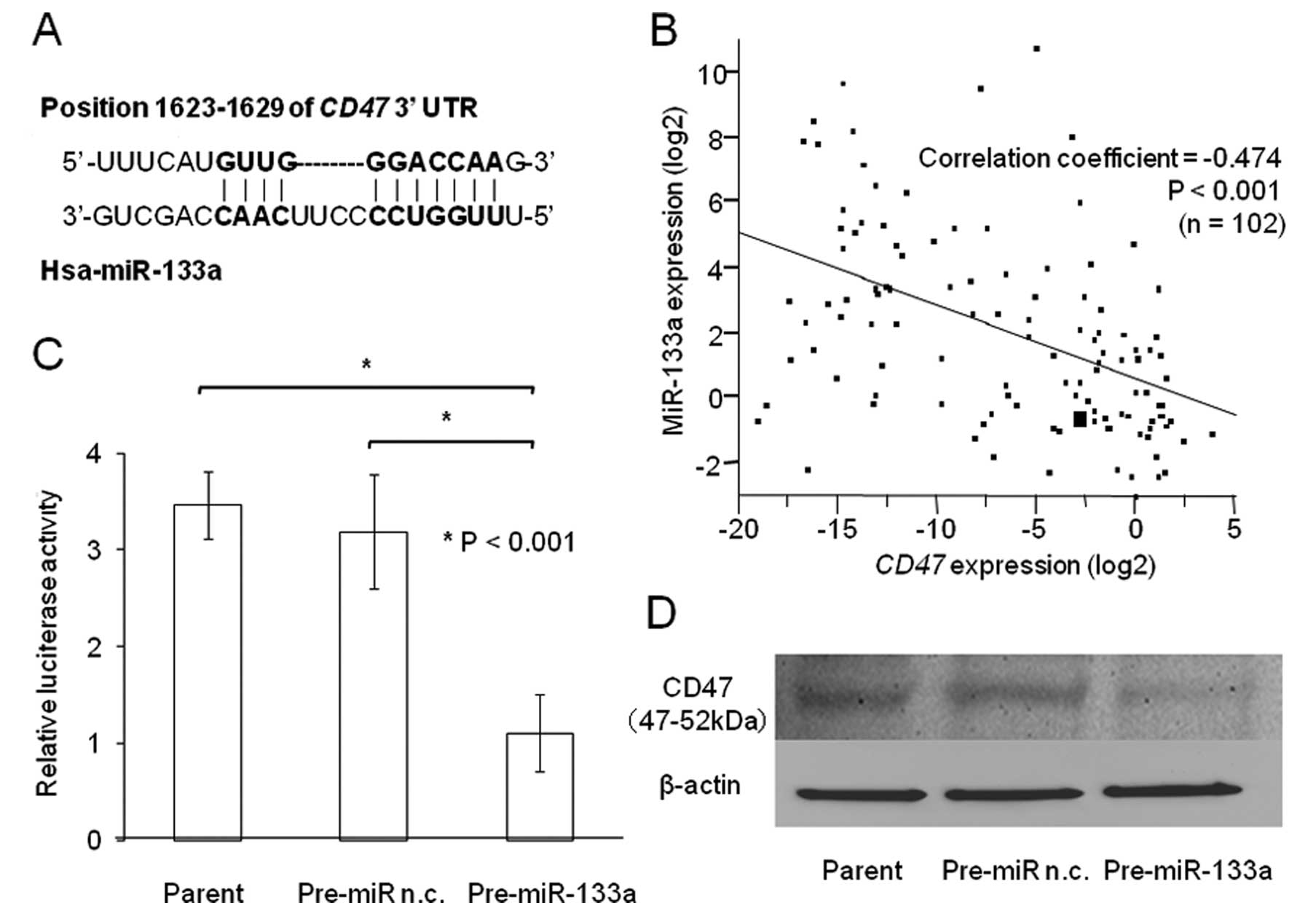
miR-133a target sequences in the 3′-UTR region of
CD47 (Fig. 3A) were predicted using
TargetScan (release 5.1: April 2009) and amplified from the genomic
DNA of normal cells. The amplified fragment was inserted into the
XhoI restriction sites of the dual-luciferase plasmid
pmirGLO vector (Promega) using the In-Fusion® Dry-Down
PCR Cloning kit (Clontech). Plasmid sequences were confirmed by
sequencing using the following primers: sense,
5′-GCAAGATCGCCGTGTAATTCTAG-3′; anti-sense,
5′-AGAGGCCTCAGCAGGTCATA-3′.
Luciferase assay
TE-8 cells were seeded in a 96-well plate and
co-transfected with 0.2 μg of pmirGLO vecter, 100 nmol/l of
Pre-miR-133a and 0.5 μl of Lipofectamine RNAiMAX (Invitrogen) in 50
μl of Opti-MEM Reduced-Serum Medium (Invitrogen). Pre-miR Negative
Control #1 was used as a control. Twenty-four hours following
transfection, the activities of the firefly and Renilla luciferases
in cell lysates were measured using the Dual-Glo®
Luciferase Assay System (Promega) and the Fluoroskan Ascent FL
(Thermo Fischer Scientific). Firefly luciferase activity was
normalized to Renilla luciferase activity. All transfection
experiments were conducted in triplicate.
Protein expression analysis
Western blot analysis was used to confirm the
expression of CD47 in Pre-miR-133a transfected cells. Total protein
was extracted from TE-8 cells 48 h after transfection using
PROPREP™ protein extraction solution (Intron Biotechnology, Inc.).
Total protein (40 μg) was electrophoresed in an Any kD™
Mini-PROTEAN® TGX™ Precast Gel (Bio-Rad Laboratories,
Hercules, CA, USA) and then electrotransferred to a Hybond-enhanced
nitrocellulose membrane (Amersham Pharmacia Biotech) at 200 mA for
180 min at 4°C. The membrane was blocked with 5% skim milk and CD47
protein was detected using CD47 mouse monoclonal antibody (Santa
Cruz Biotechnology, Santa Cruz, CA) diluted 1:100. β-actin mouse
monoclonal antibody (clone AC-74; Sigma) diluted 1:1000 served as a
control. Bands on the membrane were detected using Retiga-4000R and
QCapture Pro 6.0, an enhanced chemiluminescence detection system,
according to the manufacturer’s instructions (QImaging).
In vivo assay
Forty-eight hours following transfection with
Pre-miR-133a or Pre-miR Negative Control #1, 6-week-old female
BALB/c nude mice received subcutaneous injections of
4×106 of TE-8 cells. The tumor volume was determined by
caliper measurements at day 14 and calculated using the formula:
Volume = S × S × L/2, where S is the short length of the tumor in
mm and L is the long length of the tumor in mm.
Statistical analysis
Differences between two groups were estimated using
the Student’s t-test, the χ2 test, and the
repeated-measures ANOVA. Kaplan-Meier curves were generated for
disease-specific and overall survival, and statistical significance
was determined using the log-rank test. Univariate and multivariate
survival analyses were carried out using the Cox proportional
hazards regression model. A probability value of <0.05 was
considered significant. All statistical analyses were performed
using the JMP5.0 software (SAS Institute Inc.).
Results
Association between CD47 expression and
clinicopathological findings in ESCC patients
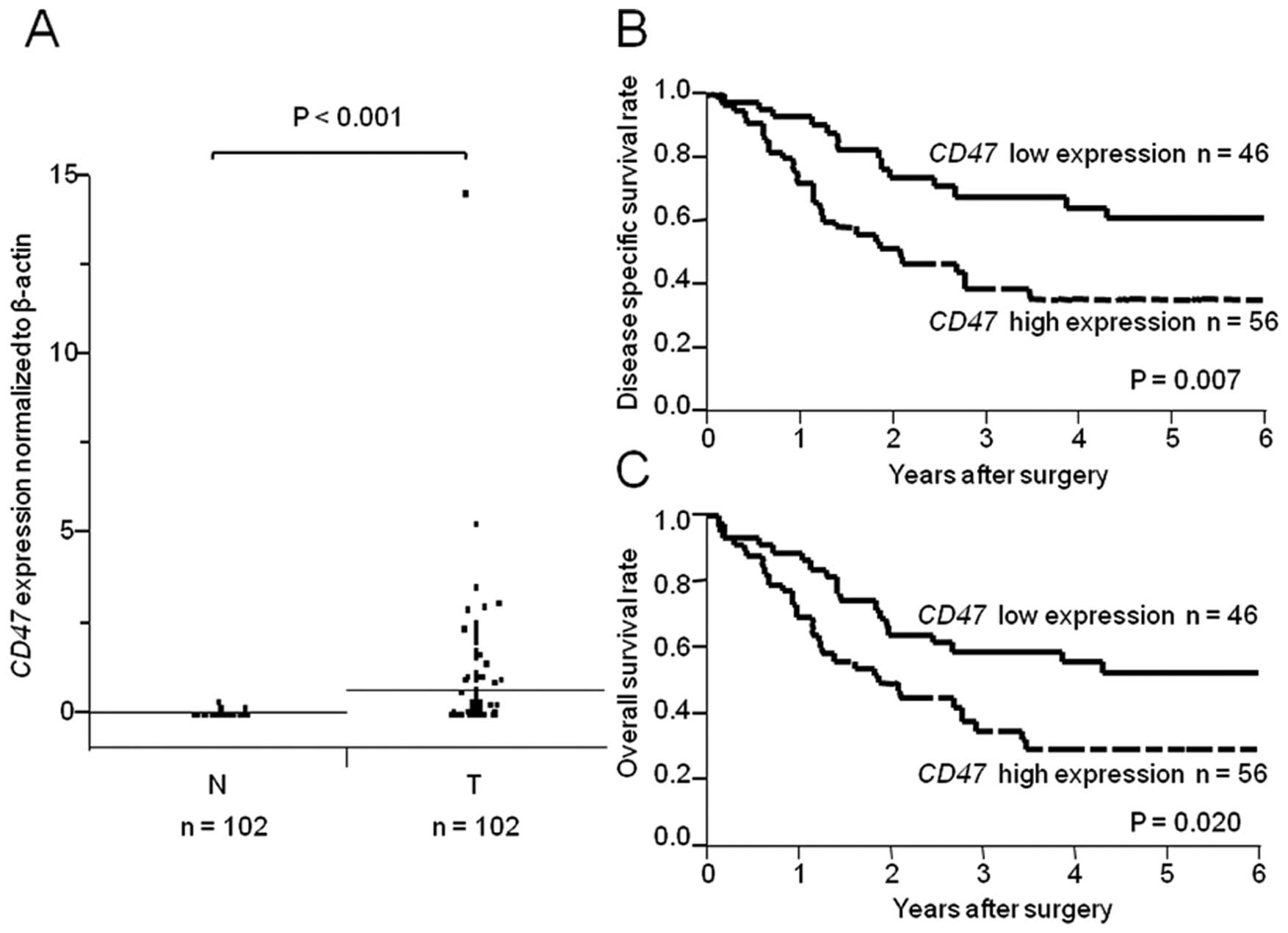
CD47 expression was higher in a subset of
ESCC samples compared with corresponding non-cancerous tissues
(Fig. 1A; P<0.001). We divided
the 102 ESCC patients into two groups according to the level of
CD47 expression in their cancerous tissue. The cut-off point
was the median level of CD47 expression in non-cancerous
tissues. The correlation between CD47 expression and the
clinicopathologic characteristics of patients is shown in Table I. The CD47 high expression
group (n=56) showed more extensive lymph node metastasis than the
low expression group (n=46; P=0.049). However, no significant
differences were observed regarding gender, age, tumor depth,
distant metastasis, lymphatic invasion, venous invasion, or TNM
stage.
 | Table IRelationship between CD47
expression and clinicopathological features. |
Table I
Relationship between CD47
expression and clinicopathological features.
| CD47
expression, n | |
|---|
|
| |
|---|
| Factors | Low (n=46) | High (n=56) | P-value |
|---|
| Gender |
| Male | 40 | 49 | 0.935 |
| Female | 6 | 7 | |
| Age (years) (mean ±
SD) | 65.0±8.0 | 64.8±7.9 | 0.864 |
| Tumor depth |
| T1 | 12 | 7 | 0.248 |
| T2 | 7 | 6 | |
| T3 | 23 | 37 | |
| T4 | 4 | 6 | |
| Lymph node
metastasis |
| N0 | 20 | 14 | 0.049a |
| N1 | 26 | 42 | |
| Distant
metastasis |
| M0 | 41 | 46 | 0.316 |
| M1 | 5 | 10 | |
| Lymphatic
invasion |
| Negative | 40 | 54 | 0.074 |
| Positive | 6 | 2 | |
| Venous
invasion |
| Negative | 34 | 47 | 0.214 |
| Positive | 12 | 9 | |
| Tumor stage |
| I | 8 | 4 | 0.148 |
| II | 17 | 16 | |
| III | 16 | 23 | |
| IV | 5 | 13 | |
Prognostic significance of CD47
expression in ESCC patients
The 5-year disease-specific survival rates of ESCC
patients in the CD47 high expression group were
significantly lower than those in the low CD47 expression
group (high expression, 35.4%; low expression, 60.8%; P=0.007;
Fig. 1B). The 5-year overall
survival rates of ESCC patients in the high expression group were
also significantly lower than those in the low expression group
(high expression, 29.4%; low expression, 52.6%; P=0.020; Fig. 1C). Univariate analysis of
disease-specific survival revealed that the relative level of tumor
depth, lymph node metastasis, distant metastasis, lymphatic
invasion, and CD47 expression were prognostic factors
(Table II). Multivariate analysis
of the five factors found to be significant in univariate analysis
showed that high expression of CD47 (P=0.045), tumor depth
(P=0.042) and lymph node metastasis (P=0.019; Table II) are independent prognostic
factors of poor survival.
 | Table IIResults of univariate and
multivariate analysis of clinicopathological factors affecting
disease-specific survival rates following surgery. |
Table II
Results of univariate and
multivariate analysis of clinicopathological factors affecting
disease-specific survival rates following surgery.
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|---|
| Clinicopathological
variable | RR | 95% CI | P-value | RR | 95% CI | P-value |
|---|
| Gender
(male/female) | 1.092 | 0.673–1.615 | 0.671 | - | - | - |
| Age
(<65/≥65) | 1.065 | 0.7965–1.427 | 0.671 | - | - | - |
| Depth of tumor
invasion (T1/T2,3,4) | 2.019 | 1.278–3.698 | 0.001a | 1.665 | 1.018–3.143 | 0.042a |
| Lymph node
metastasis (N0/N1) | 1.941 | 1.356–2.958 | <0.001a | 1.584 | 1.073–2.488 | 0.019a |
| Distant metastasis
(M0/M1) | 1.662 | 1.153–2.304 | 0.008a | 1.284 | 0.881–1.810 | 0.185 |
| Lymphatic invasion
(negative/positive) | 2.354 | 1.098–9.913 | 0.023a | 1.092 | 0.444–4.810 | 0.870 |
| Venous invasion
(negative/positive) | 1.459 | 0.985–2.369 | 0.060 | - | - | - |
| miR-133a expression
(high/low) | 1.117 | 0.835–1.499 | 0.455 | - | - | - |
| CD47 expression
(low/high) | 1.515 | 1.118–2.097 | 0.007a | 1.371 | 1.007–1.908 | 0.045a |
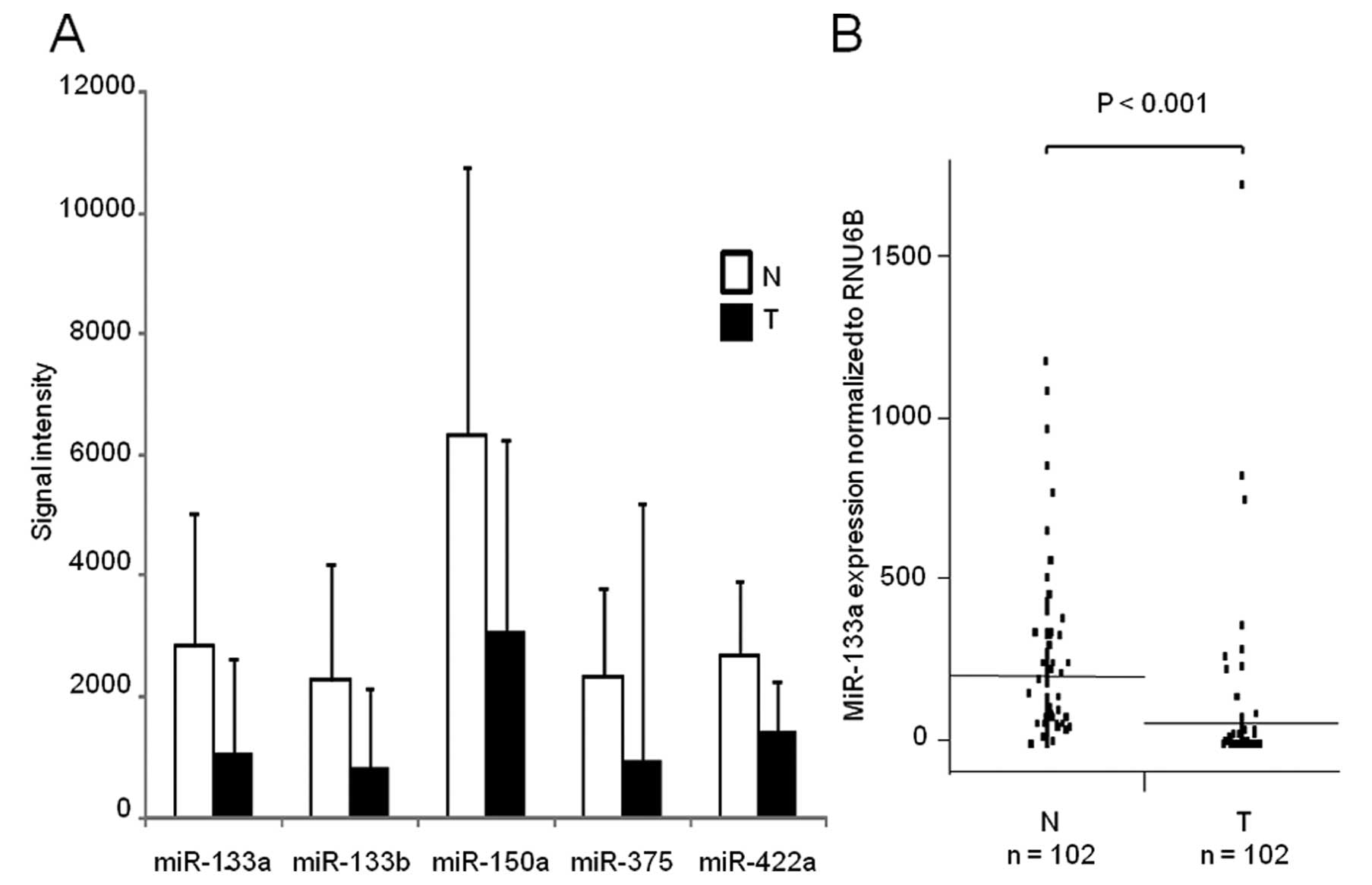
Re-analysis of GSE6188 and miR-133a
expression in ESCC
In search for CD47 regulatory miRNAs
downregulated in ESCC, we found five downregulated miRNAs during
re-analysis of a public microarray database (Fig. 2A). Among them, miR-133a expression
was validated in 102 clinical samples of ESCC and adjacent
non-cancerous tissues. The expression of miR-133a was significantly
lower in cancerous tissues compared to non-cancerous tissues
(P<0.001; Fig. 2B).
miR-133a directly binds the 3′-UTR of
CD47 and regulates its protein expression in ESCC
miR-133a was predicted to be an in silico
regulator of CD47 by TargetScan 5.0 (30) (Fig.
3A). We found an inverse correlation between miR-133a and CD47
expression in 102 clinical samples of ESCC. High levels of miR-133a
were associated with low CD47 expression (P<0.001;
Fig. 3B). The luciferase reporter
assay demonstrated that the luminescence intensity was
significantly decreased in Pre-miR-133a transfectants (Fig. 3C; P<0.001), suggesting that
miR-133a has actual target sites in the 3′-UTR of CD47 mRNA
in ESCC. The transient transfection of Pre-miR-133a also repressed
CD47 protein levels in ESCC (Fig.
3D).
Tumor suppressive activity of miR-133a in
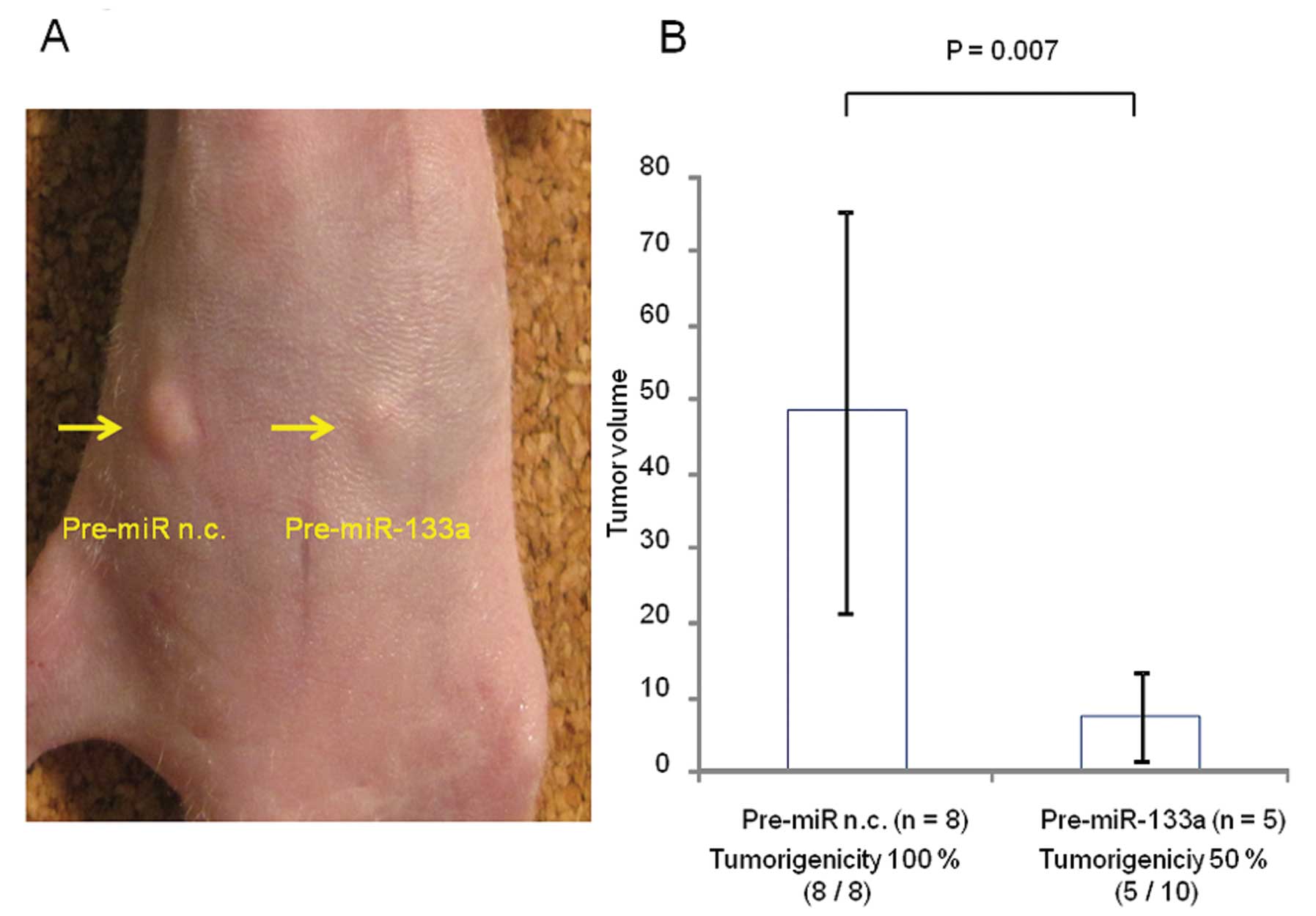
vivo
A mouse xenograft model was used to evaluate the
tumor suppressive activity of miR-133a (Fig. 4A). Pre-miR-133a strongly inhibited
the tumorigenic potential of TE-8 cells in vivo (5 of 10;
50%) compared with the negative control (8 of 8; 100%) (P=0.036,
Fig. 4B). Moreover, Pre-miR-133a
significantly inhibited tumor volume growth on Day 14 after
injection (P=0.007; Fig. 4B).
Discussion
The expression level of miR-133a in primary ESCC was
lower than in corresponding normal esophageal tissues, a finding
which is consistent with the results of a previous microarray
expression study (21). Because
tumor suppressive miRNAs are usually underexpressed in tumors, it
has been suggested that miR-133a functions as a tumor suppressor.
Indeed, this study revealed that miR-133a inhibited tumor growth in
an ESCC xenograft mouse model. Kano et al demonstrated that
transfection of miR-133a inhibits cell proliferation and cell
invasion in ESCC through the regulation of FSCN1, which is
usually increased in tumors compared to normal epithelium (31). FSCN1 overexpression is
significantly associated with poor prognosis in ESCC (32). These in vivo and in
vitro findings suggest that miR-133a contributes to decreases
in cell proliferation. Furthermore, our data revealed that miR-133a
inhibits tumor xenograftment, suggesting that miR-133a could play a
key role in tumorigenesis in ESCC.
Luciferase assays supported the premise of a
specific interaction between miR-133a and the 3′-UTR of
CD47, a protein that inhibits phagocytosis. The negative
modulatory effect of miR-133a was substantiated further by a strong
inverse relationship between the expression levels of CD47
mRNA and miR-133a. The significant reduction of CD47 protein
in an ESCC cell line following Pre-miR-133a transfection suggests
that miR-133a directly regulates CD47 expression. In
contrast, it has been demonstrated that hematopoietic cells from
CD47 knockout mice were rapidly cleared from the bloodstream
by macrophages in syngeneic wild-type mice (7,33).
Chao et al also reported that monoclonal antibody against
CD47 enabled phagocytosis of acute lymphoblastic leukemia
cells by macrophages in vitro and inhibited tumor
engraftment in vivo (13).
These findings suggest that the rapid rejection of xenograft cells
by recipient macrophages could have resulted from the critical role
of CD47 in self protection. These studies support the idea
that miR-133a contributes to anti-tumorigenesis in ESCC through the
regulation of CD47 expression. To the best of our knowledge,
this is the first study describing the clinical significance of
CD47 and its regulation by miR-133a in ESCC.
The expression of CD47 is significantly
correlated with lymph node metastasis, the main cause of death in
most cancer patients. In particular, lymph node metastasis leads to
poor prognosis after surgical resection in ESCC patients (34). However, tumor immunosurveillance is
a well-established mechanism for the regulation of tumor
progression (11), and macrophages
in the lymph node sinus are the front-line endogenic defense
against metastasis (35). Asano
et al showed that macrophages phagocytose dead tumor cells
transported via lymphatic flow and subsequently cross-present tumor
antigens to CD8+ T cells, resulting in activation of the
antitumor immunity system (36).
Therefore, ESCC characterized by high CD47 expression may have the
ability to evade phagocytosis, and after acquiring invasiveness, to
participate in the promotion of lymph node metastasis. Multivariate
Cox proportional hazard regression analysis revealed that
CD47 expression had a significantly worse prognostic impact
(P=0.045) on survival of ESCC patients independent of tumor depth
(P=0.042) and lymph node metastasis (P=0.019). Novel biomarkers
that are capable of precisely identifying high risk ESCC patients
may provide the tools for determining appropriate therapeutic
strategies. Our data indicate that CD47 could be of value as
a novel prognostic marker for the identification of aggressive
ESCC.
Although miRNAs show signs of being a new class of
targets for therapeutic applications (37), the development of effective and safe
approaches for sequence-specific antagonism of miRNAs in
vivo remains a significant scientific and therapeutic challenge
(38,39). On another front, CD47 was
identified as a therapeutic antibody target in acute myeloid
leukemia, non-Hodgkin’s lymphoma, bladder cancer and acute
lymphoblastic leukemia (12,13,40–42).
However, until now, the regulation of CD47 expression is not
well understood. The identification of miR133a-mediated CD47
regulation might provide a promising new therapeutic strategy in
treating ESCC.
In conclusion, our data indicate that CD47 is
an independent prognostic marker that is regulated by miR-133a,
which may function as a tumor suppressor in ESCC. This correlation
could provide new insight into the mechanism of cancer progression
and a promising candidate for a novel therapeutic target in
ESCC.
Acknowledgements
We thank Ms. Masako Shin for her excellent technical
assistance.
References
|
1
|
Enzinger PC and Mayer RJ: Esophageal
cancer. N Engl J Med. 349:2241–2252. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Parkin DM, Bray F, Ferlay J and Pisani P:
Global cancer statistics, 2002. CA Cancer J Clin. 55:74–108. 2005.
View Article : Google Scholar
|
|
3
|
Malthaner RA, Wong RK, Rumble RB and Zuraw
L: Neoadjuvant or adjuvant therapy for resectable esophageal
cancer: a systematic review and meta-analysis. BMC Med. 2:352004.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Gebski V, Burmeister B, Smithers BM, Foo
K, Zalcberg J and Simes J: Survival benefits from neoadjuvant
chemoradiotherapy or chemotherapy in oesophageal carcinoma: a
meta-analysis. Lancet Oncol. 8:226–234. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Tepper J, Krasna MJ, Niedzwiecki D, et al:
Phase III trial of trimodality therapy with cisplatin,
fluorouracil, radiotherapy, and surgery compared with surgery alone
for esophageal cancer: CALGB 9781. J Clin Oncol. 26:1086–1092.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Shinohara M, Ohyama N, Murata Y, et al:
CD47 regulation of epithelial cell spreading and migration, and its
signal transduction. Cancer Sci. 97:889–895. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Oldenborg PA, Zheleznyak A, Fang YF,
Lagenaur CF, Gresham HD and Lindberg FP: Role of CD47 as a marker
of self on red blood cells. Science. 288:2051–2054. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Okazawa H, Motegi S, Ohyama N, et al:
Negative regulation of phagocytosis in macrophages by the
CD47-SHPS-1 system. J Immunol. 174:2004–2011. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
van den Berg TK and van der Schoot CE:
Innate immune ‘self’ recognition: a role for CD47-SIRPalpha
interactions in hematopoietic stem cell transplantation. Trends
Immunol. 29:203–206. 2008.
|
|
10
|
Oldenborg PA, Gresham HD, Chen Y, Izui S
and Lindberg FP: Lethal autoimmune hemolytic anemia in
CD47-deficient nonobese diabetic (NOD) mice. Blood. 99:3500–3504.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Jaiswal S, Chao MP, Majeti R and Weissman
IL: Macrophages as mediators of tumor immunosurveillance. Trends
Immunol. 31:212–219. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Majeti R, Chao MP, Alizadeh AA, et al:
CD47 is an adverse prognostic factor and therapeutic antibody
target on human acute myeloid leukemia stem cells. Cell.
138:286–299. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Chao MP, Alizadeh AA, Tang C, et al:
Therapeutic antibody targeting of CD47 eliminates human acute
lymphoblastic leukemia. Cancer Res. 71:1374–1384. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Nagahara M, Mimori K, Kataoka A, et al:
Correlated expression of CD47 and SIRPA in bone marrow and in
peripheral blood predicts recurrence in breast cancer patients.
Clin Cancer Res. 16:4625–4635. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Junker A, Krumbholz M, Eisele S, et al:
MicroRNA profiling of multiple sclerosis lesions identifies
modulators of the regulatory protein CD47. Brain. 132:3342–3352.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Valencia-Sanchez MA, Liu J, Hannon GJ and
Parker R: Control of translation and mRNA degradation by miRNAs and
siRNAs. Genes Dev. 20:515–524. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Bartel DP: MicroRNAs: genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Bartel DP: MicroRNAs: target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lu J, Getz G, Miska EA, et al: MicroRNA
expression profiles classify human cancers. Nature. 435:834–838.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Volinia S, Calin GA, Liu CG, et al: A
microRNA expression signature of human solid tumors defines cancer
gene targets. Proc Natl Acad Sci USA. 103:2257–2261. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Guo Y, Chen Z, Zhang L, et al: Distinctive
microRNA profiles relating to patient survival in esophageal
squamous cell carcinoma. Cancer Res. 68:26–33. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Childs G, Fazzari M, Kung G, et al:
Low-level expression of microRNAs let-7d and miR-205 are prognostic
markers of head and neck squamous cell carcinoma. Am J Pathol.
174:736–745. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wong TS, Liu XB, Wong BY, Ng RW, Yuen AP
and Wei WI: Mature miR-184 as potential oncogenic microRNA of
squamous cell carcinoma of tongue. Clin Cancer Res. 14:2588–2592.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Nohata N, Hanazawa T, Kikkawa N, et al:
Caveolin-1 mediates tumor cell migration and invasion and its
regulation by miR-133a in head and neck squamous cell carcinoma.
Int J Oncol. 38:209–217. 2011.PubMed/NCBI
|
|
25
|
Chiyomaru T, Enokida H, Tatarano S, et al:
miR-145 and miR-133a function as tumour suppressors and directly
regulate FSCN1 expression in bladder cancer. Br J Cancer.
102:883–891. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Song T, Xia W, Shao N, et al: Differential
miRNA expression profiles in bladder urothelial carcinomas. Asian
Pac J Cancer Prev. 11:905–911. 2010.PubMed/NCBI
|
|
27
|
Bandres E, Cubedo E, Agirre X, et al:
Identification by real-time PCR of 13 mature microRNAs
differentially expressed in colorectal cancer and non-tumoral
tissues. Mol Cancer. 5:292006. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Missiaglia E, Shepherd CJ, Patel S, et al:
MicroRNA-206 expression levels correlate with clinical behaviour of
rhabdomyosarcomas. Br J Cancer. 102:1769–1777. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Rao PK, Missiaglia E, Shields L, et al:
Distinct roles for miR-1 and miR-133a in the proliferation and
differentiation of rhabdomyosarcoma cells. FASEB J. 24:3427–3437.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Lewis BP, Burge CB and Bartel DP:
Conserved seed pairing, often flanked by adenosines, indicates that
thousands of human genes are microRNA targets. Cell. 120:15–20.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kano M, Seki N, Kikkawa N, et al: miR-145,
miR-133a and miR-133b: Tumor suppressive miRNAs target FSCN1 in
esophageal squamous cell carcinoma. Int J Cancer. 127:2804–2814.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Hashimoto Y, Ito T, Inoue H, et al:
Prognostic significance of fascin overexpression in human
esophageal squamous cell carcinoma. Clin Cancer Res. 11:2597–2605.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wang H, Madariaga ML, Wang S, Van Rooijen
N, Oldenborg PA and Yang YG: Lack of CD47 on non-hematopoietic
cells induces split macrophage tolerance to CD47null cells. Proc
Natl Acad Sci USA. 104:13744–13749. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Kuwano H, Nakajima M, Miyazaki T and Kato
H: Distinctive clinicopathological characteristics in esophageal
squamous cell carcinoma. Ann Thorac Cardiovasc Surg. 9:6–13.
2003.PubMed/NCBI
|
|
35
|
Nagata H, Arai T, Soejima Y, Suzuki H,
Ishii H and Hibi T: Limited capability of regional lymph nodes to
eradicate metastatic cancer cells. Cancer Res. 64:8239–8248. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Asano K, Nabeyama A, Miyake Y, et al:
CD169-positive macrophages dominate antitumor immunity by
crosspresenting dead cell-associated antigens. Immunity. 34:85–95.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Soifer HS, Rossi JJ and Saetrom P:
MicroRNAs in disease and potential therapeutic applications. Mol
Ther. 15:2070–2079. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Elmen J, Lindow M, Schutz S, Lawrence M,
et al: LNA-mediated microRNA silencing in non-human primates.
Nature. 452:896–899. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Silvestri P, Di Russo C, Rigattieri S, et
al: MicroRNAs and ischemic heart disease: towards a better
comprehension of pathogenesis, new diagnostic tools and new
therapeutic targets. Recent Pat Cardiovasc Drug Discov. 4:109–118.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Chan KS, Espinosa I, Chao M, et al:
Identification, molecular characterization, clinical prognosis, and
therapeutic targeting of human bladder tumor-initiating cells. Proc
Natl Acad Sci USA. 106:14016–14021. 2009. View Article : Google Scholar
|
|
41
|
Chao MP, Alizadeh AA, Tang C, et al:
Anti-CD47 antibody synergizes with rituximab to promote
phagocytosis and eradicate non-Hodgkin lymphoma. Cell. 142:699–713.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Chao MP, Alizadeh AA, Tang C, et al:
Therapeutic antibody targeting of CD47 eliminates human acute
lymphoblastic leukemia. Cancer Res. 71:1374–1384. 2010. View Article : Google Scholar : PubMed/NCBI
|