Introduction
Colorectal cancer is common in Brazil. In the year
2012, 14,180 new cases of colon and rectum cancer were expected to
occur in men and 15,960 in women. These values correspond to an
estimated risk of 15 new cases per 100,000 men and 16 cases per
100,000 women (1).
Excluding non-melanoma skin tumors, colon and rectum
cancer is the second most common cancer among men in Southeast
Brazil (22/100,000) and third in South (18/100,000) and Midwest
(14/100,000) Brazil (1). In North
Brazil (4/100,000) this cancer ranks fourth; in Northeast Brazil
(5/100,000), fifth. Among women, it is the second most common
cancer in Southeast (23/100,000) and South Brazil (20/100,000), the
third in Midwest (15/100,000) and Northeast Brazil (7/100,000), and
sixth in the North (5/100,000) (1).
Familial adenomatous polyposis (FAP) is one of the
most clearly defined and well understood inherited colorectal
cancer syndrome. It is an autosomal dominant disorder that
typically presents in the form of colorectal cancer in young adults
secondary to extensive adenomatous polyposis present in the colon
(2).
The adenomatous polyposis coli (APC) gene is
on chromosome 5q21 and displays alternative splicing in multiple
coding and noncoding regions of the DNA sequence, and the primary
transcript has 15 exons. The APC gene has 8,532 base pairs
corresponding to 2,844 amino acids, resulting in a 311.8-kDa
protein. Exon 15 has the largest extension, making up more than
three quarters of the coding region (3).
Approximately 737 APC gene mutations,
including 332 germline and 402 somatic have been identified.
APC germline mutations are responsible for the occurrence of
FAP, and somatic mutations have been associated with malignant
transformation of adenomas (4).
Almost all mutations lead to truncation of the APC protein either
by nonsense (30%) or by frameshift mutations (68%). The majority of
mutations occur within the first half of the coding sequence. In an
American study, in which 1,591 patients were studied, of the 431
pathogenic or likely pathogenic mutations, frameshift, nonsense,
splice sites and large deletion or duplication mutations
represented 43, 42, 9 and 6% of cases, respectively (5).
APC germline mutations are predominate at the
5′ end of the gene, while somatic mutations mainly occur in the
region called the mutation cluster region (MCR) between codons
1,284 and 1,580 of the APC gene. In germline mutations, two
hot spot codons have been identified; one at position 1,061 and the
second at position 1,309. In somatic mutations, two hot spots seem
to occur at position 1,309 and 1,450 (3).
Several studies have attempted to correlate specific
APC mutations with clinical phenotypes. Mutations between
codons 169 to 1,578 have been generally associated with the classic
form of FAP. Mutations between codons 1,445 and 1,578 have been
associated with desmoid tumors, whereas mutations between codons
279 to 1,309 have been correlated with the development of duodenal
polyposis (6).
Based on the findings in the literature, the
objective of the present study was to detect APC germline
mutations that affect families followed up at the Oncology Clinic
of the University of Campinas (Unicamp) and to compare the
identified mutations with clinical variables.
Materials and methods
We recruited 20 nonrelative patients at the Oncology
Service in the ‘Gastrocentro’ of the Faculty of Medical Sciences of
Unicamp. The present study included families that had two or more
successive generations affected by FAP (>100 polyps); no
polyposis colorectal cancer was present. The project was approved
by the university ethics committee (#874/2008). All patients and/or
their guardians signed an informed consent form.
Clinical variables
The clinical variables analyzed in our samples
included gender (male/female), age at diagnosis (≤41 or >41
years), smoking habits (passive smoking, smoker, non-smoker), TNM
stage (I + II vs. III + IV), Astler-Coller stage (B1 + B2 vs. C1 +
C2), degree of differentiation of adenocarcinoma (moderately
differentiated, poorly differentiated, well-differentiated).
All of the variables were evaluated by medical
specialists including special considerations to TNM stage (tumor,
lymph node, metastasis), Astler-Coller stage and degree of
differentiation of the adenocarcinoma and were evaluated taking
into account previously literature (6–10).
DNA extraction
Genomic DNA was obtained by direct extraction from
lymphocytes of peripheral blood according to standard procedures
(11). DNA samples were quantified
using the NanoVue® v1.7.2 spectrophotometer (GE
Healthcare, Chicago, IL, USA). For all analyses performed, 50 ng/μl
was used to improve the polymerase chain reaction (PCR)
technique.
DNA sequencing and analysis
To identify APC mutations, DNA fragments
containing the entire coding region and intron-exon boundaries of
the APC gene were amplified, using PCR conditions as
published by Miyoshi et al(12), Nagase et al(13) and Gómez-Fernández et
al(14), with primers as listed
in Table I. The precise gradients
for temperature and buffers providing the optimal temperature for
each fragment were determined experimentally. The PCR products
indicating heterozygosity were sequenced using the Applied
Biosystems (ABI) Prism BigDye Terminator v3.1 cycle sequencing kit
and ABI 3500XL DNA sequencer (PE Applied Biosystems, Foster City,
CA, USA), using identical conditions as previously published
(12–14). The DNA sequence was analyzed using
GeneMapper software (Applied Biosystems) or Fragment Profiler (GE
Healthcare Biosciences, Piscataway, NJ, USA).
 | Table IDescription of the oligonucleotides
used for the analysis of the APC gene. |
Table I
Description of the oligonucleotides
used for the analysis of the APC gene.
| Nucleotide
nomenclature | Sequences |
|---|
| APC_EX1_F |
5′-AACCTTATAggTCCAAgggTAg-3′ |
| APC_EX1_R |
5′-ACCTCAAgTTTACAAgAgggAA-3′ |
| APC_EX2_F |
5′-AAATACAgAATCATgTCTTgAAgT-3′ |
| APC_EX2_R |
5′-ACACCTAAAgATgACAATTTgAg-3′ |
| APC_EX3_F |
5′-gACCCAAgTggACTTTTCAgg-3′ |
| APC_EX3_R |
5′-ACAATAAACTggAgTACACAAgg-3′ |
| APC_EX4_F |
5′-gAgAAgTTTgCAATAACAACTgATg-3′ |
| APC_EX4_R |
5′-TTATCCTgAATTTTAATggATTACCT-3′ |
| APC_EX5_F |
5′-AACCTCACTCTAACTggACCAA-3′ |
| APC_EX5_R |
5′-AACAgAgCTgTAATTCATTTTATTCC-3′ |
| APC_EX6_F |
5′-ggTAgCCATAgTATgATTATTTCT-3′ |
| APC_EX6_R |
5′-CTACCTATTTTTATACCCACAAAC-3′ |
| APC_EX7_F |
5′-AAgAAAgCCTACACCATTTTTgC-3′ |
| APC_EX7_R |
5′-gATCATTCTTAgAACCATCTTgC-3′ |
| APC_EX8_F |
5′-gACACTTCATTTggAgTACCTTAACA-3′ |
| APC_EX8_R |
5′-ggCATTAgTgACCAgggTTT-3′ |
| APC_EX9_F |
5′-AgTCgTAATTTTgTTTCTAAACTC-3′ |
| APC_EX9_R |
5′-TTTgAAACATgCACTACgAT-3′ |
| APC_EX10_F |
5′-TTgCTCTTCAAATAACAAAgCAT-3′ |
| APC_EX10_R |
5′-TCCACCAgTAATTgTCTATgTCA-3′ |
| APC_EX11_F |
5′-gATgATTgTCTTTTTCCTCTTgC-3′ |
| APC_EX11_R |
5′-CTgAgCTATCTTAAgAAATACATg-3′ |
| APC_EX12_F |
5′-TgACAAAggAAgAACAgATAgCA-3′ |
| APC_EX12_R |
5′-gCAgTgAgCTgAgATTgCAC-3′ |
| APC_EX13_F |
5′-TTTCTATTCTTACTgCTAgCATT-3′ |
| APC_EX13_R |
5′-ATACACAggTAAgAAATTAggA-3′ |
| APC_EX14_F |
5′-AgggACgggCAATAggATAg-3′ |
| APC_EX14_R |
5′-ggTCTTTTTgAgAgTATgAATTCTg-3′ |
| APC_EX15A_F |
5′-TTgTTACTgCATACACATTg-3′ |
| APC_EX15A_R |
5′-CAAATATggTgAAAggACA-3′ |
| APC_EX15B_F |
5′-CCCTAgAAgCAgAATTAg-3′ |
| APC_EX15B_R |
5′-TTCTTCTAAgTgCATTTC-3′ |
| APC_EX15C_F |
5′-CATggAAgAAgTgTCAgC-3′ |
| APC_EX15C_R |
5′-TTCTATTATgTgTTTgggTC-3′ |
| APC_EX15D_F |
5′-CACAgAATgAAAgATggg-3′ |
| APC_EX15D_R |
5′-gAAggTgTggACgTATTC-3′ |
| APC_EX15E_F |
5′-gAAACgTCATgTggATCAgC-3′ |
| APC_EX15E_R |
5′-TggCAATCgAACgACTCTC-3′ |
| APC_EX15F_F |
5′-CCTAgAACCAAATCCAgCAgAC-3′ |
| APC_EX15F_R |
5′-gTTggCATggCAgAAATAATAC-3′ |
| APC_EX15G_F |
5′-AgATgCTTgCTggACCTg-3′ |
| APC_EX15G_R |
5′-TTgCCACggAAAgTACTC-3′ |
| APC_EX15H_F |
5′-TCTTgCAgAATgCATTAATT-3′ |
Statistical analysis
Statistical analyses were performed using the
Statistical Package for the Social Sciences (SPSS) v.17.0 from
SPSS, Inc., Chicago, IL, USA (http://www.spss.com) by Fisher’s exact test and
χ2 test, considering α=0.05. To improve the data
presentation, the odds ratio was calculated to variables to
demonstrate the association between the clinical variables, TNM and
APC germline mutation identified.
Results
In the descriptive analysis, the average age at
diagnosis of the patients was 42.85 years (±6.892), and the age
range was from 29 to 55 years. Of the 20 patients, 18 (90%) were
Caucasian and 2 (10%) were not Caucasian; 14 (70%) were females and
6 (30%) were males. The clinical data are summarized in Table II. The frequency of cases for each
stage according to the TNM system was 2 (10%), 6 (30%), 6 (30%) and
6 (30%), respectively, for stage I, II, III and IV. The frequency
of cases for each stage according to the Astler-Coller system was 2
(10.5%), 5 (26.4%), 2 (10.5%) and 10 (52.6%), respectively, for
stages B1, B2, C1 and C2. A smoking habit was observed in 8
patients, 2 (10%) were occasional smokers and 6 (30%) were smokers;
the remaining patients (60%) were non-smokers. Two patients (10%)
had well-differentiated adenocarcinoma, 15 (75%) had moderately
differentiated and 3 (15%) had poorly differentiated
adenocarcinoma.
 | Table IIThe familial adenomatous polyposis
patients relating to gender, race, age at diagnosis, staging (TNM
and Astler-Coller), smoking habit, degree of differentiation of
adenocarcinoma and APC gene genotype. |
Table II
The familial adenomatous polyposis
patients relating to gender, race, age at diagnosis, staging (TNM
and Astler-Coller), smoking habit, degree of differentiation of
adenocarcinoma and APC gene genotype.
| Patient | Gender | Race | Age at diagnosis
(years) | Staging | Smoking habit | Degree of
differentiation in adenocarcinoma | Genotype | Mutation
deleterious/not deleterious | Extra-colonic
manifestations |
|---|
|
|---|
| TNM | Astler-Coller |
|---|
| 1 | F | C | 37 | IV | C1 | NS | MD | Glu1309X | D | Small intestine and
duodenal polyps |
| 2 | F | C | 47 | III | B2 | S | MD | Ser 932 X | D | Duodenal
polyps |
| 3 | F | NC | 31 | IV | C2 | NS | MD | Tyr935X | D | Gastric polyps |
| 4 | F | C | 40 | II | C2 | S | MD | Arg657Arga/c.3927_3931 delAAAGA | ND/D | Small intestine
polyps |
| 5 | F | C | 45 | IV | B2 | S | MD | - | - | - |
| 6 | M | C | 41 | II | B1 | S | MD | Ile606Ilea | ND | - |
| 7 | M | C | 36 | I | C2 | NS | PD | Gln1291X | D | Small intestine and
duodenal polyps |
| 8 | F | C | 29 | III | C2 | NS | MD | Gly2502Ser | D | Gastric polyps |
| 9 | M | C | 47 | II | C2 | S | MD | Glu1317Gln | D | Osteoma jaw |
| 10 | F | C | 41 | III | B2 | NS | MD | Leu629X | D | Duodenal
polyps |
| 11 | F | C | 52 | II | B2 | NS | PD | Asn1037Asna/Tyr935X | ND/D | - |
| 12 | F | NC | 40 | I | C2 | NS | WD | Thr934Thra | ND | - |
| 13 | F | C | 44 | III | C1 | NS | MD |
c.3183_3186delACAA | D | Small intestine
polyps |
| 14 | M | C | 36 | IV | B2 | NS | MD |
c.3183_3187delACAAA | D | Small intestine
polyps |
| 15 | F | C | 55 | IV | C2 | S | MD | Arg 876X | D | - |
| 16 | M | C | 49 | II | B1 | PS | MD | Lys939Lysa/Tyr951Tyra | ND/ND | Gastric polyps |
| 17 | F | C | 50 | II | NR | NS | WD | Glu892X | D | - |
| 18 | F | C | 41 | III | C2 | NS | PD | Gly974Glya/c.3927_3931delAAAGA | ND/D | - |
| 19 | F | C | 47 | III | C2 | NS | MD | Lys1454Glu | D | - |
| 20 | M | C | 49 | IV | C2 | PS | MD | Leu1564X | D | Duodenal
polyps |
For the determined mutant alleles, 16 (40%) were
deleterious and 7 (17.5%) were not deleterious. Associations were
analyzed by correlating the TNM stage with the clinical variables,
and the data are shown in Table
III. The same associations were analyzed between Astler-Coller
stage and the clinical variables as described in the Table IV.
 | Table IIIAssociation of clinical variables of
colorectal cancer according to TNM stage. |
Table III
Association of clinical variables of
colorectal cancer according to TNM stage.
| TNM stage | | | | |
|---|
|
| | | | |
|---|
| Variable | I and II n (%) | III and IV n
(%) | Total | P-value | OR | CI (5–95%) |
|---|
| Gender | | | | | | |
| Female | 4 (28.6) | 10 (71.4) | 14 | 0.274 | 0.219 | 0.014–2.242 |
| Male | 4 (66.7) | 2 (33.3) | 6 | | 1 | - |
| Race | | | | | | |
| Caucasian | 7 (38.9) | 11 (61.1) | 18 | 1 | 0.615 | 0.007–57.02 |
| Not Caucasian | 1 (50) | 1 (50) | 2 | | 1 | - |
| Presence of
deleterious allele | | | | | | |
| Presence | 5 (31.3) | 11 (68.8) | 16 | 0.048 | 0.086 |
0.001–0.984 |
| Absence | 6 (85.7) | 1 (14.3) | 7 | | 1 | - |
| Degree of
differentiation of adenocarcinoma | | | | | | |
| WD | 2 (100) | - | 2 | - | - | - |
| MD | 4 (26.7) | 11 (73.3) | 15 | 0.116 | 0.205 | 0.001–1.457 |
| PD | 2 (66.7) | 1 (33.3) | 3 | 0.688 | 3.424 | 0.149–235.6 |
| Age at diagnosis
(years) | | | | | | |
| ≤41 | 4 (40) | 6 (60) | 10 | 1 | 1 | 0.119–8.417 |
| >41 | 4 (40) | 6 (60) | 10 | | 1 | - |
| Astler-Coller
stage | | | | | | |
| B1 and B2 | 3 (42.9) | 4 (57.1) | 7 | 1 | 1.468 | 0.142–14.66 |
| C1 and C2 | 4 (33.3) | 8 (66.7) | 12 | | 1 | - |
| Smoking habit | | | | | | |
| NS | 4 (33.3) | 8 (66.7) | 12 | 0.777 | 0.518 | 0.056–4.451 |
| S and PS | 4 (50) | 4 (50) | 8 | | 1 | - |
 | Table IVAssociation of colorectal cancer
clinical variables according to Astler-Coller stage. |
Table IV
Association of colorectal cancer
clinical variables according to Astler-Coller stage.
| Astler-Coller | | | | |
|---|
|
| | | | |
|---|
| Variable | B1 and B2
(no.) | C1 and C2
(no.) | Total | P-value | OR | CI (5–95%) |
|---|
| Gender | | | | | | |
| Female | 4 | 9 | 13 | 0.617 | 0.465 | 0.04–5.09 |
| Male | 3 | 3 | 6 | | 1 | - |
| Race | | | | | | |
| Caucasian | 7 | 10 | 17 | 0.509 | - | - |
| Not Caucasian | 0 | 2 | 2 | | - | - |
| Presence of
deleterious allele | | | | | | |
| Presence | 5 | 10 | 15 | 0.376 | 0.393 |
0.04–3.36 |
| Absence | 4 | 3 | 7 | | 1 | - |
| Degree of
differentiation of adenocarcinoma | | | | | | |
| WD | 0 | 1 | 1 | - | - | - |
| MD | 6 | 9 | 15 | 0.607 | 1.933 | 0.12–122.2 |
| PD | 1 | 2 | 3 | 0.841 | 0.841 | 0.01–19.64 |
| Age at diagnosis
(years) | | | | | | |
| ≤41 | 3 | 7 | 10 | 0.650 | 0.554 | 0.05–5.03 |
| >41 | 4 | 5 | 9 | | 1 | - |
| TNM stage | | | | | | |
| I and II | 3 | 4 | 7 | 1 | 0.554 | 0.05–5.03 |
| III and IV | 4 | 8 | 12 | | 1 | - |
| Smoking habit | | | | | | |
| NS | 3 | 8 | 11 | 0.377 | 0.396 | 0.04–3.67 |
| S and PS | 4 | 4 | 8 | | 1 | - |
For the deleterious mutations detected, we found a
prevalence of nonsense mutations, with 9 (45%) mutant alleles. In 4
(20%) patients small deletions were noted, while 3 (15%) patients
had missense mutations, and 3 (15%) patients had only neutral
polymorphisms and 1 (5%)patient had no mutations found in the
exons. In 60% of patients, extra-colonic manifestations were
present; the most common being gastric polyps, duodenal and in the
small bowel (Table II).
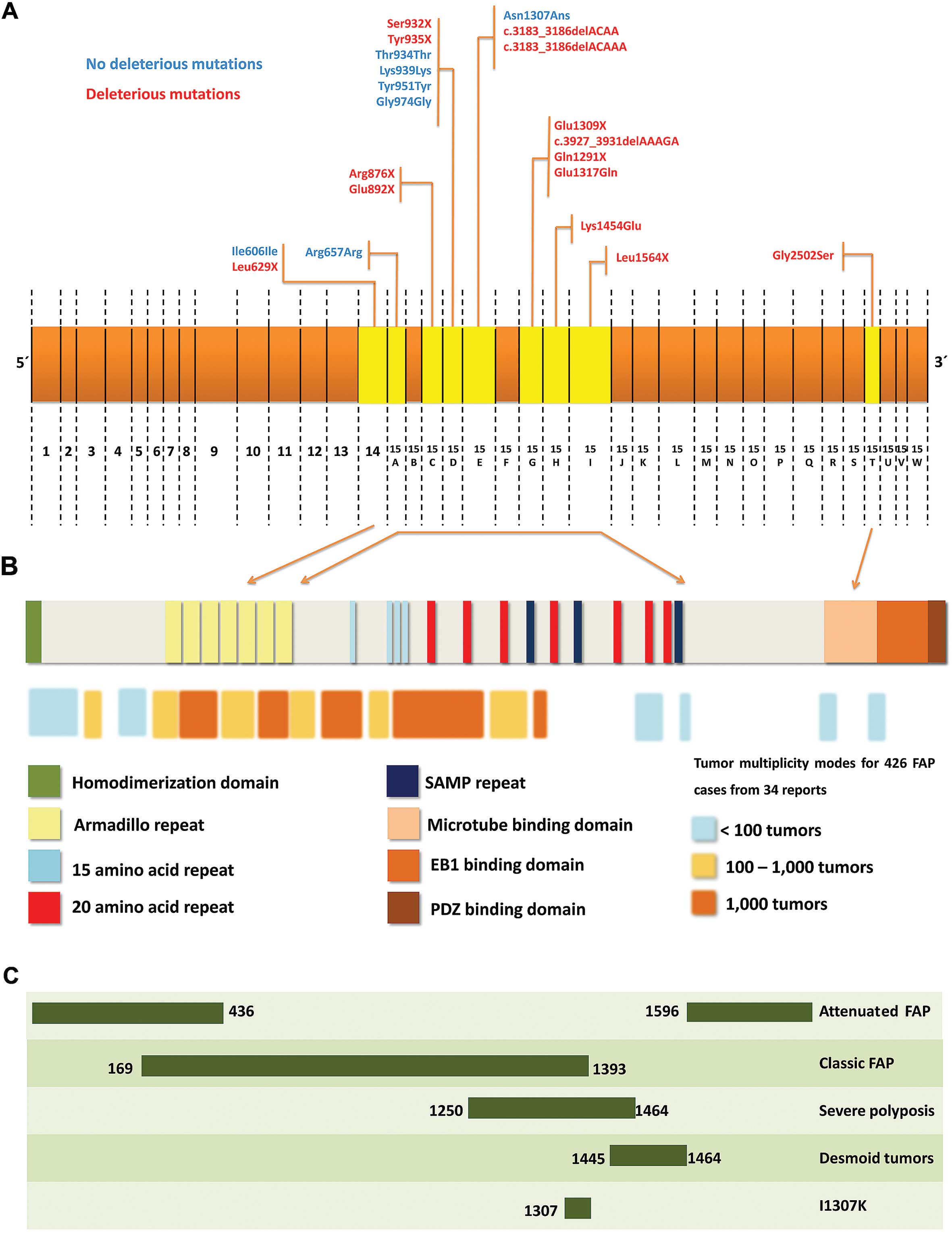
In Figs. 1 and
2, the APC gene and all
mutations identified were described in details. In the same
figures, the protein structure is described, considering the
principal mutation sites and their association with FAP.
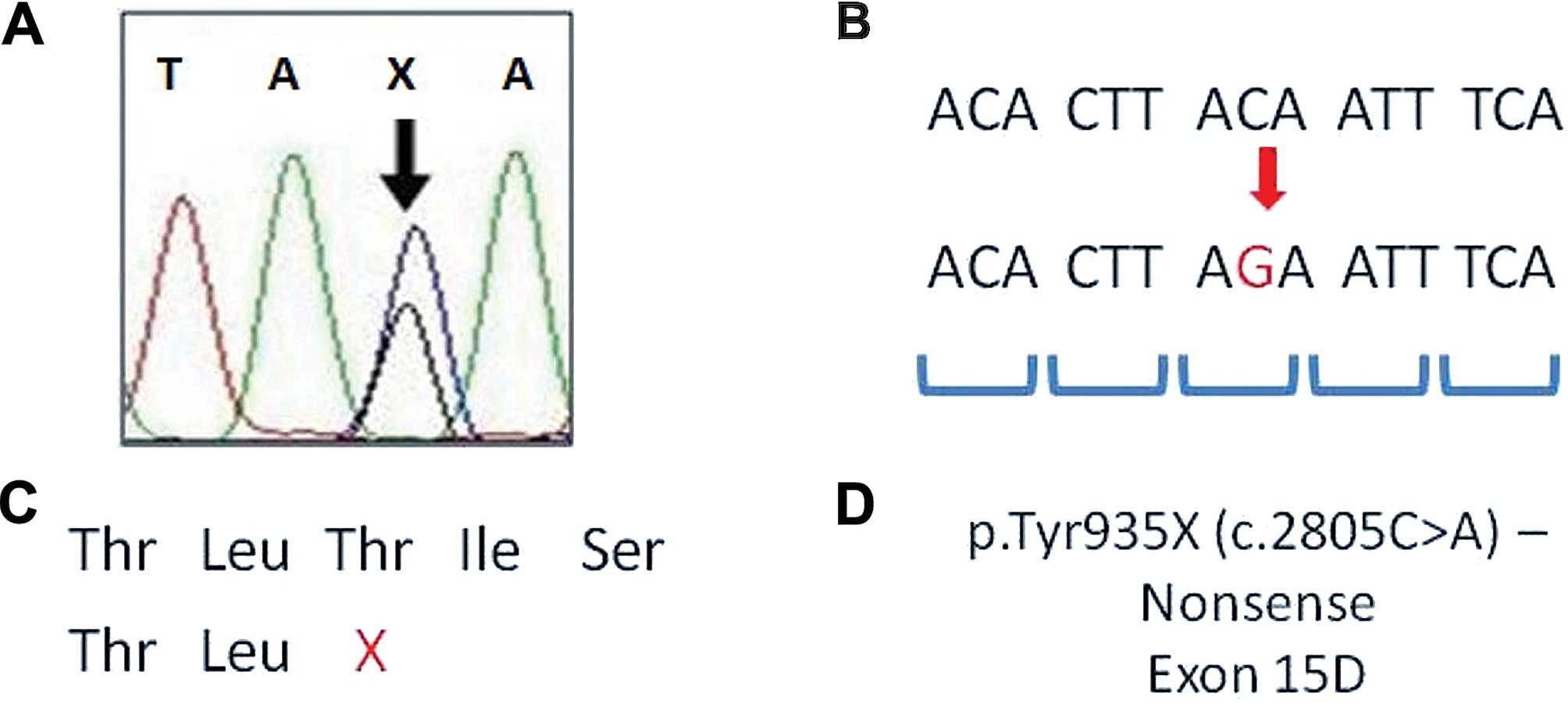 | Figure 1Representation of adenomatous
polyposis coli (APC) mutation analysis. (A) DNA sequencing
electropherogram of p.Tyr935X of APC gene; X, stop codon
mutation. (B) DNA sequence with an alteration (red sequence -
change of C to G, stop codon mutation); codons are shown in blue.
(C) Amino acid sequence of protein and the stop codon in the APC
partial amino acid sequence; X, stop codon in Thr amino acid. (D)
Mutation description - identification, type of mutation,
localization of mutation in APC gene. Thr, threonine; Tyr,
tyrosine; A, adenine, T, thymine; C, cytosine; G, guanine. Fig. 1
was adapted from previous studies (15–17). |
Associations between the clinical variables and the
identified APC germline mutations could not be calculated as
the sample size was small and some of the mutations were not
deleterious.
Discussion
The high molecular heterogeneity in the APC
gene was consistent with other studies in FAP patients (12,18).
Mutations c.3927_3931delAAAGA and pTyr935X were found in 2
patients. The c.3927_3931delAAAGA mutation occurs in exon 15 and
leads to formation of a stop codon at position 1,312. It is the
most frequent mutation in the APC gene. Its frequency varies
from 0% in southwest Spain to 2.4% in the Australian population, 5%
in the Dutch population, 7% in the Israeli population, and up to
16% in Italian FAP patients (19–22).
The pTyr935X mutation is a nonsense alteration of exon 15 that
exchanges cytosine for adenine.
In our sample, we found a predominance of nonsense
mutations (45% of the patients), followed by frameshift mutations
(20% of patients). Among the 6 (30%) patients with neutral
mutations, missense mutations occurred in more than 1 patient. We
found the missense mutation, Gly2502Ser. According to Azzopardi
et al(18), who studied 691
patients with colorectal adenomas and 969 healthy individuals
(individuals investigated for cystic fibrosis), this mutation can
be found in individuals with or without adenoma, leaving a doubt as
to whether this mutation is deleterious.
The mutation Glu1317Gln is described in the
literature as being deleterious (23–27),
although other studies considered it to be not deleterious.
Azzopardi et al(18) found
this mutation in both healthy subjects and in adenoma patients.
However, we need further monitoring and analysis of these
individuals with the family to gain a better understanding of this
result.
For the variety of mutations, we were unable to
determine a correlation between the clinical variables and the
mutations detected. It is necessary to expand the sample to support
such analysis. Yet, following analysis of the correlation of the
presence of deleterious mutations and TNM and Astler-Coller stage,
we found a positive correlation with the presence of deleterious
mutations, demonstrating a more severe disease. Patients with
deleterious mutations had an OR, 0.086 (IC =0.001–0.984); TNM stage
I + II in comparison with III + IV, when compared with the patients
with no deleterious mutations identified.
In conclusion, our study demonstrated the molecular
heterogeneity of APC germline mutations in FAP and the
difficulty in performing molecular diagnostics in a Brazilian
population, since there were no mutations noted with a higher
prevalence. Thus, molecular diagnostics requires further detailed
evaluation, which, however is hampered by the presence of neutral
mutations, and these mutations are still debatable in many
populations of the world.
Acknowledgements
We thank FAPESP for the financial support and the
Laboratório de Genética Molecular (http://www.laboratoriomultiusuario.com.br) for the
possibility of the present study.
References
|
1
|
National Cancer Institute (INCA). Estimate
2012: Cancer Incidence in Brazil. 2011, http://www.inca.gov.br/estimativa/2012/index.asp?ID=5.
Accessed February 25, 2013
|
|
2
|
Leblanc R: Familial adenomatous polyposis
and benign intracranial tumors: a new variant of Gardner’s
syndrome. Can J Neurol Sci. 27:341–346. 2000.PubMed/NCBI
|
|
3
|
Dundar M, Caglayan AO, Saatci C, et al:
How the 11307K adenomatous polyposis coli gene variant contributes
in the assessment of risk of colorectal cancer, but not stomach
cancer, in a Turkish population. Cancer Genet Cytogenet. 177:95–97.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
The APC mutations database - UMD.
http://www.umd.be/APC/.
Accessed February 25, 2013
|
|
5
|
Kerr SE, Thomas CB, Thibodeau SN, et al:
APC germline mutations in individuals being evaluated for familial
adenomatous polyposis: a review of the Mayo Clinic experience with
1591 consecutive tests. J Mol Diagn. 15:31–43. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zeichner SB, Raj N, Cusnir M, et al: A de
novo germline APC mutation (3927del5) in a patient with familial
adenomatous polyposis: case report and literature review. Clin Med
Insights Oncol. 6:315–323. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Astler VB and Coller FA: The prognostic
significance of direct extension of carcinoma of the colon and
rectum. Ann Surg. 139:846–852. 1954. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Way LW and Doherty GM: Cirurgia:
Diagnóstico e Tratamento. 11th edition. Guanabara Koogan; Rio de
Janeiro: 2004, (In Portuguese).
|
|
9
|
Towsend CM, Beauchamp RD, Evers BM and
Mattox KL: Sabiston, Tratado de Cirurgia: A Base Biológica da
Prática Cirúrgica Moderna. Elsevier; Rio de Janeiro: 2005, (In
Portuguese).
|
|
10
|
World Health Organisation. Histological
Typing of Intestinal Tumours. International Histological
Classification of Tumours. (15)WHO; Geneva: 1976
|
|
11
|
Sambrook J, Fritsch EF and Maniatis T:
Molecular Cloning: A Laboratory Manual. Cold Spring Harbor
Laboratory Press; New York: 1989
|
|
12
|
Miyoshi Y, Ando H, Nagase H, et al:
Germ-line mutations of the APC gene in 53 familial adenomatous
polyposis patients. Proc Natl Acad Sci USA. 89:4452–4456. 1992.
View Article : Google Scholar
|
|
13
|
Nagase H and Nakamura Y: Mutations of the
APC (adenomatous polyposis coli) gene. Hum Mutat. 2:425–434. 1993.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Gómez-Fernández N, Castellví-Bel S,
Fernández-Rozadilla C, et al: Molecular analysis of the APC and
MUTYH genes in Galician and Catalonian FAP families: a different
spectrum of mutations? BMC Med Genet. 10:572009.PubMed/NCBI
|
|
15
|
Goss KH and Groden J: Biology of the
adenomatous polyposis coli tumor suppressor. J Clin Oncol.
18:1967–1979. 2000.PubMed/NCBI
|
|
16
|
Amos-Landgraf JM, Kwong LN, Kendziorski
CM, et al: A target-selected Apc-mutant rat kindred enhances the
modeling of familial human colon cancer. Proc Natl Acad Sci USA.
104:4036–4041. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Half E, Bercovich D and Rozen P: Familial
adenomatous polyposis. Orphanet J Rare Dis. 4:222009. View Article : Google Scholar
|
|
18
|
Azzopardi D, Dallosso AR, Eliason K, et
al: Multiple rare nonsynonymous variants in the adenomatous
polyposis coli gene predispose to colorectal adenomas. Cancer Res.
68:358–363. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Gavert N, Yaron Y, Naiman T, et al:
Molecular analysis of the APC gene in 71 Israeli families: 17 novel
mutations. Hum Mutat. 19:6642002. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ruiz-Ponte C, Vega A, Carracedo A and
Barros F: Mutation analysis of the adenomatous polyposis coli (APC)
gene in northwest Spanish patients with familial adenomatous
polyposis (FAP) and sporadic colorectal cancer. Hum Mutat.
18:3552001. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Schnitzler M, Koorey D, Dwight T, et al:
Frequency of codon 1061 and codon 1309 APC mutations in Australian
familial adenomatous polyposis patients. Hum Mutat. (Suppl 1):
S56–S57. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Varesco L, Gismondi V, James R, et al: APC
gene mutations in Italian familial polyposis coli patients. Cancer
Detect Prev. 17:279–281. 1993.PubMed/NCBI
|
|
23
|
Laken SJ, Petersen GM, Gruber SB, et al:
Familial colorectal cancer in Ashkenazim due to a hypermutable
tract in APC. Nat Genet. 17:79–83. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lamlum H, Al Tassan N, Jaeger E, et al:
Germline APC variants in patients with multiple colorectal
adenomas, with evidence for the particular importance of E1317Q.
Hum Mol Genet. 9:2215–2221. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Frayling IM, Beck Ne, Ilyas M, et al: The
APC variants I1307K and E1317Q are associated with colorectal
tumors, but not always with a family history. Proc Natl Acad Sci
USA. 95:10722–10727. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Gryfe R, Di Nicola N, Lal G, et al:
Inherited colorectal polyposis and cancer risk of the APC I1307K
polymorphism. Am J Hum Genet. 64:378–384. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Hahnloser D, Petersen GM, Rabe K, et al:
The APC E1317Q variant in adenomatous polyps and colorectal
cancers. Cancer Epidemiol Biomarkers Prev. 12:1023–1028.
2003.PubMed/NCBI
|