Introduction
Oral tongue squamous cell carcinoma (OTSCC), the
most common type of oral cancer, exhibits increased incidence and
poor prognosis (1,2). OTSCC is significantly more aggressive
than other forms of oral cancer, as it has a propensity for rapid
local invasion, metastasis and a high recurrence rate (3,4). The
prognosis for patients with OTSCC has not strikingly improved over
the past 3 decades, even with combined treatment involving surgery,
chemotherapy and radiation; the 5-year overall survival rate of
OTSCC is ~50–60% (2,5–7).
Therefore, it is critical to develop novel biomarkers for
predicting prognosis and for establishing targeted treatments for
patients with OTSCC.
BATF2, also known as SARI (suppressor of AP-1,
regulated by IFN) and a member of the BATF subfamily of bZIP
proteins, was cloned and identified in 2008. The BATF2 gene is
located at 11q12–13. Steady-state BATF2 mRNA expression was
detected in multiple lineage-specific normal cells, but not in
their transformed/tumorigenic counterparts (8). Furthermore, overexpression of BATF2
was found to inhibit proliferation and induce apoptosis in cancer
cells but not in normal cells (8).
In contrast, BATF2 downregulation was found to promote tumor
proliferation and metastasis in lung adenocarcinoma (9). Recently, decreased expression of BATF2
was found to be associated with poor prognosis in hepatocellular
carcinoma (HCC) (10) and
colorectal carcinoma (11). Thus,
it is likely that BATF2 functions as a tumor-suppressor in cancer
development; however, no data regarding BATF2 expression and its
correlation with OTSCC are available.
In the present study, we analyzed the expression of
BATF2 in OTSCC using quantitative PCR, western blotting and
immunohistochemistry and investigated the relationship between its
expression and the clinicopathological features of the OTSCC
patients. We also evaluated the potential prognostic value of BATF2
in the postoperative survival of OTSCC patients. Our data showed
that BATF2 plays an important role in the development of OTSCC and
may be considered as a candidate tumor-suppressor and a prognostic
marker for patients with OTSCC.
Materials and methods
Patients and clinical tissue
specimens
The present study was approved by the Ethics
Committee of The Third Affiliated Hospital of Kunming Medical
University, and informed consent was obtained from all participants
prior to enrollment. A total of 16 paired OTSCC tissues and
adjacent non-tumor tissues (distance from the tumor of >2 cm)
were collected from OTSCC patients who had undergone surgical
resection at The Third Affiliated Hospital of Kunming Medical
University between May 2010 and August 2010. The fresh tissues were
immediately immersed in RNAlater (Ambion, Inc., Austin, TX, USA)
after surgical resection, stored at 4°C overnight to allow thorough
penetration of the tissue and then frozen at −80°C until RNA and
protein extraction was performed. An additional 202
paraffin-embedded OTSCC samples, which were pathologically and
clinically diagnosed between January 2000 and December 2005 at The
Third Affiliated Hospital of Kunming Medical University, were
collected for immunohistochemistry. These samples were from 112
males and 90 females, with a median age of 53 years (ranging from
21 to 78 years), and none of these patients received radiotherapy
or chemotherapy prior to surgery. The clinicopathological features
and BATF2 expression levels for the 202 patients are shown in
Table I. The histological
differentiation of the samples was determined according to the
criteria of the World Health Organization. The tumor (T)
classification, node (N) classification and clinical
tumor-node-metastasis (TNM) stage were assessed according to the
TNM classification of the American Joint Committee on Cancer (AJCC)
(12).
 | Table ICorrelation between the expression of
BATF2 and the clinicopathological variables of the 202 patients
with OTSCC. |
Table I
Correlation between the expression of
BATF2 and the clinicopathological variables of the 202 patients
with OTSCC.
| Clinicopathological
variables | BATF2 expression n
(%) | P-value |
|---|
|
|---|
| No/low | High |
|---|
| Gender | | | 0.943 |
| Male | 69 (61.6) | 43 (38.4) | |
| Female | 55 (61.1) | 35 (38.9) | |
| Age (years) | | | 0.513 |
| ≤50 | 53 (57.8) | 37 (42.1) | |
| >50 | 71 (63.5) | 41 (36.5) | |
| Tobacco history | | | 0.429 |
| Smoker | 44 (57.3) | 32 (42.7) | |
| Nonsmoker | 80 (63.5) | 46 (36.5) | |
| Clinical TNM
stage | | | 0.198 |
| I | 34 (52.3) | 31 (47.7) | |
| II | 52 (65.8) | 27 (34.2) | |
| III | 27 (61.4) | 17 (38.6) | |
| IV | 11 (78.6) | 3 (21.4) | |
| T classification | | | 0.162 |
| T1–2 | 108 (59.7) | 73 (40.3) | |
| T3–4 | 16 (76.2) | 5 (23.8) | |
| N classification | | | 0.866 |
| N0 | 93 (60.8) | 60 (39.2) | |
| N+ | 31 (63.3) | 18 (36.7) | |
| Histological
differentiation | | | 0.002 |
| Well | 79 (54.1) | 67 (45.9) | |
| Moderate | 37 (92.5) | 10 (7.5) | |
| Poor | 8 (88.9) | 1 (11.1) | |
Cell lines and cell culture
A normal tongue epithelial cell line (NTEC1) was
established by culturing normal tongue squamous epithelium from a
non-tumor patient in keratinocyte/serum-free medium (Invitrogen
Life Technologies, Carlsbad, CA, USA). OTSCC cell lines were
purchased from the American Type Culture Collection (ATCC,
Manassas, VA, USA) (CAL-27, SCC-25 and SCC-9 cells) or from the
Committee of the Type Culture Collection of the Chinese Academy of
Sciences (Shanghai, China) (TCA8113 and TSCCA cells). All cells
were cultured in DMEM/F12 (Invitrogen Life Technologies)
supplemented with 10% fetal bovine serum (HyClone, Logan, UT, USA),
penicillin (100 U/ml) and streptomycin (100 U/ml) at 37°C in a
humidified 5% CO2 incubator.
Quantitative (q)PCR
Total RNA from the cell lines and human tissues was
extracted using TRIzol reagent (Invitrogen Life Technologies).
After reverse transcription of the total RNA, the first-strand cDNA
was used as a template for detecting the expression of BATF2. qPCR
was performed with an ABI PRISM 7900HT sequence detection system.
The housekeeping gene GAPDH was used as an internal control to
normalize the expression levels of BATF2. The primer sequences
were: 5′-AGACCCCAAGGAGCAACA-3′ (sense), and
5′-CTTTTTCCAGAGACTCGTGCT-3′ (antisense) for BATF2, and
5′-CTCCTCCTGTTCGACAGTCAGC-3′ (sense), and
5′-CCCAATACGACCAAATCCGTT-3′ antisense for GAPDH. To ensure the
reproducibility of the results, all experiments were repeated 3
times. The data were analyzed using the comparative threshold cycle
(2−ΔΔCT) method.
Western blot analysis
Western blot analysis was performed as previously
described (10). When relevant, the
blots were probed with an anti-BATF2 mouse monoclonal antibody
(1:1,000 dilution; Abnova Corporation, Taiwan) and an anti-GAPDH
mouse monoclonal antibody (1:1,000 dilution; Santa Cruz
Biotechnology, Inc., Santa Cruz, CA, USA). The signals were
detected using enhanced chemiluminescence (ECL) (Amersham Pharmacia
Biotech, Piscataway, NJ, USA) according to the manufacturer’s
suggested protocol.
Immunohistochemical staining
In brief, paraffin-embedded tongue tissue specimens
were cut into 4-μm sections and incubated at 65°C for 1 h. All
sections were deparaffinized with xylene and rehydrated in a graded
ethanol series. After treatment with 3% H2O2
for 15 min to block the endogenous peroxidase, the sections were
submerged in EDTA antigen retrieval buffer (pH 8.0) and microwaved.
Then, the sections were treated with normal goat serum for 30 min
to reduce any nonspecific binding and were incubated with an
anti-BATF2 mouse monoclonal antibody (1:1,000; Abnova Corporation)
overnight at 4°C. After washing, the sections were incubated with a
biotinylated anti-rabbit secondary antibody, followed by further
incubation with streptavidin-horseradish peroxidase (both from
Dako) at 37°C for 30 min. To produce a color reaction,
diaminobenzidine (DAB) was used. For the negative controls, the
antibody was replaced with normal goat serum. The immunostained
samples were evaluated by two independent pathologists who did not
have knowledge of the clinical data. According to previous studies
(10,13), the intensity of the cells expressing
BATF2 was scored as follows: 0 (no staining), 1 (weak staining,
light yellow), 2 (moderate staining, yellow-brown) or 3 (strong
staining, brown). The percentage of positive staining was scored as
follows: 0 (no expression), 1 (1–25%), 2 (26–50%), 3 (51–75%) or 4
(>75%). The BATF2 expression level was calculated as the
intensity score plus the proportion score and was divided into 4
grades: − (score of 0 and 1), + (score of 2 and 3), ++ (score of 4
and 5) and +++ (score of 6 and 7). In the present study, ‘−’ and
‘+’ represented no/low expression (score of 0–3), whereas ‘++’ and
‘+++’ were regarded as high expression (score of 4–7).
Statistical analysis
Overall survival was calculated from the date of
surgery until the day of death or the last date in the medical
records on which the patient was reported to be alive. Disease-free
survival was calculated from the date of surgery until the last
follow-up date on which the patient was not found to have tumor
recurrence. A paired samples t-test was used to compare the mRNA
and protein expression of BATF2 in the paired OTSCC and adjacent
non-tumor tissue samples. A χ2 test was used to analyze
the relationship between BATF2 expression and the
clinicopathological features of the OTSCC patients. Survival curves
were plotted using the Kaplan-Meier method and compared with the
log-rank test. The Cox proportional hazard regression model was
used for univariate and multivariate analyses to explore the
influence of the clinicopathological variables and BATF2 expression
on survival. SPSS 18.0 software (SPSS, Chicago, IL, USA) was used
for all statistical analyses, and a P-value of <0.05 was
considered to indicate a significant result.
Results
BATF2 expression is decreased in the
OTSCC cell lines
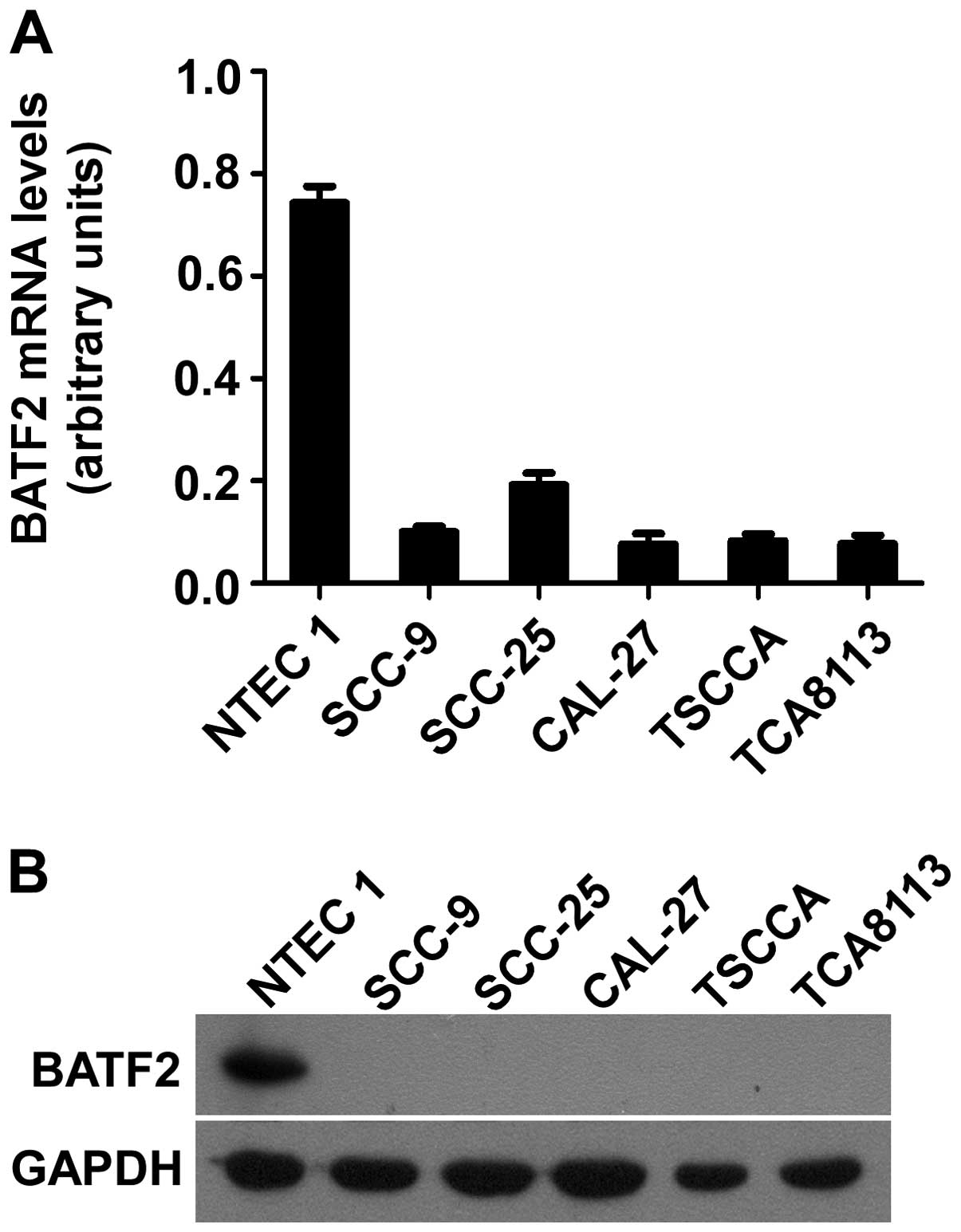
To examine the expression levels of BATF2, qPCR and
western blot analysis were conducted using the normal tongue
epithelial cell line NTEC1 and the OTSCC cell lines SCC-9, SCC-25,
CAL-27, TSCCA and TCA8113. qPCR revealed lower expression of BATF2
mRNA in all of the cancer cell lines when compared to the NTEC1
cell line (Fig. 1A). As shown in
Fig. 1B, high expression of the
BATF2 protein was observed in the NTEC1 cell line, whereas the
expression of the BATF2 protein was significantly lower, i.e., at
an undetectable level, in the OTSCC cell lines. These findings are
consistent with those from our qPCR experiment. Thus, our data
indicated that BATF2 was downregulated at both the mRNA and protein
levels in the OTSCC cell lines.
BATF2 expression is downregulated in
fresh OTSCC tissues
To determine the expression of BATF2 in OTSCC
tissues, qPCR and western blot analysis were performed using 16
OTSCC tissues and their matched adjacent non-tumor tissues. BATF2
mRNA was found to be significantly downregulated in 13 (81.25%,
13/16) of the OTSCC tissues when compared with their matched
adjacent non-tumor tissues (P=0.001) (Fig. 2A and B). Similarly, the BATF2
protein was also significantly downregulated in 14 (87.5%, 14/16)
of the tumor samples when compared with the adjacent non-tumor
tissues from the same patients (P<0.001) (Fig. 2C).
Correlation between BATF2 expression and
clinicopathological features
To verify the relationship between BATF2 and the
development of OTSCC, we examined BATF2 expression in 202
paraffin-embedded OTSCC samples and 30 adjacent non-tumor tissue
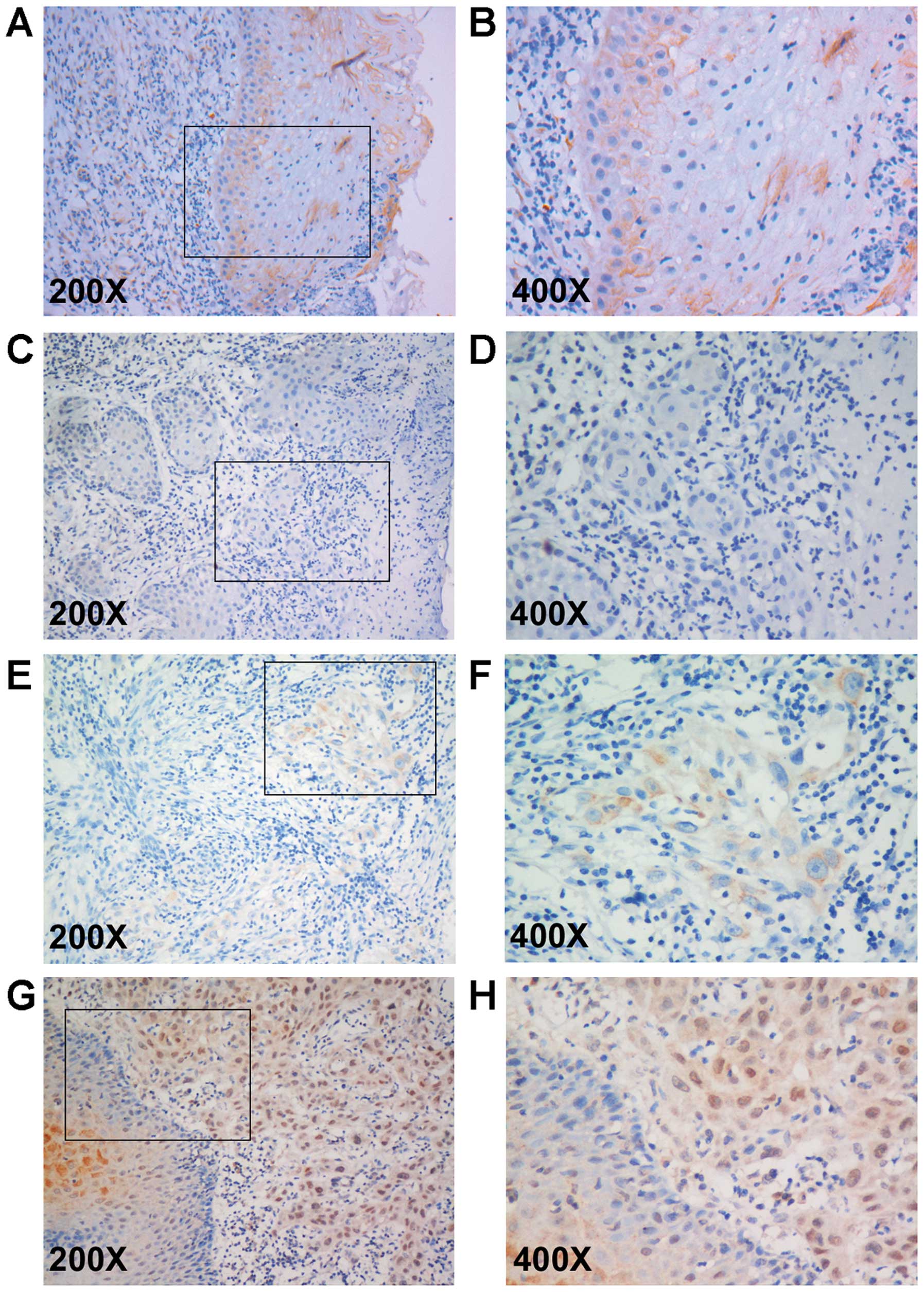
specimens by immunohistochemistry. As a result, among the 30
non-tumor tissue specimens, 18 cases (60%) showed high expression
(Fig. 3A and B), 8 cases (26.7%)
showed low expression and 4 cases (13.3%) showed negative
expression. However, among the 202 OTSCC samples, 20 cases (9.9%)
showed negative expression (Fig. 3C and
D), 104 cases (51.5%) showed low expression (Fig. 3E and F) and 78 (38.6%) cases showed
high expression (Fig. 3G and H). As
shown in Table I, the BATF2
expression level was significantly correlated with histological
differentiation (P=0.002) but not with age, gender, tobacco
history, T classification, N classification or clinical TNM
stage.
BATF2 expression and prognosis
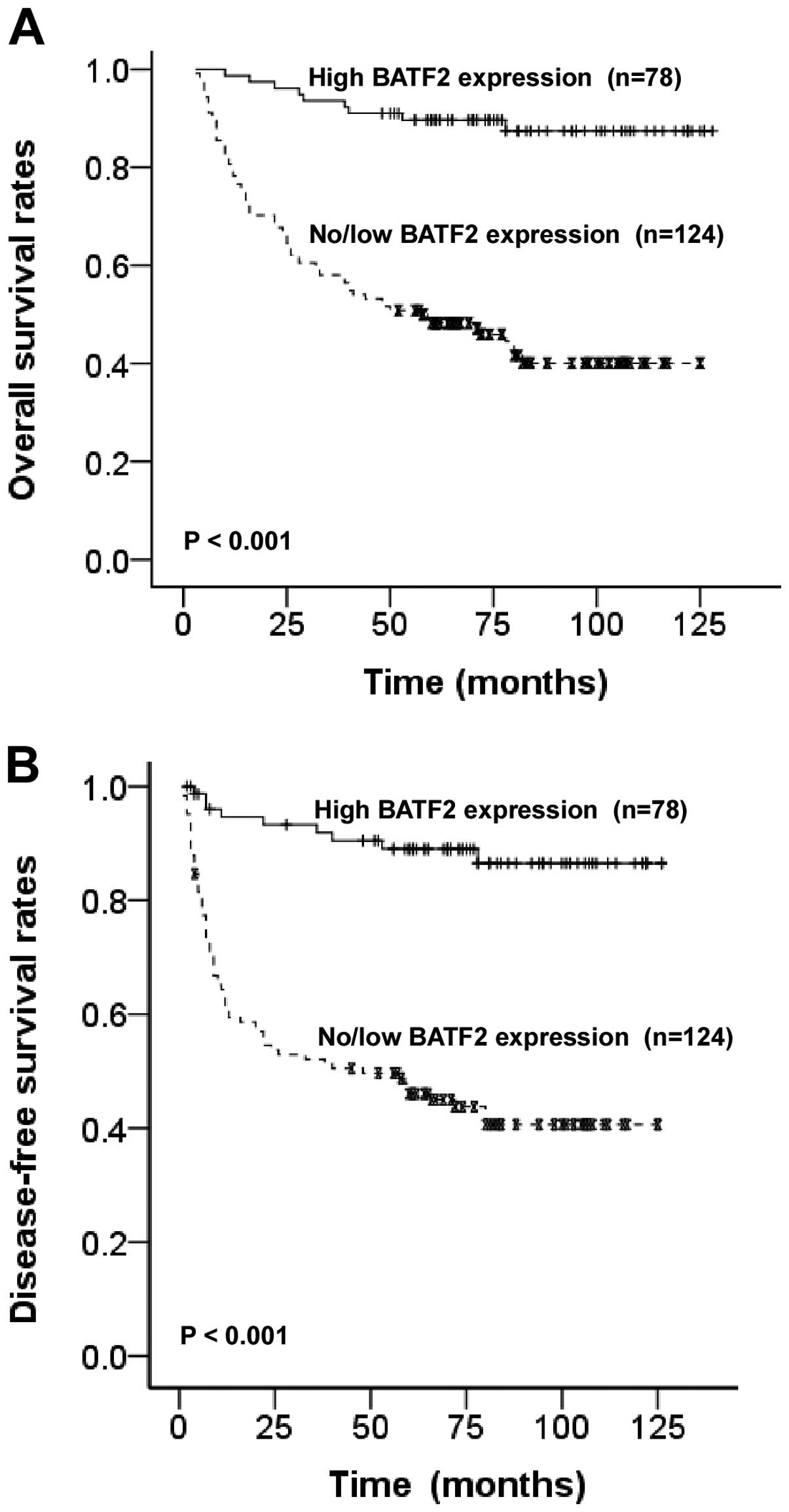
The patients with no/low BATF2 expression had a
significantly poorer 5-year overall survival rate and disease-free
survival rate than those with high BATF2 expression (50.5 vs.
89.7%, P<0.001, Fig. 4A) (42.8
vs. 86.5%, P<0.001, Fig. 4B).
Univariate Cox regression analyses demonstrated that clinical TNM
stage, histological differentiation and BATF2 expression level were
significantly associated with overall survival. Furthermore,
multivariate Cox regression analyses revealed that the above
mentioned 3 factors are independent predictors for the overall
survival of OTSCC patients (P=0.000, 0.001 and 0.001, respectively;
Table II). These findings indicate
that a high level of BATF2 expression is a biomarker for better
prognosis in patients with OTSCC.
 | Table IIUnivariate and multivariate analyses
using the Cox regression model. |
Table II
Univariate and multivariate analyses
using the Cox regression model.
| Variables | Hazard risk (95%
CI) | P-value |
|---|
| Univariate |
| Gender | 1.167
(0.856–1.589) | 0.329 |
| Age | 1.231
(0.907–1.670) | 0.182 |
| T
classification | 1.151
(0.918–1.442) | 0.223 |
| N
classification | 0.981
(0.655–1.471) | 0.928 |
| Clinical TNM
stage | 1.423
(1.095–1.848) | 0.008 |
| Histological
differentiation | 1.459
(1.186–1.793) | 0.020 |
| Expression of
BATF2 | 1.507
(1.155–1.966) | 0.002 |
| Multivariate |
| Clinical TNM
stage | 1.541
(1.363–1.743) | 0.000 |
| Histological
differentiation | 1.424
(1.158–1.750) | 0.001 |
| Expression of
BATF2 | 1.564
(1.210–2.022) | 0.001 |
Discussion
Despite advances in the early diagnosis and
management of cancer, the outcome for patients with OTSCC has not
significantly improved over the last 3 decades (2,6,7).
Therefore, it is critical to identify biomarkers and molecular
targets for these patients. Basic leucine zipper transcription
factor ATF-like (BATF), BATF2 and BATF3 belong to the activator
protein 1 (AP-1) family of transcription factors, which regulate
numerous cellular processes (14).
Su et al(8) detected
steady-state expression of BATF2 mRNA in many normal cell lines,
such as melanocytes, astrocytes, breast and prostate epithelial
cells and pancreatic mesothelial cells, but not in their malignant
counterparts. The overexpression of BATF2 was shown to induce
profound growth inhibition and apoptosis in malignant glioma,
melanoma and prostate cancer cell lines, with no effect on the
survival of the corresponding normal cells. The growth inhibitory
effect of BATF2 was further confirmed by its ability to slow the
growth rate of DU-145 prostate cancer cells injected into athymic
nude mice.
However, BATF2 expression in OTSCC has not been
reported. In the present study, the mRNA and protein expression of
BATF2 was significantly lower in the OTSCC cell lines and tissues
than in normal tongue epithelial cells or adjacent non-tumor
tissues (Figs. 1 and 2). In agreement with these results from
our qPCR and western blot analysis, little to no BATF2 expression
was observed by immunohistochemistry in 61.4% of the OTSCC samples
(124/202). Furthermore, decreased BATF2 expression was
significantly associated with poor histological differentiation in
the OTSCC tissues (Table I,
Fig. 3). In contrast to the
research of Ma et al(10),
we did not find that age or tumor size of the patients with OTSCC
was significantly correlated with the level of BATF2 expression,
indicating that decreased BATF2 expression may exert different
effects on the development of different neoplasms. Survival
analysis showed that no or low BATF2 expression was significantly
correlated with poor overall survival and disease-free survival
(P<0.001). Multivariate Cox regression analysis further
confirmed that the BATF2 expression level is an independent
prognostic factor, with a hazard risk of 1.564 (95% CI,
1.210–2.022). All of these results support the hypothesis that
BATF2 functions as a tumor-suppressor gene in the development of
OTSCC.
Nevertheless, the mechanism by which BATF2
expression is downregulated in solid tumors and the relationship
between its decreased expression and poor prognosis are not fully
understood. Mutation of tumor-suppressor genes, particularly in
exons, is one of the most notable features that may lead to gene
inactivation. In the study performed by Ma et al(10), no mutation was detected in any of
the 3 exons of the BATF2 gene in 5 HCC cell lines and 8 HCC tumor
tissues. Wang et al(9)
reported that loss of BATF2 expression initiates the
epithelial-mesenchymal transition, which is visualized by
repression of E-cadherin and upregulation of vimentin in lung
adenocarcinoma cell lines and clinical lung adenocarcinoma
specimens. Using a human lung xenograft mouse model, the knockdown
of endogenous BATF2 in human carcinoma cells was found to lead to
the development of multiple lymph node metastases by modulating the
(GSK)-3β-β-catenin signaling pathway. Huang et al(15) demonstrated that a BCR-ABL kinase
inhibitor or siRNA specific to BCR-ABL upregulated BATF2 mRNA
expression in human leukemia cells. Both JAK/STAT and RAS/MAPK
signaling inhibitors (AG490 and PD98059, respectively) upregulated
BATF2 mRNA expression, while the PI3K/AKT pathway inhibitor
LY294002 had no such effect. Therefore, this group reported that
BATF2 mRNA expression was suppressed by BCR-ABL through the
downstream RAS/MAPK and JAK/STAT signaling pathways in human
leukemia cells. Dash et al(16) found that BATF2 selectively
suppressed the transcription of CCN1, a secretory integrin-binding
protein that regulates angiogenesis, cell adhesion, migration,
proliferation, survival and apoptosis. Hence, BATF2 inhibited
CCN1-induced anchorage-independent growth and invasion in breast
cancer, malignant glioma and metastatic melanoma cells by
inhibiting the activation of MAP kinase and the PI3/AKT kinases.
The AP-1 transcription factor BATF3 is required for the homeostatic
development of classical CD8α+ dendritic cells, which
prime CD8 T-cell responses against intracellular pathogens.
Recently, Tussiwand et al(17) identified an alternative pathway that
results from the molecular compensation of BATF3 by BATF2, which is
induced by cytokines in response to infection. Whether this pathway
is related to carcinogenesis remains elusive.
In summary, BATF2 expression was decreased in most
of the cases of OTSCC analyzed in the present study, and lower
BATF2 expression was found to be related to poor histological
differentiation and poor prognosis in patients with OTSCC,
demonstrating that BATF2 may serve as a tumor-suppressor gene in
the development of OTSCC and that BATF2 may be used as a prognostic
biomarker and therapeutic target for OTSCC. However, the exact
mechanism by which the expression of BATF2 is decreased and how
BATF2 functions in the development of OTSCC remain to be
explored.
Acknowledgements
The present study was partly supported by grants
from the National Natural Science Foundation of China (nos.
30960444 and 81260402), and the Special Foundation of High Levels
of Health Technical Personnel Training in Yunnan Province (no.
D-201243).
References
|
1
|
Jemal A, Siegel R, Xu J and Ward E: Cancer
statistics, 2010. CA Cancer J Clin. 60:277–300. 2010. View Article : Google Scholar
|
|
2
|
Shiboski CH, Schmidt BL and Jordan RC:
Tongue and tonsil carcinoma: increasing trends in the U.S.
population ages 20–44 years. Cancer. 103:1843–1849. 2005.PubMed/NCBI
|
|
3
|
Franceschi D, Gupta R, Spiro RH and Shah
JP: Improved survival in the treatment of squamous carcinoma of the
oral tongue. Am J Surg. 166:360–365. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Yuen AP, Lam KY, Chan AC, et al:
Clinicopathological analysis of elective neck dissection for N0
neck of early oral tongue carcinoma. Am J Surg. 177:90–92. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Myers JN, Elkins T, Roberts D and Byers
RM: Squamous cell carcinoma of the tongue in young adults:
increasing incidence and factors that predict treatment outcomes.
Otolaryngol Head Neck Surg. 122:44–51. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wen Y, Dai X, Wang C, et al: A
retrospective clinical study of 6539 cases of malignant
oral-maxillofacial tumors. Hua Xi Kou Qiang Yi Xue Za Zhi.
19:296–299. 2001.(In Chinese).
|
|
7
|
Goldstein DP, Bachar GY, Lea J, et al:
Outcomes of squamous cell cancer of the oral tongue managed at the
Princess Margaret Hospital. Head Neck. 35:632–641. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Su ZZ, Lee SG, Emdad L, et al: Cloning and
characterization of SARI (suppressor of AP-1, regulated by
IFN). Proc Natl Acad Sci USA. 105:20906–20911. 2008.
|
|
9
|
Wang C, Su Y, Zhang L, et al: The function
of SARI in modulating epithelial-mesenchymal transition and lung
adenocarcinoma metastasis. PLoS One. 7:e380462012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ma H, Liang X, Chen Y, et al: Decreased
expression of BATF2 is associated with a poor prognosis in
hepatocellular carcinoma. Int J Cancer. 128:771–777. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Liu ZB, Yang Y, Ye XG, Wang L, Tian PY and
Zhang YY: Expression and significance of SARI and CCN1 in human
colorectal carcinomas. Zhonghua Yi Xue Za Zhi. 91:2397–2401.
2011.(In Chinese).
|
|
12
|
Edge SB, Byrd DR, Compton CC, Fritz AG,
Greene FL and Trotti A: AJCC Cancer Staging Manual. 7th edition.
Springer; New York: 2010
|
|
13
|
Soumaoro LT, Uetake H, Higuchi T, Takagi
Y, Enomoto M and Sugihara K: Cyclooxygenase-2 expression: a
significant prognostic indicator for patients with colorectal
cancer. Clin Cancer Res. 10:8465–8471. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Murphy TL, Tussiwand R and Murphy KM:
Specificity through cooperation: BATF-IRF interactions control
immune-regulatory networks. Nat Rev Immunol. 13:499–509. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Huang Q, Yang Y, Li X and Huang S:
Transcription suppression of SARI (suppressor of AP-1, regulated by
IFN) by BCR-ABL in human leukemia cells. Tumour Biol. 32:1191–1197.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Dash R, Su ZZ, Lee SG, et al: Inhibition
of AP-1 by SARI negatively regulates transformation progression
mediated by CCN1. Oncogene. 29:4412–4423. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Tussiwand R, Lee WL, Murphy TL, et al:
Compensatory dendritic cell development mediated by BATF-IRF
interactions. Nature. 490:502–507. 2012. View Article : Google Scholar : PubMed/NCBI
|