Introduction
Conventional lipoma is a benign tumor composed of
mature fat cells, most frequently occurring between the ages of 40
and 60 years (1,2). Lipomas may occur anywhere in the body,
including inside bones and parenchymatous organs. The prognosis is
excellent regardless of whether they are superficial or
deep-seated, although intramuscular lipomas with an infiltrative
growth pattern may recur locally (3). Lipomas have been extensively analyzed
by chromosome banding, through which aberrations have been noted in
approximately 83% of the cases (4).
The most common alteration is structural rearrangement of
chromosome region 12q13–15 (75% of the cases with clonal
aberrations), resulting in transcriptional upregulation of the
HMGA2 gene, which causes tumorigenesis. Other characteristic
cytogenetic findings include deletions of 13q (15–20%),
supernumerary ring chromosomes or giant marker chromosomes (6%) and
rearrangements of band 6p21–23 (5%), which harbors the HMGA1
gene (5–7). In 10% of the cases, none of these
characteristic aberrations is noted, suggesting that conventional
lipomas sometimes develop through alternate molecular routes. One
uncommon, but recurrent, cytogenetic finding is structural
rearrangement of chromosome arm 15q, usually without concomitant
rearrangement of 12q (5). In order
to assess whether such 15q rearrangements might target genes of
importance for lipomagenesis, we selected seven cases for further
fluorescence in situ hybridization (FISH) and/or single
nucleotide polymorphism (SNP) and gene expression array studies.
Apart from mapping the breakpoints in 15q, we also assessed the
status of the HMGA2 gene using FISH or quantitative
real-time PCR (qRT-PCR), in order to investigate its cryptic
involvement in the tumorigenesis of this group of lipomas.
Materials and methods
Samples
Seven conventional lipomas with structural
rearrangement of 15q were selected among the 550 lipomas studied at
the Department of Clinical Genetics, Lund Univerity (Lund, Sweden),
from 1984. Data concerning patient gender and age, tumor location
and karyotype are documented in Table
I. All cases were analyzed by chromosome banding after
short-term culturing according to standard methods. Karyotypes were
described according to ISCN (2013). All samples were obtained after
informed consent, and the study was approved by the regional ethics
committee of Lund University (Dnr 2011/289).
 | Table IClinical and cytogenetic information
for seven conventional lipomas with rearrangement of chromosome arm
15q. |
Table I
Clinical and cytogenetic information
for seven conventional lipomas with rearrangement of chromosome arm
15q.
| Case | Location | Karyotype | Age
(years)/gender | No. of
recurrences |
|---|
| 1 | Axilla, deep |
46,XY,del(15)(q13q21) | 60/M | 2 |
| 2 | Arm,
subcutaneous |
46,XX,der(4)t(4;15)(p16;q22),t(5;9)(q22;q32),ins(8;13)
(q24;q34q14),
add(15)(q15),add(16)(q13),der(20)t(16;20)(q13;q12) | 42/F | 0 |
| 3 | Neck,
subcutaneous |
46,XY,−5,der(6)del(6)(p?)
t(6;15)(q11;q12–15),−10,der(15)
t(6;15)(?;q15–21),+der(?)t(?;6)(?;q?)x2,+mar | 36/M | 0 |
| 4 | Neck,
subcutaneous |
46,Y,t(X;15;11)(q22;q22;q23) | 41/M | 0 |
| 5 | Shoulder, deep |
47,XY,der(6)t(6;15)(q15;q15),−15,+2r | 72/M | 0 |
| 6 | Arm, deep |
46,XY,der(12)ins(12;15)(q1?1;q12q21),t(15;17)(q12–21;q2?),
del(15)(q1?2),der(17)t(15;17)(q2?;q2?3) | 59/M | 0 |
| 7 | Back, deep |
46,XX,t(6;15)(q13;q22) | 48/F | 0 |
SNP array analysis
Cases 1 and 3–5 were analyzed by SNP arrays to
detect global copy number aberrations. DNA was extracted from fresh
frozen tumor biopsies using the DNeasy Tissue kit, according to the
manufacturer’s instructions (Qiagen, Valencia, CA, USA) and
afterwards hybridized onto the Illumina Human OmniQuad version 1.0
BeadChip (Illumina, Inc., San Diego, CA, USA) containing 1.2
million markers, following standard protocols supplied by the
manufacturer. SNP positions were based on the NCBI36/hg18 sequence
assembly. Data analysis was performed using the GenomeStudio
software 1.6.1 (Illumina), detecting imbalances by visual
inspection. Constitutional copy number variations were excluded
querying the Database of Genomic Variants (http://projects.tcag.ca/cgi-bin/variation) (8).
Gene expression array analysis
RNA of good quality was extracted from cases 1–4 and
hybridized onto Affymetrix Human Gene 1.0 ST Arrays (Affymetrix,
Santa Clara, CA, USA) as previously described (9). Twenty-five conventional lipomas
without detectable 15q rearrangements were included as controls.
Gene expression data were normalized, background-corrected and
summarized by using the Robust Multichip Analysis algorithm
implemented in the Expression Console version software 1.1
(Affymetrix). Gene expression levels were subsequently compared
between cases and controls. All genes located in the region 24–60
Mb in 15q were evaluated using a t-test, and P-values were adjusted
for multiple testing by Benjamini-Hochberg false discovery rate
(FDR) correction (Qlucore Omics Explorer; Qlucore AB, Lund,
Sweden). Genes with a P-value <0.05 and an FDR <0.2 were
considered to be significantly altered.
FISH analysis
FISH analyses were performed on all seven cases. To
pinpoint the breakpoints in 15q, we performed a chromosome walking
with bacterial artificial chromosome (BAC) probes. To assess the
involvement of HMGA2, we performed a break-apart assay with
the BAC probes RP11-299L9 and RP11-427K2, specific for the 5′-part
and the 3′-part of the gene, respectively. Whole chromosome paint
(WCP) probes were used to detect chromosomal rearrangements and,
when applicable, to discriminate tumor cells from normal cells.
Probes and slides were prepared and analyzed as previously
described (10).
qRT-PCR
qRT-PCR was used to study the expression level of
the HMGA2 gene in cases 1 and 4, which showed no
rearrangement at FISH analysis. To detect differences in the
expression levels of HMGA2 5′- and 3′-ends, consistent with
an intragenic rearrangement, a TaqMan gene expression assay was
performed using probes Hs00171569_m1 (exons 1–2), and Hs00971725_m1
(exons 4–5) as previously described (11). ACTB was used as endogenous
controls. Conventional lipomas showing a t(3;12), with an
HMGA2/LPP fusion, resulting in overexpressing the 5′-part of
HMGA2 (control 1) and a t(5;12), resulting in overexpression
of the entire HMGA2 gene (control 2), were used as
additional controls. The qRT-PCR analysis was performed as
previously described (11).
Results
Results from G-banding and SNP array analyses are
shown in Tables I and II, respectively. Case 1 had a deletion
spanning 15q12-q21.2 as the sole cytogenetic anomaly; the same
karyotype was also found in two local recurrences occurring 2 and 3
years after surgery for the primary tumor. SNP array analysis
mapped the deletion at 25.572–50.200 Mb, in agreement with the FISH
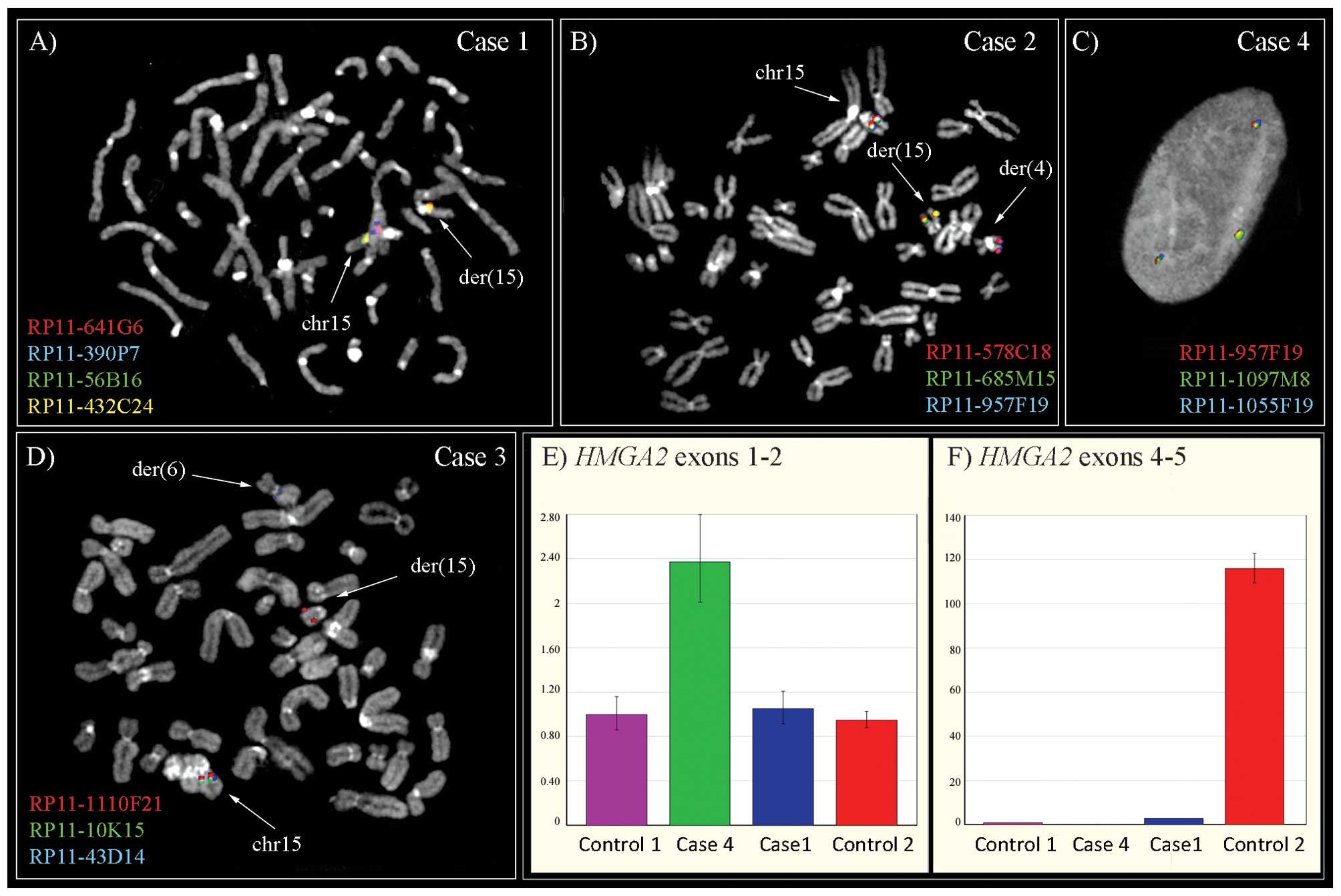
results (Fig. 1). No rearrangement
of the HMGA2 gene was noted at FISH analysis, while qRT-PCR
showed overexpression of the 5′-part (Fig. 1). The remaining six lipomas showed
involvement of 15q15-q22 in translocations with different partners
(Table I). Three of these were also
analyzed by SNP array. Case 3 had numerous hemizygous deletions,
including the region 37.940–38.104 Mb in 15q; this deletion was
confirmed and better mapped by FISH at 37.892–38.104 Mb, disclosing
an unbalanced t(6;15) (Fig. 1). The
SNP data also revealed deletion of the 5′-part of HMGA2 and
of 13q14.11-q14.3. In case 4, the SNP array analysis disclosed a
deletion in 11q23.1 as the single imbalance. FISH analysis mapped
the breakpoint on chromosome 15 at 47.331–47.441 Mb (15q21.1),
defined by split signals for BAC probes RP11-957F19 and
RP11-1097M8. While FISH analysis revealed a seemingly intact
HMGA2 locus, qRT-PCR revealed overexpression of the 5′-part
of the gene (Fig. 1). In case 5,
SNP array analysis identified amplification of 12q, including the
HMGA2 locus, and a hemizygous deletion in band 15q21.3 at
52.663–52.685 Mb. FISH revealed that the translocation breakpoint
was close to the centromere of 15q. Three cases were studied only
by FISH. Case 2 had a breakpoint in 15q21.1, at 47.174–47.270 Mb,
defined by split signals for BAC probe RP11-578C18. In addition,
this case had a deletion of the 3′-part of HMGA2. In case 6,
we mapped the breakpoint in 15q to 50.425–58.884 Mb and also found
that the 3′-part of the HMGA2 gene was deleted. In case 7,
we mapped the breakpoint in chromosome 15 at 57.643–57.692 Mb, in
the region between BAC probes RP11-1030I22 and RP11-1030O17. In
addition, this case showed deletion of the 3′-part of HMGA2.
Thus, we found no clustering of breakpoints in 15q, and the region
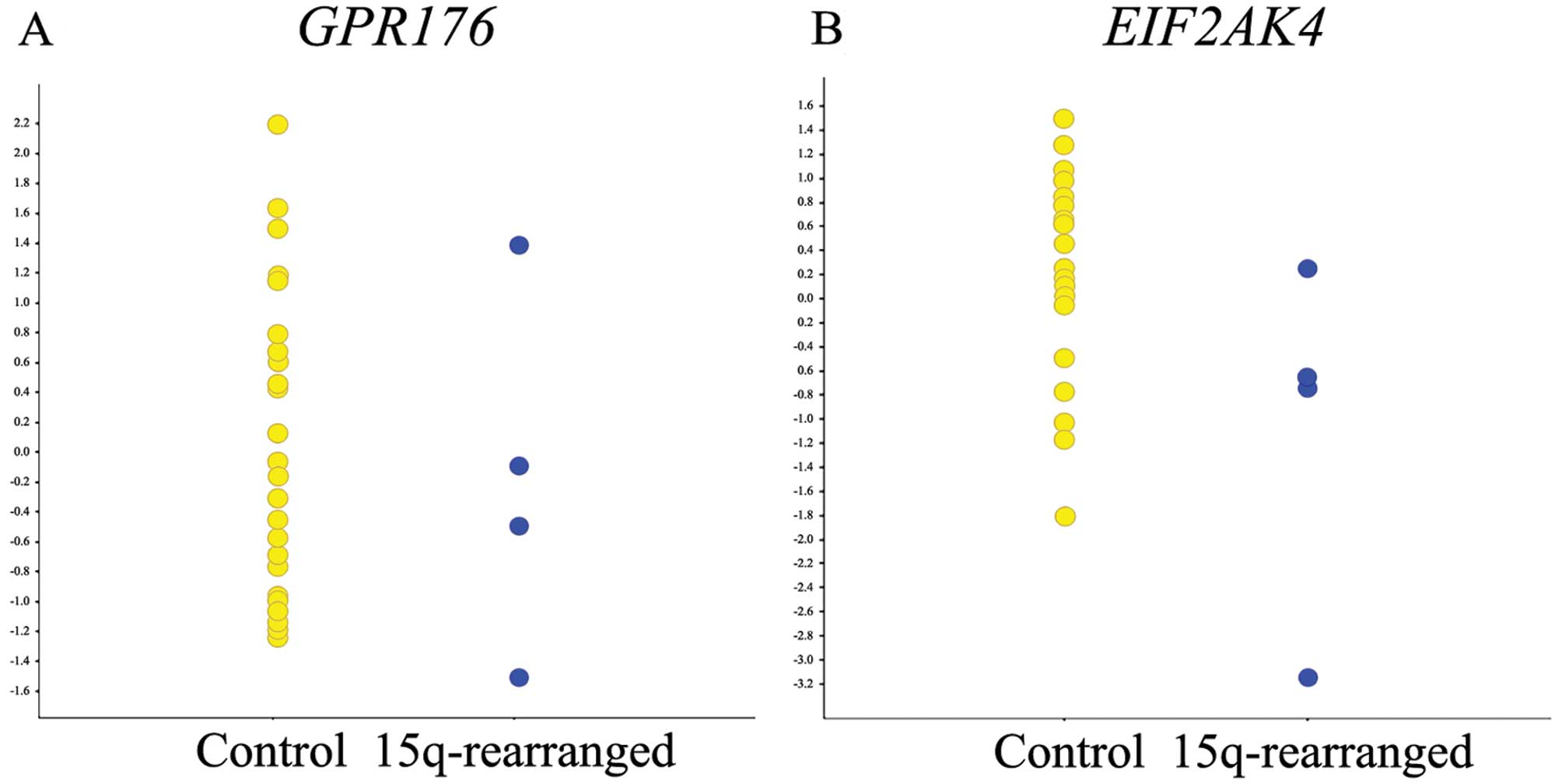
37.892–38.104 Mb in 15q was the only recurrently deleted region,
detected in two cases. This aberration affected two genes,
GPR176 and EIF2AK4, neither of which showed different
expression levels in lipomas with a 15q rearrangement when compared
to the control tumors (Fig. 2).
Instead, HMGA2 was involved in all seven cases, being either
partially deleted or split or cryptically deregulated.
 | Table IITable showing SNP array results and
the HMGA2 involvement for the studied cases. |
Table II
Table showing SNP array results and
the HMGA2 involvement for the studied cases.
| Case | SNP array
analysis | Method used to detect
involvement of HMGA2/Statusa |
|---|
|
|---|
| Band | SNP array
results | Position
NCBI36/hg18 |
|---|
| 1 | 15q12-q21.2 | del |
chr15:25572122-50200608 | qRT-PCR/+ |
| 2 | No data | FISH/+ |
| 3 | 1p36.21 | del |
chr1:15681459-15842337 | SNP array/+ |
| 3p25.3-p25.2 | del |
chr3:11356973-11747815 | |
| 3p24.1 | del |
chr3:27448147-27774038 | |
| 3p21.1 | del |
chr3:53122926-53244203 | |
| 3p21.1 | del |
chr3:53321422-53504180 | |
| 3p14.3 | del |
chr3:57068780-57212261 | |
| 3p13 | del |
chr3:73056422-73357684 | |
| 3p13 | del |
chr3:73911714-74117031 | |
| 5q31.3 | del |
chr5:141412453-141542868 | |
| 6q23.2 | del |
chr6:132493337-132685728 | |
| 7p14.1 | del |
chr7:42550790-42560558 | |
| 12q14.2 | del |
chr12:62387591-62966981 | |
| 12q14.3 | del |
chr12:64366833-64519077 | |
| 13q14.11-q14.3 | del |
chr13:43391527-49535788 | |
| 13q14.3 | del |
chr13:50535906-50596640 | |
| 15q15.1 | del |
chr15:37940000-38104900 | |
| 4 | 11q23.1 | LOH |
chr11:112486970-112561533 | qRT-PCR/+ |
| 5 | 11q22.1 | del |
chr11:98871868-98888063 | SNP array/+ |
| 12q13.3-14.1 | amp |
chr12:56343030-56776958 | |
| 12q14.1 | amp |
chr12:57322409-57774332 | |
| 12q14.1 | amp |
chr12:59587140-60060029 | |
| 12q14.1 | amp |
chr12:61364051-61459825 | |
| 12q14.3 | amp |
chr12:64385997-66074782 | |
| 12q15-q21.2 | amp |
chr12:67333542-76053035 | |
| 12q21.2 | amp |
chr12:76430478-76593563 | |
| 12q21.33 | amp |
chr12:89855182-89977501 | |
| 12q21.33 | amp |
chr12:90132557-90489984 | |
| 12q21.33 | amp |
chr12:90489984-90721904 | |
| 12q21.33 | amp |
chr12:90730613-91063729 | |
| 12q21.33 | amp |
chr12:91077361-91291882 | |
| 12q21.33 | amp |
chr12:91330328-91419845 | |
| 12q22 | amp |
chr12:94480909-94526441 | |
| 12q22 | amp |
chr12:94527237-94720773 | |
| 12q22 | amp |
chr12:94722197-94812991 | |
| 12q22 | amp |
chr12:95037274-95273987 | |
| 12q23.1 | amp |
chr12:95290705-96148524 | |
| 12q23.1 | amp |
chr12:97080720-97094264 | |
| 12q23.1 | amp |
chr12:97103285-97198077 | |
| 12q23.1 | amp |
chr12:97226786-97479566 | |
| 12q23.1 | amp |
chr12:97494248-97672573 | |
| 12q23.1 | amp |
chr12:97678033-98155350 | |
| 12q23.1 | amp |
chr12:98162407-98648233 | |
| 12q23.1 | amp |
chr12:98676267-98802171 | |
| 12q23.2 | amp |
chr12:100721860-101242139 | |
| 12q23.2 | amp |
chr12:102081102-102169706 | |
| 15q21.3 | del |
chr15:52663504-52685923 | |
| 6 | No data | FISH/+ |
| 7 | No data | FISH/+ |
Discussion
Previous genetic analyses of conventional lipomas
have identified several different molecular subgroups. The most
prominent one is characterized by aberrant transcriptional
upregulation of HMGA2, either of the whole gene or of its
5′-part, as a driver mutation (11). Minor subsets instead develop through
upregulation of the closely related HMGA1 gene or through
deletion of one or more genes in 13q; the exact pathogenetic
mechanism(s) involved in the latter cases remain unidentified
(5). Even when combining the
results of cytogenetic and molecular genetic analyses, deregulation
of these genes cannot, however, account for the development of all
conventional lipomas. In the present study, we investigated the
possibility of the existence of yet another pathway, involving a
locus on chromosome arm 15q. Prompted by the finding of an
interstitial deletion of 15q as the sole cytogenetic aberration in
multiple samples from one conventional lipoma (case 1), we mapped
the breakpoints in 15q in seven cases that at G-banding analysis
had shown various rearrangements of this chromosome arm, but no
detectable involvement of 12q14.3. Neither FISH nor SNP array data
indicated a shared breakpoint, excluding that they result in a
recurrent fusion gene or in transcriptional upregulation of a
specific target gene. Moreover, we found concomitant rearrangement
of the HMGA2 locus in all seven cases, disclosing its
primary role in the tumorigenesis also of this subgroup of lipomas,
and thus revealing 15q rearrangements as secondary changes.
Nevertheless, local recurrences are exceedingly rare for
conventional lipomas, and when they occur they often have features
of a misdiagnosed atypical lipoma, such as supernumerary ring
chromosomes, large size and location in the thigh (4); the lipoma of case 1 recurred twice,
always with the morphology of a conventional lipoma. Thus, it can
be speculated that certain 15q rearrangements, such as the large
interstitial deletion in this case, could have an impact on the
growth and recurrence potential of lipomas, functioning as a
cooperative mutation for lipomagenesis. The only other lipoma
showing a deletion overlapping with the one characterized in case
1, was case 3. The shared deleted region at 37.892–38.104 Mb was
found to contain the 5′-part of GPR176, encoding a G
protein-coupled receptor involved in responses to hormones, growth
factors and neurotransmitters (12), and the 5′-part of the
EIF2AK4, encoding a kinase that acts in response to varied
cellular stresses (13). However,
global gene expression analysis did not identify either of these
genes as a potential target and neither has been implicated in
adipocytic tumorigenesis.
Although 15q-rearrangements appear to be secondary
to HMGA2 deregulation, several of the deletions on
chromosome 15, as the one observed in case 1, could be of relevance
for growth characteristics, increasing the risk for local
recurrence in conventional lipomas.
Acknowledgements
We acknowledge the assistance with the microarray
analyses from the Swegene Centre for Integrative Biology at Lund
University. The present study was supported by the Swedish Cancer
Society, the Swedish Research Council and the Royal Physiographic
Society in Lund.
Abbreviations:
|
FISH
|
fluorescence in situ
hybridization
|
|
SNP
|
single nucleotide polymorphism
|
|
qRT-PCR
|
quantitative real-time PCR
|
|
FDR
|
false discovery rate
|
|
BAC
|
bacterial artificial chromosome
|
|
WCP
|
whole chromosome paint
|
References
|
1
|
Rydholm A and Berg NO: Size, site and
clinical incidence of lipoma. Factors in the differential diagnosis
of lipoma and sarcoma. Acta Orthop Scand. 54:929–934. 1983.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Nielsen GP and Mandahl N: Lipoma. WHO
Classification of Tumours of Soft Tissue and Bone. Fletcher CDM,
Bridge JA, Hogendoorn PCW and Mertens F: IARC Press; Lyon: pp.
20–21. 2013
|
|
3
|
Colella G, Biondi P, Caltabiano R, Vecchio
GM, Amico P and Magro G: Giant intramuscular lipoma of the tongue:
a case report and literature review. Cases J. 2:79062009.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Billing V, Mertens F, Domanski HA and
Rydholm A: Deep-seated ordinary and atypical lipomas:
histopathology, cytogenetics, clinical features, and outcome in 215
tumours of the extremity and trunk wall. J Bone Joint Surg Br.
90:929–933. 2008. View Article : Google Scholar
|
|
5
|
Bartuma H, Hallor KH, Panagopoulos I, et
al: Assessment of the clinical and molecular impact of different
cytogenetic subgroups in a series of 272 lipomas with abnormal
karyotype. Genes Chromosomes Cancer. 46:594–606. 2007. View Article : Google Scholar
|
|
6
|
Mandahl N, Bartuma H, Magnusson L,
Isaksson M, Macchia G and Mertens F: HMGA2 and MDM2
expression in lipomatous tumors with partial, low-level
amplification of sequences from the long arm of chromosome 12.
Cancer Genet. 204:550–556. 2011. View Article : Google Scholar
|
|
7
|
Nishio J: Contributions of cytogenetics
and molecular cytogenetics to the diagnosis of adipocytic tumors. J
Biomed Biotechnol. 2011:5240672011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Iafrate AJ, Feuk L, Rivera MN, et al:
Detection of large-scale variation in the human genome. Nat Genet.
36:949–951. 2004. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Nord KH, Magnusson L, Isaksson M, et al:
Concomitant deletions of tumor suppressor genes MEN1 and
AIP are essential for the pathogenesis of the brown fat
tumor hibernoma. Proc Natl Acad Sci USA. 107:21122–21127. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Jin Y, Möller E, Nord KH, et al: Fusion of
the AHRR and NCOA2 genes through a recurrent
translocation t(5;8)(p15;q13) in soft tissue angiofibroma results
in upregulation of aryl hydrocarbon receptor target genes. Genes
Chromosomes Cancer. 51:510–520. 2012.
|
|
11
|
Bartuma H, Panagopoulos I, Collin A, et
al: Expression levels of HMGA2 in adipocytic tumors
correlate with morphologic and cytogenetic subgroups. Mol Cancer.
8:362009.
|
|
12
|
Hata S, Emi Y, Iyanagi T and Osumi T: cDNA
cloning of a putative G protein-coupled receptor from brain.
Biochim Biophys Acta. 1261:121–125. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Berlanga JJ, Santoyo J and De Haro C:
Characterization of a mammalian homolog of the GCN2 eukaryotic
initiation factor 2α kinase. Eur J Biochem. 265:754–762.
1999.PubMed/NCBI
|