Introduction
Gastric cancer (GC) remains the fourth most commonly
diagnosed cancer and is the second leading cause of
cancer-associated mortality worldwide (1). The carcinogenesis of GC is complicated
and involves the dysregulation of oncogenes and tumor suppressors
(2). Many molecules are responsible
for GC tumorigenesis, and despite advances in surgical and
chemotherapeutic interventions, the overall 5-year survival for GC
patients remains low (30%) and the recurrence rate high (3–5). Thus,
identifying new biomarkers or factors that improve the diagnosis of
GC prior to tumorigenesis and predict prognosis responsible for
cancer recurrence in patients with GC is crucial.
A number of microRNA (miRNA) genes have been
identified in the human genome and it has been suggested that at
least 50% of all protein-encoding genes are regulated by miRNA
(6). Mature miRNAs are 21–22
nucleotides in size and affect the post-translational expression of
genes by interacting with complementary target sites within the
3′-untranslated region of the messenger RNA (7). Recent findings have shown the role of
miRNAs in a variety of basic biological and pathological processes
(8) and the association of miRNA
signatures with human diseases has been established (9). miR-377 can have a critical role in the
pathophysiology of diabetic nephropathy (10) and human nucleus pulposus cells
(11). miR-377 was responsible for
metabolizing the excess heme generated during hemolysis (12). miR-377 may be required for
glioblastoma multiforme (GBM) development and serve as a
therapeutic target for the treatment of GBM (13). However, the expression of miR-377 in
GC and its prognostic values remain to be elucidated.
In the present study, we evaluated the clinical
significance of miR-377. The expression level of miR-377 was
overexpressed in GC tissues and cell lines. Furthermore, the
correlation between the expression level of miR-377 and
clinicopathological characteristics was analyzed. The prognostic
value of miR-377 on the prognosis of GC patients was also
estimated. Further analyses showed that the overexpression of
miR-377 promoted GC cell proliferation. Moreover, p53, PTEN and
TIMP1 were identified as direct targets of miR-377.
Materials and methods
Patients and specimens
This study was approved by the Medical Ethics
Committee of Jiangsu Cancer Hospital (Nanjing, China) and written
informed consent was obtained from all of the patients. The
specimens were handled and made anonymous according to the ethical
and legal standards. Paired tissue specimens (tumor and adjacent
normal mucosa) from 102 patients with GC were obtained and
histologically confirmed by a pathologist at Jiangsu Cancer
Hospital and the First People’s Hospital of Yunnan Province from
January, 2007 to December, 2010. The samples were derived from
patients who had not received adjuvant treatment including
radiotherapy or chemotherapy prior to surgery in order to eliminate
potential treatment-induced changes to gene expression profiles.
After excision, the tissue specimens were immediately frozen in
liquid nitrogen for subsequent analysis. The specimens were stained
with hematoxylin and eosin and examined histopathologically. The
sections that consisted of >80% carcinoma cells were used to
prepare the total RNA. Clinicopathological information, including
gender, age, tumor size, depth of invasion, lymph node invasion,
distant metastasis, TNM stage, tumor differentiation and early
recurrence was available for all the patients.
Cell culture
The human HGC-27, MKN-45, BGC-823, AGS, SGC-7901,
MGC80-3, NCI-N87 and SNU-1 gastric cancer cell lines, and GES-1
normal gastric cell line were obtained from the American Type
Culture Collection (ATCC; Manassas, VA, USA). MKN-45, BGC-823, AGS,
SGC-7901 and SNU-1 cells were grown in Dulbecco’s modified Eagle’s
medium (DMEM) and HGC-27, MGC80-3, NCI-N87 and GES-1 were grown in
RPMI-1640. In each case, the medium was supplemented with 15% fetal
calf serum (FCS), 100 U/ml penicillin and 100 U/ml streptomycin.
The cells were incubated at 37°C in a humidified incubator
containing 5% CO2.
Reverse transcriptase-quantitative PCR
(RT-qPCR)
Approximately 40 mg of tissue sample or harvested
cells (2×106) was homogenized in 1 ml TRIzol reagent
(Invitrogen, Carlsbad, CA, USA), according to the manufacturer’s
instructions. The purity and concentration of RNA were determined
using a NanoDrop 1000 spectrophotometer (Thermo Fisher scientific,
Wilmington, DE, USA). RNA (2 µg) was reverse transcribed
from each sample to produce cDNA using the PrimeScript RT reagent
kit (Takara Biotechnology, Dalian, China). Quantitative PCR was
employed to determine the relative expression level of target genes
using the SYBR Premix ExTaq II kit (Takara) on the 7300 Real-Time
PCR systems (Applied Biosystems, Carlsbad, CA, USA). The PCR
cycling profile was denatured at 95°C for 30 sec, followed by 40
cycles of annealing at 95°C for 5 sec and extension at 60°C for 34
sec. Small nucleolar RNA U6 was used as an internal standard for
normalization. The cycle threshold (CT) value was calculated. The
2−ΔCT (ΔCT =CTmiR-377−CTU6RNA)
method was used to quantify the relative amount of miR-377.
miRNA mimics, plasmids and
transfection
The miR-377 mimic, miR-377 mutant (miR-377-mut) and
miR-377-inhibitor (miR-377-in) were purchased from the shanghai
GenePharma, Co. (Shanghai, China), together with the negative
control (miR-control). The transfection of miRNAs (50 nM) was
performed using X-tremeGENE (Roche, Reinach, Switzerland) according
to the manufacturer’s instructions. The 3′-UTR of p53, PTEN and
TIMP1 was PCR-amplified from MKN-45 genomic DNA and cloned
downstream of the luciferase gene in the pGL vector (Promega,
Madison, WI, USA). For the reporter assay, the cells were cultured
in 96-well plates and transfected with luciferase reporters (50 ng)
and 50 nM of miR-control, miR-377 mimics or miR-377-mut. After 48
h, luciferase activity was measured using the dual-luciferase
reporter system (Promega). The Renilla activity was used as
an internal control. Each transfection was performed in
triplicate.
MTT assay
The cells were seeded in 96-well plates
(2×103/well) 24 h after transfection. MTT (Beyotime,
Haimen, China) was added to each well and the cells were cultured
for 4 h at 37°C. The reaction was stopped by 150 µl DMSO and
the optical density at 490 nm was detected on a microplate reader
(Thermo Fisher scientific, Kalamazoo, MI, USA).
Colony formation assay
Five hundred of each transfected cells were plated
in 6-well plates and cultured for 14 days without any disturbance.
The cells were stained with 0.5% crystal violet for 1 h at 37°C.
visible colonies were counted in four different fields and the mean
value was calculated.
Cell cycle analysis
The cells were seeded into 6-well plates with a
density of 1×105 cells/well after transfection and
maintained in DMEM/RPMI-1640 containing 15% FCS. Cultured cells
were trypsinized after 48 h and fixed with 70% ethanol at 4°C
overnight before being stained with propidium iodide (PI)
(Invitrogen). DNA contents were detected using an LSRII flow
cytometer (BD Biosciences, san Jose, CA, USA). Data were analyzed
by Flow Jo (Tree Star, Ashland, OR, USA). TargetScan, miRanda and
DIANA were used to determine the putative human protein-coding gene
targets of miR-377.
Luciferase reporter assays
MKN-45 cells were co-transfected with miR-377 or
control or the mutated 3′-UTR (Mut) of p53, PTEN and TIMP1. After
48 h, the cells were collected and luciferase activity was assayed
using the dual-luciferase assay system (Promega).
Protein isolation and western blot
analysis
Total proteins were extracted with RIPA lysis buffer
with proteinase/phosphatase inhibitors (Beyotime). The lysate was
separated by 10% sodium dodecyl sulfate polyacrylamide gel
electrophoresis and the gel was blotted onto a PVDF membrane
(Bio-Rad, Hercules, CA, USA). The membrane was blocked in 1% BSA
and then incubated with one of the following antibodies: anti-p53
(Cellsignal, no. 2527), anti-PTEN (Cellsignal, no. 5384),
anti-TIMP1 (Cellsignal, no. 8946) or anti-GAPDH (Cellsignal, no.
2118). Goat radish peroxidase-conjugated anti-rabbit IgG
(Cellsignal, no. 2975) were incubated as the secondary antibodies.
Subsequent visualization was detected using a SuperSignal West
Femto Maximum Sensitivity Substrate (Thermo Fisher scientific).
Statistical analysis
Data are presented as means ± SD. A comparison of
the level of miR-377 expression between GC and adjacent normal
tissue was performed using the Wilcoxon test. The correlation
between the expression of miR-377 and clinicopathological
characteristics was assessed using a two-sample Student’s t-test.
The postoperative survival rate was analyzed using the Kaplan-Meier
method and differences in survival rates were assessed with the
log-rank test. A Cox proportional hazards analysis was performed to
calculate the hazard ratio (HR) and the 95% confidence interval
(CI) to determine the association between miR-377 expression and
survival. In addition, a multivariate Cox regression was performed
to adjust for other covariates. The tests were two-tailed and
results with P<0.05 were considered statistically significant.
Statistical analyses were performed using SPSS 19.0 software (SPSS,
Chicago, IL, USA).
Results
miR-377 was overexpressed in human GC
tissues and cell lines
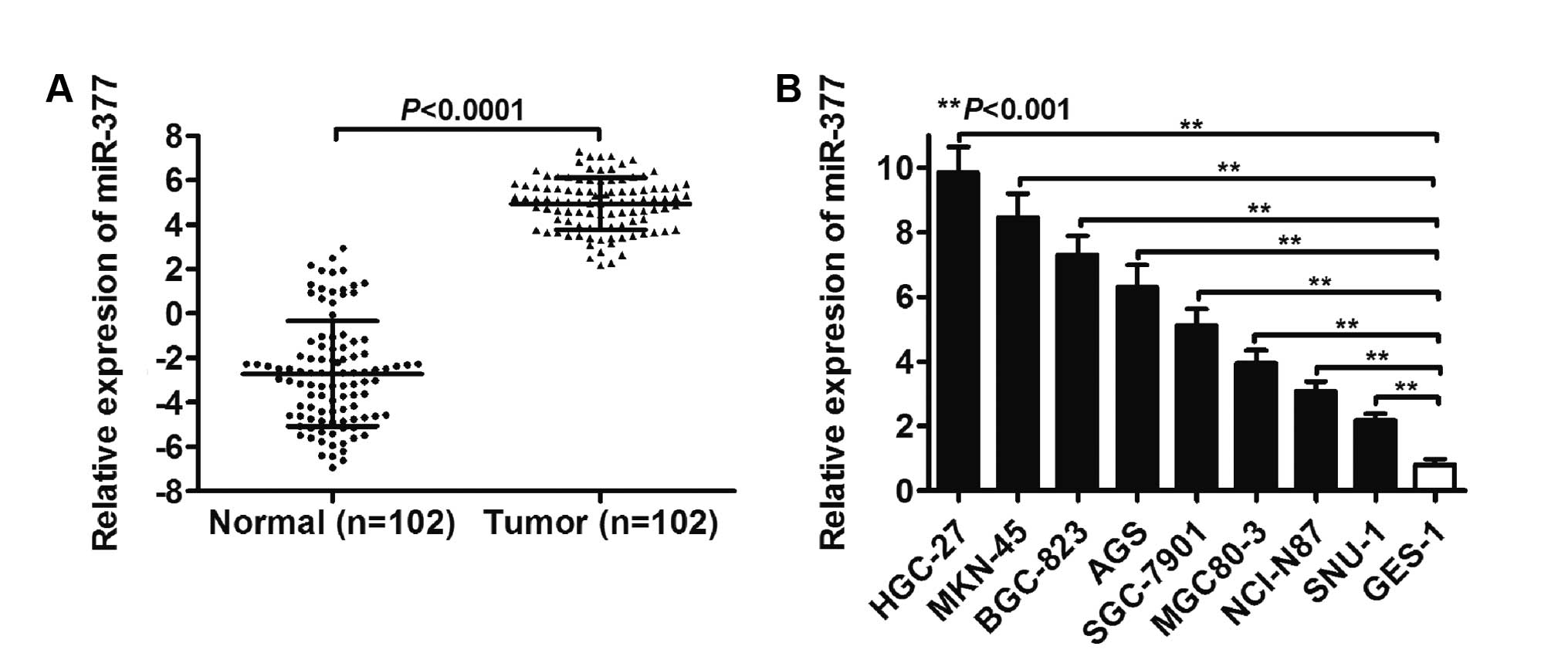
As shown in Fig. 1A,
the overall expression level of miR-377 was determined by RT-qPCR
in 102 paired normal and GC tissues. A significantly different
expression level of miR-377 between the normal and tumor groups was
identified (P<0.0001). Furthermore, miR-377 expression in tumor
tissues showed elevated levels of miR-377 compared to the
corresponding normal tissues, with an average increase of
9.62-fold. Levels of miR-377 in the BGC-823, AGS, SGC-7901,
MGC80-3, NCI-N87 and SNU-1 GC cell lines HGC-27, MKN-45, were also
higher than that in the normal GES-1 cell line (P<0.001,
Fig. 1B).
Correlations between miR-377 expression
and clinicopatho- logical characteristics
To identify the clinical relevance of miR-377
expression in GC, the correlations between miR-377 expression and
clinicopathological parameters such as gender, age, tumor size,
depth of invasion, lymph node invasion, distant metastasis, TNM
stage, tumor differentiation and early recurrence were examined
(Table I). Of the 102 GC patients,
82 cases were included in the miR-377-high group and the remaining
20 cases were included in the miR-377-low group. As shown in
Table I, the results demonstrated
that a high expression of miR-377 was significantly correlated with
distant metastasis (P=0.034), TNM stage (P=0.030) and early
recurrence (P=0.044). However, there was no significant association
between miR-377 expression and other clinicopathological
chacteristics, such as gender, age and tumor size, depth of
invasion, lymph node invasion and tumor differentiation
(P>0.05).
 | Table IClinicopathological parameters of GC
patients and the correlation with miR-377 expression. |
Table I
Clinicopathological parameters of GC
patients and the correlation with miR-377 expression.
| Characteristics | No. | miR-377 expression
| P-value |
|---|
| Low | High |
|---|
| Gender | | | | 0.328 |
| Male | 67 | 15 | 52 | |
| Female | 35 | 5 | 30 | |
| Age (years) | | | | 0.936 |
| <60 | 62 | 12 | 50 | |
| ≥60 | 40 | 8 | 32 | |
| Tumor size (cm) | | | | 0.174 |
| <4.5 | 63 | 15 | 48 | |
| ≥4.5 | 39 | 5 | 34 | |
| Depth of
invasion | | | | 0.942 |
| T1,T2 | 21 | 4 | 17 | |
| T3,T4 | 81 | 16 | 65 | |
| Lymph node
invasion | | | | 0.085 |
| Absent | 78 | 18 | 60 | |
| Present | 24 | 2 | 22 | |
| Distant
metastasis | | | | 0.034 |
| Absent | 40 | 12 | 28 | |
| Present | 62 | 8 | 54 | |
| TNM stage | | | | 0.030 |
| I,II | 35 | 11 | 24 | |
| III,IV | 67 | 9 | 58 | |
| Tumor
differentiation | | | | 0.258 |
| Well,
moderate | 60 | 14 | 46 | |
| Poor,
mucinous | 42 | 6 | 36 | |
| Early
recurrence | | | | 0.044 |
| No | 41 | 12 | 29 | |
| Yes | 61 | 8 | 53 | |
Prognostic value of miR-377
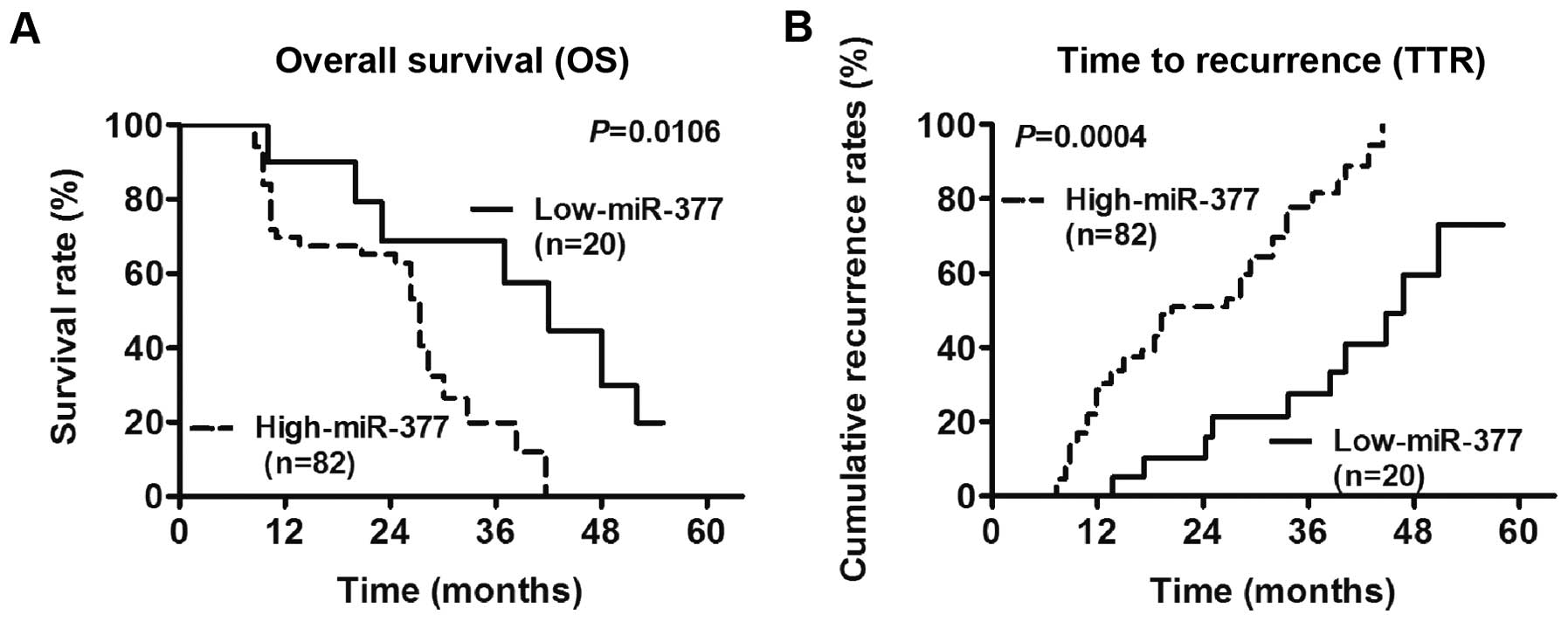
The results showed that miR-377 was a prognostic
factor for overall survival (OS) and time to recurrence (TTR) in
GC. The Log-rank test results showed that the 3- and 5-year OS
rates in the group with miR-377 upregulation were significantly
lower than those in the group with miR-377 downregulation
(P=0.0106) (Fig. 2A). Similarly,
the cumulative TTR rates in the miR-377 upregulation group were
significantly higher than those in the miR-377 downregulation group
(P=0.0004) (Fig. 2B). Furthermore,
the Cox regression analysis suggested that distant metastasis and
miR-377 expression status were independent factors that affected OS
(P=0.027, HR=1.62, 95% CI=1.13–2.78), while miR-377 expression
status was an independent factor for TTR (P=0.022, HR=2.14, 95%
CI=0.87-4.42) (Table II).
Collectively, these data suggested that miR-377 was an independent
prognostic biomarker in GC.
 | Table IIUnivariate and multivariate analyses
of prognostic variables of overall survival and time to recurrence
in GC patients. |
Table II
Univariate and multivariate analyses
of prognostic variables of overall survival and time to recurrence
in GC patients.
| Variables | Univariate analysis
| Multivariate
analysis
|
|---|
| HR | 95% CI | P-value | HR | 95% CI | P-value |
|---|
| Overall
survival | 3.56 | 3.28–4.34 | 0.713 | | | |
| Gender (male vs.
female) | 2.77 | 2.65–3.98 | 0.248 | | | |
| Age (<60 vs.
≥60) | 2.81 | 2.36–3.51 | 0.057 | | | |
| Tumor size
(<4.5 vs. ≥4.5) | 2.15 | 1.76–3.01 | 0.323 | | | |
| Depth of invasion
(T1,T2 vs. T3,T4) | 1.88 | 1.45–3.54 | 0.082 | | | |
| Lymph node
invasion (absent vs. present) | 2.55 | 1.76–4.22 | 0.013 | 1.73 | 1.26–2.63 | 0.091 |
| Distant metastasis
(absent vs. present) | 1.39 | 0.88–3.19 | 0.025 | 1.82 | 1.04–3.15 | 0.016 |
| TNM stage (I,II
vs. III,IV) | 1.14 | 0.63–1.79 | 0.014 | 0.96 | 0.65–1.61 | 0.065 |
| Tumor
differentiation | 1.37 | 1.01–2.63 | 0.419 | | | |
| (Well, moderate
vs. poor, mucinous) | | | | | | |
| Early recurrence
(no vs. yes) | 1.19 | 0.88–2.69 | 0.035 | 1.65 | 1.17–3.05 | 0.043 |
| miR-377 expression
(low vs. high) | 1.48 | 1.09–2.43 | 0.006 | 1.62 | 1.13–2.78 | 0.027 |
| Time to
recurrence | | | | | | |
| Gender (male vs.
female) | 2.08 | 1.86–2.83 | 0.216 | | | |
| Age (<60 vs.
≥60) | 2.68 | 2.38–3.59 | 0.971 | | | |
| Tumor size
(<4.5 vs. ≥4.5) | 4.02 | 3.21–4.46 | 0.317 | | | |
| Depth of invasion
(T1,T2 vs. T3,T4) | 2.94 | 2.62–3.94 | 0.462 | | | |
| Lymph node
invasion (absent vs. present) | 3.27 | 2.95–4.09 | 0.013 | 2.97 | 1.89–3.32 | 0.093 |
| Distant metastasis
(absent vs. present) | 2.38 | 0.52–4.25 | 0.045 | 3.71 | 3.39–4.67 | 0.232 |
| TNM stage (I,II
vs. III,IV) | 1.92 | 0.67–4.29 | 0.018 | 2.51 | 2.19–3.33 | 0.307 |
| Tumor
differentiation | 1.56 | 0.18–3.08 | 0.054 | | | |
| (Well, moderate
vs. poor, mucinous) | | | | | | |
| Early recurrence
(no vs. yes) | 1.23 | 0.54–2.61 | 0.027 | 2.03 | 0.17–3.85 | 0.041 |
| miR-377 expression
(low vs. high) | 0.77 | 0.41–2.87 | 0.012 | 2.14 | 0.87–4.42 | 0.022 |
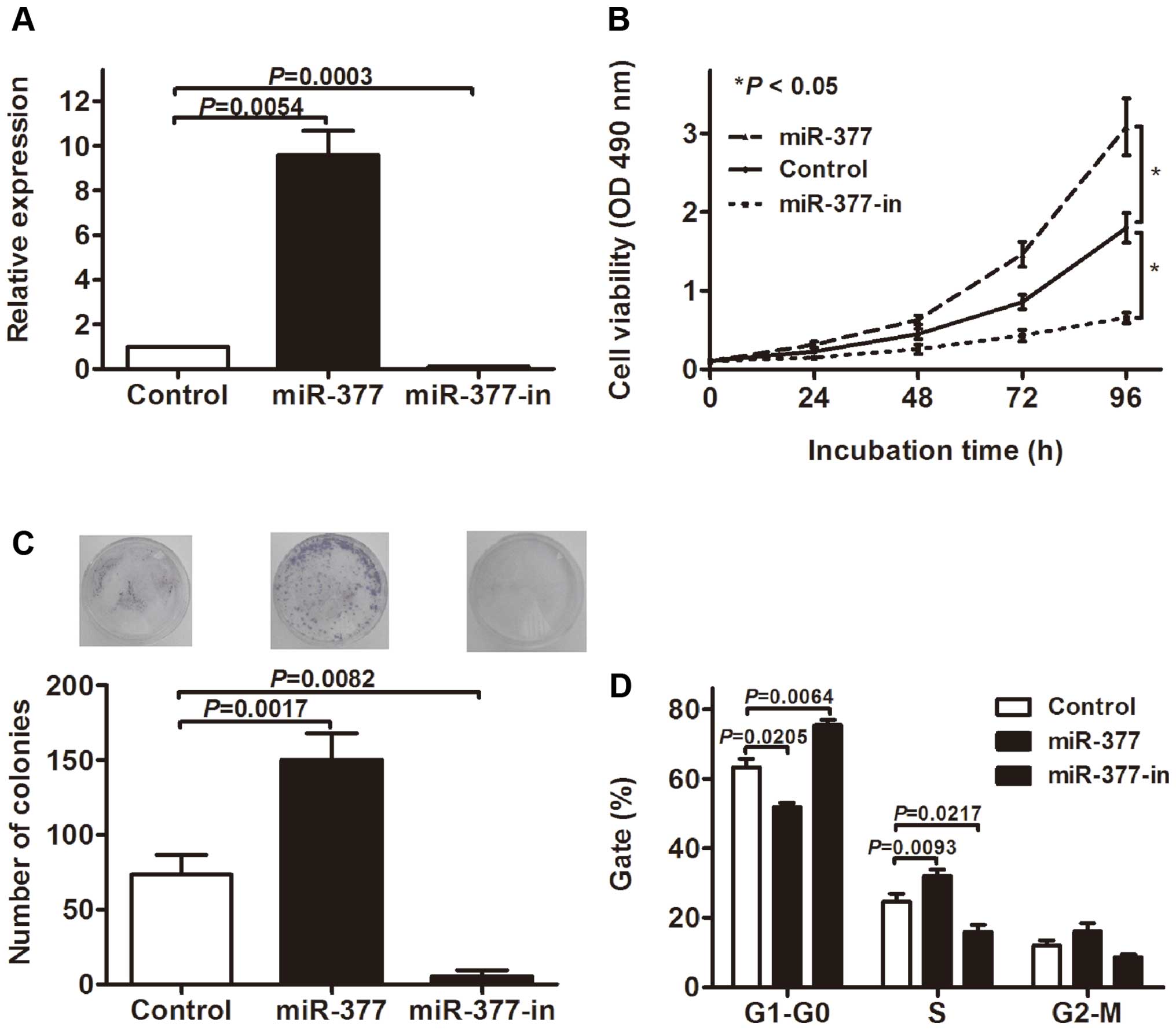
miR-377 promotes GC cell
proliferation
To examine the role of miR-377 in GC tumorigenesis,
we examined the effect of miR-377 overexpression and inhibition on
the proliferation of MKN-45 GC cell lines. The cells were
transfected with miR-377 mimic (miR-377), miR-377 inhibitor
(miR-377-in) or the miR scramble control oligonucleotides
(Control). RT-qPCR showed that miR-377 was significantly increased
in the cells transfected with miR-377 mimics and decreased in the
miR-377 inhibitor group compared with the control (Fig. 3A). The MTT assay showed that the
overexpression of miR-377 significantly promoted the proliferation
of MKN-45 cells, whereas the inhibition of miR-377 suppressed cell
proliferation (Fig. 3B). The colony
formation assay was performed to further confirm the effect of
miR-377 on GC cell proliferation, and data indicated that the
overexpression of miR-377 significantly increased colony numbers in
MKN-45 cell cultures, whereas the knockdown of miR-377 expression
obviously decreased colony formation (Fig. 3C). Furthermore, we assessed the cell
cycle by flow cytometry. As shown in Fig. 3D, the transfection of miR-377
decreased the percentage of cells in G1 peak but increased that in
the s peak. Similarly, miR-377-in led to cell cycle arrests in
MKN-45 cells. Collectively, these results showed that miR-377
promotes GC cell growth.
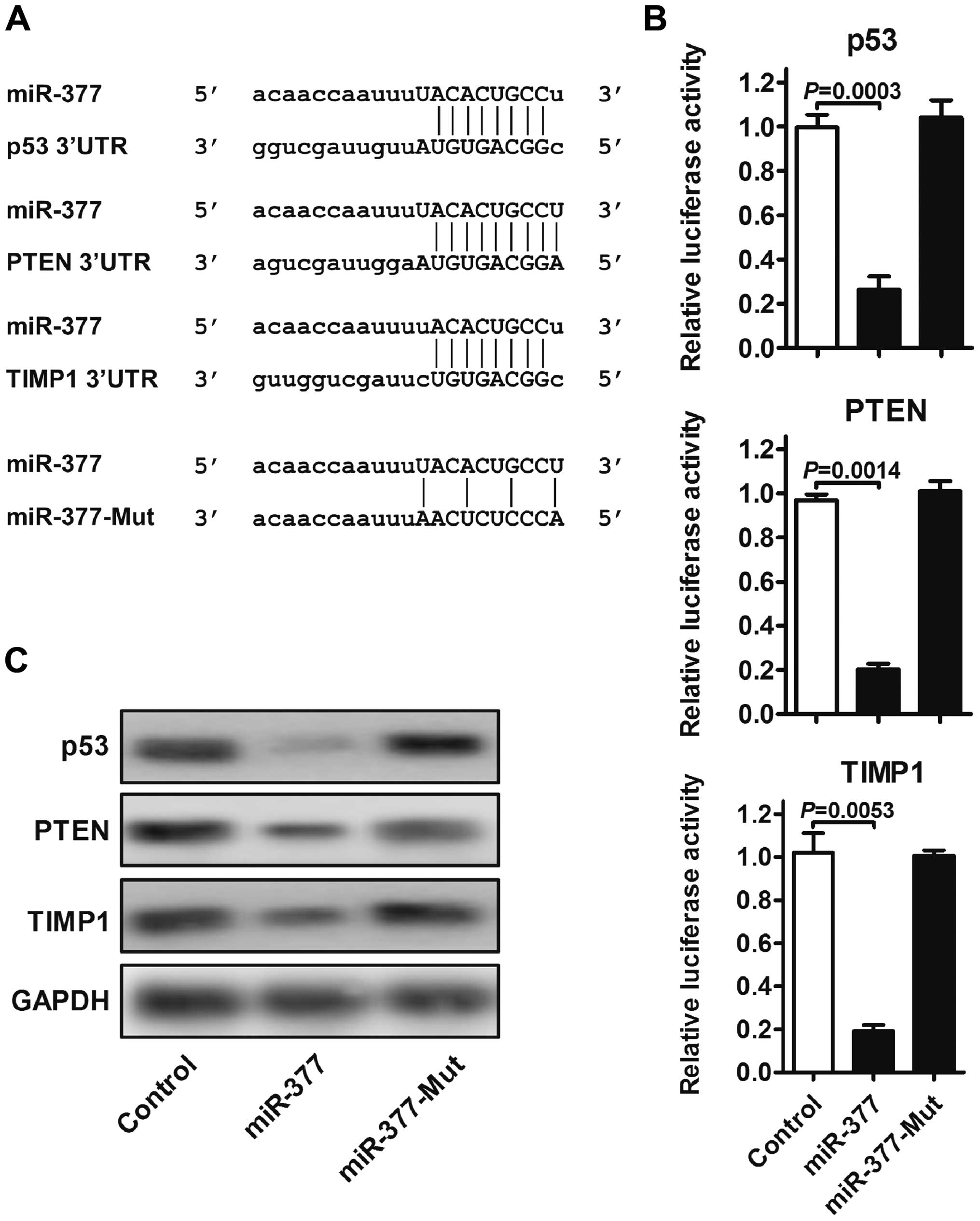
miR-377 directly targets p53, PTEN and
TIMP1
To gain insight into the biological implications of
miR-377 on GC tumorigenesis, we used TargetScan, miRanda and DIANA
for the putative human protein-coding gene targets of miR-377. The
tumor-suppressor genes p53, PTEN and TIMP1 were predicted to have
miR-377-binding elements in their 3′-UTRs with high fidelity scores
(Fig. 4A). To assess whether the
predicted miR-377-binding sites in the 3′-UTR of the three target
genes were responsible for miR-377 regulation, we cloned the 3′-UTR
regions downstream of a luciferase reporter gene and co-transfected
these vectors together with control, miR-377 and miR-377-mut into
MKN-45 cells, respectively. The luciferase activity of cells
transfected with miR-377 was significantly decreased compared with
the control. However, the mutation of miR-377 clearly abrogated the
repression of the luciferase activity (Fig. 4B). To investigate whether miR-377
regulated these targets, the protein expression levels of p53, PTEN
and TIMP1 were assessed by western blot analysis. As shown in
Fig. 4C, the overexpression of
miR-377 markedly altered the protein levels of p53, PTEN and TIMP1,
whereas the protein expression of these target proteins was not
affected by mutant miR-377. These results suggested that p53, PTEN
and TIMP1 are direct targets of miR-377 and these three targets may
mediate the promotive effect of miR-377 on tumorigenesis.
Discussion
GC is one of the most common and lethal cancers,
with a high relapse rate (1,14).
Therefore, it is essential to develop novel, prognostic factors and
therapeutic strategies. The outcome of GC patients is determined
primarily by the presence or absence of metastasis (15). Thus, identification of the precise
molecular mechanisms that modulate malignant transformation is
necessary. Identification of miRNA molecular profiles associated
with the prognosis of patients with GC only shed light on the
elucidation of the underlying biological mechanisms involved in the
development or progression of the disease, and provide the
opportunity to identify novel targets for GC diagnosis and
treatment (16). To the best of our
knowledge, we have shown for the first time that miR-377 was
frequently upregulated in GC tissues than in their normal adjacent
tissues. Furthermore, the results suggest that miR-377 is involved
in the progression of GC. Of note, we found that a high expression
of miR-377 was a significant predictor of OS and TTR.
Previous findings suggested that miR-377 plays a
vital role in stress responses during pathological conditions, such
as diabetes and aging (9,10,17).
Overexpression of miR-377 has been observed in human mesangial
cells, and was associated with an increased expression of the
matrix protein, fibronectin, which is accumulated in excess in
diabetic nephropathy (10). In the
present study, we first confirmed that miR-377 was frequently
upregulated in GC tissues than in their normal adjacent mucosa, and
identified a high miR-377 expression as a valid factor associated
with advanced tumor stages. These data suggest the potential of
miR-377 to serve as a molecular target for GC therapy, especially
for tumors with a high potency of metastasis.
The correlation between the miR-377 expression level
and clinicopathological values of GC remains unclear. In the
present study, we first investigated the relationship of miR-377
expression with clinicopathological characteristics and prognosis.
The miR-377 expression was found to be associated with distant
metastasis, TNM stage and early TTR, suggesting that miR-377 was
involved in the tumorigenesis, development, progression and
metastasis of GC. Of note, miR-377 expression was significantly
associated with the OS and TTR of GC patients. In support of this,
the Kaplan-Meier analysis of overall survival showed that patients
whose tumors with a higher miR-377 expression had a significantly
worse OS and TTR, indicating that a high miR-377 level is a
biomarker of poor prognosis for GC patients. Moreover, the Cox
proportional hazards model showed that miR-377 was an independent
marker of poor OS and TTR of the known clinical prognostic
indicators including distant metastasis, TNM stage and early TTR.
Therefore, constitutes a molecular prognostic marker for GC
patients, identifying higher risk of death in patients. Thus, good
candidates are to receive a more aggressive treatment. To the best
of our knowledge, this is the first report to describe the clinical
significance of miR-377 in TTR and prognosis of GC patients.
To examine the significance of miR-377 upregulation
in GC, several assays were performed. First, we conducted a cell
growth assay on MKN-45 cells to assess whether the differential
expression of miR-377 affected proliferation. We found that the
upregulation of miR-377 expression significantly promoted
proliferation and that inhibition of miR-377 suppressed cell
proliferation. When assessing the cell cycle of MKN-45 cells by
flow cytometry, it was found that miR-377-mimic-transfected cells
yielded a population with more cells in the S-phase, and fewer
cells in G1-phase, and that inhibition of miR-377 led to cell cycle
arrest. The S-phase is the period in which a cell performs DNA
replication and the G1/S transition is a major checkpoint in the
regulation of the cell cycle (18).
Our data suggest that miR-377 affects mechanisms that control the
G1/S transition in GC cells.
To investigate the downstream effects of miR-377
overexpression in GC cells, we developed a luciferase-reporter
system to assess whether miR-377 can regulate known tumor
suppressors and found that miR-377 targets p53, PTEN and TIMP1. p53
is an important tumor-suppressor gene and responds to diverse
cellular stresses to regulate the expression of target genes,
especially inducing cell cycle arrest and apoptosis (19). Mutations in p53 are associated with
a variety of human cancers (20).
The p53 has been previously reported to be downregulated in GC
tissues, and restoring its expression inhibits growth and reduces
clonogenic activity in GC cell lines (21). The result is consistent with our
observations that MKN-45 cells have a downregulated p53 expression
caused by miR-377, and increased proliferation ability. PTEN has
been identified as a tumor suppressor that is mutated in a large
number of cancers at high frequency (22). The protein encoding this gene is
phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase (23). It negatively regulates the
intracellular levels of phosphatidylinositol-3,4,5-trisphosphate in
cells and negatively regulates the AKT/PKB signaling pathway
(24). Thus, it seems likely that
our observation of increased cell numbers in the S-phase and
decreased numbers in the G1-phase was caused by the modulation of
PTEN by miR-377. TIMP1 has been demonstrated to be a suppressor of
tumorigenesis and progression (25). It is a natural inhibitor of the
matrix metalloproteinases (MMPs), which are able to promote cell
proliferation and may also have an anti-apoptotic function
(26). Dysregulation of TIMP1 has
been found in several types of cancer (27). In the present study, TIMP1 was found
to be downregulated by miR-377 and when miR-377-overexpressing
cells exhibited increased growth activity. This result suggests
that the dysregulation of TIMP1 by miR-377 is a mechanism
associated with this process. The present study provides
confounding evidence that an upregulated expression of miR-377 in
GC modulates core mechanisms in the control of cell cycle
progression and proliferation, by inhibiting the expression of
tumor suppressor p53, PTEN and TIMP1. The differential expression
of miR-377 in GC can potentially be utilized in diagnostic
applications and therapeutic interventions.
In conclusion, to the best of our knowledge, the
results of the present study have, for the first time, demonstrated
that miR-377 was overexpressed in GC tissue and cell lines and
associated with tumorigenesis and poor prognosis. The present study
also demonstrated that miR-377 was an independent prognostic factor
of patients with GC. Of note, the upregulated expression of miR-377
in GC modulates core mechanisms in the control of proliferation and
cell cycle progression, by inhibiting expression of tumor
suppressor p53, PTEN and TIMP1. The differential expression of
miR-377 in GC can be a candidate therapeutic target and a potential
biomarker for the diagnosis and prognosis in GC.
Acknowledgments
The present study was supported by grants from the
Agency of Jiangsu Province Science and Technology (no. 2013035) and
the Research Office of Jiangsu Cancer Hospital (no. ZK201401).
References
|
1
|
Gibson CJ, Britton KA, Miller AL and
Loscalzo J: Clinical problem-solving. Out of the blue. N Engl J
Med. 370:1742–1748. 2014.PubMed/NCBI
|
|
2
|
Wu WKK, Lee CW, Cho CH, Fan D, Wu K, Yu J
and Sung JJ: MicroRNA dysregulation in gastric cancer: A new player
enters the game. Oncogene. 29:5761–5771. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Song JH and Meltzer SJ: MicroRNAs in
pathogenesis, diagnosis, and treatment of gastroesophageal cancers.
Gastroenterology. 143:35–47. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lin X, Yang M, Xia T and Guo J: Increased
expression of long noncoding RNA ABHD11-AS1 in gastric cancer and
its clinical significance. Med Oncol. 31:422014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Shen L, Shan YS, Hu HM, Price TJ, Sirohi
B, Yeh KH, Yang YH, Sano T, Yang HK, Zhang X, et al: Management of
gastric cancer in Asia: Resource-stratified guidelines. Lancet
Oncol. 14:e535–e547. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Chitwood DH and Timmermans MC: Small RNAs
are on the move. Nature. 467:415–419. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Morris KV, Chan SW, Jacobsen SE and Looney
DJ: Small interfering RNA-induced transcriptional gene silencing in
human cells. Science. 305:1289–1292. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Cai H, Yuan Y, Hao YF, Guo TK, Wei X and
Zhang YM: Plasma microRNAs serve as novel potential biomarkers for
early detection of gastric cancer. Med Oncol. 30:4522013.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yu X, Luo L, Wu Y, Yu X, Liu Y, Yu X, Zhao
X, Zhang X, Cui L, Ye G, et al: Gastric juice miR-129 as a
potential biomarker for screening gastric cancer. Med Oncol.
30:3652013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wang Q, Wang Y, Minto AW, Wang J, Shi Q,
Li X and Quigg RJ: MicroRNA-377 is up-regulated and can lead to
increased fibronectin production in diabetic nephropathy. FAsEB J.
22:4126–4135. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wen ZL, Huang W, Feng YL, Wang Y, Liang J,
Cai W, Kang K, Chang D, Zhu P, Milliard RW, et al: MicroRNA-377
regulates angiogenesis by targeting vEGF: Implications for
mesenchymal stem cells based therapy in ischemic heart disease.
Circulation. 128. 2013
|
|
12
|
Li N, Liu Q, Su Q, Wei C, Lan B, Wang J,
Bao G, Yan F, Yu Y, Peng B, et al: Effects of legumain as a
potential prognostic factor on gastric cancers. Med Oncol.
30:6212013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Orditura M, Galizia G, Sforza V,
Gambardella V, Fabozzi A, Laterza MM, Andreozzi F, Ventriglia J,
Savastano B, Mabilia A, et al: Treatment of gastric cancer. World J
Gastroenterol. 20:1635–1649. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Vicentini C, Fassan M, D’Angelo E, Corbo
V, Silvestris N, Nuovo GJ and Scarpa A: Clinical application of
microRNA testing in neuroendocrine tumors of the gastrointestinal
tract. Molecules. 19:2458–2468. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Beckman JD, Chen C, Nguyen J, Thayanithy
V, Subramanian V, Steer CJ and Vercellotti GM: Regulation of heme
oxygenase-1 protein expression by miR-377 in combination with
miR-217. J Biol Chem. 286:3194–3202. 2011. View Article : Google Scholar :
|
|
16
|
Zhang R, Luo H, Wang S, Chen W, Chen Z,
Wang HW, Chen Y, Yang J, Zhang X, Wu W, et al: MicroRNA-377
inhibited prolife ration and invasion of human glioblastoma cells
by directly targeting specificity protein 1. Neuro Oncol.
16:1510–1522. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Tsirimonaki E, Fedonidis C, Pneumaticos
SG, Tragas AA, Michalopoulos I and Mangoura D: PKCε signalling
activates ERK1/2, and regulates aggrecan, ADAMTs5, and miR377 gene
expression in human nucleus pulposus cells. PLoS One. 8:e820452013.
View Article : Google Scholar
|
|
18
|
Bertoli C, Skotheim JM and de Bruin RAM:
Control of cell cycle transcription during G1 and S phases. Nat Rev
Mol Cell Biol. 14:518–528. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
19
|
Duffy MJ, Synnott NC, McGowan PM, Crown J,
O’Connor D and Gallagher WM: p53 as a target for the treatment of
cancer. Cancer Treat Rev. 40:1153–1160. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Shetzer Y, Solomon H, Koifman G,
Molchadsky A, Horesh S and Rotter V: The paradigm of mutant
p53-expressing cancer stem cells and drug resistance.
Carcinogenesis. 35:1196–1208. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kubota E, Williamson CT, Ye R, Elegbede A,
Peterson L, Lees-Miller SP and Bebb DG: Low ATM protein expression
and depletion of p53 correlates with olaparib sensitivity in
gastric cancer cell lines. Cell Cycle. 13:2129–2137. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Song MS, Salmena L and Pandolfi PP: The
functions and regulation of the PTEN tumour suppressor. Nat Rev Mol
Cell Biol. 13:283–296. 2012.PubMed/NCBI
|
|
23
|
Hollander MC, Blumenthal GM and Dennis PA:
PTEN loss in the continuum of common cancers, rare syndromes and
mouse models. Nat Rev Cancer. 11:289–301. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Carnero A, Blanco-Aparicio C, Renner O,
Link W and Leal JFM: The PTEN/PI3K/AKT signalling pathway in
cancer, therapeutic implications. Curr Cancer Drug Targets.
8:187–198. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Bjerre C, Vinther L, Belling KC, Würtz SØ,
Yadav R, Lademann U, Rigina O, Do KN, Ditzel HJ, Lykkesfeldt AE, et
al: TIMP1 overexpression mediates resistance of MCF-7 human breast
cancer cells to fulvestrant and down-regulates progesterone
receptor expression. Tumour Biol. 34:3839–3851. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lu YC, Chang JT, Liao CT, Kang CJ, Huang
SF, Chen IH, Huang CC, Huang YC, Chen WH, Tsai CY, et al:
OncomiR-196 promotes an invasive phenotype in oral cancer through
the NME4-JNK-TIMP1-MMP signaling pathway. Mol Cancer. 13:2182014.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Bunatova K, Pesta M, Kulda V, Topolcan O,
Vrzalova J, Sutnar A, Treska V, Pecen L and Liska V: Plasma TIMP1
level is a prognostic factor in patients with liver metastases.
Anticancer Res. 32:4601–4606. 2012.PubMed/NCBI
|