Introduction
Regulation of gene expression is predominantly
directed by alternative pre-mRNA splicing. A fine-tuned balance of
factors is vital to the subtle distinction in the determination of
splicing patterns with crucial biological consequences. Alterations
in alternative splicing patterns result in functional changes in
gene products and may account for both the cause or consequence of
various human diseases (1).
Cancer-associated changes in splicing processes accompanying the
development and progression of malignant phenotypes have been
identified in numerous studies (2,3).
Various pre-mRNAs originating from aberrant splicing most likely
predispose cancer and are under consideration to serve as
therapeutic targets (2), e.g. tumor
suppressors (4), angiogenic factors
(5–7) or apoptosis inhibitors (8,9).
Specific pre-mRNAs from alternatively spliced genes have been shown
to contribute to acquisition of drug-resistant phenotypes in
reaction to anticancer chemotherapies (10).
The proto-oncogene recepteur d’origine nantais (RON,
MST1R) encodes a transmembrane tyrosine kinase receptor for
macrophage-stimulating protein (MSP) and plays a crucial role in
various tumor biological processes, such as cell motility,
adhesion, proliferation, apoptosis and epithelial-to-mesenchymal
transition (EMT) (11). The 185-kDa
RON mature protein is characterized by a disulfide-linked
extracellular α (40 kDa) and a transmembrane β chain (145 kDa) and
is organized in heterodimers with unique functional features of the
MET protein family to control cell dissociation and invasion of
extracellular matrices (scattering) (12,13).
The multifunctional factor is involved in several oncogenic
pathways, such as PI-3 kinase/Akt, β-catenin, MAP kinase and JNK
(14), where functional diversity
originates from various alternative RON splice variants (13). In addition to mature RON, six
isoforms emerging from alternative pre-mRNA splicing, protein
truncation or alternative transcription start have been identified
to date (Table I) (13,15).
 | Table IRON splicing isoforms and functional
implications. |
Table I
RON splicing isoforms and functional
implications.
| RON variant | Mechanism | Tyrokinase
activity | Cellular
localization | Functional
implication | Refs. |
|---|
| RONΔ170 | Splicing exon 19
deletion | − | Membrane | Inhibitor of
oncogenic activities | (13,30) |
| RONΔ165 | Splicing in-frame
deletion in exon 11 | + | Accumulation of
single-chain pro-RONΔ165 in cytoplasm | Motile and invasive
phenotype | (11–13,18,28,30,31) |
| RONΔ160 | Splicing in-frame
deletion in exon 5–6 | + | Membrane | Tumor growth,
increase in β-catenin target gene expression, metastasis | (13,17,18,30–33) |
| RONΔ155 | Splicing combined
deletion of exons 5, 6 and 11 | + | Cytoplasm | Tumor growth,
similarly to RONΔ160 | (13,18,30,31) |
| RONΔ110 | Proteolytic
truncation | + | Cell surface,
cytoplasm | RON-overexpressing
cancer cells, tumorigenicity | (13,30,33) |
| RONΔ55 | Alternative
transcription start in exon 11 | + | Membrane,
cytoplasm | Cell transformation
and migration | (13,30) |
Expression of alternative RON isoforms Δ160 and Δ165
are clearly correlated to tumor progression processes (12,16–18).
Altered RON expression profiles represent an auspicious target for
therapeutic approaches, such as chemical inhibitors or specific
antibodies against cancer-associated RON protein variants (13).
Ovarian cancer is diagnosed in 200,000 women every
year worldwide. It accounts for 20% of all cancer-related deaths
among women. In ~90% of cases, development of ovarian cancer is a
sporadic event, in 10% there is a genetic disposition [breast and
ovarian cancer syndrome (BOC) or Lynch II syndrome] (19). Malignant transformation in ovarian
cancer is supported by several molecular changes such as
upregulation of RTKs (receptor tyrosine kinases), mutation of tumor
suppressor genes and increased activity of proteolytic enzymes.
Ferrandina and colleagues (20) investigated the prognostic relevance
of RON protein expression levels in ovarian cancer patients by
immunohistochemical analysis pointing out a specific significant
link of platinum resistance and RON expression. The correlation of
RON-associated chemoresistance against cisplatin was also shown
in vitro (21).
Splicing factors belonging to the family of SR
proteins (SRp20, SRp40, SRp55, SRp75 and ASF/SFRS1) and hTra2-β1
play a crucial role in constitutive and alternative splicing and
are involved in post-transcriptional processes such as mRNA nuclear
export, nonsense-mediated decay and translation (22,23).
In breast cancer as well as in ovarian cancer a specific induction
of different splicing factors has been observed (24,25).
YB-1 is a member of the family of cold-shock proteins and acts as a
splicing and transcription factor. In ovarian cancer,
overexpression of YB-1 is associated with chemotherapy drug
resistance and serves as a negative prognostic factor (26,27).
The functional role of RON in ovarian carcinogenesis
remains unclear. In the present study, we performed a targeted
analysis of expression of specific RON variants in primary ovarian
cancer and investigated its potential correlation to
clinicopathological parameters. Since RON alternative splicing is
governed by splicing factor activity (13,28) we
also performed correlative analysis of expression levels of several
splicing factors with known involvement in RON mRNA processing.
Materials and methods
Patients and tissue samples
The present study is based on a multi-center cohort
of 43 patients who received surgical treatment for primary
epithelial ovarian cancer between 2003 and 2009. Tissues were
sampled during primary surgery at the Departments of Obstetrics and
Gynecology of the university Medical Center Freiburg, Charité
Campus Virchow-Klinikum Berlin, Medical Center Bayreuth, university
Medical Center Greifswald, university Medical Center Vienna and the
Oncological Institute of Moldavia and stored at the Tumor Bank
Ovarian Cancer (TOC). Specimens were freshly frozen in liquid
nitrogen and immediately stored at −80°C until further analysis.
Clinicopathological parameters were documented in the TOC database
including age, date of initial diagnosis and surgery, FIGO and TNM
stage, grade, histological subtype, tissue sample localization,
post-surgical therapies and follow-up data. Forty-five samples from
43 patients were collected and analyzed. Usually one tumor specimen
was obtained from each patient (41 patients). In two patients 2
tumor samples of different origin (omentum majus/left ovary and
omentum majus/uterus) were collected. In addition, 4
non-pathological ovarian tissue specimens were obtained at surgery
performed at the Department of Obstetrics and Gynecology of the
university Medical Center Freiburg and were histologically
classified at the Institute of Pathology. All patients provided
written consent and the study was approved by the Ethics Review
Committee of the university Medical Center Freiburg, Germany, in
accordance with human rights for patients in research.
Statistical analysis
The patient cohort (N=43) was subgrouped for
statistical analysis in regard to age, FIGO stage, TNM stage,
histological subtype, grade, localization of tumor tissue sampling
and resistance to platinum-derived chemotherapy (Table II). Statistical analysis was
performed using SPSS software (version 20.0; SPSS, Inc., Chicago,
IL, USA). Correlations were tested with the Spearman’s correlation
coefficient (rS) and the Mann-Whitney U test. A P-value
<0.05 was considered to indicate a statistically significant
result.
 | Table IICharacteristics of the ovarian cancer
patients evaluated in the study (N=43). |
Table II
Characteristics of the ovarian cancer
patients evaluated in the study (N=43).
| No. of
patients | Percentage (%) |
|---|
| Age (years) | | |
| <45 | 7 | 16.3 |
| 45–60 | 19 | 44.2 |
| ‰>60 | 16 | 37.2 |
| N/O | 1 | 2.3 |
| FIGO stage | | |
| I | 9 | 20.9 |
| II | 3 | 7.0 |
| III | 24 | 55.8 |
| IV | 4 | 9.3 |
| N/O | 3 | 7.0 |
| Grade | | |
| 1 | 6 | 14.0 |
| 2 | 13 | 30.2 |
| 3 | 17 | 39.5 |
| N/O | 7 | 16.3 |
| Histological
subtype | | |
| Serous | 33 | 76.7 |
| Mucinous | 4 | 9.3 |
| Others | 4 | 9.3 |
| N/O | 2 | 4.7 |
RNA isolation and PCR methods
Total RNA was isolated by TRIzol® reagent
(Invitrogen™, Life Technologies™ GmbH, Thermo Fischer Scientific,
Darmstadt, Germany) method according to the manufacturer’s protocol
after homogenization of tissue specimens by
Ultra-Turrax® (IKA Werke, Staufen, Germany). RNA quality
and quantity was determined by densitometry. Reverse transcription
of 2 μg purified RNA into cDNA was performed using MMLV-RT
(Promega GmbH, Mannheim, Germany), RiboLock RNase inhibitor
(Fermentas GmbH, Thermo Fischer Scientific, St. Leon-Roth,
Germany), and random hexamer primers (New England BioLabs GmbH,
Frankfurt, Germany) followed by either real-time PCR using
QuantiFast SYBR-Green PCR Master Mix (Qiagen, Hilden, Germany) or
conventional RT-PCR Taq polymerase (Genaxxon Bioscience
GmbH, Ulm, Germany) and gene-specific primers. For best possible
precision only singular PCR was performed. Isoforms with known
oncologic potential, such as isoforms with exon 5 and 6 skip
(potential RONΔ165) and exon 11 skip (potential RONΔ160) were
investigated. A PCR spanning exons 4 to 12 was not performed since
estimated amplicon sizes (>1,000 bp) were too big for accurate
separation. Expression levels of housekeeping gene RPS18 served as
the comparative value. The sequence of primers and expected
amplicon sizes were: for real-time PCR: RPS18-S, 5′-TAC TCA ACA CCA
ACA TCG ATG GGC-3′ and -AS, 5′-GCT TTC CTC AAC ACC ACA TGA GCA-3′;
HIF-1α-S, 5′-CGT TCC TTC GAT CAG TTG TC-3′ and -AS, 5′-TCA GTG GTG
GCA GTG GTA GT-3′; RON standard-S, 5′-TGA GGT CAA GGA TGT GCT GA-3′
and -AS, 5′-GTG ACT TGA TGG CAC ATT GG-3′; SRp75-S, 5′-TAA GGG CTA
CGG GAA GAT CC-3′ and -AS: 5′-CCA CAA AGG TCT TTG CCA TT-3′;
SRp55-S, 5′-GAC GGC TAC AGC TAC GGA AG-3′ and -AS, 5′-CCA ACT GCA
CCG ACT AGA AA-3′; SRp40-S, 5′-TCC AAG GGA TGC AGA TGA TGC TGT-3′
and -AS, 5′-TAT CGT CCT CTA CCT CTT CCA CCT-3′; SRp20-S 5′-GAA GAC
TCA TCG GAG CGT GT-3′ and -AS, 5′-TGT TGC CAT TGT TTC CAA GA-3′;
htra2-β1-S, 5′-ATG ACC AGC AGT CTA GGC GTT CAA-3′ and -AS, 5′-ATC
CTA CGC CCA TCA AGC TCC ATT-3′ and YB-1-S, 5′-TGG TTC AAT GTA AGG
AAC GGA-3′ and -AS, 5′-ACT GCG AAG GTA CTT CCT GG-3′; ASF/SF2-S,
5′-CGT GTT CTA CAA ATA CGG CG-3′ and -AS, 5′-ACC CAT CGT AAT CAT
AGC CG-3′; for RT-PCR: RONΔ165 (exon 11 skipping) S 5′-ACC TAG TTC
CAC TGA AGC CT-3′ and -AS, 5′-ACC AGT AGC TGA AGA CCA GT-3′ (339
and 147 bp); RONΔ160 (exon 5–6 skipping) S, 5′-CAC TGC CCA CCT AAG
CTT AC-3′ and -AS, 5′-CTG GTG CCT ACA GAC AGA CT-3′ (445, 285, 279
and 119 bp). RT-PCR products were subsequently analyzed by gel
electrophoresis.
Protein isolation and western blot
analysis
Protein isolation was performed in parallel to RNA
extraction by TRIzol® reagent method according to the
manufacturer’s protocol after homogenization of tissue specimens by
Ultra-Turrax®. Protein concentrations were determined with the BCA™
protein assay kit (Pierce, Thermo Fischer Scientific, Rockford, IL,
USA). Protein samples were separated by 10% SDS-PAGE and
transferred onto polyvinylidene difluoride (PVDF) membranes (Carl
Roth GmbH & Co. KG, Karlsruhe, Germany) by using a Bio-Rad
Trans-Blot Cell (Bio-Rad Laboratories GmbH, Munich, Germany).
Membranes were soaked in blocking solution [phosphate-buffered
saline (PBS) with 3% dry non-fat milk, 0.1% Tween-20] for 1 h at
room temperature and incubated with the primary polyclonal rabbit
IgG antibody anti-RON β (Santa Cruz Biotechnology, Inc., Santa
Cruz, CA, USA; dilution 1:500) in blocking buffer overnight at 4°C.
After washing, the membranes were further incubated with
horseradish peroxidase-conjugated goat anti-rabbit IgG secondary
antibody (Jackson ImmunoResearch Europe Ltd., Suffolk, UK; dilution
1:7,500) in blocking buffer for 2 h at room temperature. Finally
the immunocomplexes were made visible by an enhanced
chemiluminescence western blot detection kit (Pierce) and exposed
to X-ray film (Fujifilm Europe GmbH, Düsseldorf, Germany).
Results
Expression of oncogenic RON variants in
primary ovarian cancer
Forty-five tumor samples of primary ovarian cancer
and 4 non-pathological ovarian tissue samples were analyzed in
regard to the expression profiles of total RON (all isoforms). In
addition, the expression of the alternatively spliced RON isoforms
with exon skip 5–6 (potential RONΔ160 and RONΔ155) and exon skip 11
(potential RONΔ165) bearing a known oncogenic potential was
investigated in particular.
High levels of RON expression were detected in all
45 ovarian tumor tissues with statistical significance (105.94±6.46
vs. 0.00, P=0.001). The origin of the tumor specimens did not
influence the expression levels of RON; tissue samples of primary
tumors (N=25) as well as metastases (N=20) showed high levels of
RON expression (105.29±5.45 vs. 106.70±7.46, P=0.337). In contrast,
all non-pathological (physiological) ovarian tissue samples (N=4)
were characterized by no detectable RON expression (Fig. 1 and Table III). Alternative splicing of RON
exon 5/6 or exon 11 was noted in 39 of the 45 tumor samples
(86.67%) whereas 6 tumor samples showed no alternative splicing of
RON (13.33%). Notably, the non-pathological ovarian tissues did not
express any RON mRNA variants.
 | Table IIIRON expression in physiological
ovary, primary ovarian cancer tumor and metastatic tissues. |
Table III
RON expression in physiological
ovary, primary ovarian cancer tumor and metastatic tissues.
| N | RON standard
(median) | SD | P-value |
|---|
| Tumor | 25 | 105.29 | 5.45 | |
| Metastasis | 20 | 106.70 | 7.46 | 0.337 |
| Tumor | 25 | 105.29 | 5.45 | |
| Normal tissue | 4 | 0.00 | 0.00 | 0.002 |
| Metastasis | 20 | 106.70 | 7.46 | |
| Normal tissue | 4 | 0.00 | 0.00 | 0.002 |
|
Tumor/metastasis | 45 | 105.94 | 6.46 | |
| Normal tissue | 4 | 0.00 | 0.00 | 0.001 |
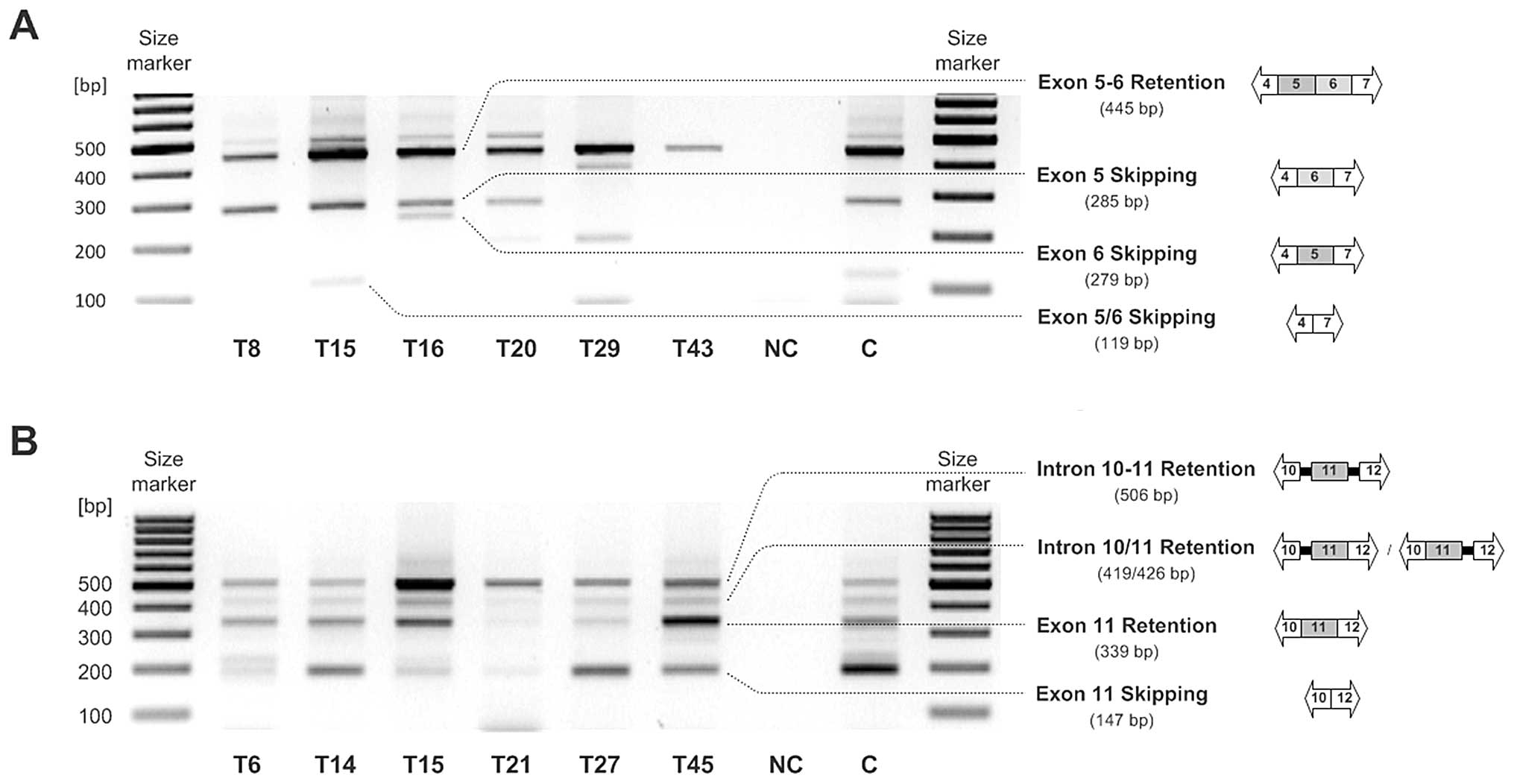
RT-PCR analysis employing RON exon 4 to 7 spanning
primers resulted in the detection of three different amplicons.
Full length RON (445 bp) expression was found in most malignant
ovarian tissues (39 of 45; 86.67%). The expected potential RONΔ160
or RONΔ155 mRNA (119 bp), characterized by exclusion of exon
cassette 5-6, was observed in a fraction of tumor samples only (11
of 45; 24.40%). The third RT-RCR amplicon with estimated 280 bp was
detected at a higher frequency in the ovarian cancer tissue
specimens (29 of 45; 64.44%). This RON mRNA could have been either
characterized by solitary exon 5 (284 bp) or exon 6 exclusion (297
bp), whereas subsequent sequence analysis clearly identified the
exon 5 skipping mRNA variant (284 bp) (Fig. 2A). In the metastatic tissues,
expression of RON mRNA variants with exclusion of exon cassette 5-6
was more frequent than that in the primary tumors (35 vs. 16%),
whereas in the primary tumor samples, expression of the exon 5
skipping mRNA variant (285 bp) was observed more often (76 vs.
50%).
Expression analysis of RON variants with exon 11
skipping (potential RONΔ165) via RT-PCR was performed using RON
exon 10 to 12 spanning primers and resulted in the detection of
conventionally spliced RON including exon 11 (339 bp) as well as an
exon 11-skipping RON variant (192 bp) with varying intensity in the
majority of the ovarian tumor tissues (37 of 45; 82.22%). In the
metastatic tissues, expression of RON variants with exclusion of
exon 11 was more frequent than that in the primary tumor tissues
(90 vs. 76%). Additionally, another amplicon (509 bp) occurred in
almost all analyzed malignant tissues (44 of 45; 97.78%). This mRNA
originated from intron 10–11 and intron 11–12 retention. Sequencing
analysis revealed that this PCR product does not encode for a
biologically active protein, since premature termination codons
(PTCs) are introduced (Fig. 2b).
PTCs most likely constitute RNA degradation via nonsense-mediated
decay.
Quantitative RON protein expression analysis
(western blot analysis) employed a specific antibody directed
against the C-terminus of RON. Non-pathological ovarian tissues
showed expression of two RON protein isoforms only, in fact
unprocessed pre-RON (190 kDa) and ‘short form RON’ (RONΔ55 with 55
kDa). Ovarian tumor specimens also expressed pre-RON and RONΔ55,
plus a variety of additional protein isoforms, e.g. RONΔ155 (125
kDa β-chain), RONΔ160 (135 kDa, β-chain), RONΔ165 (165 kDa, single
chain) (data not shown). Only slightly differing molecular weights
of the different isoforms made a precise quantification
impossible.
In the ovarian tumor samples, a significant
correlation between total RON splicing and the expression of
splicing factors ASF/SFRS1 (P=0.035), SRp40 (P=0.015), SRp55
(P=0.015) and SRp75 (P=0.039) was detected. More detailed analysis
comparing tumor samples with alternative splicing of RON exon 5/6
or exon 11 (N=39) to tumor samples without alternative splicing of
RON exon 5/6 or exon 11 (N=6) revealed different correlations to
splicing factor expression. While tumor samples with alternative
splicing events of RON were characterized by significant
correlations of RON expression and the expression of certain
splicing factors (SRp40, P=0.047; SRp55, P=0.005; SRp75, P=0.019),
no significant correlations in tumor samples lacking RON
alternative splicing were found, except for SRp40 (P=0.005).
Notably, tumor tissues with alternative splicing of exon 5/6
(potential RONΔ16 0, N=11) showed no correlation of RON expression
to splicing factors. In contrast, in the tumor samples with
alternative splicing of exon 11 (potential RONΔ165, N=37),
significant correlations of RON expression to SRp40 (P=0.033),
SRp55 (P=0.05) and SRp75 (P=0.028) were determined (Table IV).
 | Table IVRON and splicing factor
expression. |
Table IV
RON and splicing factor
expression.
| RON expression | All tumor samples
(N=45) | Alternative RON
splicing (N=39) | No alternative RON
splicing (N=6) | Alternative
splicing Exon 11 (N=37) | Alternative
splicing Exon 5/6 (N=11) |
|---|
| ASF/SFRS1 | 0.035 | 0.074 | 0.544 | 0.070 | 0.555 |
| SRp20 | 0.648 | 0.385 | 0.266 | 0.488 | 0.401 |
| SRp40 | 0.015 | 0.047 | 0.005 | 0.033 | 0.979 |
| SRp55 | 0.015 | 0.005 | 0.623 | 0.005 | 0.689 |
| SRp75 | 0.039 | 0.019 | 0.872 | 0.028 | 0.537 |
| Tra2ß | 0.979 | 0.793 | 0.397 | 0.984 | 0.370 |
| Yb-1 | 0.119 | 0.090 | 0.872 | 0.119 | 0.631 |
Correlation of RON expression to
molecular and clinicopathological features
A significant correlation of FIGO stage and grade
(P=0.014) as well as of FIGO stage and platin-resistance (P=0.034)
was observed in the tumor samples in general. Corresponding results
were observed in the tumor samples with RON alternative splicing
events (FIGO stage/grading, P=0.031;
FIGO-stage/platinum-resistance, P=0.028). Tumor samples without
alternative splicing of RON showed no correlations to the
clinicopathological parameters.
Significant correlations of RON expression and
clinicopathological parameters such as age, FIGO stage, grade,
histological subtype or resistance to platinum-derived chemotherapy
were not detected in the analyzed tumor samples in general as well
as in any subgroup (e.g. tumor samples with RON alternative
splicing) (Table V). An exceptional
significant correlation of RON expression with grade (P=0.043) was
found in the metastatic issues.
 | Table VCorrelation of RON expression to
clinicopathological parameters. |
Table V
Correlation of RON expression to
clinicopathological parameters.
| RON expression | All tumor samples
(N=45) | Primary tumor
(N=25) | Metastasis
(N=20) | Alternative RON
splicing (N=39) | No alternative RON
splicing (N=6) | Alternative
splicing Exon 11 (N=37) | Alternative
splicing Exon 5/6 (N=11) |
|---|
| Age | 0.205 | 0.141 | 0.972 | 0.247 | 0.544 | 0.320 | 0.331 |
| FIGO stage | 0.663 | 0.728 | 0.242 | 0.560 | 0.864 | 0.321 | 0.766 |
| Grade | 0.579 | 0.217 | 0.043 | 0.638 | 0.638 | 0.506 | 0.689 |
| Histology | 0.489 | 0.799 | 0.446 | 0.721 | 0.158 | 0.670 | 0.915 |
| Platin
resistance | 0.638 | 0.586 | 0.158 | 0.312 | | 0.312 | |
| FIGO/grading | 0.014 | 0.030 | 0.611 | 0.031 | 0.239 | 0.010 | 0.846 |
| FIGO/platin | 0.034 | 0.063 | 0.704 | 0.028 | | 0.028 | |
Expression of splicing factors in the
ovarian tumor specimens
To elucidate the potential role of intrinsic
splicing factors within malignant cells, we investigated the
expression patterns of those factors, known to be involved in the
regulation of RON mRNA processing.
Expression levels of splicing factors ASF/SFRS1,
htra2-β1, YB-1, SRp20, SRp40, SRp55 and SRp75 were evaluated in the
ovarian tumor samples (N=45) and non-pathological ovarian tissue
specimens (N=4) by real-time-RT-PCR analysis. In comparison to the
non-pathological tissues, the expression of all investigated
splicing factors showed a significant upregulation in the ovarian
tumor samples (Table VI).
 | Table VIExpression of splicing factors in the
physiological ovarian and ovarian cancer tissues. |
Table VI
Expression of splicing factors in the
physiological ovarian and ovarian cancer tissues.
| Tumor (N=45) | Physiological
(N=4) | P-value |
|---|
| ASF/SFRS1 | 98.82±3.20 | 0.00 | 0.001 |
| htra2-β1 | 117.35±3.82 | 0.00 | 0.001 |
| YB-1 | 114.51±5.02 | 100.50±2.50 | 0.001 |
| SRp20 | 115.26±7.66 | 95.00±1.26 | 0.001 |
| SRp40 | 110.38±4.25 | 107.50±1.71 | 0.024 |
| SRp55 | 123.21±3.55 | 73.00±2.28 | 0.001 |
| SRp75 | 122.26±4.51 | 91.00±1.82 | 0.001 |
Except for SRp20 (118.00±7.86 vs. 113.75±6.72,
P=0.027) no difference in expression levels of the splicing factors
in the primary tumor samples (N=25) vs. metastases (N=20) were
evident.
In tumor samples with alternative RON splicing
various significant splicing factor interactions were observed
(e.g. SRp55/SRp75; P<0.001). Notably, this phenomenon was not
detected in the tumor samples lacking RON alternative splicing
(Table VII).
 | Table VIIExpression of splicing factors in
ovarian cancer. |
Table VII
Expression of splicing factors in
ovarian cancer.
| | ASF/SFRS1 | SRp20 | SRp40 | SRp55 | SRp75 | Tr2β |
|---|
| All tumor
samples | SRp20 | 0.691 | | | | | |
| SRp40 | 0.064 | 0.037 | | | | |
| SRp55 | 0.002 | 0.489 | 0.000 | | | |
| SRp75 | 0.291 | 0.235 | 0.000 | 0.000 | | |
| Tra2β | 0.166 | 0.000 | 0.004 | 0.198 | 0.039 | |
| Yb-1 | 0.413 | 0.411 | 0.000 | 0.000 | 0.000 | 0.049 |
| With alternative
splicing | SRp20 | 0.501 | | | | | |
| SRp40 | 0.077 | 0.194 | | | | |
| SRp55 | 0.000 | 0.886 | 0.000 | | | |
| SRp75 | 0.165 | 0.194 | 0.000 | 0.000 | | |
| Tra2β | 0.290 | 0.000 | 0.015 | 0.152 | 0.013 | |
| Yb-1 | 0.165 | 0.376 | 0.000 | 0.000 | 0.000 | 0.027 |
| Without alternative
splicing | SRp20 | 0.787 | | | | | |
| SRp40 | 0.704 | 0.208 | | | | |
| SRp55 | 0.156 | 0.397 | 0.787 | | | |
| SRp75 | 0.623 | 0.623 | 0.704 | 0.544 | | |
| Tra2β | 0.208 | 0.397 | 0.266 | 0.468 | 0.468 | |
| Yb-1 | 0.072 | 0.872 | 0.872 | 0.787 | 0.468 | 0.544 |
Statistical correlation analysis in regard to
splicing factor expression and clinicopathological features showed
a significant correlation of YB-1 (P<0.001), SRp40 (P=0.046),
SRp55 (P=0.002) and SRp75 (P=0.002) with FIGO stage I and II (N=12)
compared to FIGO stage III and IV (N=30). YB-1 also showed a
significant correlation to platinum resistance (P=0.042). Other
significant correlations of splicing factor expression and
clinicopathological parameters such as age, grade or histologic
subtype were not observed.
Discussion
RON and its splice variants play an important role
in various tumor biological processes such as cell motility,
adhesion, proliferation, apoptosis and epithelial-to-mesenchymal
transition (EMT) (11). In the
present study, we analyzed the expression of RON and specific
alternatively spliced RON variants with known oncologic potential
in ovarian cancer tumor samples. In addition, the expression levels
of splicing factors which are involved in the regulation of
alternative splicing of RON were investigated.
In all of the ovarian tumor samples (N=45), high
levels of RON expression were detected, whereas in the
non-pathological ovarian tissues (N=4) RON expression was absent
(P=0.001). This observation supports the findings of Maggiora et
al (29) who described
overexpression of RON in ovarian carcinomas but only rarely
detectable RON expression in normal ovarian tissue. There was no
difference in RON expression regarding the origin of the sample
(primary tumor vs. metastasis). Alternatively spliced RON variants
with exclusion of exon 11 (potential RONΔ165) were present in the
majority of the analyzed tumor samples (82.22%) whereas the
alternatively spliced RON variant with exclusion of exon 5/6
(potential RONΔ160 or RONΔ155) was detectable in 24.40% of the
tumor samples only. Expression of alternatively spliced RON
variants was more frequent in metastases than in the primary tumors
(exon 5/6 skip, 35 vs. 16%; exon 11 skip, 90 vs. 76%). The findings
at the mRNA level were confirmed by western blot analysis at the
protein level. While the non-pathological ovarian tissues only
demonstrated pre-RON and RONΔ55 expression, ovarian cancer
specimens expressed a variety of RON isoforms (including pre-RON,
RONΔ55, RONΔ160 and RONΔ165).
Since alternatively spliced RON variants with
exclusion of exon 11 (potential RONΔ165) are present more often in
ovarian tumor samples than other alternatively spliced RON
variants, these isoforms may play a more important role in ovarian
cancer development and progression than others. Elevated expression
levels of alternatively spliced RON isoforms in metastases in
comparison to primary tumors might indicate that alterations in RON
mRNA processing not only occur during tumor development, but are
most likely an accompanying phenomenon in gradational ovarian
cancer progression. Significant correlations of RON mRNA expression
(neither overall RON expression nor expression of the investigated
alternatively spliced RON variants) to clinicopathological
parameters such as FIGO stage, grade or platinum resistance were
not found in the present study. These findings are consistent for
the most part with an immunohistochemical study of Ferrandina et
al (20), who observed no
correlations of RON expression to age, FIGO stage and histology. In
the present study significant correlations were found for grade 3
and higher incidence of immunohistochemical RON expression as well
as a significant inverse correlation of RON expression and survival
in the subgroup of patients with platinum-resistant ovarian cancer
recurrence.
To elucidate the role of intrinsic splicing factors
in alternative splicing in ovarian cancer, we also analyzed the
expression of splicing factors known to be involved in RON mRNA
processing.
All investigated splicing factors ASF/SFRS1,
htra2-β1, YB-1, SRp20, SRp40, SRp55 and SRp75 showed a significant
upregulation in the ovarian cancer samples when compared to the
non-pathological ovarian tissues. Except for SRp20, no differences
in splicing factor expression were noted in the primary tumors and
metastases. Significant correlations of RON expression and splicing
factors were noted in the ovarian cancer samples in general as well
as in samples with known alternative splicing events of RON. These
groups could also be characterized by significant interactions of
splicing factors. In contrast in the tumor specimens without
alternatively spliced RON mRNA neither correlations of RON to
splicing factor expression nor significant splicing factor
interactions were observed. These findings emphasize the crucial
role of the investigated splicing factors for RON mRNA processing.
Notably, alternative splicing of RON exon 5/6 does not appear to be
regulated by the investigated splicing factors since there were no
correlations observed for RON expression and splicing factor
expression in the tumor samples exhibiting alternative exon 5/6
splicing.
Several splicing factors were correlated to FIGO
stage. A significant inverse correlation of YB-1, SRp40, SRp55 and
SRp75 to FIGO stage I and II vs. FIGO stage III and IV was found in
the present study. YB-1 was also correlated to platinum-resistance.
These findings are consistent with other previously published
studies (26,27).
In conclusion, our findings account for an essential
regulatory interplay of splicing factor-driven alterations in the
RON alternative splicing pattern with subsequent tumor biological
consequences in the development and progression of ovarian cancer.
To the best of our knowledge, this is the first study describing
significant changes in splicing factor expression in the regulation
of RON alternative splicing in ovarian cancer patients. The
observed essential interplay of different splicing factors in the
mRNA processing of RON in ovarian cancer might extend the options
for new therapeutic approaches not only to alternatively spliced
RON isoforms but also to the involved regulatory splicing
factors.
References
|
1
|
Skotheim RI and Nees M: Alternative
splicing in cancer: Noise, functional, or systematic? Int J Biochem
Cell Biol. 39:1432–1449. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Tazi J, Bakkour N and Stamm S: Alternative
splicing and disease. Biochim Biophys Acta. 1792:14–26. 2009.
View Article : Google Scholar
|
|
3
|
Venables JP: Unbalanced alternative
splicing and its significance in cancer. BioEssays. 28:378–386.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Huiping C, Kristjansdottir S, Bergthorsson
JT, Jonasson JG, Magnusson J, Egilsson V and Ingvarsson S: High
frequency of LOH, MSI and abnormal expression of FHIT in gastric
cancer. Eur J Cancer. 38:728–735. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Cheung N, Wong MP, Yuen ST, Leung SY and
Chung LP: Tissue-specific expression pattern of vascular
endothelial growth factor isoforms in the malignant transformation
of lung and colon. Hum Pathol. 29:910–914. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Elias AP and Dias S: Microenvironment
changes (in pH) affect VEGF alternative splicing. Cancer
Microenviron. 1:131–139. 2008. View Article : Google Scholar
|
|
7
|
Hirschfeld M, Jaeger M, Buratti E, Stuani
C, Grueneisen J, Gitsch G and Stickeler E: Expression of
tumor-promoting Cyr61 is regulated by hTRA2-2401 and acidosis. Hum
Mol Genet. 20:2356–2365. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Krieg A, Mahotka C, Krieg T, Grabsch H,
Müller W, Takeno S, Suschek CV, Heydthausen M, Gabbert HE and
Gerharz CD: Expression of different survivin variants in gastric
carcinomas: First clues to a role of survivin-2b in tumour
progression. Br J Cancer. 86:737–743. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Vegran F, Boidot R, Oudin C, Riedinger JM
and Lizard-Nacol S: Distinct expression of Survivin splice variants
in breast carcinomas. Int J Oncol. 27:1151–1157. 2005.PubMed/NCBI
|
|
10
|
Eblen ST: Regulation of chemoresistance
via alternative messenger RNA splicing. Biochem Pharmacol.
83:1063–1072. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ghigna C, De Toledo M, Bonomi S, Valacca
C, Gallo S, Apicella M, Eperon I, Tazi J and Biamonti G:
Pro-metastatic splicing of Ron proto-oncogene mRNA can be reversed:
Therapeutic potential of bifunctional oligonucleotides and indole
derivatives. RNA Biol. 7:495–503. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Collesi C, Santoro MM, Gaudino G and
Comoglio PM: A splicing variant of the RON transcript induces
constitutive tyrosine kinase activity and an invasive phenotype.
Mol Cell Biol. 16:5518–5526. 1996.PubMed/NCBI
|
|
13
|
Lu Y, Yao HP and Wang MH: Multiple
variants of the RON receptor tyrosine kinase: Biochemical
properties, tumorigenic activities, and potential drug targets.
Cancer Lett. 257:157–164. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Danilkovitch-Miagkova A: Oncogenic
signaling pathways activated by RON receptor tyrosine kinase. Curr
Cancer Drug Targets. 3:31–40. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wang MH, Wang D and Chen YQ: Oncogenic and
invasive potentials of human macrophage-stimulating protein
receptor, the RON receptor tyrosine kinase. Carcinogenesis.
24:1291–1300. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang MH, Kurtz AL and Chen Y:
Identification of a novel splicing product of the RON receptor
tyrosine kinase in human colorectal carcinoma cells.
Carcinogenesis. 21:1507–1512. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Xu XM, Zhou YQ and Wang MH: Mechanisms of
cytoplasmic β-catenin accumulation and its involvement in
tumorigenic activities mediated by oncogenic splicing variant of
the receptor originated from Nantes tyrosine kinase. J Biol Chem.
280:25087–25094. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhou YQ, He C, Chen YQ, Wang D and Wang
MH: Altered expression of the RON receptor tyrosine kinase in
primary human colorectal adenocarcinomas: Generation of different
splicing RON variants and their oncogenic potential. Oncogene.
22:186–197. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lynch HT, Casey MJ, Lynch J, White TE and
Godwin AK: Genetics and ovarian carcinoma. Semin Oncol. 25:265–280.
1998.PubMed/NCBI
|
|
20
|
Ferrandina G, Martinelli E, Petrillo M,
Prisco MG, Zucconi A, Santaguida S, Zannoni G, Scambia G and
Ferlini C: Prognostic role of the recepteur d’origine nantais (RON)
expression in ovarian cancer patients. Gynecol Oncol. 111:237–243.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Prislei S, Mariani M, Raspaglio G,
Mozzetti S, Filippetti F, Ferrandina G, Scambia G and Ferlini C:
RON and cisplatin resistance in ovarian cancer cell lines. Oncol
Res. 19:13–22. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Huang Y and Steitz JA: SRprises along a
messenger’s journey. Mol Cell. 17:613–615. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zahler AM, Lane WS, Stolk JA and Roth MB:
SR proteins: A conserved family of pre-mRNA splicing factors. Genes
Dev. 6:837–847. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Fischer DC, Noack K, Runnebaum IB,
Watermann DO, Kieback DG, Stamm S and Stickeler E: Expression of
splicing factors in human ovarian cancer. Oncol Rep. 11:1085–1090.
2004.PubMed/NCBI
|
|
25
|
Stickeler E, Kittrell F, Medina D and
Berget SM: Stage-specific changes in SR splicing factors and
alternative splicing in mammary tumorigenesis. Oncogene.
18:3574–3582. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Fujii T, Seki N, Namoto-Matsubayashi R,
Takahashi H, Inoue Y, Toh U, Kage M and Shirouzu K: YB-1 prevents
apoptosis via the mTOR/STAT3 pathway in HER-2-overexpressing breast
cancer cells. Future Oncol. 5:153–156. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kamura T, Yahata H, Amada S, Ogawa S,
Sonoda T, Kobayashi H, Mitsumoto M, Kohno K, Kuwano M and Nakano H:
Is nuclear expression of Y box-binding protein-1 a new prognostic
factor in ovarian serous adenocarcinoma? Cancer. 85:2450–2454.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ghigna C, Giordano S, Shen H, Benvenuto F,
Castiglioni F, Comoglio PM, Green MR, Riva S and Biamonti G: Cell
motility is controlled by SF2/ASF through alternative splicing of
the Ron protooncogene. Mol Cell. 20:881–890. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Maggiora P, Lorenzato A, Fracchioli S,
Costa B, Castagnaro M, Arisio R, Katsaros D, Massobrio M, Comoglio
PM and Flavia Di Renzo M: The RON and MET oncogenes are
co-expressed in human ovarian carcinomas and cooperate in
activating invasiveness. Exp Cell Res. 288:382–389. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wang MH, Yao HP and Zhou YQ: Oncogenesis
of RON receptor tyrosine kinase: A molecular target for malignant
epithelial cancers. Acta Pharmacol Sin. 27:641–650. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Camp ER, Liu W, Fan F, Yang A, Somcio R
and Ellis LM: RON, a tyrosine kinase receptor involved in tumor
progression and metastasis. Ann Surg Oncol. 12:273–281. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Chen YQ, Zhou YQ, Angeloni D, Kurtz AL,
Qiang XZ and Wang MH: Overexpression and activation of the RON
receptor tyrosine kinase in a panel of human colorectal carcinoma
cell lines. Exp Cell Res. 261:229–238. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Ma Q, Zhang K, Guin S, Zhou YQ and Wang
MH: Deletion or insertion in the first
immunoglobulin-plexin-transcription (IPT) domain differentially
regulates expression and tumorigenic activities of RON receptor
tyrosine kinase. Mol Cancer. 9:3072010. View Article : Google Scholar : PubMed/NCBI
|