Introduction
MicroRNAs (miRNAs), a type of non-coding RNAs, play
important regulatory roles in gene expression by binding to the
3′-untranslated region (3′UTR) of target mRNAs and lead to mRNA
degradation or translation inhibition (1). miRNAs are aberrantly expressed in
different types of tumors and play act as tumor suppressors or
oncogenes depending on their target genes (2–4). The
roles of miRNAs in tumor include cell growth, death, autophagy,
apoptosis, angiogenesis and differentiation, metastasis and therapy
resistance in tumor progression (5–7).
Breast cancer is one of the leading causes of cancer
mortality among women worldwide (8). A number of miRNAs appear to be
involved in breast cancer progression (5–7).
miR-519d is reported as a negative regulator of tumor progression
in chondrosarcoma (9), osteosarcoma
(10), ovarian cancer (11) and hepatocellular carcinoma (12,13).
However, its expression and cell function in breast cancer remains
to be determined.
Signal transducer and activator of transcription 3
(STAT3) is a key cytoplasmic transcription factor that is usually
activated by cytokine receptors and tyrosine kinase in the cells
(14). STAT3 plays an important
role in early embryo genesis and the regulation of various types of
cells (15–16). The role of STAT3 in breast cancer
has been widely studied and aberrant activation of STAT3 can also
promote breast cancer formation and progression (15–16).
STAT3 activation is frequently observed in primary breast cancers
and is associated with poor prognosis and invasiveness (15–16).
It has been shown that miRNAs regulate STAT3 expression in cancer
cells (17), but its regulation by
miRNAs needs to be further elucidated in breast cancer.
In the present study, we examined the potential role
of miR-519d in breast cancer. The miRNA prediction software
indicated that STAT3 may be the downstream target of miR-519d.
Furthermore, the correlation between the expression of miR-519d and
STAT3 in breast cancer samples was explored.
Materials and methods
Patients and tumor tissues
Human breast cancer tissues and matched normal
adjacent breast specimens from the patients diagnosed with breast
cancer were obtained at the Shengjing Hospital of China Medical
University between 2009 and 2013. The diagnosis was based on
pathological evidence and the specimens were immediately
snap-frozen and stored at −80°C. None of the patients received
chemotherapy or radiotherapy before the surgical excision. All the
patients provided written informed consent for the use of their
tissues. The study protocol was approved by the Ethics Committee of
Shengjing Hospital.
Cell lines and culture
Human MCF7, BT474, SKBR3 and MDA231 breast cancer
cell lines were primarily obtained from the American Type Culture
Collection (ATCC; Manassas, VA, USA) and were maintained in the
medium according to the instructions from ATCC. MCF10A cells were
cultured in Dulbecco's modified Eagle's medium (DMEM)/F12 with 10%
fetal bovine serum (FBS) and antibiotics (100 U/ml penicillin and
100 μg/ml streptomycin sulfate, EGF and insulin). The cells
were grown in a humidified incubator at 37°C with 5%
CO2.
Quantitative RT-PCR
Total RNA was isolated from breast cancer tissues or
cells using TRIzol (Invitrogen-Life Technologies, Carlsbad, CA,
USA). miR-519d and U6 were polyadenylated using a poly-A polymerase
based First-Strand Synthesis kit (Takara Bio, Japan) following the
manufacturer's instructions. To quantify the STAT3 and GAPDH mRNA
levels, 1 μg of total RNA was subjected to First-Strand cDNA
synthesis using a PrimeScript RT Reagent kit (Takara). The qPCR was
performed using SYBR-Green PCR master mix (Takara) on the ABI
7500HT system. U6 or GAPDH were used as an endogenous control. All
the primers were ordered from Invitrogen. The relative fold
expression was calculated with the 2−ΔΔCT method. The
RT-qPCR reactions were run in triplicate (18).
Transfection
miR-519d, anti-miR-519d and their controls were
designed and synthesized by RiboBio Co. (Guangzhou, China). The
cells were transfected with miRNA using Lipofectamine 2000
(Invitrogen-Life Technologies). Briefly, breast cancer cells were
seeded in 12-well plates and grown until they reached 40%
confluence prior to transfection, and RNA and protein were
extracted 48 h later. The final concentration of miR-519d mimic and
anti-miR-519d was 50 nM. Lentiviral miR-519d (LV-miR-519d) and
empty lentiviral vector (LV-miR-control) were constructed by
Genechem Co. (Shanghai, China) and were infected into the breast
cancer cells according to the manufacturer's instructions.
MTT assay
An MTT assay was employed to detect the growth of
breast cancer cells and the growth curve was delineated. Briefly,
2.5×103 cells/well were seeded in a 96-well plate and
allowed to adhere. At different time-points, 20 μl of the
MTT solution was added to each well (5 mg/ml and 0.5% MTT) and the
cells were continued to culture for 4 h. After the incubation, the
supernatant was discarded and 150 μl dimethyl sulfoxide was
added to each well, and the culture plate was agitated at low speed
for 10 min until the crystal dissolved completely. The ELISA reader
was used to measure the absorbance at 570 nm.
Apoptosis assay
For apoptosis analysis, the cells were collected,
washed twice with cold phosphate-buffered saline and resuspended in
binding buffer at a cell density of 1×106/ml. The cells
were then stained with Annexin V-FITC and propodium iodide
according to the manufacturer's instructions. The signal was
obtained by a FACSCalibur flow cytometer (BD Biosciences, San Jose,
CA, USA) and was analyzed with CellQuest software.
Cell cycle analysis
MCF7 or SKBR3 cells were transfected with miR-519 or
miR-control and the cells were collected for cell cycle analysis.
The cells were fixed in 70% ethanol overnight at −20°C, and treated
with DNA staining solution. Cell cycle analysis was performed using
flow cytometry.
Matrigel invasion assay
MCF7 or SKBR3 cells were transfected with miR-519 or
miR-control and were collected for the invasion assay using a
Matrigel-coated chamber (24-well plates, 8-mm pore size; Corning
Inc., Corning, NY, USA). Briefly, breast cancer cells transfected
with miR-519 or miR-control (5×104) were seeded in the
upper chamber at 37°C with the medium with 0.1 and 20% FBS in the
lower wells. After 48 h, the chambers were removed and the
non-invading cells were then wiped with cotton swabs. Invaded cells
at the bottom of the membrane were stained with 0.1% crystal violet
and counted under microscopic observation with five fields.
Western blot analysis
Breast cancer cells were collected after 48 h
treatment with 50 nM miR-519d mimic or anti-miR-519d and the
controls. Protein extraction, SDS-PAGE gel electrophoresis and
western blotting were performed as previously described. Several
different primary antibodies were used including STAT3 (Cell
Signaling Technology, Danvers, MA, USA) and GAPDH (Santa Cruz
Biotechnology, Inc., Santa Cruz, CA, USA). Secondary antibody
incubations were performed for 2 h at room temperature and the
protein bands were visualized on X-ray film using an enhanced
chemiluminescent ECL substrate.
Luciferase reporter assay
A total of 1×104 cells were seeded in
96-well plates in 200 μl medium. A total of 100 ng wild-type
or mutant reporter constructs were co-transfected combined with 50
nM miR-519d or miR-control using the Lipofectamine 2000
transfection reagent into the breast cancer cells according to the
manufacturer's instruction. After 48 h, luciferase activity was
measured with the Dual-Luciferase reporter assay system (Promega,
Madison, WI, USA). Firefly luciferase activity was then normalized
to the corresponding Renilla luciferase activity.
Breast cancer xenograft model
Tumorigenicity of 1×107 cells in mammary
fat pads of 4-week-old female nude mice (BALB/c-nude) was assessed.
The tumor size was measured with a vernier caliper every five days
and the tumor volumes were calculated using the formula: (length ×
width2)/2mm3. The use of nude mice complied
with the guide for the use of Laboratory Animals and the study was
approved by the Animal Care and Use Committee of Shengjing
Hospital.
Statistical analysis
The results are presented as mean ± standard
deviation (SD). Statistical significance was determined using the
Student's t-test. P<0.05 was considered statistically
significant.
Results
The expression of miR-519d is lower in
breast cancer patients and cells
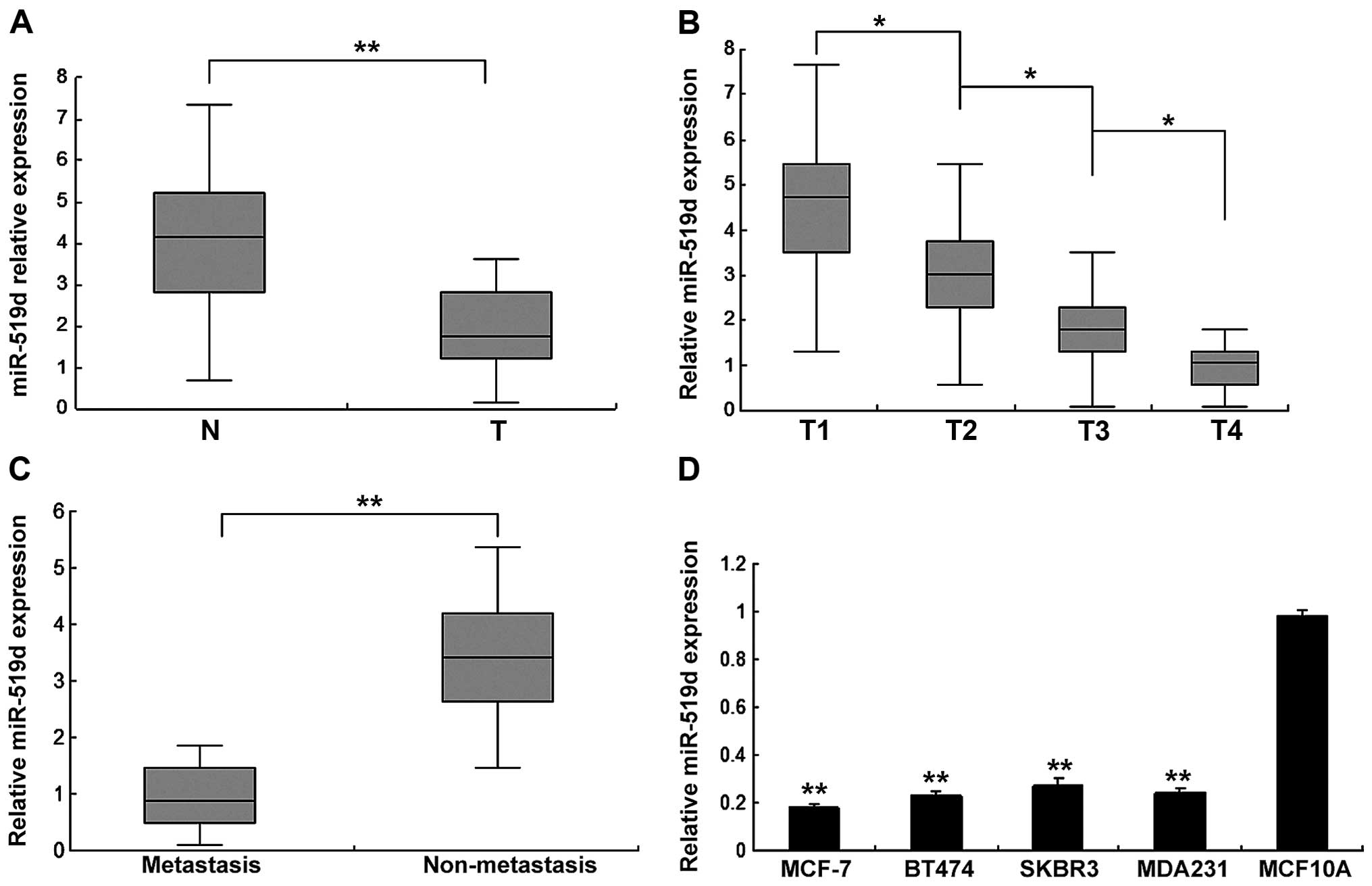
Firstly, we examined the expression of miR-519d in
human breast cancer specimens by quantitative RT-PCR (RT-qPCR). The
expression levels of miR-519d were analyzed and they were
downregulated in breast cancer tissues compared with normal samples
(Fig. 1A). Cancer tissues were
classified as T1, T2, T3 and T4, and we found that miR-519d
expression was higher in stage T1, whereas stages T2, T3 and T4 had
lower levels, showing a significant correlation of miR-519d with T
stage of breast cancer (Fig. 1B).
Breast cancer tissues were classified as non-metastasis and
metastasis, and the result showed that miR-519d expression in the
metastasis group was lower than that of the non-metastasis group
(Fig. 1C). miR-519d was lower in
human ovarian cancer cells compared with normal cells (Fig. 1D). These results suggested that
miR-519d may function as a negative regulator in the progression of
human breast cancer.
miR-519d suppresses breast cancer cell
proliferation
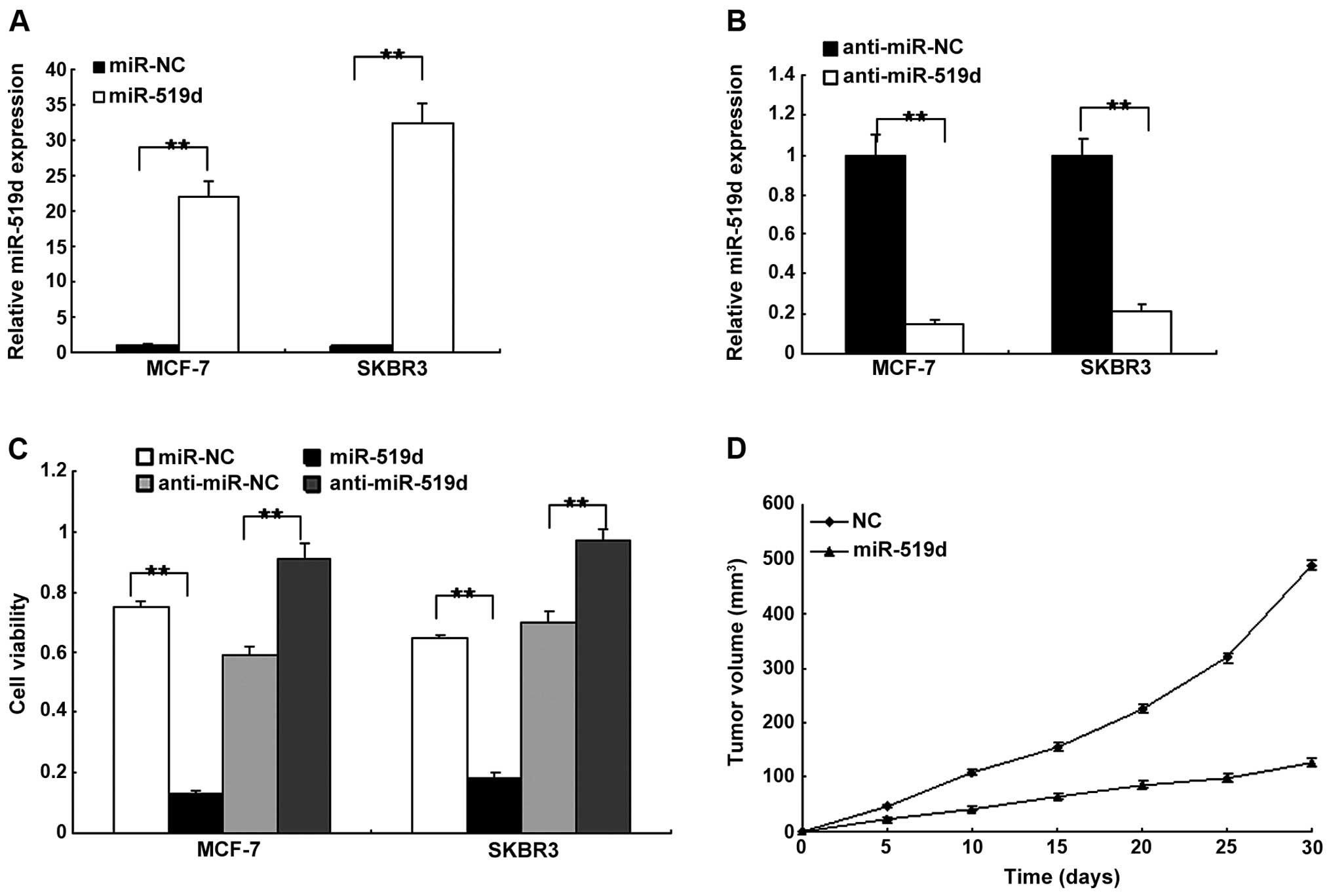
To examine the cell function of miR-519d in breast
cancer cells, the MCF7 and SKBR3 cells were transfected with
miR-519d mimics or the control, and the miR-519d was restored in
the two cell lines (Fig. 2A).
miR-519d expression was decreased in MCF7 and SKBR3 cells with
anti-miR-519d (Fig. 2B). Using the
MTT assay, cell proliferation was examined. The results indicated
that miR-519d inhibited cell proliferation while deletion of
miR-519 promoted cell proliferation in the MCF7 and SKBR3 cells
(Fig. 2C). To confirm that miR-519d
inhibited tumor growth, in vivo tumor models were
established by implanting the MCF7 cells transfected with
lentivirus-mediated miRNA-519d or negative control. Tumors with
miR-519d-transfected cells grew significantly more gradually than
the control (Fig. 2D). These
results indicated that miR-519d suppressed breast cancer growth
in vitro and in vivo.
miR-519d leads to cell apoptosis and
changes cell cycle distribution in breast cancer cells
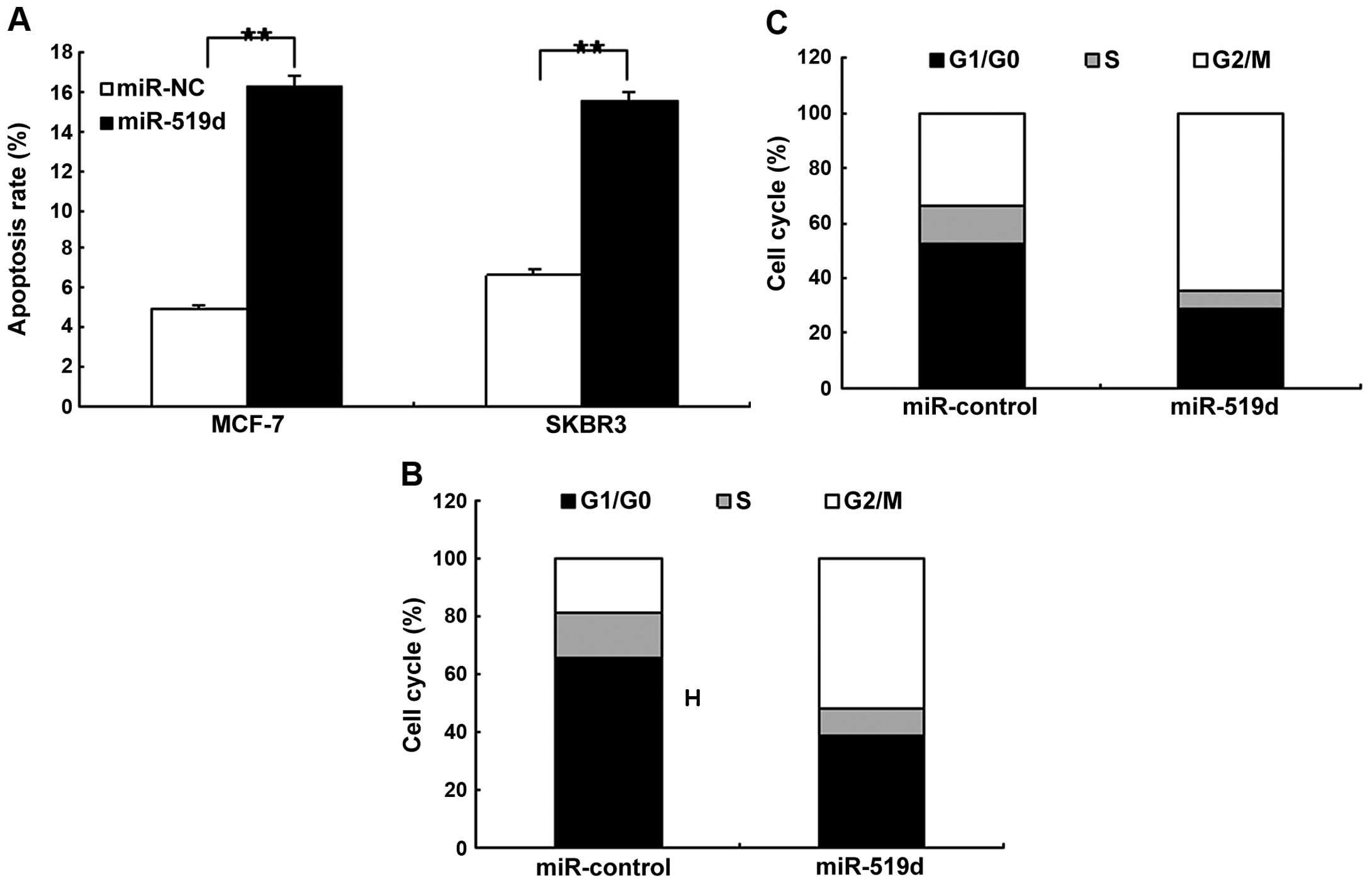
We determined whether miR-519d promoted cancer cell
apoptosis or induced changes to the cell cycle distribution. MCF7
and SKBR3 cells were transfected with miR-519d mimics to observe
cell apoptosis and the cell cycle using flow cytometry. It was
found that the apoptotic rate increased significantly in MCF7 and
SKBR3 cells compared to the control (Fig. 3A). Results from the cell cycle
analysis showed that miR-519d reduced the proportion of G0/G1 and
G2/M phases significantly and increased the proportion of S phase
in the MCF7 and SKBR3 cells when compared with the controls
(Fig. 3B and C).
miR-519d inhibits invasion of the breast
cancer cells
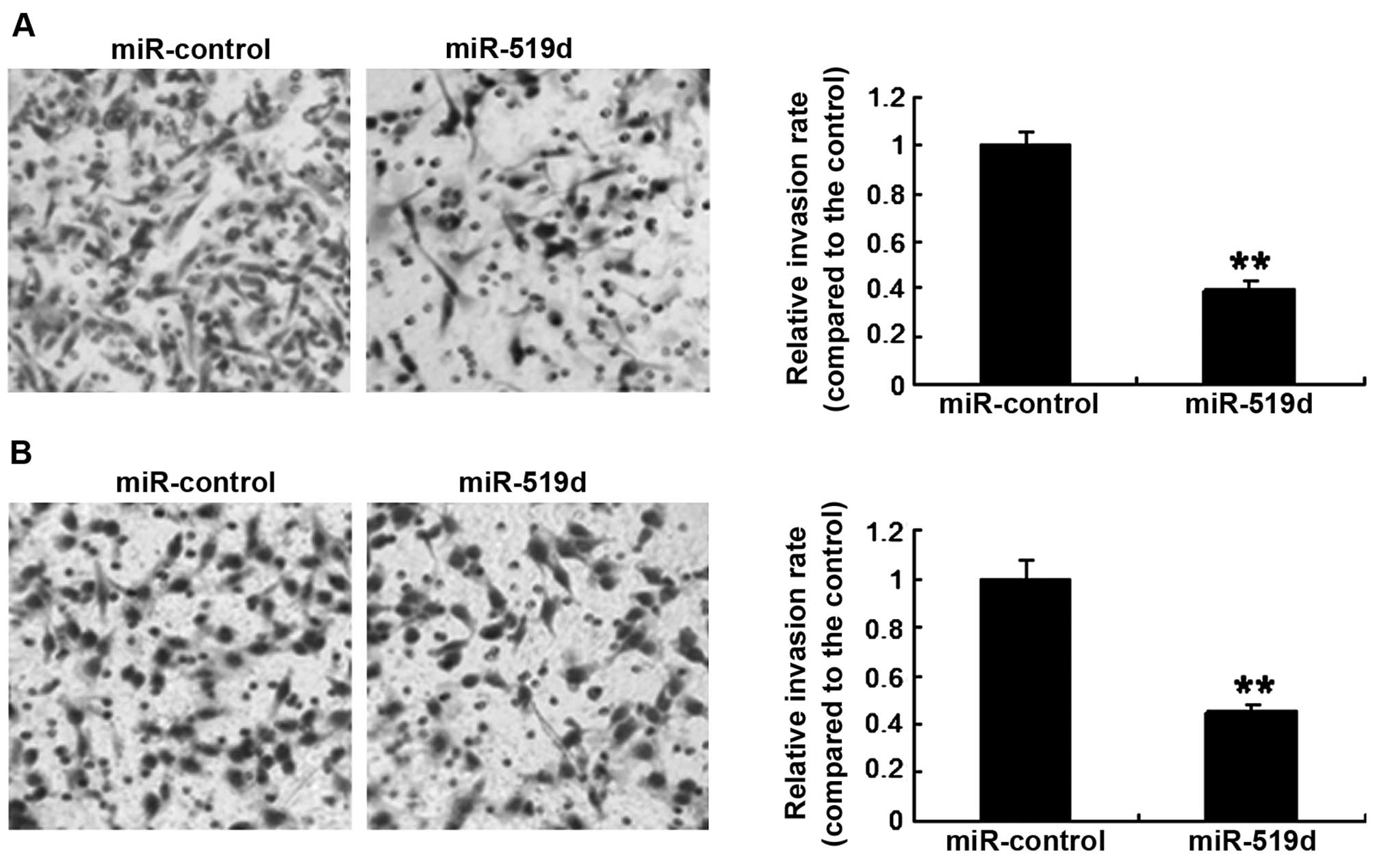
Breast cancer cell invasiveness was assayed by a
Transwell system. MCF-7 cells were transfected with the miR-519d
mimics or the control, and it was found that the invasion was
markedly lower in the cells with miR-519d overexpression than the
cells with miR-control (Fig. 4A).
Similar resutls were obtained for the SKBR3 cell line (Fig. 4B). The results showed that miR-519d
inhibited breast cancer cell invasion.
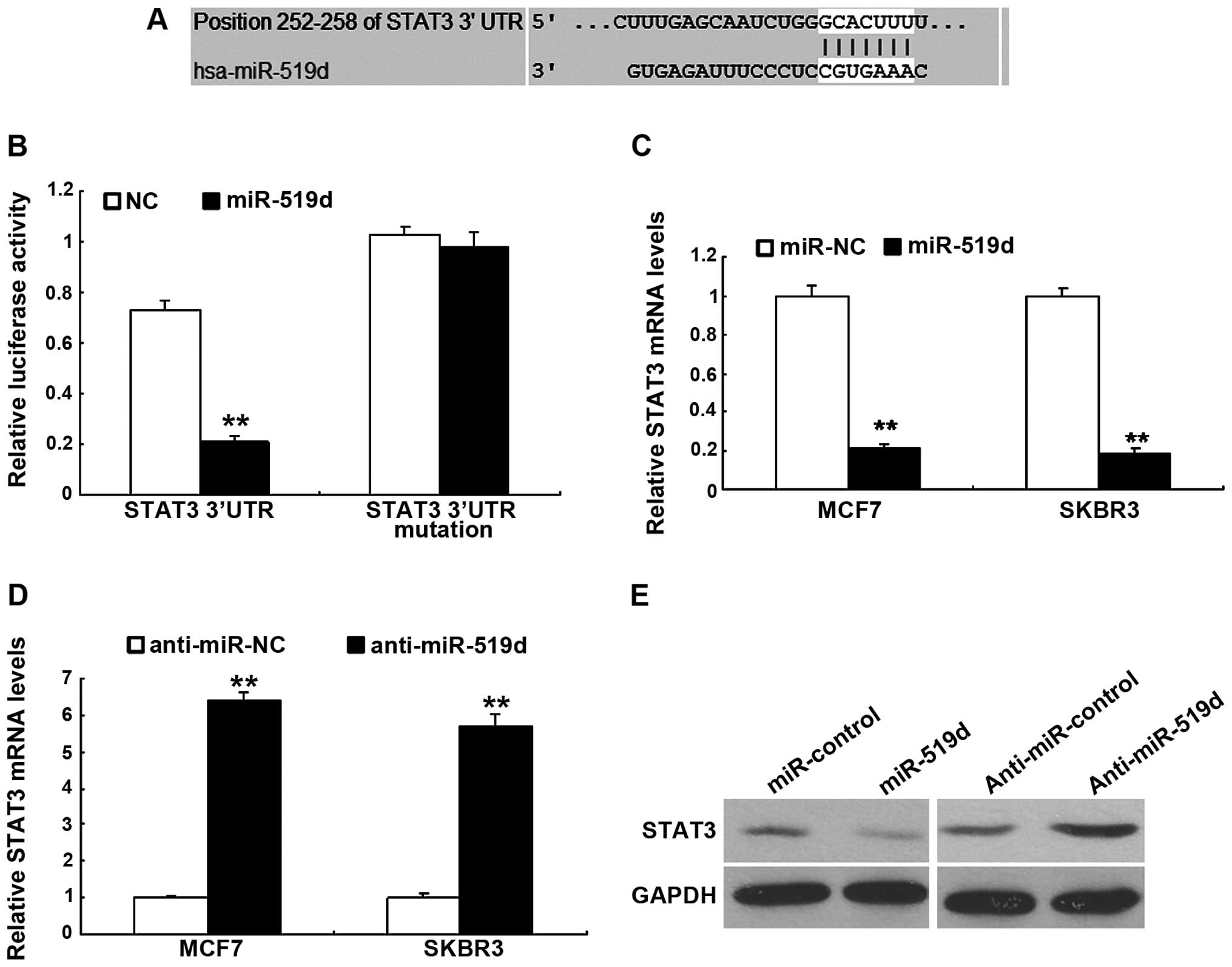
miR-519d downregulates STAT3 expression
in the breast cancer cells
To investigate the molecular mechanism that miR-519d
regulates cell function including cell proliferation and metastasis
in breast cancer cells, we used a TargetScan software that
predicted the putative target genes of miR-519d. Our analysis
identified that STAT3 was one of the potential targets for miR-519d
(Fig. 5A). Results from the
luciferase activity assay showed that the miR-519d significantly
decreased the luciferase activity of with the wild-type 3′UTR, but
not the mutant 3′UTR vector (Fig.
5B). miRNA regulates gene expression at the transcriptional and
translational levels. However, whether miR-519d downregulated STAT3
mRNA or protein remained to be determined. MCF-7 and SKBR3 cells
were transfected with miR-519d to assess STAT3 mRNA using RT-qPCR
and it was found that miR-519d led to an obvious decrease in STAT3
mRNA (Fig. 5C) and the protein
(Fig. 5D). By contrast,
transfection of anti-miR-519d resulted in an upregulation in STAT3
expression as determined by western blot analysis (Fig. 5E).
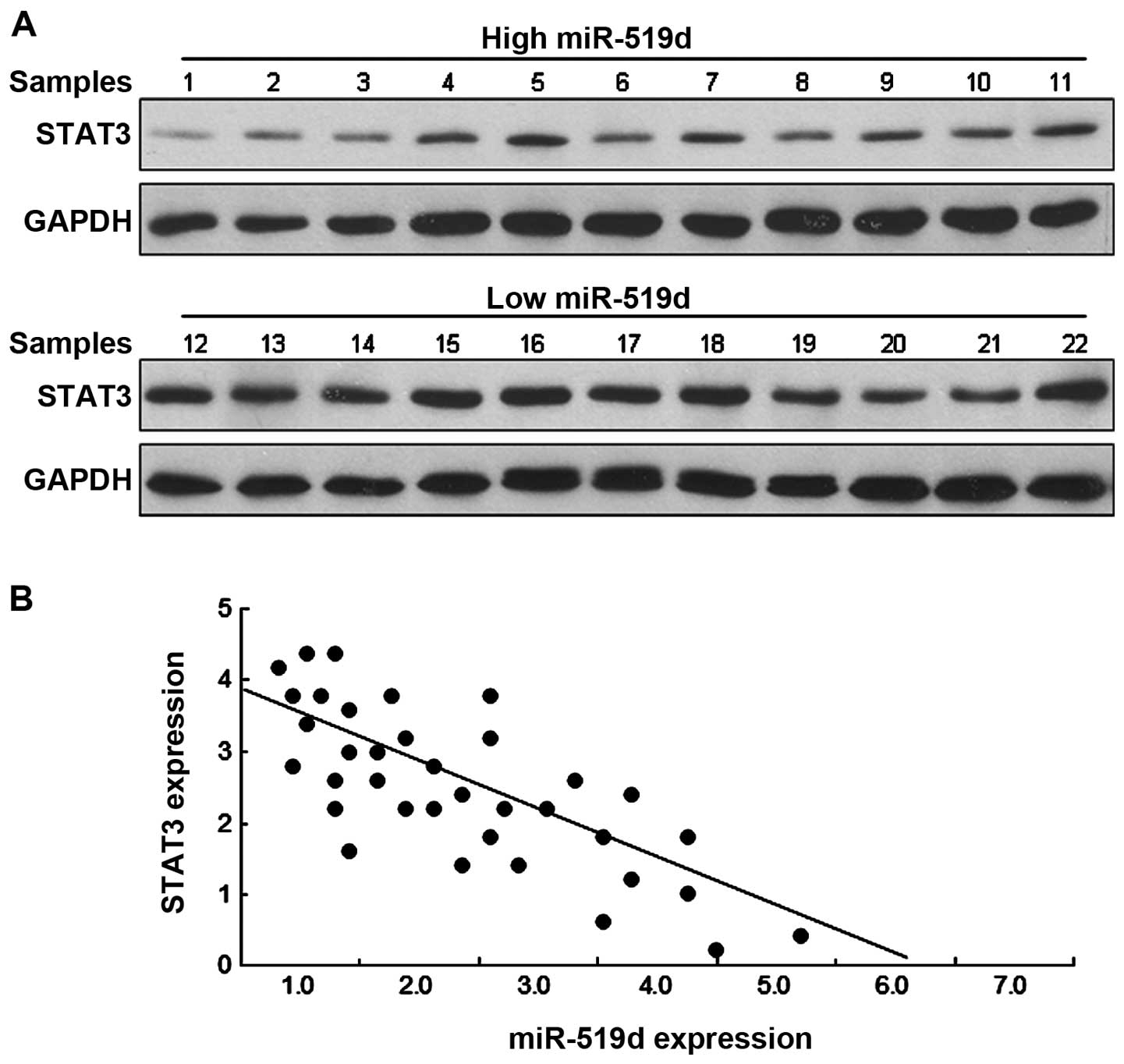
miR-519d is negatively correlated to
STAT3 in breast cancer tissues
The above data identified that STAT3 is a direct
target gene of miR-519d. We then determined whether there is any
relationship between STAT3 and miR-519d in breast cancer tissues.
Western blot analysis revealed that the breast cancer tissues with
a low miR-519d expression had a higher STAT3 protein level while
those with a high miR-519d expression had a lower STAT3 protein
level (Fig. 6A). As shown in
Fig. 6B, there was a significant
inverse relationship between STAT3 and miR-519d expression levels
in breast cancer tissues.
Discussion
In recent years, a number of miRNAs that play
important roles in breast cancer progression and may be critical
diagnostic markers have been identified. One of these is miR-519,
which is reported in osteosarcoma (9), ovarian cancer (10) and HCC (11–12).
Previous findings have shown that miR-519d is a potential tumor
suppressor. However, little is known with regard to the molecular
mechanisms involved in the miR-519d modulation in the process of
breast cancer. In the present study, we found that miR-519d was
frequently downregulated in breast cancer tissues and cell lines.
We also found that it suppressed breast cancer growth and invasion,
induced apoptosis and cell cycle arrest and its expression was
negatively correlated with STAT3 expression in breast cancer.
miR-519d belongs to the chromosome 19 miRNA cluster
(C19MC), the largest human miRNA cluster (12). It has been shown that miR-519d plays
roles as a negative regulator in tumors. For example, miR-519d in
human osteosarcoma is downregulated and then promotes metastasis
(10). miR-519d represses ovarian
cancer cell proliferation and enhances cisplatin-mediated
cytotoxicity in vitro by targeting XIAP (11). miR-519d suppresses cell growth in
the hepatocellular carcinoma targeting MKi67 (13). In our study, miR-519d was found to
significantly suppress the proliferation of breast cancer cells by
induction of apoptosis and cell cycle arrest at S phase.
Furthermore, the growth of xenograft tumors was significantly
inhibited after being transfected with miR-519d. These results
suggest that miR-519d might act as an inhibitor in the progression
of breast tumorigenesis.
STAT3 is regulated by miRNAs in cancer-like miR-130b
(18), miR-874 (19), miR-124 (20), miR-200 (20), miR-1181 (21) and miR-7 (22). In the present study, we found STAT3
was verified as a new target gene of miR-519d and its mRNA and
protein levels were inhibited in breast cancer cells by miR-519d.
Breast cancer tissues were classified as low and high miR-519d
expression. The results indicated that expression of miR-519d and
STAT3 was negatively correlated.
In conclusion, the results of the present study have
demonstrated that the low expression of miR-519d was associated
with TNM staging and metastasis of breast cancer. This study also
shows that miR-519d plays an important role in the regulation of
breast cancer cell proliferation and invasion by downregulating
STAT3. Our results indicate that miR-519d is a novel biomarker that
can be used to predict the prognosis and progression of breast
cancer.
References
|
1
|
Vislovukh A, Vargas TR, Polesskaya A and
Groisman I: Role of 3′-untranslated region translational control in
cancer development, diagnostics and treatment. World J Biol Chem.
5:40–57. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
van Rooij E and Kauppinen S: Development
of microRNA therapeutics is coming of age. EMBO Mol Med. 6:851–864.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zhu J, Zheng Z, Wang J, Sun J, Wang P,
Cheng X, Fu L, Zhang L, Wang Z and Li Z: Different miRNA expression
profiles between human breast cancer tumors and serum. Front Genet.
5:1492014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Siegel R, Ma J, Zou Z and Jemal A: Cancer
statistics, 2014. CA Cancer J Clin. 64:9–29. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Corcoran C, Friel AM, Duffy MJ, Crown J
and O'Driscoll L: Intracellular and extracellular microRNAs in
breast cancer. Clin Chem. 57:18–32. 2011. View Article : Google Scholar
|
|
6
|
Melo SA and Esteller M: Dysregulation of
microRNAs in cancer: Playing with fire. FEBS Lett. 585:2087–2099.
2011. View Article : Google Scholar
|
|
7
|
Greene SB, Herschkowitz JI and Rosen JM:
Small players with big roles: microRNAs as targets to inhibit
breast cancer progression. Curr Drug Targets. 11:1059–1073. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Le Quesne J and Caldas C: Micro-RNAs and
breast cancer. Mol Oncol. 4:230–241. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tsai CH, Tsai HC, Huang HN, Hung CH, Hsu
CJ, Fong YC, Hsu HC, Huang YL and Tang CH: Resistin promotes tumor
metastasis by down-regulation of miR-519d through the AMPK/p38
signaling pathway in human chondrosarcoma cells. Oncotarget.
6:258–270. 2015.
|
|
10
|
Tsai HC, Su HL, Huang CY, Fong YC, Hsu CJ
and Tang CH: CTGF increases matrix metalloproteinases expression
and subsequently promotes tumor metastasis in human osteosarcoma
through down-regulating miR-519d. Oncotarget. 5:3800–3812.
2014.PubMed/NCBI
|
|
11
|
Pang Y, Mao H, Shen L, Zhao Z, Liu R and
Liu P: MiR-519d represses ovarian cancer cell proliferation and
enhances cisplatin-mediated cytotoxicity in vitro by targeting
XIAP. Onco Targets Ther. 7:587–597. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Fornari F, Milazzo M, Chieco P, Negrini M,
Marasco E, Capranico G, Mantovani V, Marinello J, Sabbioni S,
Callegari E, et al: In hepatocellular carcinoma miR-519d is
up-regulated by p53 and DNA hypomethylation and targets CDKN1A/p21,
PTEN, AKT3 and TIMP2. J Pathol. 227:275–285. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hou YY, Cao WW, Li L, Li SP, Liu T, Wan
HY, Liu M, Li X and Tang H: MicroRNA-519d targets MKi67 and
suppresses cell growth in the hepatocellular carcinoma cell line
QGY-7703. Cancer Lett. 307:182–190. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Haura EB, Turkson J and Jove R: Mechanisms
of disease: Insights into the emerging role of signal transducers
and activators of transcription in cancer. Nat Clin Pract Oncol.
2:315–324. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Walker SR, Xiang M and Frank DA: Distinct
roles of STAT3 and STAT5 in the pathogenesis and targeted therapy
of breast cancer. Mol Cell Endocrinol. 382:616–621. 2014.
View Article : Google Scholar
|
|
16
|
Clevenger CV: Roles and regulation of stat
family transcription factors in human breast cancer. Am J Pathol.
165:1449–1460. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Haghikia A, Hoch M, Stapel B and
Hilfiker-Kleiner D: STAT3 regulation of and by microRNAs in
development and disease. JAK-STAT. 1:143–150. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhao G, Zhang JG, Shi Y, Qin Q, Liu Y,
Wang B, Tian K, Deng SC, Li X, Zhu S, et al: Correction: miR-130b
is a prognostic marker and inhibits cell proliferation and invasion
in pancreatic cancer through targeting STAT3. PLoS One.
8:e738032013. View Article : Google Scholar
|
|
19
|
Zhang X, Tang J, Zhi X, Xie K, Wang W, Li
Z, Zhu Y, Yang L, Xu H and Xu Z: miR-874 functions as a tumor
suppressor by inhibiting angiogenesis through STAT3/VEGF-A pathway
in gastric cancer. Oncotarget. 6:1605–1617. 2015.PubMed/NCBI
|
|
20
|
Xiao Y, Wang J, Yan W, Zhou Y, Chen Y,
Zhou K, Wen J, Wang Y and Cai W: Dysregulated miR-124 and miR-200
expression contribute to cholangiocyte proliferation in the
cholestatic liver by targeting IL-6/STAT3 signalling. J Hepatol.
62:889–896. 2015. View Article : Google Scholar
|
|
21
|
Jiang J, Li Z, Yu C, Chen M, Tian S and
Sun C: MiR-1181 inhibits stem cell-like phenotypes and suppresses
SOX2 and STAT3 in human pancreatic cancer. Cancer Lett.
356:962–970. 2015. View Article : Google Scholar
|
|
22
|
Zhang H, Cai K, Wang J, Wang X, Cheng K,
Shi F, Jiang L, Zhang Y and Dou J: MiR-7, inhibited indirectly by
lincRNA HOTAIR, directly inhibits SETDB1 and reverses the EMT of
breast cancer stem cells by downregulating the STAT3 pathway. Stem
Cells. 32:2858–2868. 2014. View Article : Google Scholar : PubMed/NCBI
|