Introduction
Fibroadenomas (FA) and phyllodes tumors (PT) are
fibroepithelial tumors of the breast with stromal and epithelial
components. It has been suggested that the insulin-like growth
factor 2 (IGF2) gene is associated with the pathogenesis of
breast fibroepithelial tumors (1).
The IGF2 gene is located within the IGF2/H19
imprinted gene cluster on chromosome 11p15 and is expressed
predominantly from the paternal allele (2–4).
IGF2 promotes the growth and proliferation of cells in many
different tissues (5) and is highly
expressed in various types of tumors (6–8)
including fibroepithelial tumors of the breast (1). As reported in studies of Wilms'
tumors, loss of IGF2 imprinting (LOI) manifested by
biallelic gene expression results in IGF2 overexpression and
subsequent tumor formations (9,10).
Occurrence of LOI in almost all types of cancers such as colorectal
cancer (33–88%) (11–13), prostate cancer (83%) (14) and breast cancer (0–60%) (15–18)
has been frequently reported, but the association of LOI with
IGF2 expression has not been clearly established yet.
Imprinted genes are associated with CpG-rich regions
which have allele-specific DNA methylation and are known as
differentially methylated regions (DMRs) (15). Loss of methylation at the human
IGF2 DMR0 has been reported to be associated with
IGF2 LOI in several malignancies (2,6,19,20)
including breast tumors (15,21).
DMR0 methylation levels are significantly lower in cancer tissues
(29–31%) than in normal tissues (45–51%) (6,15,20),
but IGF2 mRNA expression is not always related to LOI or
DMR0 methylation status (7,15,17),
suggesting that aberrant IGF2 expression may well be induced
by other unknown mechanisms.
MED12 mutations have recently been identified
in fibroepithelial tumors of the breast (22), that is, in 47–59% of FAs and 67–80%
of PTs (22–25) and were detected specifically in the
stromal cells of these tumors. MED12 mutations were first
described in a study of leiomyomas of the uterus, and enhanced
expression of IGF2 has been found in leiomyomas with
MED12 mutations (26),
implicating that MED12 mutation is associated with
IGF2 expression also in fibroepithelial tumors of the
breast.
In the present study, we therefore first
investigated whether LOI occurs in FAs and PTs and is associated
with IGF2 expression since these important issues have not
been properly addressed yet. Furthermore, we studied the impact of
DMR0 methylation or MED12 mutation on LOI or IGF2
expression, respectively.
Materials and methods
Patient samples
For this study 58 FAs and 27 PTs from 52 and 24
female patients, respectively, were analyzed. Clinicopathological
characteristics of these tumors are shown in Table I. All these tumors are the same as
those analyzed in our previous study on MED12 mutations
(23). The patients underwent
tumorectomy or mastectomy between 1993 and 2005. Five patients had
synchronous multiple FAs, one patient synchronous FA and PT, one
patient metachronous FA and PT, and two patients metachronous
multiple PTs. Histological diagnosis of all tumors was confirmed by
two pathologists (E.M. and J.I.). This study was approved by the
Osaka university Research ethics Committee.
 | Table IImprinting status of IGF2 in
fibroadenomas and phyllodes tumors. |
Table I
Imprinting status of IGF2 in
fibroadenomas and phyllodes tumors.
|
Characteristics | Imprinting status
| P-value |
|---|
| LOI (−) | LOI (+) | Total |
|---|
| Tumor
histology | | | | |
| Fibroadenomas | 12 | 13 | 25 | 0.224 |
| Phyllodes
tumors | 5 | 12 | 17 | |
| Fibroadenomas | | | | |
| Age
(years)a | 22 (18–63) | 30 (16–48) | | 0.465 |
| Tumor size
(mm)a | 27 (13–55) | 29 (13–98) | | 0.723 |
| Histological
type | | | | |
|
Intracanalicular | 8 | 8 | 16 | 0.923 |
|
Pericanalicular | 2 | 3 | 5 | |
| Organoid | 2 | 2 | 4 | |
| Mastopathic | 0 | 0 | 0 | |
| Phyllodes
tumors | | | | |
| Age
(years)a | 50 (21–52) | 44 (13–61) | | 0.632 |
| Tumor size
(mm)a | 25 (17–72) | 43 (15–220) | | 0.333 |
| Histological
grade | | | | |
| Benign | 5 | 8 | 13 | 0.070 |
| Borderline | 0 | 4 | 4 | |
| Malignant | 0 | 0 | 0 | |
DNA extraction from formalin-fixed
paraffin-embedded (FFPE) tumor tissues
For DNA extraction, three to eight 10-µm
sections per tumor were cut from the FFPE tumor tissues and mounted
onto a polyethylene napthalate (PEN) membrane slide (Leica
Microsystems GmbH, Wetzlar, Germany). The FFPE tumor tissues slides
were stained with hematoxylin after deparaffinization and the tumor
area was macrodissected with a scalpel and with stereoscopic
assistance. Genomic DNA from the paraffin sections was extracted
and purified using the QIAamp DNA FFPE kit (Qiagen, Valencia, CA,
USA) and 1 µg of genomic DNA was subjected to sodium
bisulfite treatment with the EpiTect Bisulfite kit (Qiagen).
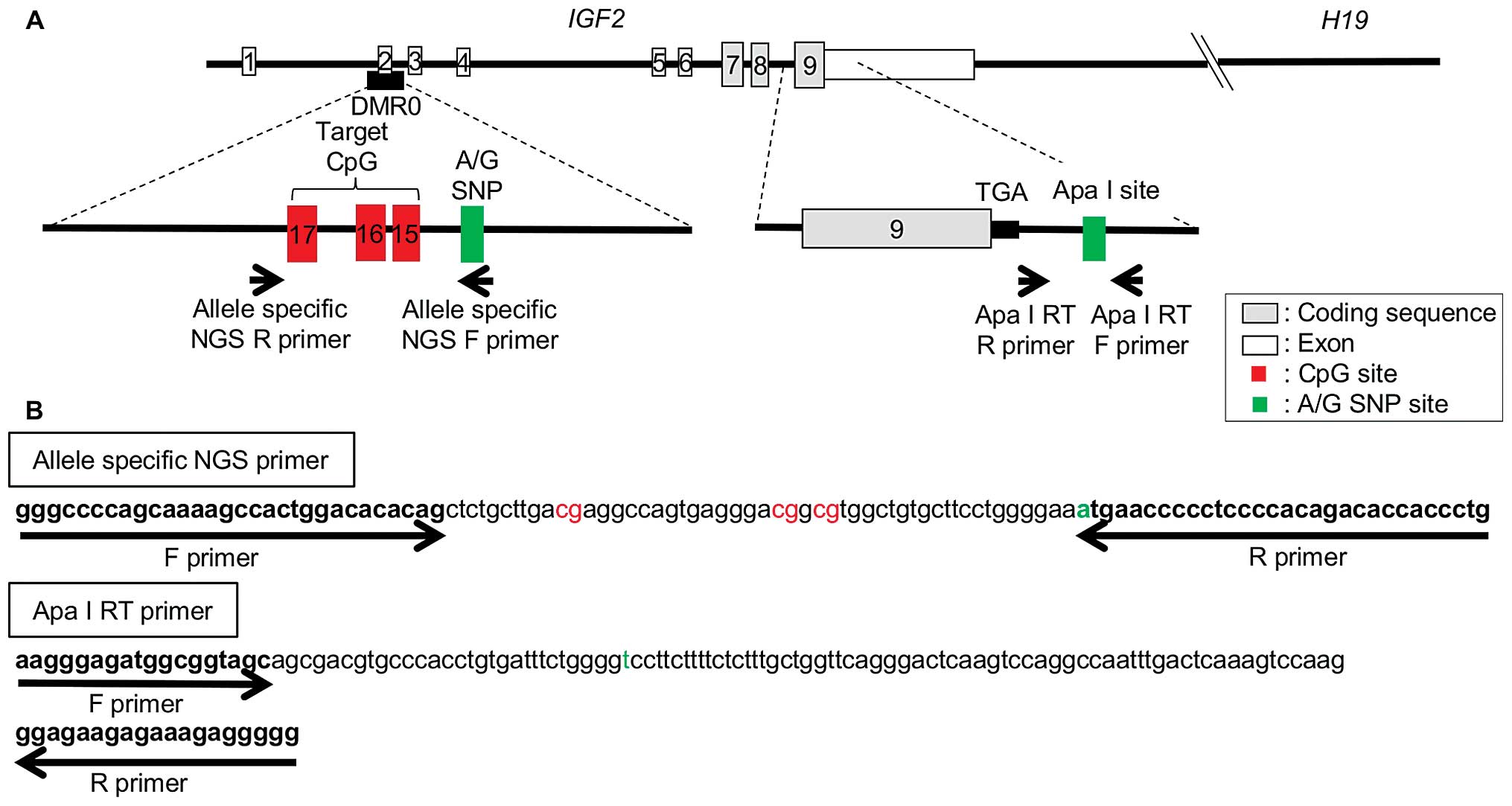
Quantitative IGF2 DMR0 methylation
analysis using NGS
Since it has been reported that IGF2 LOI
correlates with hypomethylation of three CpGs (CpG 15–17) included
in DMR0, we performed target sequencing of this region by means of
next-generation sequencing (NGS). For allele-specific
pyrosequencing, two primer sets were designed, which recognized the
A or G allele of the rs3741210 polymorphism near the three targeted
CpGs. The following NGS primers were designed for IGF2 DMR0
allele-specific methylation: forward
5′-GGGCCCCAGCAAAAGCCACTGGACACACAG-3′, reverse for A-allele
5′-CAGGGTGGTGTCTGTGGGGAGGGGGTTCAT-3′ and reverse for G-allele
5′-CAGGGTGGTGTCTGTGGGGAGGGGGTTCAC-3′ (amplicon size = 111 bp;
Fig. 1). A total volume of 30
µl contained 4 µl of bisulfite DNA and 0.4 µM
of each primer. PCR was carried out using Takara Ex Taq®
Hot Start Version (Takara Bio Inc., Shiga, Japan). PCR conditions
were as follows: initial denaturing at 95°C for 15 min; 40 cycles
at 95°C for 30 sec, at 56°C for 30 sec, at 72°C for 30 sec, and a
final extension at 72°C for 10 min.
The PCR amplicons were purified with the QIAquick
PCR Purification kit (Qiagen). The samples amplified with both A-
and G-allele specific PCR were defined as informative for an A/G
polymorphism. Twenty-four FAs and 13 PTs were subjected to
methylation analysis. The NGS methylation assay was performed by
using the GS Junior system (Roche Diagnostics, Basel, Switzerland)
according to the manufacturer's instructions, and data were
analyzed with the GS Amplicon Variant Analyzer (AVA) software
(version 2.7; Roche Diagnostics). The methylation ratio was
calculated by dividing the number of cytosines by that of the total
reads at each CpG site. The average methylation ratio of the three
CpG sites was then calculated and we defined one allele showing a
higher methylation ratio as the paternal allele and the other
allele showing a lower methylation ratio as the maternal
allele.
Selection of samples with informative Apa
I polymorphism
IGF2 imprinting status was determined by
assaying for Apa I polymorphism (rs680) within the IGF2 exon
9 by means of restriction digestion of PCR products obtained by
using the following Apa I RT primers: forward
5′-AAGGGAGATGGCGGTAGC-3′ and reverse 5′-CCCCCTCTTTCTCTTCTCC-3′
(amplicon size = 129 bp; Fig. 1). A
total volume of 20 µl contained 2 µl of genomic DNA
and 0.4 µM of each primer. PCR was carried out using Takara
Ex Taq Hot Start Version (Takara Bio Inc.). PCR conditions were as
follows: initial denaturing at 95°C for 15 min; 40 cycles at 95°C
for 30 sec, at 61°C for 30 sec, at 72°C for 30 sec, and a final
extension at 72°C for 10 min. Five microliters of each of the PCR
products was digested with 15 units of Apa I at 37°C for 60 min
(Takara Bio Inc.). The A allele (not digested by Apa I) produced
129 bp and the G allele (digested by Apa I) produced 80 and 49 bp.
Samples which showed heterozygous A/G at the Apa I polymorphism
were defined as informative and selected for RNA isolation.
RNA extraction from FFPE tissues
For RNA extraction, two to four 10-µm
sections were removed from the FFPE tumor tissues. After
deparaffinization, staining and macrodissection as detailed above,
RNA was extracted and purified with the RNeasy FFPE kit (Qiagen).
Reverse transcription from RNA to cDNA was performed with the
ReverTra Ace qPCR RT kit (Toyobo, Osaka, Japan). The incubation was
as follows: 37°C for 20 min; 98°C for 5 min.
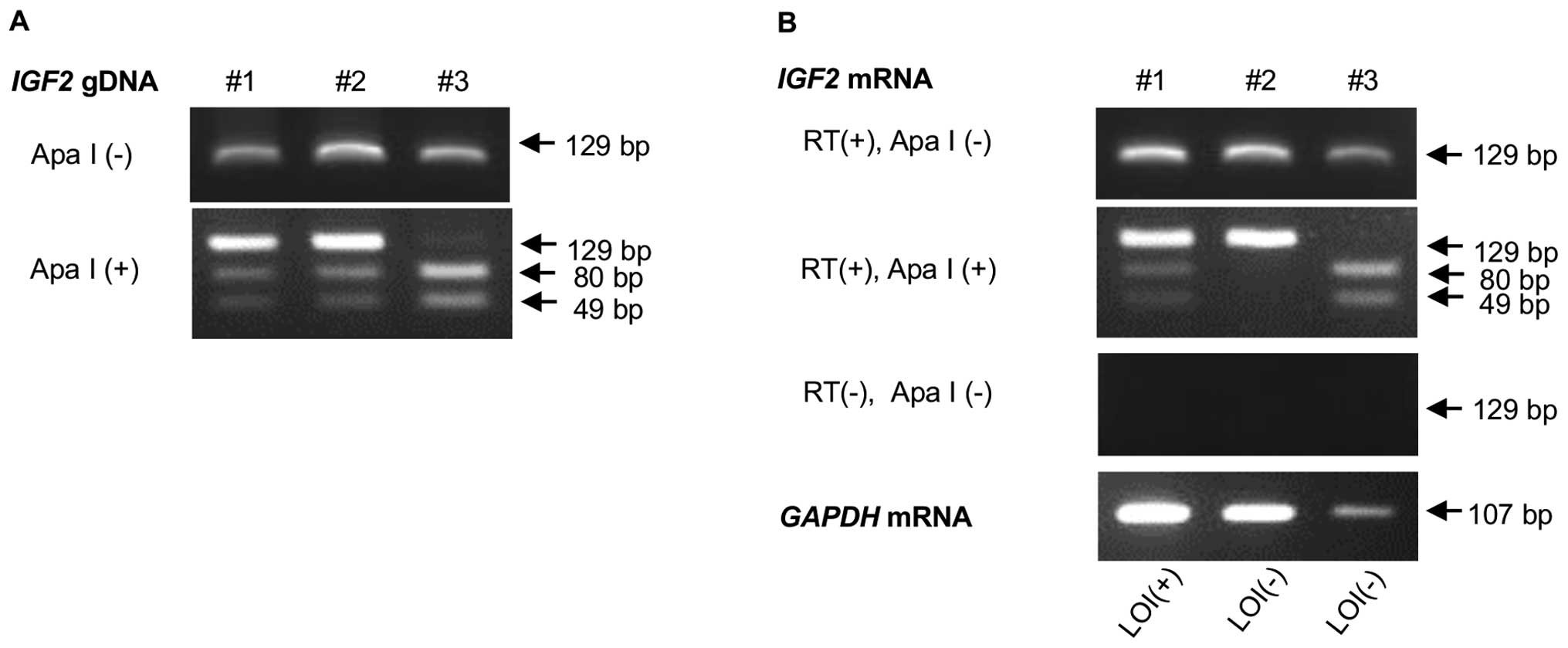
Analysis of IGF2 imprinting status
RT-PCR was used to analyze RNAs from the informative
specimens from 25 FAs and 17 PTs for allele-specific Apa I site
polymorphism. PCR and Apa I digestion were performed as detailed
above. A specimen was identified as LOI when two bands were clearly
visible on a gel with a ratio of at least 1:4 as previously
described (Fig. 2) (15).
Real-time RT-PCR
Quantitative mRNA expression was measured by using
the Light cycler 480 Real-time PCR System (Roche Applied Science,
Mannheim, Germany) at 95°C (10 min), followed by 50 cycles at 95°C
(15 sec) and at 60°C (60 sec), and finally one cycle at 50°C (10
sec). IGF2 and glyceraldehyde 3-phosphate dehydrogenase
(GAPDH) TaqMan® Gene Expression Assays (assay
identification numbers: Hs01005963_m1 and Hs02758991_g1; Applied
Biosystems, Foster City, CA, USA) were used for the real-time qPCR
assay. The expression of IGF2 was normalized to that of
GAPDH, and each assay was performed in duplicate.
IGF2 expression analysis using laser
microdissection (LMD)
A 10-µm section was cut from each of the FFPE
tumor tissues of three FAs and three PTs and mounted onto a
polyethylene napthalate (PEN) membrane slide (Leica Microsystems
GmbH), which was stained with hematoxylin after deparaffinization.
One area of the epithelium or stroma was selected and dissected
with the laser microdissection system LMD7000 (Leica). Each LMD
specimen was automatically collected by gravity into the cap of a
microdissection tube. The LMD specimens were then subjected to RNA
extraction with the RNeasy FFPE kit (Qiagen). Reverse
transcription, IGF2 imprinting analysis and real-time RT-PCR
were performed as described above.
IGF2 expression analysis of matched
normal breast tissues
Twelve normal breast tissues adjacent to the tumors
could be obtained for six FAs and six PTs. These tissues were
macrodissected and RNA was extracted with the RNeasy FFPE kit.
Reverse transcription, IGF2 imprinting analysis and
real-time RT-PCR were performed as described above.
Correlation with IGF2 expression and
MED12 mutation status
The 23 FAs and 17 PTs which were identified as
informative for Apa I polymorphisms were subjected to MED12
mutation analysis. Since mutation in the MED12 exon 2
reportedly correlates with IGF2 overexpression, we used NGS
for target sequencing of this exon. In brief, DNA from the paraffin
sections was amplified by means of PCR using the following primers:
forward 5′-AACAACTAAACGCCGCTTTC-3′ and reverse
5′-ATGCTCATCCCCAGAGACAG-3′; amplicon, 98 bp. Primers were designed
to cover 93% of MED12 mutations of FAs as previously
reported by Lim et al (22).
The PCR amplicons were purified with NucleoSpin Gel and PCR
Clean-Up (Macherey-Nagel, Düren, Germany). NGS was performed with
the GS Junior system and the data were analyzed with GS AVA
software, which can detect mutations with a minimum read count of
two and a minimum read percentage of 0.25%. Variant allele
frequencies were calculated for each position and a threshold of 5%
was used to characterize candidate variants based on the findings
by Lim et al (22).
Statistics
Associations of the clinicopathological
characteristics with IGF2 imprinting status were evaluated
by means of the chi-square test or Fisher exact test. Differences
in DMR0 methylation ratio or IGF2 mRNA expression were
assessed with the t-test. All statistical analyses were two-sided
and P<0.05 was considered to be significant.
Results
Imprinting status of IGF2 in FAs and
PTs
A total of 58 FAs and 27 PTs were subjected to Apa I
polymorphism analysis and 25 FAs (43%) and 17 PTs (63%) were
identified as heterozygous (A/G) for this polymorphism and used for
the subsequent analysis. The frequency of LOI was higher for PTs
(71%, 12/17) than for FAs (52%, 13/25) although the difference was
not statistically significant. However, the imprinting status of
IGF2 in FAs and PTs was not associated with age, tumor size,
histological type or histological grade (Table I).
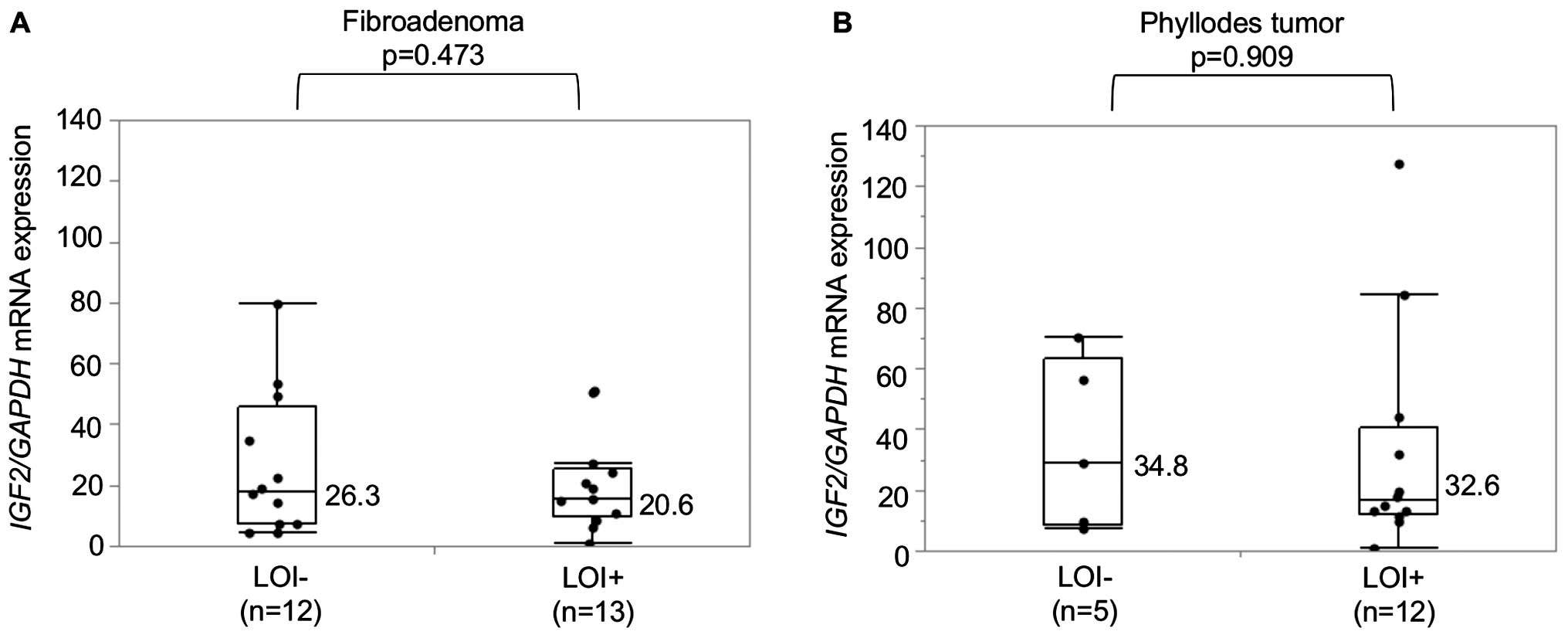
Relationship between LOI and IGF2 mRNA
expression
IGF2 mRNA expression in 25 FAs and 17 PTs was
determined by means of qRT-PCR and showed no significant difference
between LOI positive (LOI+) and LOI negative
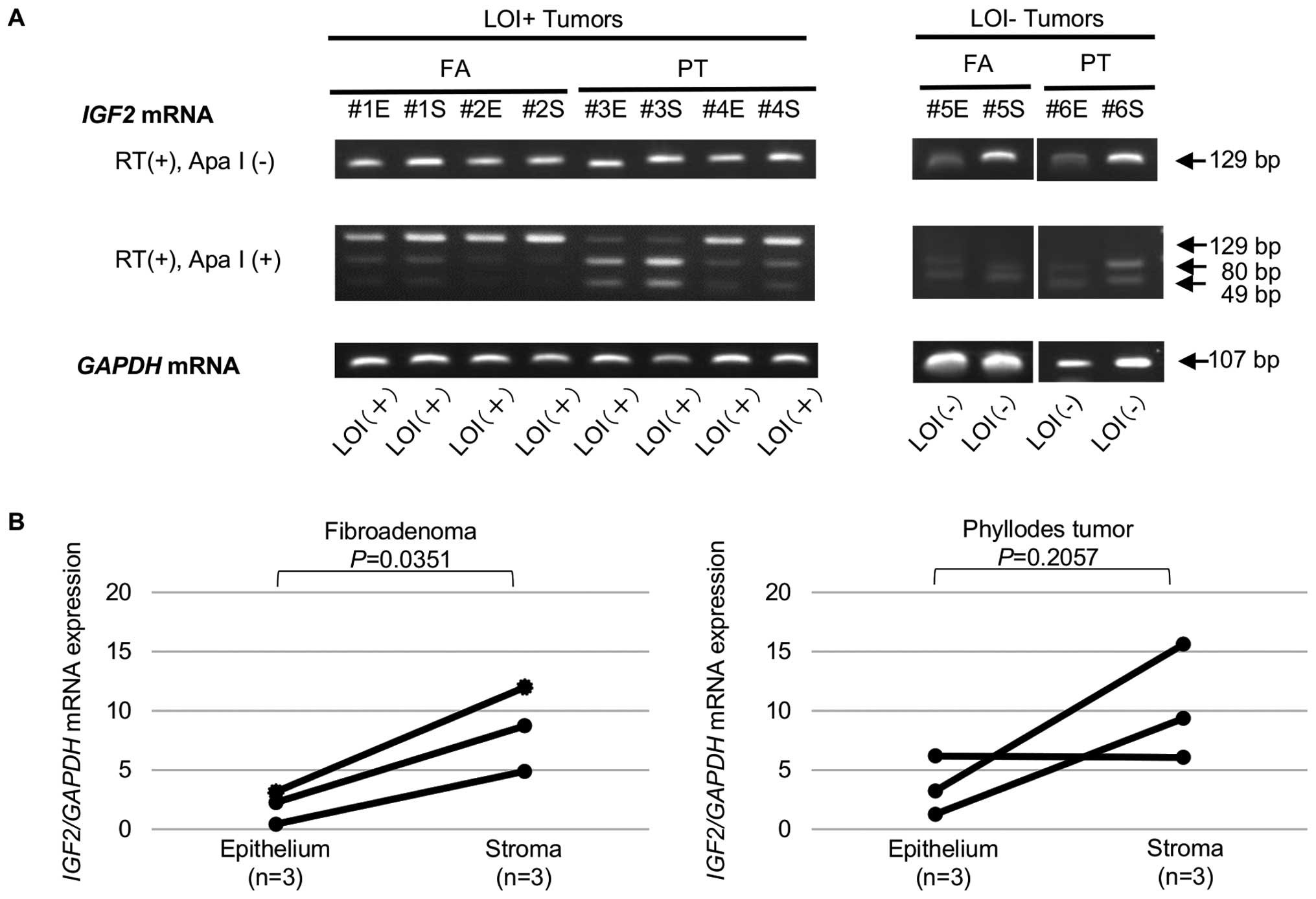
(LOI−) tumors in either FAs or PTs (Fig. 3). The epithelial cells and stromal
cells were then separately obtained from the six tumors (three FAs
and three PTs) with the aid of LMD and subjected to the LOI assay.
The IGF2 imprinting status of the epithelial cells and the
stromal cells in each tumor was identical, i.e., three tumors
contained LOI+ epithelial cells and LOI+
stromal cells and three tumors LOI− epithelial cells and
LOI− stromal cells (Fig.
4A). On the contrary, IGF2 mRNA expression in most
tumors (three FAs and two PTs) was upregulated in the stromal cells
in comparison with that in the epithelial cells (Fig. 4B).
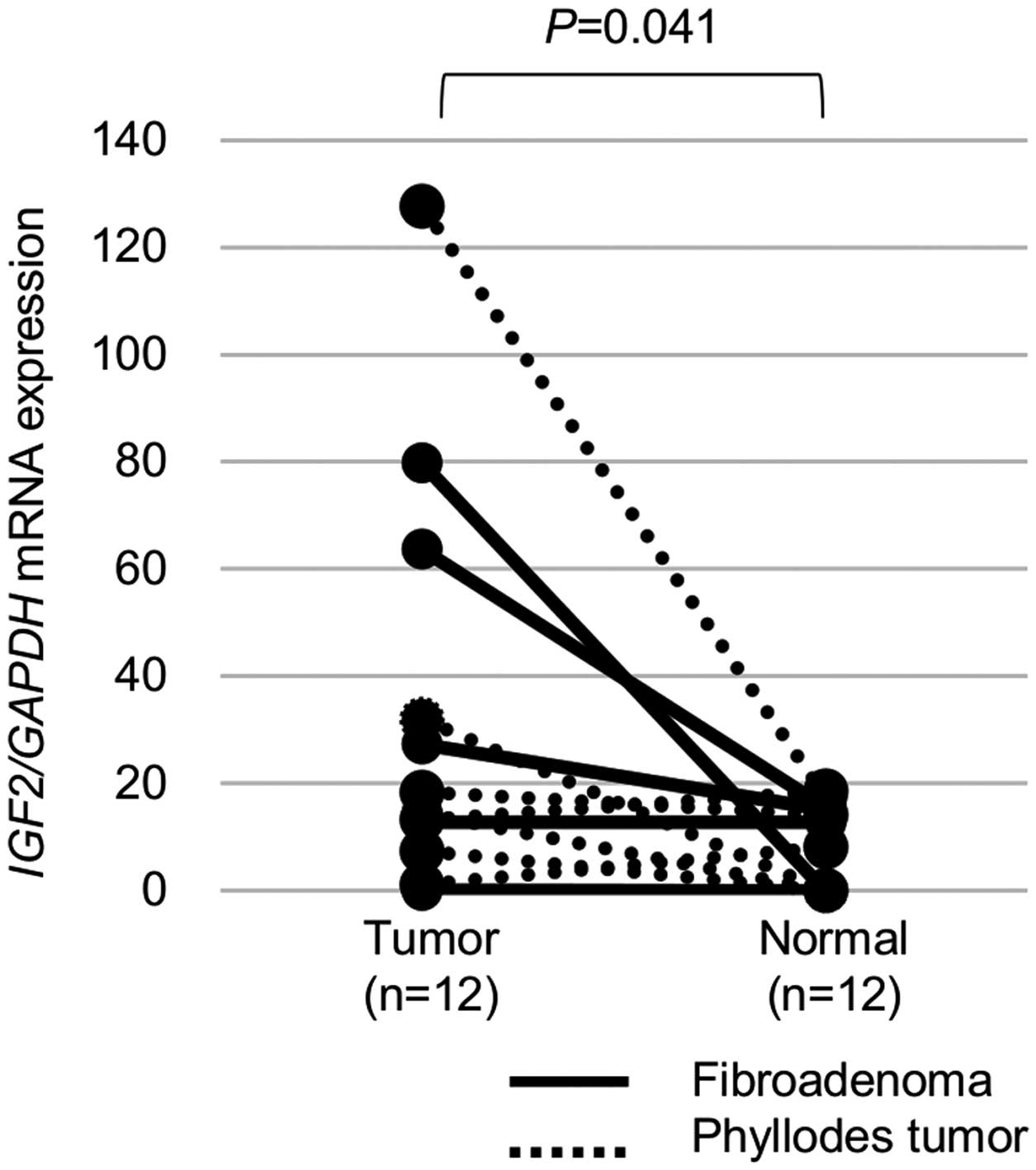
We also analyzed LOI and IGF2 mRNA expression
in the normal tissues surrounding the tumors (6 FAs and 6 PTs). LOI
was observed in four (33%) of the 12 normal tissues, while
IGF2 mRNA expression was significantly lower in the normal
tissues than in the tumors (P=0.041, Fig. 5).
Correlation between IGF2 mRNA expression
and MED12 mutation
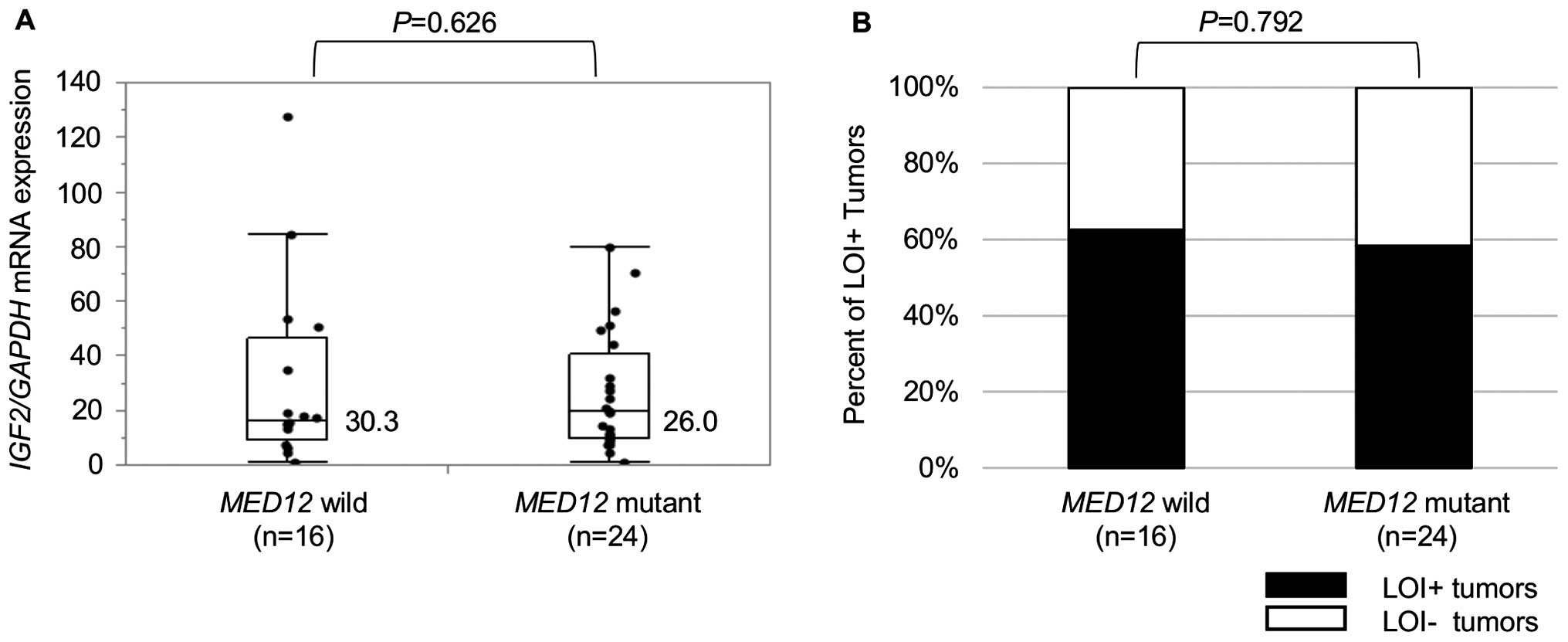
Of the 40 tumors (23 FAs and 17 PTs), 24 (12 FAs and
12 PTs) harbored MED12 mutations. No significant difference
in IGF2 mRNA expression was observed between the tumors with
and without MED12 mutations (Fig. 6A). In addition, MED12
mutation status showed no correlation with IGF2 imprinting
status (Fig. 6B).
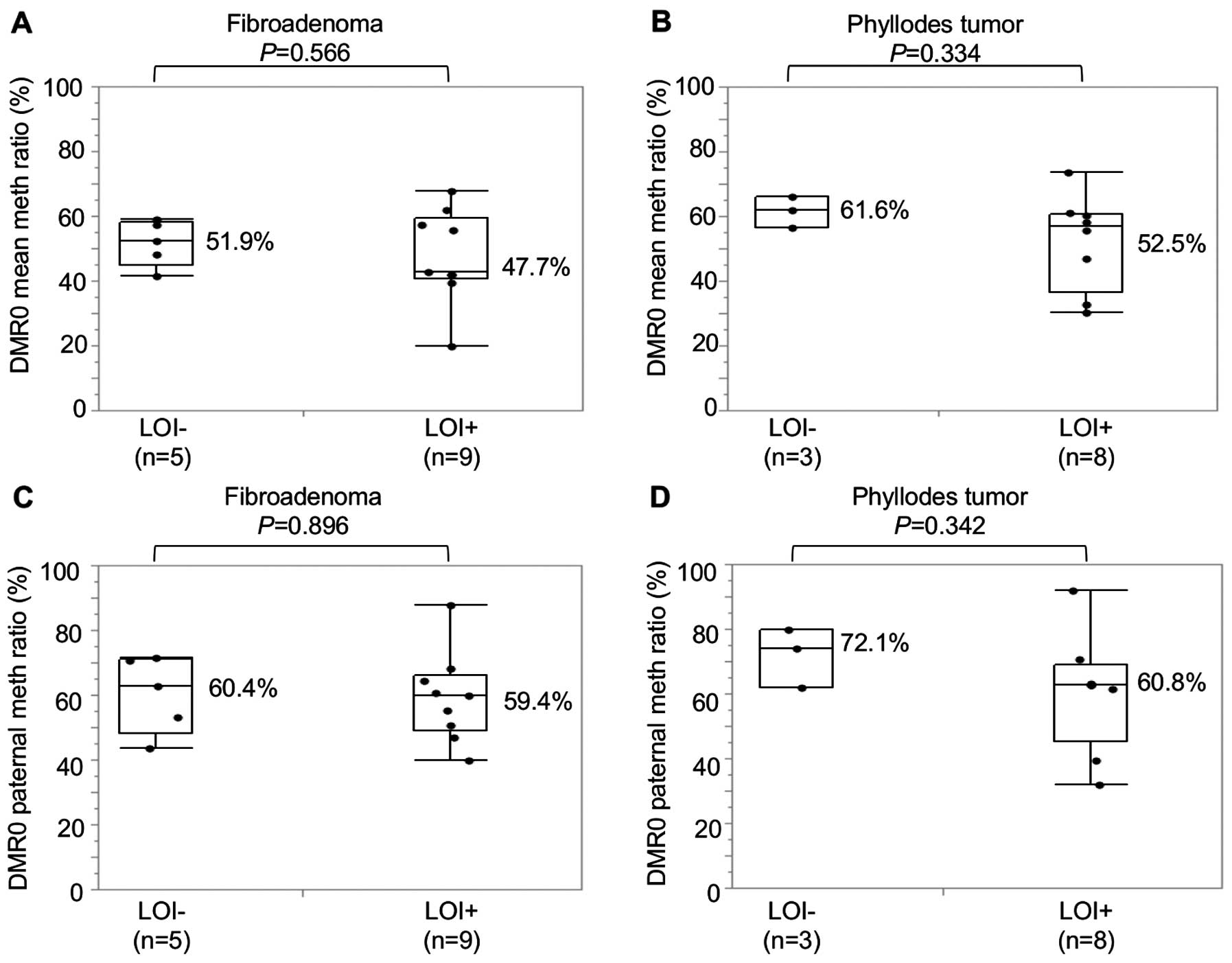
Relationship between LOI and IGF2 DMR0
methylation
The relationship between LOI and DMR0 methylation
was examined in 14 FAs and 11 PTs which were polymorphic for both
rs680 in exon 9 and rs3741210 in DMR0. There was no significant
difference in DMR0 methylation ratios of either allele or a
paternal allele between LOI+ and LOI− tumors
(Fig. 7).
Discussion
In the present study, LOI was observed in 52%
(13/25) of FAs and 71% (12/17) of PTs but the IGF2 mRNA
expression was not associated with LOI, in contrast to the two
classical studies on Wilms' tumors, which reported that IGF2
mRNA expression was two times higher in LOI+ than
LOI− tumors (9,10). Although Kaneda et al reported
that IGF2 was upregulated in LOI+ intestinal
crypts in mice (27), no
association of LOI with IGF2 mRNA upregulation has been
reported by other investigators in studies of several types of
tumors including breast cancer (7,17,28).
We therefore considered it unlikely that LOI plays an important
role in the upregulation of IGF2 mRNA in the majority of
tumors including FAs and PTs.
We were able to confirm that IGF2 mRNA
expression is significantly upregulated in FAs and PTs in
comparison with surrounding normal tissues as previously described
(7,21,29). A
separate analysis of IGF2 mRNA expression in the epithelial
cells and stromal cells obtained by means of LMD demonstrated that
IGF2 mRNA expression is more upregulated in the stromal than
the epithelial cells. This observation seems to be consistent with
the fact that, although FAs and PTs consist of both epithelial and
stromal components, they essentially stem from overgrowth of the
stromal cells since the stromal cells, but not the epithelial
cells, harbor MED12 mutations (3,23,25).
Thus, it is speculated that IGF2 plays a definite and
significant role in the pathogenesis of these tumors even though
the mechanism of its upregulation is not related to LOI.
Noteworthy, the imprinting status of the epithelial
cells and stromal cells in each tumor was identical, i.e., every
tumor consisted of either LOI+ epithelial cells and
LOI+ stromal cells or LOI− epithelial cells
and LOI− stromal cells. It has been reported that LOI
can be induced by treatment with butyrate, which modifies histone
acetylation (5). Furthermore,
oxidative stress reportedly induces NF-κB binding to the
CCCTC-binding factor (CTCF) promoter, which then leads to reduced
CTCF expression, loss of CTCF binding to the ICR and IGF2
LOI (30). The epithelial cells of
FAs and PTs often exhibit hyperplastic change, so that we speculate
that a certain factor produced from the stromal cells might
stimulate the proliferation of the epithelial cells, and such a
factor might also be implicated in the induction of LOI in the
epithelial cells.
Consistent with previous observations regarding
colon cancer, esophageal cancer and breast cancer that LOI was
detected not only in tumor tissues but also in the matched normal
tissues (28,31,32),
we identified LOI in 33% of the normal tissues surrounding the FAs
or PTs. It has been reported that the presence of LOI in normal
tissues can be a risk factor for the development of colon cancer
(33,34). Since LOI in the normal tissues from
the healthy controls was not analyzed in the present study, it
remains as yet unknown whether the presence of LOI in normal tissue
represents a risk for the development of FAs or PTs.
Di Tommaso et al reported that leiomyomas
with MED12 mutations expressed significantly higher levels
of IGF2 mRNA (26). However,
we could not detect any significant association of MED12
mutations with the upregulation of IGF2 mRNA levels,
indicating that MED12 mutations perform a different role in
the pathogenesis of leiomyomas and of FAs or PTs. DMR0
hypomethylation is reportedly associated with LOI in colorectal
cancers (19) but not in other
types of cancer (6,15). Since a significant association
between LOI status and DMR0 methylation levels could not be
demonstrated in our study, DMR0 methylation is unlikely to play an
important role in the development of LOI in FAs and PTs.
In conclusion, we were able to demonstrate that
IGF2 mRNA expression is upregulated in the stromal cells in
FAs and PTs, but that LOI of IGF2 is not implicated in this
upregulation. Furthermore, MED12 mutations were found not to
be associated with induction of LOI or IGF2 mRNA
upregulation, while DMR0 hypomethylation was found to be unlikely
to play a significant role in the induction of LOI. Further studies
are thus needed to clarify the role of LOI and the mechanism of
IGF2 mRNA upregulation in the pathogenesis of FAs and
PTs.
Abbreviations:
|
FA
|
fibroadenoma
|
|
PT
|
phyllodes tumor
|
References
|
1
|
Sawyer EJ, Hanby AM, Poulsom R, Jeffery R,
Gillett CE, Ellis IO, Ellis P and Tomlinson IP: Beta-catenin
abnormalities and associated insulin-like growth factor
overexpression are important in phyllodes tumours and fibroadenomas
of the breast. J Pathol. 200:627–632. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Reik W, Dean W and Walter J: Epigenetic
reprogramming in mammalian development. Science. 293:1089–1093.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Nordin M, Bergman D, Halje M, Engström W
and Ward A: Epigenetic regulation of the Igf2/H19 gene cluster.
Cell Prolif. 47:189–199. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Murrell A, Ito Y, Verde G, Huddleston J,
Woodfine K, Silengo MC, Spreafico F, Perotti D, De Crescenzo A,
Sparago A, et al: Distinct methylation changes at the IGF2-H19
locus in congenital growth disorders and cancer. PLoS One.
3:e18492008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Shin JH, Li RW, Gao Y, Bickhart DM, Liu
GE, Li W, Wu S and Li CJ: Butyrate induced IGF2 activation
correlated with distinct chromatin signatures due to histone
modification. Gene Regul Syst Bio. 7:57–70. 2013.PubMed/NCBI
|
|
6
|
Murata A, Baba Y, Watanabe M, Shigaki H,
Miyake K, Ishimoto T, Iwatsuki M, Iwagami S, Yoshida N, Oki E, et
al: IGF2 DMR0 methylation, loss of imprinting, and patient
prognosis in esophageal squamous cell carcinoma. Ann Surg Oncol.
21:1166–1174. 2014. View Article : Google Scholar
|
|
7
|
Cheng YW, Idrees K, Shattock R, Khan SA,
Zeng Z, Brennan CW, Paty P and Barany F: Loss of imprinting and
marked gene elevation are 2 forms of aberrant IGF2 expression in
colorectal cancer. Int J Cancer. 127:568–577. 2010. View Article : Google Scholar
|
|
8
|
Giani C, Cullen KJ, Campani D and
Rasmussen A: IGF-II mRNA and protein are expressed in the stroma of
invasive breast cancers: An in situ hybridization and
immunohistochemistry study. Breast Cancer Res Treat. 41:43–50.
1996. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Steenman MJ, Rainier S, Dobry CJ, Grundy
P, Horon IL and Feinberg AP: Loss of imprinting of IGF2 is linked
to reduced expression and abnormal methylation of H19 in Wilms'
tumour. Nat Genet. 7:433–439. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ravenel JD, Broman KW, Perlman EJ, Niemitz
EL, Jayawardena TM, Bell DW, Haber DA, Uejima H and Feinberg AP:
Loss of imprinting of insulin-like growth factor-II (IGF2) gene in
distinguishing specific biologic subtypes of Wilms tumor. J Natl
Cancer Inst. 93:1698–1703. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Cui H, Horon IL, Ohlsson R, Hamilton SR
and Feinberg AP: Loss of imprinting in normal tissue of colorectal
cancer patients with microsatellite instability. Nat Med.
4:1276–1280. 1998. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Takano Y, Shiota G and Kawasaki H:
Analysis of genomic imprinting of insulin-like growth factor 2 in
colorectal cancer. Oncology. 59:210–216. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang FR, He XB, Yang YH and Xie W: The
expression and imprinting status of insulin-like growth factor 2
gene in colorectal cancer. Zhonghua Yi Xue Yi Chuan Xue Za Zhi.
20:31–34. 2003.In Chinese. PubMed/NCBI
|
|
14
|
Jarrard DF, Bussemakers MJ, Bova GS and
Isaacs WB: Regional loss of imprinting of the insulin-like growth
factor II gene occurs in human prostate tissues. Clin Cancer Res.
1:1471–1478. 1995.PubMed/NCBI
|
|
15
|
Ito Y, Koessler T, Ibrahim AE, Rai S,
Vowler SL, Abu-Amero S, Silva AL, Maia AT, Huddleston JE,
Uribe-Lewis S, et al: Somatically acquired hypomethylation of IGF2
in breast and colorectal cancer. Hum Mol Genet. 17:2633–2643. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yun K, Soejima H, Merrie AE, McCall JL and
Reeve AE: Analysis of IGF2 gene imprinting in breast and colorectal
cancer by allele specific-PCR. J Pathol. 187:518–522. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yballe CM, Vu TH and Hoffman AR:
Imprinting and expression of insulin-like growth factor-II and H19
in normal breast tissue and breast tumor. J Clin Endocrinol Metab.
81:1607–1612. 1996.PubMed/NCBI
|
|
18
|
McCann AH, Miller N, O'Meara A, Pedersen
I, Keogh K, Gorey T and Dervan PA: Biallelic expression of the IGF2
gene in human breast disease. Hum Mol Genet. 5:1123–1127. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Cui H, Onyango P, Brandenburg S, Wu Y,
Hsieh CL and Feinberg AP: Loss of imprinting in colorectal cancer
linked to hypomethylation of H19 and IGF2. Cancer Res.
62:6442–6446. 2002.PubMed/NCBI
|
|
20
|
Baba Y, Nosho K, Shima K, Huttenhower C,
Tanaka N, Hazra A, Giovannucci EL, Fuchs CS and Ogino S:
Hypomethylation of the IGF2 DMR in colorectal tumors, detected by
bisulfite pyrosequencing, is associated with poor prognosis.
Gastroenterology. 139:1855–1864. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Shetty PJ, Movva S, Pasupuleti N,
Vedicherlla B, Vattam KK, Venkatasubramanian S, Ahuja YR and Hasan
Q: Regulation of IGF2 transcript and protein expression by altered
methylation in breast cancer. J Cancer Res Clin Oncol. 137:339–345.
2011. View Article : Google Scholar
|
|
22
|
Lim WK, Ong CK, Tan J, Thike AA, NG CC,
Rajasegaran V, Myint SS, Nagarajan S, Nasir ND, McPherson JR, et
al: Exome sequencing identifies highly recurrent MED12 somatic
mutations in breast fibroadenoma. Nat Genet. 46:877–880. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Mishima C, Kagara N, Tanei T, Naoi Y,
Shimoda M, Shimomura A, shimazu K, Kim SJ and Noguchi S: Mutational
analysis of MED12 in fibroadenomas and phyllodes tumors of the
breast by means of targeted next-generation sequencing. Breast
Cancer Res Treat. 152:305–312. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Cani AK, Hovelson DH, McDaniel AS, Sadis
S, Haller MJ, Yadati V, Amin AM, Bratley J, Bandla S, Williams PD,
et al: Next-gen sequencing exposes frequent MED12 mutations and
actionable therapeutic targets in phyllodes tumors. Mol Cancer Res.
13:613–619. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yoshida M, Sekine S, Ogawa R, Yoshida H,
Maeshima A, Kanai Y, Kinoshita T and Ochiai A: Frequent MED12
mutations in phyllodes tumours of the breast. Br J Cancer.
112:1703–1708. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Di Tommaso S, Tinelli A, Malvasi A and
Massari S: Missense mutations in exon 2 of the MED12 gene are
involved in IGF-2 overexpression in uterine leiomyoma. Mol Hum
Reprod. 20:1009–1015. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kaneda A, Wang CJ, Cheong R, Timp W,
Onyango P, Wen B, Iacobuzio-Donahue CA, Ohlsson R, Andraos R,
Pearson MA, et al: Enhanced sensitivity to IGF-II signaling links
loss of imprinting of IGF2 to increased cell proliferation and
tumor risk. Proc Natl Acad Sci USA. 104:20926–20931. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhao R, DeCoteau JF, Geyer CR, Gao M, Cui
H and Casson AG: Loss of imprinting of the insulin-like growth
factor II (IGF2) gene in esophageal normal and adenocarcinoma
tissues. Carcinogenesis. 30:2117–2122. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Hubertus J, Lacher M, Rottenkolber M,
Müller-Höcker J, Berger M, Stehr M, von Schweinitz D and Kappler R:
Altered expression of imprinted genes in Wilms tumors. Oncol Rep.
25:817–823. 2011. View Article : Google Scholar
|
|
30
|
Yang B, Wagner J, Damaschke N, Yao T,
Wuerzberger-Davis SM, Lee MH, Svaren J, Miyamoto S and Jarrard DF:
A novel pathway links oxidative stress to loss of insulin growth
factor-2 (IGF2) imprinting through NF-κB activation. PLos One.
9:e880522014. View Article : Google Scholar
|
|
31
|
Cui H: Loss of imprinting of IGF2 as an
epigenetic marker for the risk of human cancer. Dis Markers.
23:105–112. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
van Roozendaal CE, Gillis AJ, Klijn JG,
van Ooijen B, Claassen CJ, Eggermont AM, Henzen-Logmans SC,
Oosterhuis JW, Foekens JA and Looijenga LH: Loss of imprinting of
IGF2 and not H19 in breast cancer, adjacent normal tissue and
derived fibroblast cultures. FEBS Lett. 437:107–111. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Woodson K, Flood A, Green L, Tangrea JA,
Hanson J, Cash B, Schatzkin A and Schoenfeld P: Loss of
insulin-like growth factor-II imprinting and the presence of
screen-detected colorectal adenomas in women. J Natl Cancer Inst.
96:407–410. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Cui H, Cruz-Correa M, Giardiello FM,
Hutcheon DF, Kafonek DR, Brandenburg S, Wu Y, He X, Powe NR and
Feinberg AP: Loss of IGF2 imprinting: A potential marker of
colorectal cancer risk. Science. 299:1753–1755. 2003. View Article : Google Scholar : PubMed/NCBI
|