Introduction
Renal cell carcinomas (RCCs) are the most common
cancers that originate from the renal parenchyma, and account for
approximately 90% of all adult kidney malignancies (1). The major histologic subtype of RCC is
clear cell RCC (ccRCC) that represents 70–80% of RCCs, followed by
papillary (10–20%), chromophobe (5%) and collecting duct (1%)
subtypes (2). Globally, the
incidence of kidney cancer is approximately 270,000 new cases and
116,000 deaths, annually (3).
Despite considerable improvements in the diagnosis and treatment of
RCCs in recent years, nearly 20–30% of all patients present with
metastasis at the time of initial diagnosis, and approximately 20%
of RCC patients who undergo nephrectomy may suffer recurrence or
metastasis of the disease (4). For
these patients, the prognosis is extraordinarily poor and median
survival is not more than one year (5). Therefore, it is of great necessity to
investigate the underlying molecular mechanisms of the
tumorigenesis and progression of RCC. Identifying more novel
biomarkers is still required to promote early diagnosis, targeted
therapy and prognosis evaluation.
Nucleolar and spindle-associated protein 1 (NUSAP1)
is a recently identified protein with a molecular weight of 55 kDa
that plays a crucial role in spindle microtubule organization
(6). NUSAP1 exhibits a cell
cycle-dependent localization and is selectively expressed in
proliferating cells. Its mRNA and protein expression levels reach a
peak at the transition of G2 to mitosis and then rapidly
decline after cell division (7).
The depletion and overexpression of NUSAP1 in cells result in
abnormal chromosome segregation, aberrant spindle assembly,
defective cytokinesis, G2/M arrest and microtubule
bundling, respectively (7,8).
In addition to playing an essential role in mitosis,
NUSAP1 has recently attracted broad attention for its involvement
in cancers. Previous studies have shown that elevated expression of
NUSAP1 is correlated with malignancies, including pancreatic
adenocarcinoma, melanoma, glioblastoma, hepatocellular carcinoma,
and prostate cancer (9–13). NUSAP1 has also been associated with
the aggressiveness of meningioma (11), high risk and poor outcome in breast
cancers (14,15). In contrast, a study of childhood ALL
revealed that the expression level of NUSAP1 was decreased in
patients presenting with a poor prognosis (16). These findings suggest a critical
role of NUSAP1 in the initiation and progression of human
cancers.
To date, rare research has been conducted on the
expression and clinical significance of NUSAP1 in RCCs. In the
present study, the expression of NUSAP1 was firstly evaluated in
both ccRCC tissues and RCC cell lines by reverse
transcription-polymerase chain reaction (RT-PCR), western blot (WB)
assay and immunohistochemistry (IHC) assays. Then, we
immunohistochemically analyzed its expression and correlation with
clinicopathological characteristics of the ccRCC patients. The
present study demonstrated that there was a close relationship
between NUSAP1 expression and the prognosis of ccRCC patients.
Furthermore, we investigated the biological behavior of human RCC
cells after downregulation of NUSAP1 expression in vitro,
which suggested that overexpression of NUSAP1 was associated with
cell migration, proliferation and invasion of RCC.
Materials and methods
Patients and tissue specimens
For the RT-PCR analysis, 38 pairs of ccRCC tissue
specimens and matched adjacent normal tissues were collected from
patients who underwent radical nephrectomy at the Second Affiliated
Hospital of Anhui Medical University (Anhui, China) between March
2010 and August 2012. In addition, we prepared 124
paraffin-embedded ccRCC samples for immunohistochemical analysis
from Zhujiang Hospital of Southern Medical University (Guangzhou,
China) between January 2006 and February 2010. Each case was
histologically confirmed as ccRCC and with a verification of no
preoperative chemotherapy or radiotherapy. The patients enrolled in
the immunohistochemical analysis had been followed up until
February 2015 with a median follow-up period of 51.5 months (5–60
months). Clinicopathological data of these patients including age,
gender, tumor size, Fuhrman grade, clinical stage, lymphatic and
distant metastasis (Table I) were
gathered from well-documented medical records. Tumor stage and
grade were classified according to the American Joint Commission on
Cancer (AJCC) tumor, nodes and metastasis (TNM) system and Fuhrman
criteria. The patients enrolled in the present study had given
written informed consent, and the study was approved by the Ethics
Committee of The Second Affiliated Hospital of Anhui Medical
University and Zhujiang Hospital of Southern Medical
University.
 | Table IRelationship between NUSAP1 expression
and clinicopathological characteristics of the ccRCC patients. |
Table I
Relationship between NUSAP1 expression
and clinicopathological characteristics of the ccRCC patients.
| Variable | Cases n (%) | NUSAP1 expression
| χ2 | P-value |
|---|
| High n (%) | Low n (%) |
|---|
| Gender | | | | 0.108 | 0.743 |
| Male | 80 (64.5) | 43 (53.8) | 37 (46.2) | | |
| Female | 44 (35.5) | 25 (56.8) | 19 (43.2) | | |
| Age (years) | | | | 3.016 | 0.082 |
| ≤50 | 43 (34.5) | 19 (44.2) | 24 (55.8) | | |
| >50 | 81 (65.5) | 49 (60.5) | 32 (39.5) | | |
| Fuhrman grade | | | | 18.243 | <0.001a |
| G1–G2 | 84 (67.7) | 35 (41.7) | 49 (58.3) | | |
| G3–G4 | 40 (32.3) | 33 (82.5) | 7 (17.5) | | |
| Tumor size
(cm) | | | | 5.816 | 0.016a |
| ≤7.0 | 86 (69.4) | 41 (47.7) | 45 (52.3) | | |
| >7.0 | 38 (30.6) | 27 (71.1) | 11 (28.9) | | |
| Clinical stage | | | | 16.187 | <0.001a |
| I–II | 94 (75.8) | 42 (44.7) | 52 (55.3) | | |
| III–IV | 30 (24.2) | 26 (86.7) | 4 (13.3) | | |
| Lymph node
metastasis | | | | 3.015 | 0.082 |
| No | 108 (87.1) | 56 (51.9) | 52 (48.1) | | |
| Yes | 16 (12.9) | 12 (75.0) | 4 (25.0) | | |
| Distant
metastasis | | | | 5.199 | 0.023a |
| No | 111 (89.5) | 57 (51.4) | 54 (48.6) | | |
| Yes | 13 (10.5) | 11 (84.6) | 2 (15.4) | | |
Cell culture and transfection
Four human RCC cell lines (786-O, A704, ACHN and
A498) and an immortalized normal human proximal tubule epithelial
cell line HK-2 were purchased from the Cell Bank of the Type
Culture Collection of the Chinese Academy of Sciences (Shanghai,
China) and the American Type Culture Collection (ATCC; Rockville,
MD, USA), respectively. HK-2 cells were cultured in K-SFM medium,
while RCC cells were cultured in Dulbecco's modified Eagle's medium
(DMEM), both supplemented with 10% fetal bovine serum (FBS) and 1%
penicillin-streptomycin (both from Gibco, Carlsbad, CA, USA). All
cells were cultured in a sterile incubator under the condition of
5% CO2 at 37°C. The small interfering RNA (siRNA)
targeting NUSAP1 (si-NUSAP1) and scrambled siRNA (si-NC, as
negative control) were purchased from GenePharma (Shanghai, China).
The sequence designed for si-NUSAP1 was:
5′-GCACCAAGAAGCUGAGAAUTTAUUCUCAGCUUCUUGGUGCTT-3′. RCC cell lines
786-O and A704 were transfected with either si-NUSAP1 or si-NC
using Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA, USA)
following the manufacturer's instructions. Forty-eight hours after
transfection, the mRNA and protein were harvested and analyzed.
Real-time quantitative RT-PCR
RNA isolation from the RCC cells and tissue samples
was performed using TRIzol reagent (Invitrogen) in accordance with
the manufacturer's instructions. The first-strand cDNA was
synthesized by RNA using MMLV reverse transcriptase (Takara, Otsu,
Japan) following the protocol provided. Real-time quantitative
RT-PCR (qRT-PCR) was operated using SYBR Premix Ex Taq™ kit
(Takara) on an ABI 7500 RT-PCR System (Applied Biosystems, Foster
City, CA, USA). The primer sequences designed for cDNA
amplification were as follows: NUSAP1, 5′-GAAGCTGAGAGACAGCCACT-3′
(forward), and 5′-TCTgTgAgTCAgggTCCACA-3′ (reverse); and
glyceraldehyde-3-phosphate dehydrogenase (GAPDH, a housekeeping
gene), 5′-GAAAGCCTGCCGGTGACTAA-3′ (forward), and
5′-GCCCAATACGACCAAATCAGAG-3′ (reverse). The 2−ΔΔCt
method (17) for calculating the
relative levels of NUSAP1 mRNA was applied in the present study,
and all experiments were accomplished in triplicate.
WB assay
Human RCC cells or tissue samples were lysed in RIPA
buffer containing proteinase inhibitor cocktails. After
centrifugation at 12,000 rpm for 20 min, the BCA protein assay kit
(Sigma, St. Louis, MO, USA) was used to quantify the protein
concentrations. Equivalent amounts of harvested proteins (50
µg) were separated using SDS-PAGE, and transferred onto
polyvinylidene difluoride (PVDF) membranes (Bio-Rad, Hercules, CA,
USA). After being blocked with 5% bovine serum albumin (BSA) in
Tris-buffered saline and Tween-20 (TBST) for 2 h, the membranes
were incubated overnight at 4°C with the following antibodies:
rabbit polyclonal anti-NUSAP1 (Thermo Fisher Scientific, Inc.,
Waltham, MA, USA), and goat polyclonal anti-GAPDH (Santa Cruz
Biotechnology, Santa Cruz, CA, USA). Then, the membranes were
washed and incubated with secondary antibodies at room temperature
for 1 h. Proteins were finally visualized using ECL immunoblotting
detection reagent (Biobox Biotech. Co., Ltd, Nanjing, China)
according to the manufacturer's instructions.
IHC and staining analysis
IHC was performed to determine the protein level of
NUSAP1 expression in the ccRCC and matched adjacent normal tissues
using EliVision method for IHC staining. All formalin-fixed and
paraffin-embedded samples were cut in 4-µm thick sections,
deparaffinized and dehydrated. These sections were soaked in 3%
H2O2 to block endogenous peroxides, then,
immerged in effervescent citrate buffer (10 mM, pH 6.0) and
incubated with 5% BSA for 30 min, respectively. Thereafter, the
slides were incubated overnight at 4°C with primary rabbit
polyclonal anti-NUSAP1 antibody (1:50; Thermo Fisher Scientific),
and further incubated with the secondary antibody on the next day.
The DAB kit (ZSGB-Bio, Beijing, China) was used to perform the
visualization. The slides were washed in distilled water and
counterstained with hematoxylin at the end of the staining
process.
IHC staining results were measured based on the
multiplication of staining intensity and density scores. The
intensity score was calculated according to the average intensity
of positive NUSAP1-staining cells (0, none; 1, weak; 2, moderate;
3, strong). The density score was calculated from the results of
the percentage of positive-staining cells (1, <5%; 2, 5–25%; 3,
>25–50%; 4, >50%). The overall score was finally determined,
and scores of 0–4 were defined as low expression while scores >4
were considered as high expression. The immunohistochemical
staining was evaluated by two independent observers blinded to the
clinical outcomes.
Scratch migration assay
Cell migration ability was determined by scratch
migration assay. 786-O and A704 cells transfected with the negative
control (si-NC) or si-NUSAP1 were seeded into 6-well plates at a
concentration of 5×105 cells/well and cultured
overnight. Then, the monolayer cells were scratched with a sterile
10-µl pipette tip, washed thrice with phosphate-buffered
saline (PBS) and incubated at 37°C in 5% CO2. The wound
closure was observed and photographed by a inversion fluorescence
microscope (Olympus, Tokyo, Japan) after 24 h. Each experiment was
performed in triplicate.
Cell invasion assay
Transwell invasion assay was conducted on a 24-well
Transwell chamber plates with a pore size of 8 µm. The
Transwell filter inserts were precoated with 40 µl Matrigel
(dilution at 1:3; BD Biosciences) at 37°C for 5 h. 786-O and A704
cells (1×105 cells/well) were seeded in serum-free DMEM
in the upper chamber, and the lower chamber contained DMEM with 10%
FBS. After incubation at 37°C for 24 h, the non-invaded cells were
scraped off and the invaded cells were fixed with 4%
paraformaldehyde solution and stained with 0.1% crystal violet. The
invaded cells were observed under a light microscope and counted in
five randomly chosen fields.
Cell proliferation assay
To determine cell proliferation, 786-O and A704
cells transfected with siRNA were plated in 96-well plates at a
density of 3×103 cells/well. Cell Counting Kit-8 (CCK-8;
Ding Guo Biotech. Co,. Ltd, Guangzhou, China) was applied to
quantify the cell proliferation at 0, 12, 24 and 48 h. After CCK-8
solution (10 µl) was added to each well, the cells were
incubated for 2 h at 37°C. The absorbance that represented
proliferating cell numbers was detected at 450 nm using a
microplate reader (Thermo). Each experiment was performed in
triplicate and independently repeated three times.
Flow cytometric analysis
Flow cytometric analysis was performed to determine
cell apoptosis and cell cycle distribution. The Annexin V-FITC
apoptosis detection kit (Beyotime-Bio, Shanghai, China) and
propidium iodide (PI) was used for apoptosis assay. 786-O and A704
cells transfected with siRNA were harvested after incubation for 48
h, washed twice by PBS and suspended in 195 µl binding
buffer with 5 µl Annexin V-FITC, and incubated for 10 min at
room temperature in the dark. Then, the cells were re-suspended in
190 µl binding buffer with 10 µl PI, incubated in ice
water and immediately analyzed. For cell cycle analysis, 786-O and
A704 cells were collected and washed twice with PBS, fixed in 75%
ice-cold ethanol overnight. After that, the cells were suspended in
300 µl PBS containing 20 µl RNase and incubated at
37°C for 30 min, and then 400 µl PI was added in the cell
suspension, mixed and incubated at 4°C for 30 min. The results were
analyzed by flow cytometry (BD Biosciences) and each experiment was
performed in triplicate.
Statistical analysis
All statistical analyses were analyzed using SPSS 20
statistical software (IBM, Chicago, IL, USA). Continuous data
expressed as mean ± SD were determined using the two-tailed paired
Student's t-test. The Pearson's Chi-square test was performed to
analyze the correlations between NUSAP1 expression and
clinicopathological characteristics of the ccRCC patients. The
overall survival curves were drawn according to the Kaplan-Meier
method and compared using log-rank test. P-value <0.05 was
considered to indicate a statistically significant difference.
Factors influencing the overexpression of
NUSAP1
Recently, we completed reduced representation
bisulfite sequencing (RRBS) and transcriptome sequencing of 34
pairs of RCC tissues and adjacent normal controls. In addition,
whole-genome sequencing (WGS) was performed for another 61 paired
RCC and adjacent normal tissues. All the analytical processes were
referred to in our previous study (18). To date, studies related to these
data have not yet been published.
Results
Increased expression of NUSAP1 in ccRCC
tissues and RCC cell lines
To investigate the expression of NUSAP1 at the mRNA
and protein levels in ccRCC tissues, qRT-PCR and WB assay were
applied in 38 pairs of ccRCC and matched adjacent normal tissues.
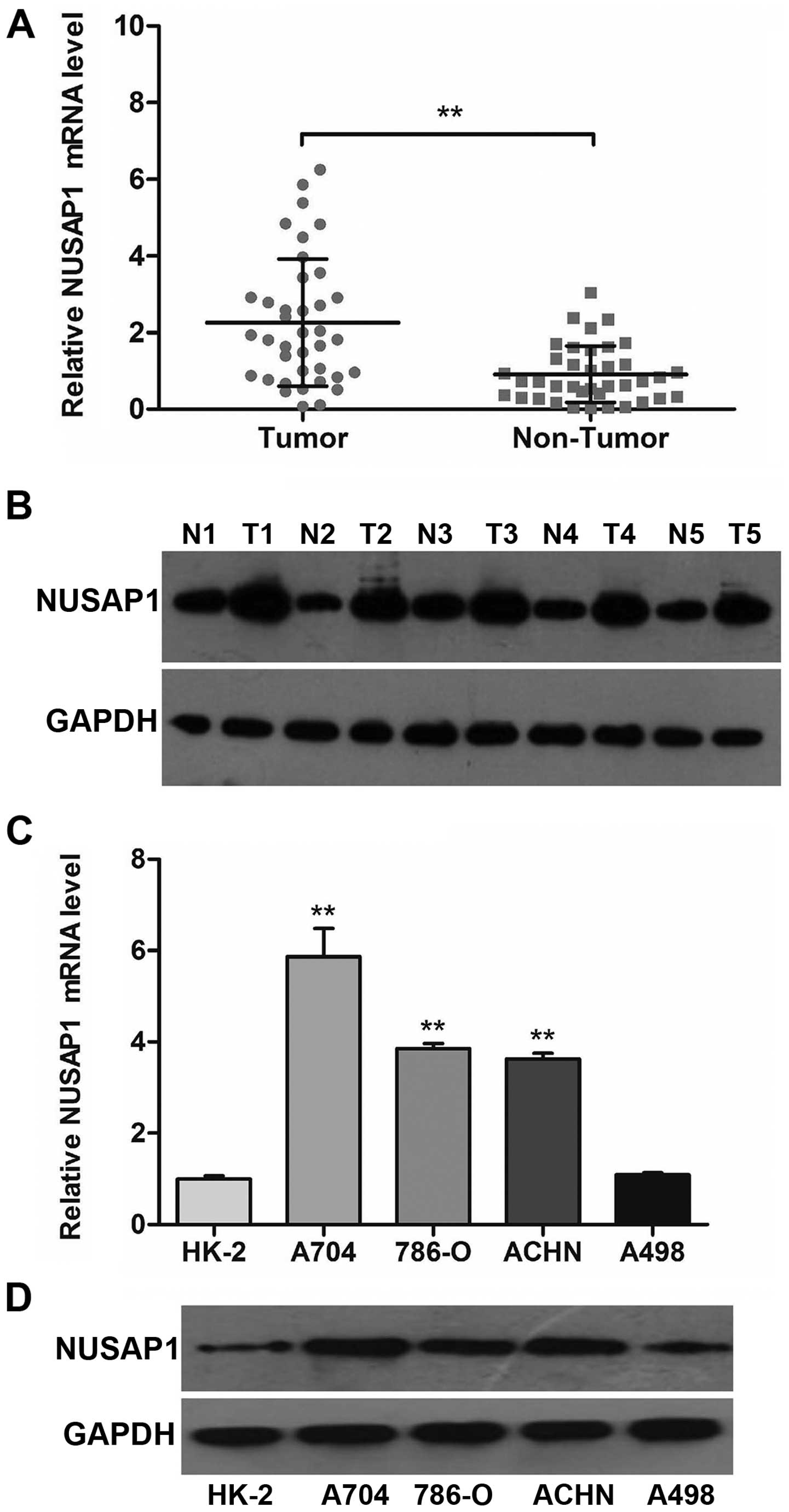
Our results revealed that NUSAP1 was over-expressed in ccRCC
tissues, when compared with levels in the matched normal tissues
(Fig. 1A; P<0.001). In line with
the mRNA data, the protein level of NUSAP1 in representative cancer
tissues was also clearly higher than that in the adjacent normal
tissues (Fig. 1B).
In addition, we determined the expression of NUSAP1
according to the above methods in five types of cell lines,
including HK-2, A704, 786-O, ACHN and A498. The data revealed that
NUSAP1 mRNA and protein levels were statistically elevated in three
RCC cell lines (A704, 786-O and ACHN) compared with that in the
HK-2 cell line (Fig. 1C and D;
P<0.001).
Immunohistochemical analysis of NUSAP1
expression in ccRCC tissues and its correlation with
clinicopathological characteristics
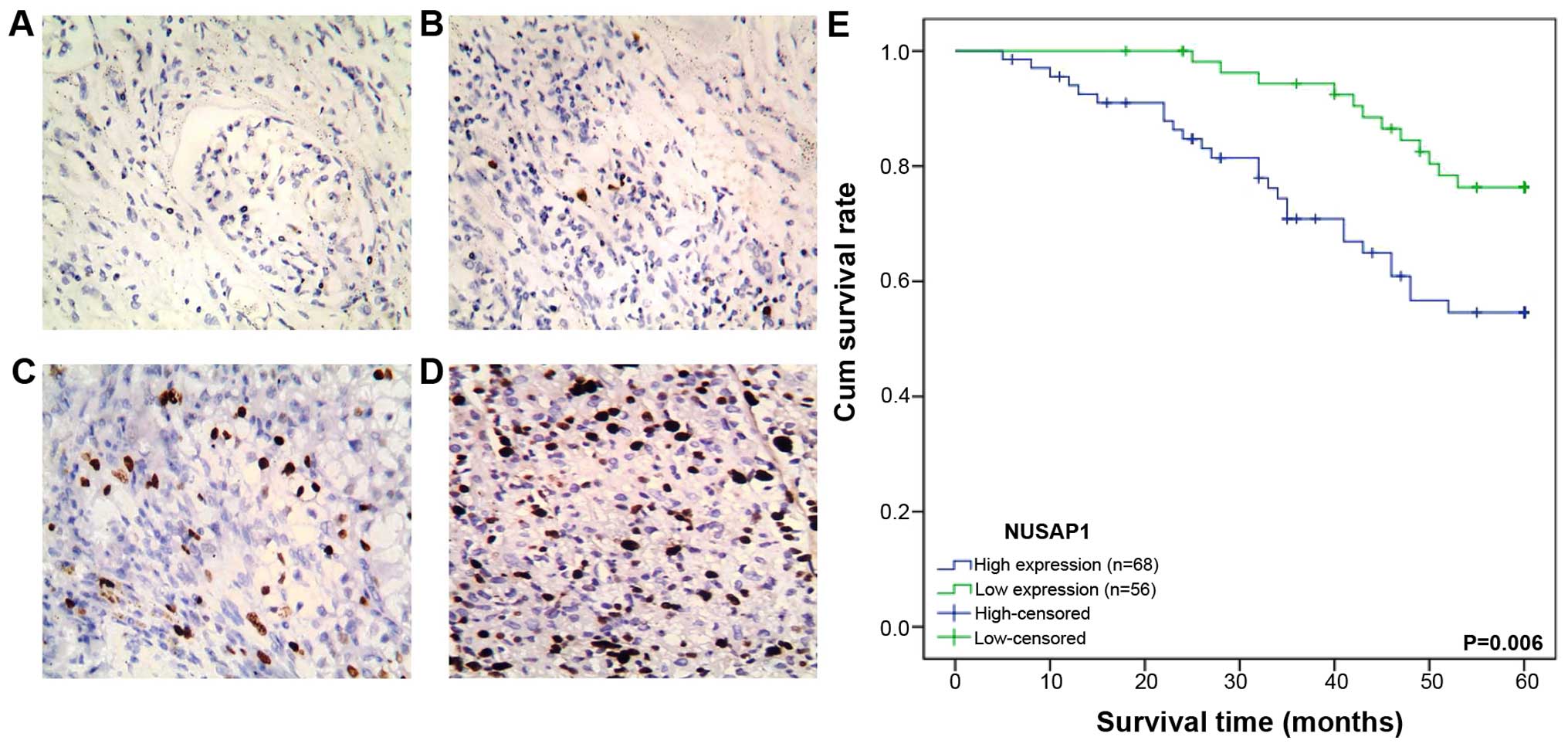
We analyzed the protein level of NUSAP1 in 124
pieces of ccRCC sections by immunohistochemical staining. A total
of 68 (54.8%) cases showed high expression of NUSAP1 while 56 cases
(45.2%) revealed low expression (Fig.
2B–D). The adjacent normal tissues exhibited either no or weak
staining of NUSAP1 (Fig. 2A). The
relationships between NUSAP1 expression and clinicopathological
characteristics of the ccRCC patients are summarized in Table I. Our data demonstrated that the
expression of NUSAP1 was significantly correlated with Fuhrman
grade (P<0.001), tumor size (P=0.016), clinical stage
(P<0.001) and distant metastasis (P=0.023), while no significant
correlation was found between NUSAP1 expression and gender, age and
lymph node metastasis (P>0.05).
NUSAP1 expression predicts the prognosis
of ccRCC patients
The association between NUSAP1 expression and the
prognosis of ccRCC patients was analyzed by Kaplan-Meier method.
The 5-year survival rate in the group of patients with high NUSAP1
expression was 61.8%, whereas it was increased to 78.6% for these
patients with low NUSAP1 expression (Fig. 2E). The log-rank test was used to
compare the correlation between survival and NUSAP1 expression in
the low and high expression groups. Our results suggested that the
patients with low NUSAP1 expression had a significantly longer
overall survival (OS) time than those with high NUSAP1 expression
(P=0.006).
NUSAP1 downregulation suppresses the
growth and aggressiveness of RCC cells in vitro
As shown in Fig. 1C and
D, three RCC cell lines (A704, 786-O and ACHN) exhibited an
obviously increased expression of NUSAP1. We thus chose A704 and
786-O cell lines to determine the biological behaviors of human RCC
cell lines following the downregulation of NUSAP1 expression in
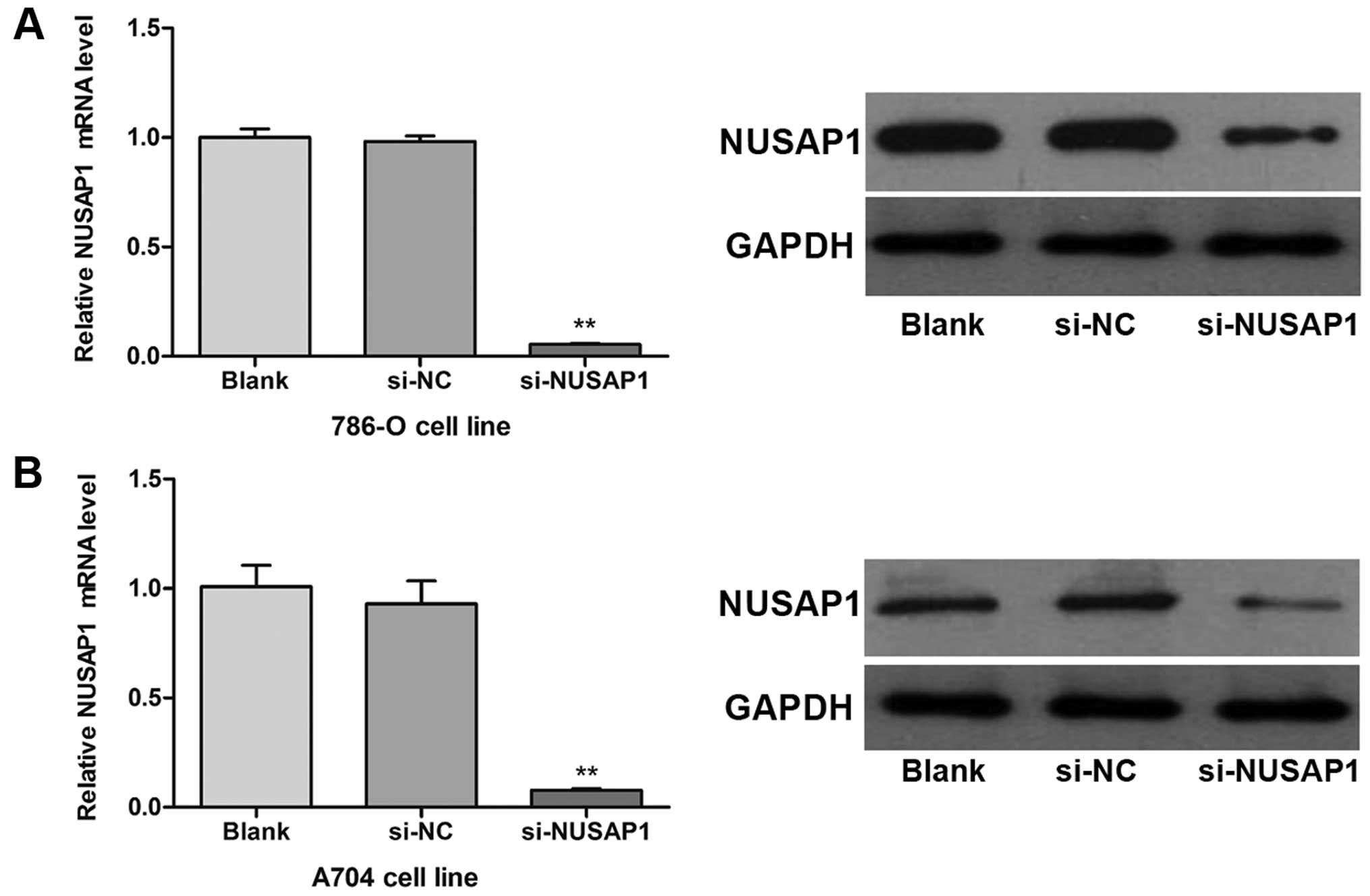
vitro. After transfection with siRNA targeting NUSAP1
(si-NUSAP1), cells exhibited a significant decrease in mRNA and
protein levels of NUSAP1 compared with those transfected with
scrambled siRNA (si-NC) (Fig. 3A and
B; P<0.001). Our results suggested that the NUSAP1
expression was efficiently downregulated in the human RCC cells
in vitro.
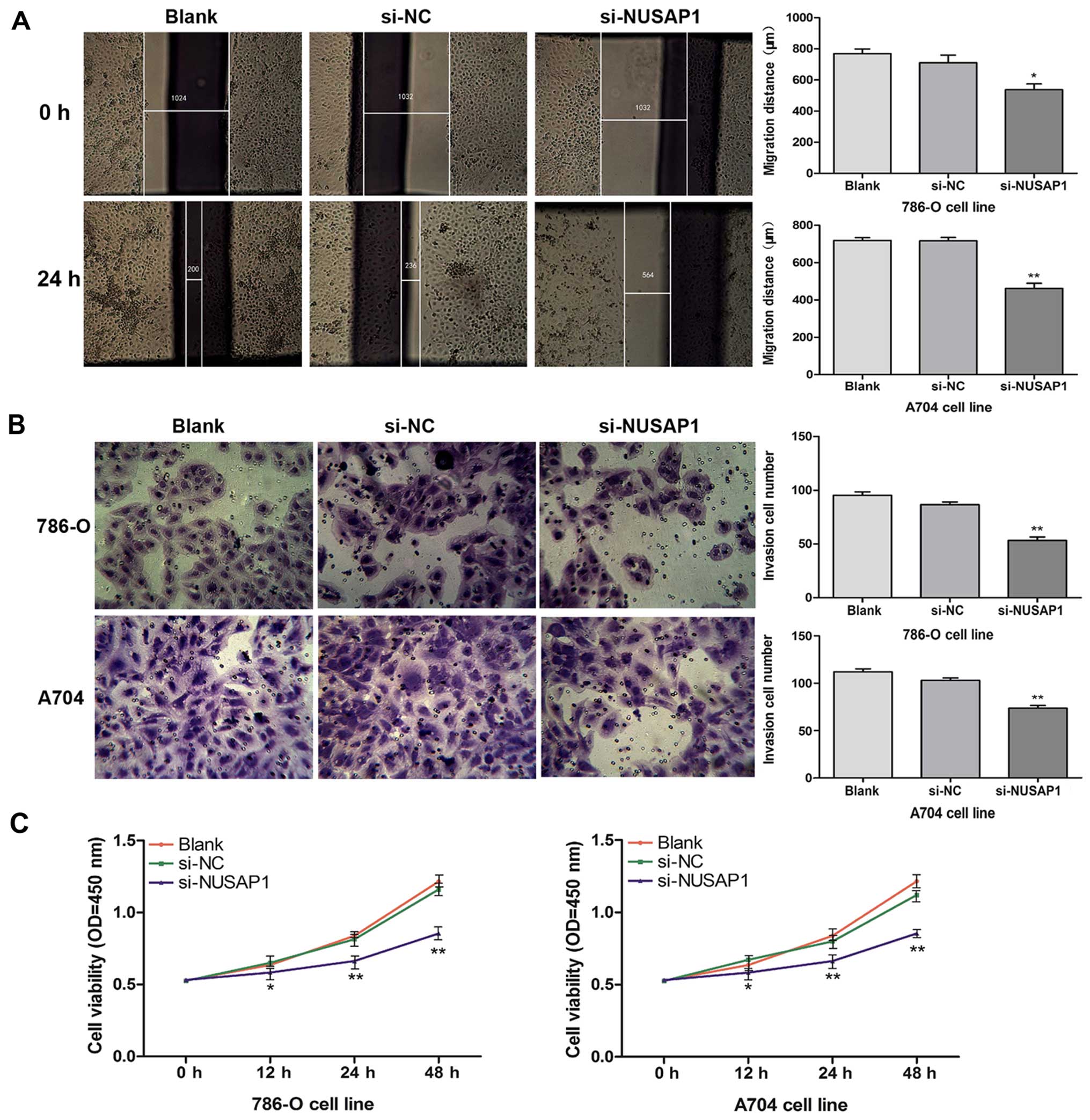
Moreover, we performed scratch migration, Transwell
invasion and CCK-8 assays to investigate the effects of the
downregulation of NUSAP1 on cell migration, invasion and
proliferation in the two RCC cell lines (A704 and 786-O),
respectively. The results showed that downregulation of NUSAP1 by
siRNA caused a significant inhibition of cell migration of the
786-O (P<0.05) and A704 (P<0.001) cell lines (Fig. 4A). In accordance with this result,
downregulation of NUSAP1 also resulted in a clear decrease in cell
invasive ability (Fig. 4B;
P<0.001). In the CCK-8 assay, we found that the proliferation
rate of the cells transfected with si-NUSAP1 was significantly
decreased compared with the rate in the cells treated with
si-NC/blank (Fig. 4C; P<0.05,
P<0.001). In summary, these results indicated that NUSAP1
expression was closely associated with cell growth and
aggressiveness of RCC.
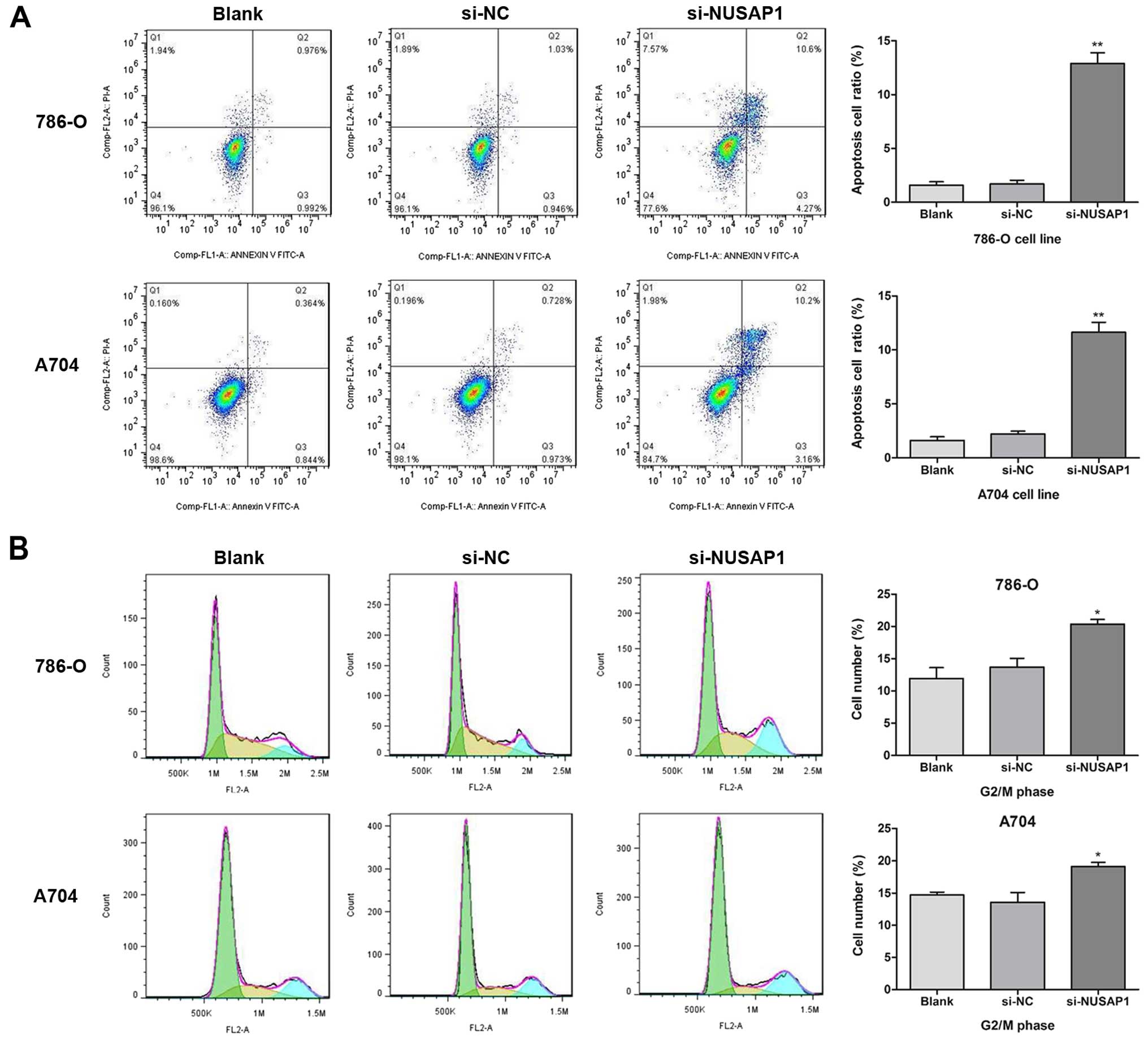
NUSAP1 downregulation induces apoptosis
and cell cycle arrest of RCC cells
To further investigate the mechanism of cell growth
inhibition by downregulation of NUSAP1 expression in RCC cells, the
cell cycle progression of 786-O and A704 cells was analyzed using
flow cytometry. As shown in Fig.
5B, after transfection with si-NUSAP1, a higher rate of
G2/M phase arrest when compared with that in the si-NC
group (mean rate, 20.37 vs. 13.69%; P<0.05) was identified in
the 786-O cells, as well as in the A704 cells (mean rate, 19.11 vs.
13.59%; P<0.05). Moreover, flow cytometric analysis showed that
there was a higher percentage of apoptotic cells in the
si-NUSAP1-transfected RCC cells, when compared with this percentage
in cells treated with si-NC (Fig.
5A; P<0.001). Therefore, our results suggest that
downregulation of NUSAP1 expression could induce apoptosis and cell
cycle arrest of RCC cells.
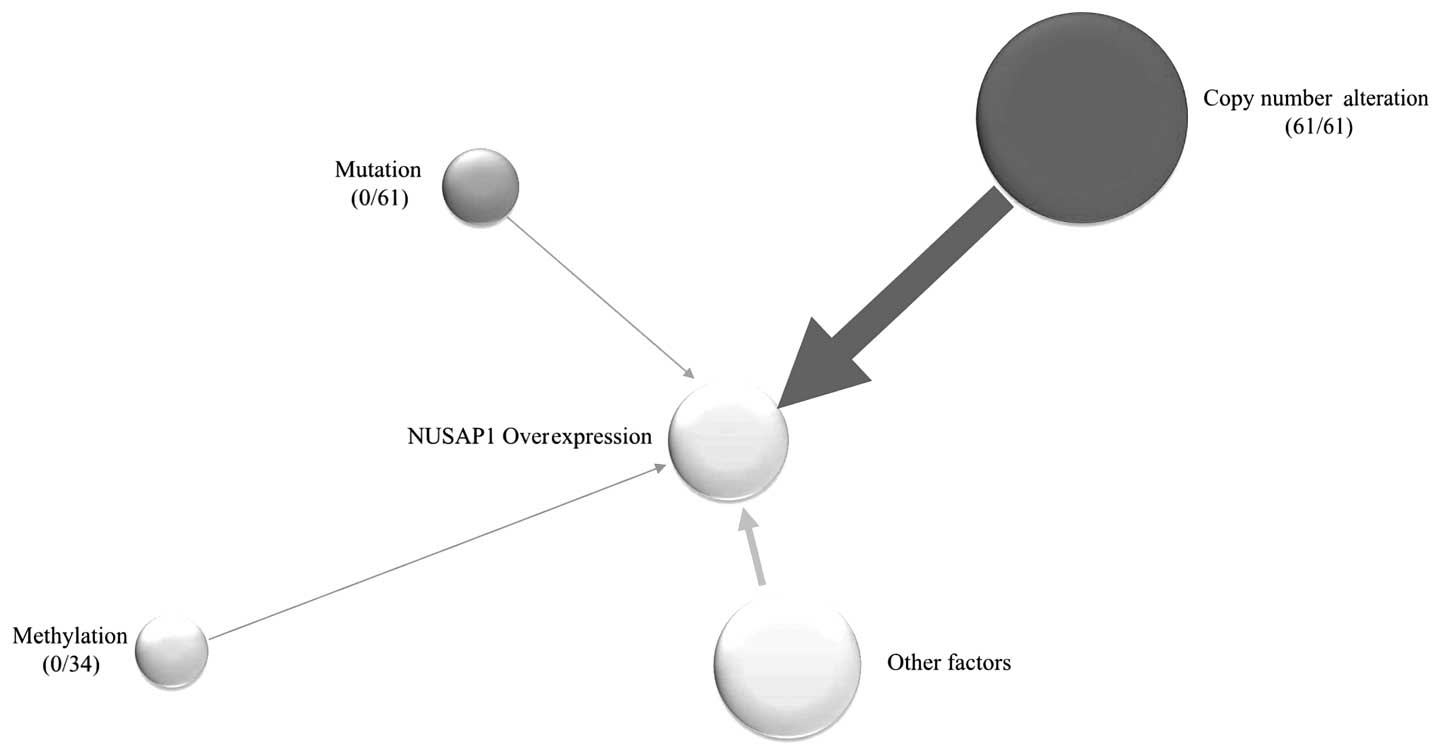
Copy number alterations (CNAs) influence
the expression of NUSAP1
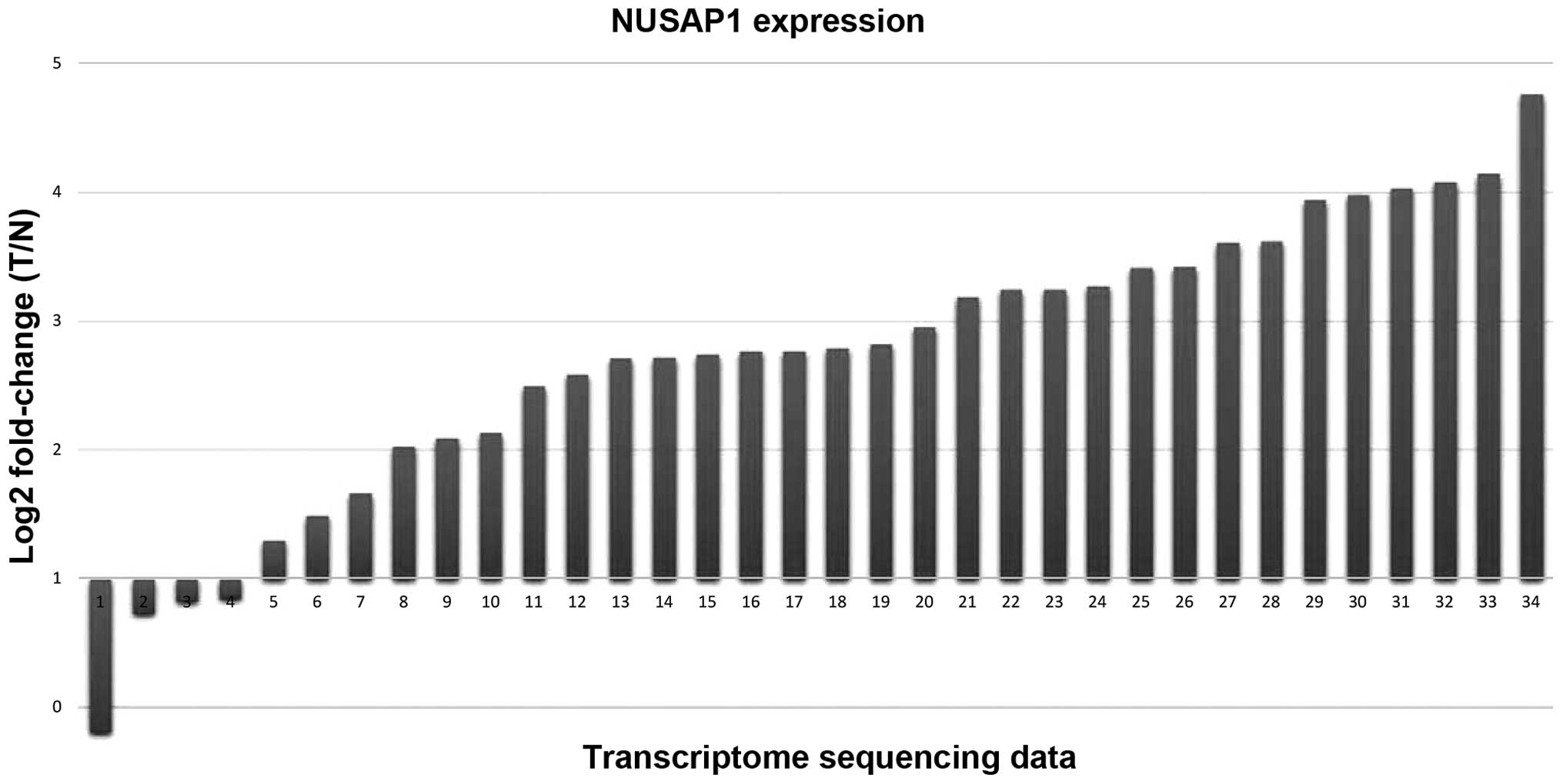
We analyzed the RBBS-seq data, and no methylation
variation of the promoter region of NUSAP1 (0/34, 0%) was
identified by quantitative analysis. However, when analyzing the
transcriptome data, we identified that NUSAP1 was overexpressed in
30 of the 34 paired RCCs relative to the adjacent normal controls
(Fig. 6). Intriguingly, we
identified that CNAs of the NUSAP1 region existed in all of the 61
paired RCCs (61/61, 100%) (Fig. 7
and Table II). Thus, we assumed
that the overexpression of NUSAP1 in RCC was mostly due to CNAs.
All of the analysis pipelines were referred to in our previous
study (18).
 | Table IIDetail information of the copy number
alterations of NUSAP1 in each tumor. |
Table II
Detail information of the copy number
alterations of NUSAP1 in each tumor.
| Gene_ID | Tumor_ID | Chr | Start | End | Np | Mean | Arm | Snvs | Ai | Median |
|---|
| NUSAP1 | s0 | chr15 | 25350206 | 102504996 | 7698 | −0.0109 | q | 55200 | 0.279345089 | 1 |
| NUSAP1 | s10 | chr15 | 38728401 | 46001801 | 728 | −0.2991 | q | 3287 | 0.315852751 | 0.807955743 |
| NUSAP1 | s100 | chr15 | 40759001 | 42050401 | 130 | 0.414 | q | 1062 | 0.275158292 | 1.327034164 |
| NUSAP1 | s102 | chr15 | 20709801 | 64638001 | 4319 | 0.1043 | q | 31445 | 0.27427683 | 1.068181818 |
| NUSAP1 | s104 | chr15 | 21360641 | 102503196 | 8030 | 0.1958 | q | 57702 | 0.284325477 | 1.142857143 |
| NUSAP1 | s106 | chr15 | 20709801 | 71496601 | 5004 | 0.2211 | q | 37642 | 0.337889277 | 1.166666667 |
| NUSAP1 | s108 | chr15 | 20709801 | 66594801 | 4514 | −0.0064 | q | 36556 | 0.303544235 | 1 |
| NUSAP1 | s110 | chr15 | 25501806 | 65942601 | 4038 | −0.0802 | q | 28232 | 0.27952482 | 0.951456311 |
| NUSAP1 | s112 | chr15 | 20709801 | 102504396 | 8092 | 0.0516 | q | 59613 | 0.289589089 | 1.03960396 |
| NUSAP1 | s120 | chr15 | 20709801 | 102502796 | 8091 | 0.1149 | q | 63673 | 0.286246703 | 1.081632653 |
| NUSAP1 | s122 | chr15 | 41332001 | 41952801 | 63 | 0.2006 | q | 374 | 0.273066667 | 1.153846154 |
| NUSAP1 | s124 | chr15 | 20709801 | 102505396 | 8092 | 0.0726 | q | 61169 | 0.289522145 | 1.051020408 |
| NUSAP1 | s126 | chr15 | 20709801 | 102503996 | 8092 | 0.1222 | q | 61144 | 0.300015989 | 1.082413584 |
| NUSAP1 | s128 | chr15 | 20709801 | 68051801 | 4660 | −0.3636 | q | 31259 | 0.465118203 | 0.775700935 |
| NUSAP1 | s14 | chr15 | 20709801 | 102507796 | 8094 | 0.0218 | q | 62011 | 0.291422215 | 1.018181818 |
| NUSAP1 | s16 | chr15 | 24729806 | 102505996 | 7759 | −0.2949 | q | 42076 | 0.740367921 | 0.815789474 |
| NUSAP1 | s18 | chr15 | 20709801 | 62136001 | 4068 | 0.0365 | q | 29248 | 0.315644532 | 1.02970297 |
| NUSAP1 | s20 | chr15 | 20709801 | 102505796 | 8092 | −0.1055 | q | 59054 | 0.300144424 | 0.932638289 |
| NUSAP1 | s22 | chr15 | 20709801 | 102502396 | 8094 | 0.0815 | q | 63784 | 0.285202245 | 1.056179775 |
| NUSAP1 | s24 | chr15 | 20709801 | 102383796 | 8082 | −0.0322 | q | 59942 | 0.308401091 | 0.978947368 |
| NUSAP1 | s26 | chr15 | 20709801 | 102504596 | 8094 | −0.0281 | q | 59679 | 0.304879746 | 0.988303694 |
| NUSAP1 | s28 | chr15 | 20709801 | 102509796 | 8095 | −0.1533 | q | 60822 | 0.30586911 | 0.9 |
| NUSAP1 | s30 | chr15 | 20709801 | 102506796 | 8094 | 0.1142 | q | 60481 | 0.298919296 | 1.081081081 |
| NUSAP1 | s32 | chr15 | 20709801 | 102506396 | 8094 | 0.0282 | q | 61255 | 0.311361302 | 1.021505376 |
| NUSAP1 | s34 | chr15 | 20709801 | 102501796 | 8094 | 0.0188 | q | 59756 | 0.297926443 | 1.012539308 |
| NUSAP1 | s36 | chr15 | 20709801 | 102028996 | 8047 | −0.0725 | q | 61481 | 0.286325104 | 0.96 |
| NUSAP1 | s38 | chr15 | 25501806 | 102507996 | 7682 | 0.0417 | q | 57184 | 0.2814581 | 1.028571429 |
| NUSAP1 | s4 | chr15 | 20709801 | 102509396 | 8084 | −0.0308 | q | 61369 | 0.304033643 | 0.97979798 |
| NUSAP1 | s40 | chr15 | 20709801 | 102510396 | 8093 | 0.0192 | q | 56548 | 0.306970417 | 1.01369863 |
| NUSAP1 | s42 | chr15 | 20709801 | 102508196 | 8095 | −0.0408 | q | 61258 | 0.2981276 | 0.978947368 |
| NUSAP1 | s44 | chr15 | 20709801 | 102504996 | 8095 | −0.047 | q | 63256 | 0.29900973 | 0.96875 |
| NUSAP1 | s46 | chr15 | 40788201 | 42078801 | 130 | −0.865 | q | 233 | 0.895454545 | 0.555555556 |
| NUSAP1 | s48 | chr15 | 20704991 | 100335961 | 15656 | 0.0227 | q | 60839 | 0.284898937 | 1.015151515 |
| NUSAP1 | s50 | chr15 | 31687371 | 100337196 | 13679 | 0.1568 | q | 49530 | 0.25923184 | 1.125 |
| NUSAP1 | s52 | chr15 | 20709801 | 102501596 | 8094 | −0.0348 | q | 59474 | 0.287460229 | 0.98757716 |
| NUSAP1 | s54 | chr15 | 20709801 | 102506796 | 8082 | −0.0881 | q | 65229 | 0.307597479 | 0.946236559 |
| NUSAP1 | s56 | chr15 | 24727406 | 96845596 | 7194 | −0.6389 | q | 37315 | 0.71711977 | 0.642857143 |
| NUSAP1 | s58 | chr15 | 20709801 | 102510396 | 8092 | 0.0182 | q | 61613 | 0.302473804 | 1.014925373 |
| NUSAP1 | s6 | chr15 | 20709801 | 102509996 | 8094 | 0.1546 | q | 60593 | 0.280029132 | 1.112359551 |
| NUSAP1 | s60 | chr15 | 20709801 | 102506196 | 8090 | 0.1564 | q | 54711 | 0.290879563 | 1.11627907 |
| NUSAP1 | s62 | chr15 | 20709801 | 102507396 | 8093 | 0.0699 | q | 62295 | 0.293077342 | 1.050505051 |
| NUSAP1 | s64 | chr15 | 20709801 | 89873396 | 6829 | −0.0218 | q | 51236 | 0.285149303 | 0.989361702 |
| NUSAP1 | s66 | chr15 | 28875606 | 60649801 | 3171 | −0.6257 | q | 23826 | 0.320973335 | 0.647887324 |
| NUSAP1 | s68 | chr15 | 20709801 | 102440796 | 8078 | −0.402 | q | 61877 | 0.338655047 | 0.75862069 |
| NUSAP1 | s70 | chr15 | 20709801 | 102503996 | 8092 | 0.0921 | q | 58793 | 0.288186153 | 1.064516129 |
| NUSAP1 | s72 | chr15 | 20709801 | 102508396 | 8086 | 0.389 | q | 62189 | 0.273379856 | 1.307692308 |
| NUSAP1 | s74 | chr15 | 20709801 | 102503996 | 8093 | −0.0213 | q | 63626 | 0.311079859 | 0.99009901 |
| NUSAP1 | s76 | chr15 | 20709801 | 93391196 | 7182 | 0.2855 | q | 52133 | 0.28283461 | 1.216981132 |
| NUSAP1 | s78 | chr15 | 20789801 | 102506196 | 8085 | −0.0992 | q | 62903 | 0.314847052 | 0.931818182 |
| NUSAP1 | s8 | chr15 | 20709801 | 102507596 | 8094 | −0.0214 | q | 63291 | 0.291604673 | 0.989130435 |
| NUSAP1 | s80 | chr15 | 20709801 | 102508196 | 8092 | 0.1063 | q | 57907 | 0.280533032 | 1.075949367 |
| NUSAP1 | s82 | chr15 | 20709801 | 90512996 | 6894 | 0.0514 | q | 51402 | 0.385165292 | 1.033333333 |
| NUSAP1 | s84 | chr15 | 20709801 | 102502396 | 8092 | −0.1288 | q | 59308 | 0.354867788 | 0.915789474 |
| NUSAP1 | s86 | chr15 | 20709801 | 102503796 | 8092 | −0.0767 | q | 61377 | 0.289897677 | 0.949494949 |
| NUSAP1 | s88 | chr15 | 20709801 | 102504196 | 8091 | −0.0986 | q | 59518 | 0.306860135 | 0.94 |
| NUSAP1 | s90 | chr15 | 25350206 | 102502196 | 7697 | 0.2473 | q | 55249 | 0.255991001 | 1.184466019 |
| NUSAP1 | s92 | chr15 | 21370441 | 50571201 | 2851 | 0.3202 | q | 21738 | 0.287522867 | 1.266055046 |
| NUSAP1 | s94 | chr15 | 20709801 | 75322201 | 5387 | −0.0493 | q | 37385 | 0.311031557 | 0.962962963 |
| NUSAP1 | s96 | chr15 | 41269201 | 42020001 | 76 | 0.0274 | q | 215 | 0.346910112 | 1.026491228 |
| NUSAP1 | s98 | chr15 | 20709801 | 102509196 | 8091 | 0.0142 | q | 58809 | 0.286709788 | 1.009615385 |
| NUSAP1 | s999 | chr15 | 20709801 | 67311401 | 4586 | −0.0995 | q | 33217 | 0.281856442 | 0.946236559 |
Discussion
Tremendous advances have been made over the last
decade in our understanding of the genetic basis and progress of a
targeted therapeutic armamentarium for RCC. As is known to all,
various conventional agents that target the vascular endothelial
growth factor (VEGF) pathway or mammalian target of rapamycin
(mTOR) may benefit renal cell carcinoma (RCC) patients (19). However, despite more options and
advances in treatment, most patients with advanced disease still
exhibit a markedly poor outcome, and we have not yet elucidated the
mechanism behind its development (20). Therefore, continued research to
identify more genes which drive the initiation and progression of
RCC is clearly warranted.
The function of NUSAP1 has been investigated in
several recent studies, which demonstrate its crucial role in cell
mitosis and tumorigenesis (6).
Although, the overexpression of NUSAP1 at the mRNA level has been
found in several types of cancers, there is limited research
concentrating on the expression and clinical significance of NUSAP1
in RCC. Our previous transcriptome sequencing data indicated that
the expression level of NUSAP1 was significantly higher in RCC
tissues than that in matched adjacent normal tissues. Methylation
of high-density CpG regions known as CpG islands (CGIs) has been
widely described as a mechanism associated with gene expression
regulation (21), whereas no
methylation change of NUSAP1 (0/34, 0%) was identified in the
present study. In addition, in a previous study using both
transcriptional profile data and CNA, they identified that genes
with differential expression may be caused by CNAs (22). To our surprise, CNAs of NUSAP1
existed in all of the 61 paired RCCs, which suggest that CNAs could
be the primary cause for the overexpression of NUSAP1 in RCC.
Next, we determined the mRNA and protein levels of
NUSAP1 expression in 38 pairs of ccRCC and matched normal tissues
and five cell lines (including four RCC cell lines and the HK-2
cell line). The qRT-PCR and western blot analyses showed that
NUSAP1 was relatively overexpressed in the RCC tissues, as well as
in three RCC cell lines (A704, 786-O and ACHN). Our data
demonstrated that an elevated expression of NUSAP1 at the
transcription and translation levels may closely correlate with the
tumorigenesis of RCC, and NUSAP1 may be a potential indicator for
RCC patients. In addition, immunohistochemical analysis was
performed to determine the association between NUSAP1 expression
and clinicopathological characteristics of the ccRCC patients. The
results showed that NUSAP1 expression was significantly associated
with the level of malignancy of ccRCC. Namely, upregulation of
NUSAP1 was associated with aggressive features of ccRCC, such as
Fuhrman grade, tumor size, clinical stage and metastasis. In
various recent studies, NUSAP1 expression was found to be
associated with the poor prognosis of patients with melanoma
(10), and NUSAP1 was also
identified as part of a malignancy-risk gene signature for breast
cancer (14). Consistent with these
studies, our survival analysis similarly revealed that
overexpression of NUSAP1 was obviously correlated with a shorter
overall survival time of ccRCC patients. These results suggest that
NUSAP1 could play an important role in the progression of RCC.
In attempting to determine the biological role of
NUSAP1 in RCC, we performed a series of functional experiments
after downregulation of NUSAP1 expression in vitro. In the
si-NUSAP1-transfected cell lines, the migration, proliferation and
invasion of RCC cells were significantly inhibited compared with
the control groups. A study by Gordon et al (23) demonstrated that knockdown of NUSAP1
by siRNA reduced the proliferation and invasion in prostate cancer
cell lines, suggested that NUSAP1 could influence tumor cell growth
and aggressiveness. Furthermore, Nie et al (24) found that NUSAP1 depletion blocked
the migration of neural crest cells in zebrafish embryos, which
indicates that its overexpression may promote cancer cell
migration. As we mentioned earlier, NUSAP1 is a microtubule-binding
protein that is selectively expressed in proliferating cells. Its
depletion causes G2/M arrest in cell cycle progression. In the
present study, we also discovered that downregulation of NUSAP1
induced apoptosis and G2/M arrest of RCC cells. In agreement with
our results, knockdown of NUSAP1 by siRNA in HeLa cells also led to
mitotic arrest and abnormal chromosome condensation (7). In addition, Vanden Bosch et al
(8) found the rapid disintegration
and small cellular fragments were present in reduced growth of
NUSAP1-null mice embryos, and presumed that lack of NUSAP1 may
result in apoptosis. Based on its crucial role in mitosis, and
NUSAP1 depletion suppressed cell growth and induced apoptosis, we
thus assumed that it could represent a novel therapeutic target for
RCC patients.
In conclusion, we demonstrated that NUSAP1
overexpression was closely related to the clinicopathological
features of RCC and predicted an unfavorable prognosis for RCC
patients. Downregulation of NUSAP1 induced cell apoptosis and
inhibited cell migration, proliferation and invasion. NUSAP1 may
thus serve as a potential prognostic indicator and a novel
therapeutic target for RCC patients. However, our research is only
a preliminary discussion on the expression and biological function
of NUSAP1 in RCC. Further studies are necessary to confirm these
findings and uncover the mechanisms of these processes.
Acknowledgments
The present study was supported by grants from the
National Natural Science Foundation of China (nos. 81301740 and
81402336).
References
|
1
|
Chow WH, Dong LM and Devesa SS:
Epidemiology and risk factors for kidney cancer. Nat Rev Urol.
7:245–257. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lam JS, Shvarts O, Leppert JT, Figlin RA
and Belldegrun AS: Renal cell carcinoma 2005: New frontiers in
staging, prognostication and targeted molecular therapy. J Urol.
173:1853–1862. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ferlay J, Shin HR, Bray F, Forman D,
Mathers C and Parkin DM: Estimates of worldwide burden of cancer in
2008: GLOBOCAN 2008. Int J Cancer. 127:2893–2917. 2010. View Article : Google Scholar
|
|
4
|
Athar U and Gentile TC: Treatment options
for metastatic renal cell carcinoma: A review. Can J Urol.
15:3954–3966. 2008.PubMed/NCBI
|
|
5
|
Gupta K, Miller JD, Li JZ, Russell MW and
Charbonneau C: Epidemiologic and socioeconomic burden of metastatic
renal cell carcinoma (mRCC): A literature review. Cancer Treat Rev.
34:193–205. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Iyer J, Moghe S, Furukawa M and Tsai MY:
What's Nu(SAP) in mitosis and cancer? Cell Signal. 23:991–998.
2011. View Article : Google Scholar
|
|
7
|
Raemaekers T, Ribbeck K, Beaudouin J,
Annaert W, Van Camp M, Stockmans I, Smets N, Bouillon R, Ellenberg
J and Carmeliet G: NuSAP, a novel microtubule-associated protein
involved in mitotic spindle organization. J Cell Biol.
162:1017–1029. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Vanden Bosch A, Raemaekers T, Denayer S,
Torrekens S, Smets N, Moermans K, Dewerchin M, Carmeliet P and
Carmeliet G: NuSAP is essential for chromatin-induced spindle
formation during early embryogenesis. J Cell Sci. 123:3244–3255.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kokkinakis DM, Liu X and Neuner RD:
Modulation of cell cycle and gene expression in pancreatic tumor
cell lines by methionine deprivation (methionine stress):
Implications to the therapy of pancreatic adenocarcinoma. Mol
Cancer Ther. 4:1338–1348. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bogunovic D, O'Neill DW, Belitskaya-Levy
I, Vacic V, Yu YL, Adams S, Darvishian F, Berman R, Shapiro R,
Pavlick AC, et al: Immune profile and mitotic index of metastatic
melanoma lesions enhance clinical staging in predicting patient
survival. Proc Natl Acad Sci USA. 106:20429–20434. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Marie SK, Okamoto OK, Uno M, Hasegawa AP,
Oba-Shinjo SM, Cohen T, Camargo AA, Kosoy A, Carlotti CG Jr, Toledo
S, et al: Maternal embryonic leucine zipper kinase transcript
abundance correlates with malignancy grade in human astrocytomas.
Int J Cancer. 122:807–815. 2008. View Article : Google Scholar
|
|
12
|
Satow R, Shitashige M, Kanai Y, Takeshita
F, Ojima H, Jigami T, Honda K, Kosuge T, Ochiya T, Hirohashi S, et
al: Combined functional genome survey of therapeutic targets for
hepatocellular carcinoma. Clin Cancer Res. 16:2518–2528. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Gulzar ZG, McKenney JK and Brooks JD:
Increased expression of NuSAP in recurrent prostate cancer is
mediated by E2F1. Oncogene. 32:70–77. 2013. View Article : Google Scholar
|
|
14
|
Chen DT, Nasir A, Culhane A, Venkataramu
C, Fulp W, Rubio R, Wang T, Agrawal D, McCarthy SM, Gruidl M, et
al: Proliferative genes dominate malignancy-risk gene signature in
histologically-normal breast tissue. Breast Cancer Res Treat.
119:335–346. 2010. View Article : Google Scholar
|
|
15
|
Lauss M, Kriegner A, Vierlinger K, Visne
I, Yildiz A, Dilaveroglu E and Noehammer C: Consensus genes of the
literature to predict breast cancer recurrence. Breast Cancer Res
Treat. 110:235–244. 2008. View Article : Google Scholar
|
|
16
|
Cario G, Fetz A, Bretscher C, Möricke A,
Schrauder A, Stanulla M and Schrappe M: Initial leukemic gene
expression profiles of patients with poor in vivo prednisone
response are similar to those of blasts persisting under prednisone
treatment in childhood acute lymphoblastic leukemia. Ann Hematol.
87:709–716. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2−ΔΔCT method. Methods. 25:402–408. 2001. View Article : Google Scholar
|
|
18
|
Huang Y, Gao S, Wu S, Song P, Sun X, Hu X,
Zhang S, Yu Y, Zhu J, Li C, et al: Multilayered molecular profiling
supported the monoclonal origin of metastatic renal cell carcinoma.
Int J Cancer. 135:78–87. 2014. View Article : Google Scholar
|
|
19
|
Srinivasan R, Ricketts CJ, Sourbier C and
Linehan WM: New strategies in renal cell carcinoma: Targeting the
genetic and metabolic basis of disease. Clin Cancer Res. 21:10–17.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Jonasch E and Motzer RJ: Ten years of
progress in renal cell carcinoma. J Natl Compr Canc Netw.
10:690–693. 2012.PubMed/NCBI
|
|
21
|
Moarii M, Boeva V, Vert JP and Reyal F:
Changes in correlation between promoter methylation and gene
expression in cancer. BMC Genomics. 16:8732015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yang Z, Zhuan B, Yan Y, Jiang S and Wang
T: Integrated analyses of copy number variations and gene
differential expression in lung squamous-cell carcinoma. Biol Res.
48:472015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Gordon CA, Gulzar ZG and Brooks JD: NUSAP1
expression is upregulated by loss of RB1 in prostate cancer cells.
Prostate. 75:517–526. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Nie J, Wang H, He F and Huang H: Nusap1 is
essential for neural crest cell migration in zebrafish. Protein
Cell. 1:259–266. 2010. View Article : Google Scholar
|