Introduction
Cutaneous malignant melanoma (CMM) is a type of
cancer that develops from melanocytes and includes four subtypes
according to clinical organization criteria: malignant nevoid
lentigo maligna melanoma (LMM), superficial spreading melanoma
(SSM), acral lentiginous melanoma (ALM), and nodular melanoma (NM).
Environmental factors, such as ultraviolet irradiation and family
heredity, have been regarded as high-risk factors for the incidence
of CMM (1). An increasing number of
studies reveal that mutation or aberrant activation of certain
signaling pathways also contributes significantly to the incidence
and progression of CMM, including the anti-oncogene p53, the
p21/cyclin D1/cyclin-dependent kinase 4 (CDK4) pathway, and the
epidermal growth factor receptor (EGFR)/Akt pathway (2,3).
The transcription factor Kruppel-like 6 (KLF6) is an
antitumor member of the KLF zinc finger DNA-binding protein family
that plays an important role in the regulation of cell
proliferation, migration, and apoptosis (4). Inactivation and downregulation of KLF6
have been regarded as an important sign of tissue carcinogenesis
(5). In many parenchymal
carcinomas, KLF6 suppresses activation of the EGFR/Akt pathway or
the p21-mediated expression of cyclin D1/CDK4, and functions as a
tumor suppressor (6–8). However, its role and underlying
mechanism in the regulation of malignant melanoma are not yet fully
understood. Several recent studies demonstrated that the expression
level of KLF6 in patients with malignant melanoma is significantly
decreased, and its level is negatively correlated with the size and
metastasis of the tumor (9–11). Thus, KLF6 is likely to play a
negative regulatory role in the development of melanoma.
MicroRNAs (miRNAs) are involved in cancer
initiation, progression, and treatment response. miR-4262 is a
newly identified miRNA and a provisional family member predicted in
human embryonic stem cells and neural precursors (12), the function of which has not been
well studied. Recent research has shown that miR-4262 expression is
aberrantly expressed in patients with prostate cancer (13). Another study on adenocarcinoma
pathogenesis indicated that miR-4262 expression was dysregulated
and had an effect on cisplatin resistance of adenocarcinoma cells
(14). These clues suggest that
miR-4262 may play a role in the regulation of cancer initiation or
progression.
In this study, we found that miR-4262 was markedly
upregulated in various types of human CMM tissues and cell lines,
displaying an opposite expression pattern to that of KLF6. We then
explored the roles of KLF6 and miR-4262 in the proliferation of CMM
cell lines, as well as their interaction.
Materials and methods
Ethics statement and sampling
The study enrolled 110 CMM patients of stages I and
II, including 30 LMM (mean age 45±9.2 years), 30 SSM (mean age
47±9.8 years), 30 ALM (mean age 46±9.5 years) and 20 NM (mean age
45±9.4 years) cases, with an equal number of males and females. The
patients had no evidence of lymph node metastasis and their tumor
diameters were ≤2 cm. Moreover, none of the patients had received
preoperative anticancer treatment. CMM tissues and matched adjacent
normal epithelial tissues were obtained from each subject by
minimally invasive surgery. The study was approved by the Ethics
Committee of The Second Affiliated Hospital of Xi'an Jiao Tong
University. The donors and their guardians were previously informed
of the experimental details and provided written consents.
Cell culture and transfection
Normal human melanocytes HACAT and foreskin
fibroblasts HFFs, as well as human CMM cell lines A375, Malme-3M,
SK-MEL-2, SK-MEL-5, and M14 were cultured in a 5% CO2
atmosphere at 37°C in Dulbecco's modified Eagle's medium (DMEM)
with 10% fetal bovine serum (FBS), and 100 mg/ml penicillin and 100
U/ml streptomycin (all from Gibco, Rockville, MD, USA). All cell
lines were purchased from the American Type Culture
Collection® (ATCC; Rockefeller, MD, USA).
When the cell density reached approximately 70%
confluency, the pcDNA-KLF6 expression vector, KLF6 siRNA, or the
single-stranded oligo miRNA inhibitors or mimics (designed and
synthesized by Gene Pharma, Shanghai, China) were transfected into
the cells with Lipofectamine® 3000 (Invitrogen,
Carlsbad, CA, USA) according to the manufacturer's
instructions.
3′-UTR luciferase reporter assay
The wild-type 3′-UTR of human KLF6 mRNA was
amplified by PCR, and a truncated 3′-UTR with deletion of the
miR-4262 targeting site was amplified by nested PCR, using primers
linked with the XhoI and NotI restriction sites at
the beginning and end, respectively. All primers were designed and
synthesized by GenScript Co., Ltd. (Nanjing, China). The PCR
products were excised with NotI and XhoI and inserted
into the psiCHECK™-2 vector (Promega, Madison, WI, USA) at the 3′
end of the Renilla gene CDS. Firefly luciferase activity was
used as the internal control. The WT/truncated 3′-UTR
dual-luciferase vectors were transfected or co-transfected with the
miR-4262 mimic into 293T cells (100% confluence; ATCC) using
X-tremeGENE 9 DNA transfection reagent (Roche, Basel, Switzerland).
The medium was changed 6 h later. Cells were incubated for another
48 h with commercial cell lysis buffer (Merck & Co., Inc.,
Whitehouse Station, NJ, USA). The luciferase activity was measured
using a luminometer (Promega) according to the manufacturer's
instructions.
Real-time qPCR
Total RNA of the tissues or cells was extracted with
TRIzol reagent (Takara, Dalian, China) according to the
manufacturer's instructions. After quality and integrity were
checked, ~1,000 ng of total RNA was used in the first-strand cDNA
synthesis reaction. The miR-4262 stem-loop primer and quantitative
primers, as well as the pre-miR-4262 quantitative primers, were
designed and produced by Invitrogen. Each individual sample was run
in triplicate wells. PCR amplification cycles were performed using
the iQTM5 Multicolor Real-Time PCR detection system (Bio-Rad
Laboratories, Inc., Hercules, CA, USA) and SYBR Premix Ex Taq II
kit (Invitrogen). The reactions were initially denatured at 95°C
for 3 min followed by 40 cycles of 95°C for 10 sec, 55°C for 30
sec, and 72°C for 20 sec. U6 RNA was used to normalize the
expression of miR-4262, and β-actin was used to normalize the
expression of other RNA transcripts. The 2−ΔΔCt method
was used to evaluate the relative expression level of the RNA
transcripts.
Western blotting
Cells were lysed in lysis buffer (Beyotime Institute
of Biotechnology, Shanghai, China) containing 1 mM PMSF. The
concentration of total protein was determined using the BCA protein
assay (Tiangen Biotech Co., Ltd., Beijing, China). Fifty micrograms
of protein in each sample was separated by 12% SDS-PAGE and then
transferred to PVDF membranes (Millipore, Boston, MA, USA) for
immunoblotting analysis. The following primary antibodies were
used: anti-KLF6 (1:300), anti-EGFR (1:400), anti-p21 (1:300),
anti-cyclin D1 (1:300), anti-CDK4 (1:200) (Abcam, Cambridge, MA,
USA), and anti-GAPDH (1:800; Santa Cruz Biotechnology, Inc., Santa
Cruz, CA, USA), which was used as the internal reference. After
incubation with the appropriate HRP-conjugated secondary antibody,
the proteins were detected using a ChemiDoc XRS imaging system and
analysis software Quantity One (Bio-Rad Laboratories, Inc.).
Detection of cell proliferation
The cell proliferation assay was performed using the
CellTiter-Blue H cell viability assay kit (Promega) and the MTT
method (Sigma-Aldrich, St. Louis, MO, USA) according to the
manufacturer's instructions.
Online website. The targeting relationship between
miR-4262 and KLF6 was evaluated by the online server TargetScan
(http://www.targetscan.org/cgi-bin/targetscan/vert_70/view_gene.gi?rs=ENST00000542957.1&taxid=9606&members=&showcnc=0&shownc=0&showncf&subset=1).
Statistical analysis. Each experiment was carried
out for a minimum of three independent replicates, and each
replicate experiment was performed in triplicate. Data are
represented as mean ± SEM. Statistics were calculated using SPSS
19.0 (SPSS Inc., Chicago, IL, USA). Multiple comparisons were
assessed by one-way ANOVA followed by Dunnett's tests. Differences
between groups were considered statistically significant at
P<0.05.
Results
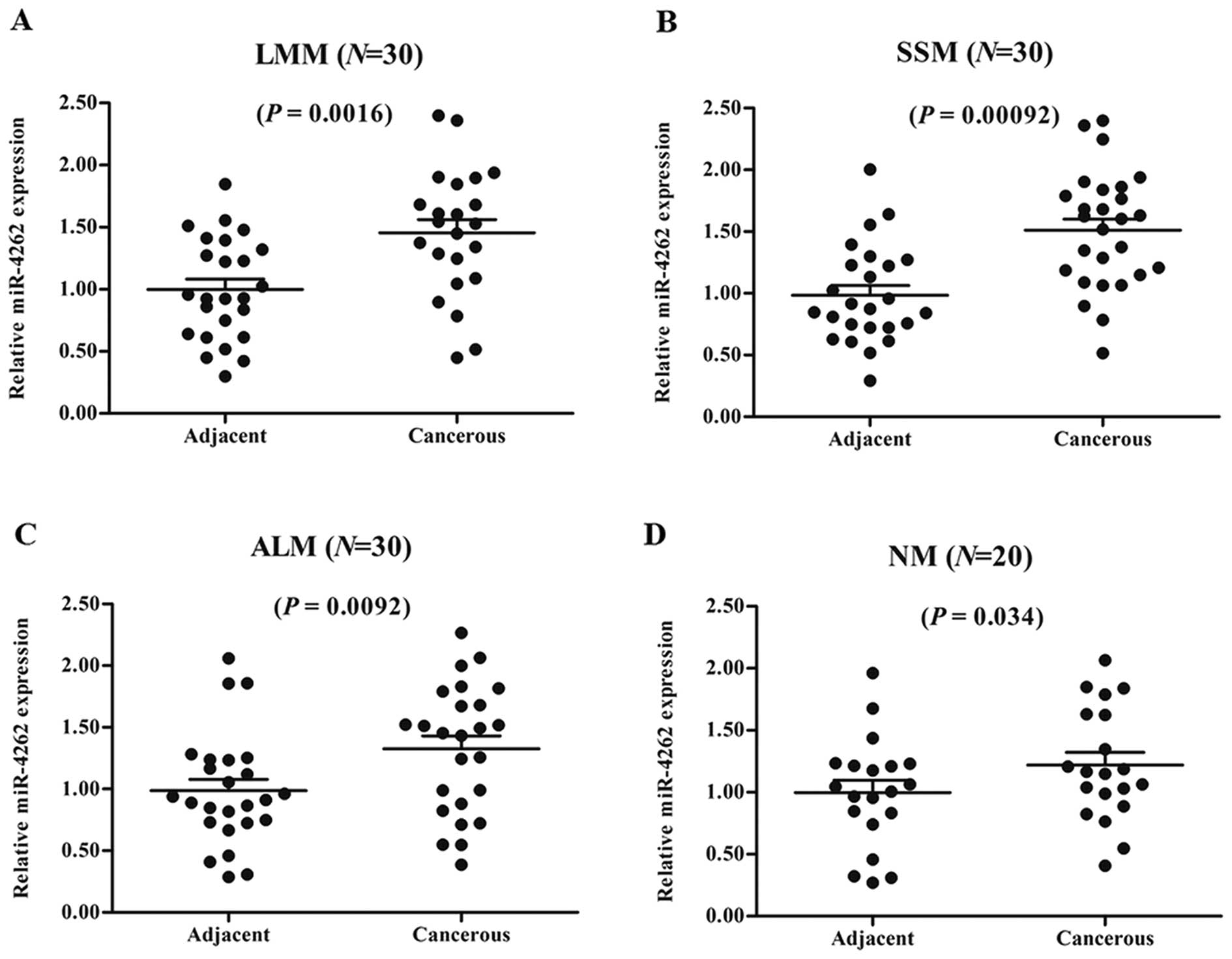
miR-4262 is upregulated in different
types of human CMM tissues and multiple human CMM cell lines
To explore the potential role of miR-4262 in the
progression of human CMM, the levels of miR-4262 were first
examined in cancerous epithelial tissues and matched adjacent
normal epithelium of the CMM patients, including 30 LMM, 30 SSM, 30
ALM, and 20 NM patients. The results showed that miR-4262
expression was upregulated in all of the cancerous tissues compared
with that noted in the matched tissues (Fig. 1A-D). Then, the levels of
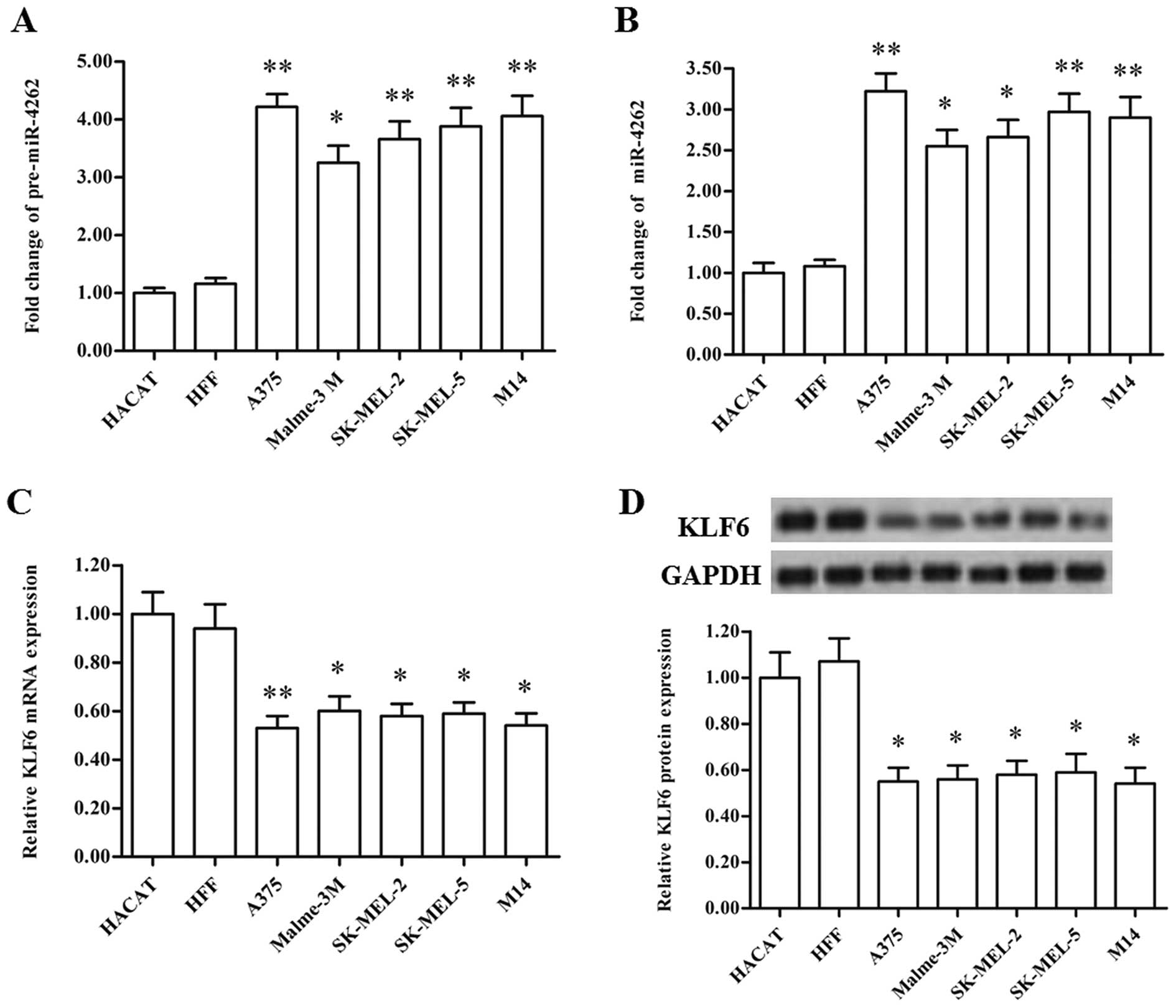
pre-miR-4262 and miR-4262 were detected in two types of normal
human melanocytes and five types of CMM cell lines. Our results
showed that, compared with the normal melanocytes, pre-miR-4262 and
miR-4262 were both robustly upregulated in the CMM cell lines
(Fig. 2A and B). These data suggest
that miR-4262 may be involved in the regulation of CMM
progression.
KLF6 is identified as a target gene of
miR-4262
Recent studies indicate that the transcription
factor KLF6 is downregulated in patients with malignant melanoma
and negatively correlated with melanoma growth and metastasis. The
expression levels of mRNA and protein were detected in the CMM cell
lines in this study. Our results showed that KLF6 mRNA and protein
were also significantly downregulated in the CMM cell lines
(Fig. 2C and D), displaying an
opposite expression pattern to that of miR-4262. Then the targeting
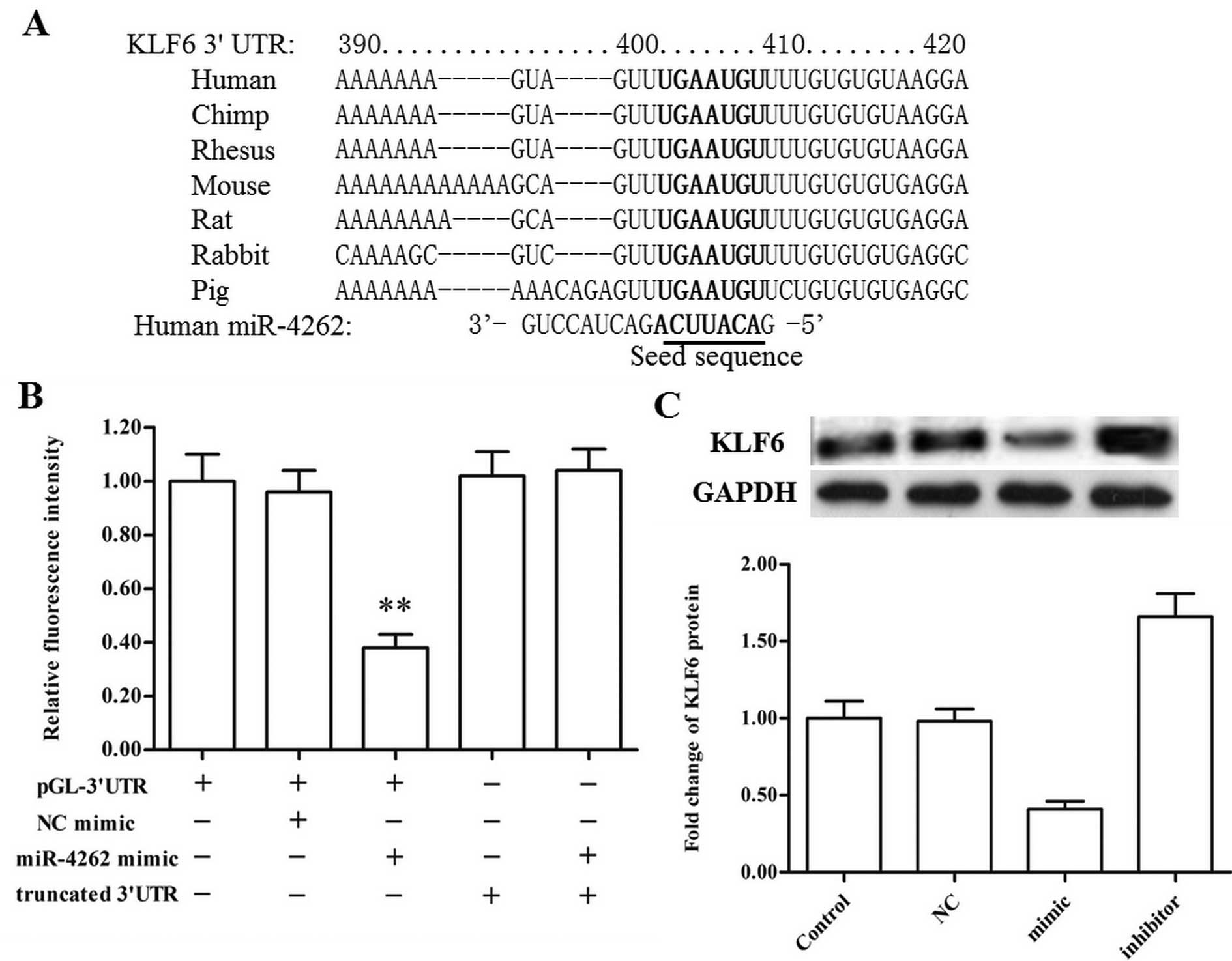
relationship between miR-4262 and KLF6 was evaluated using the
online server TargetScan. The output showed that the miR-4262 seed
sequence completely matched bases 2955–2962 of the KLF6 mRNA 3′-UTR
(Fig. 3A). 3′-UTR luciferase
reporter assay revealed that the miR-4262 mimic markedly reduced
fluorescence intensity in the psiCHECK-WT 3′-UTR group but had no
effect on that of the mutant 3′-UTR group (with a deletion of bases
390–420; Fig. 3B). Moreover, the
miR-4262 mimic and inhibitor were respectively transfected into the
CMM A375 cells, and western blotting showed that the miR-4262 mimic
sharply reduced the expression of KLF6 protein while the miR-4262
inhibitor had an opposite effect (Fig.
3C). The above data demonstrated that miR-4262 directly targets
and negatively regulates the expression of KLF6.
KLF6 suppresses the proliferation of
A375 human CMM cells
To explore the exact role of KLF6 in the
proliferation of CMM cells, a pcDNA-KLF6 expression vector and KLF6
siRNA were individually transfected into A375 human CMM cells. Cell
proliferation was evaluated using the MTT and CellTiter-Blue H
assays. The results showed that A375 cell proliferation was notably
reduced by pcDNA-KLF6 transfection and increased by the KLF6 siRNA
transfection (Fig. 4A and B).
Simultaneously, KLF6 overexpression significantly reduced the level
of the pro-tumorigenic protein EGFR, increased that of the tumor
suppressor p21, and caused a decrease in the levels of cyclin D1
and CDK4 (Fig. 4C). KLF6 siRNA
transfection had an opposite effect on the expression of the above
proteins when compared to the results from KLF6 overexpression
(Fig. 4C). These data indicate that
KLF6 has a negative effect on CMM cell proliferation.
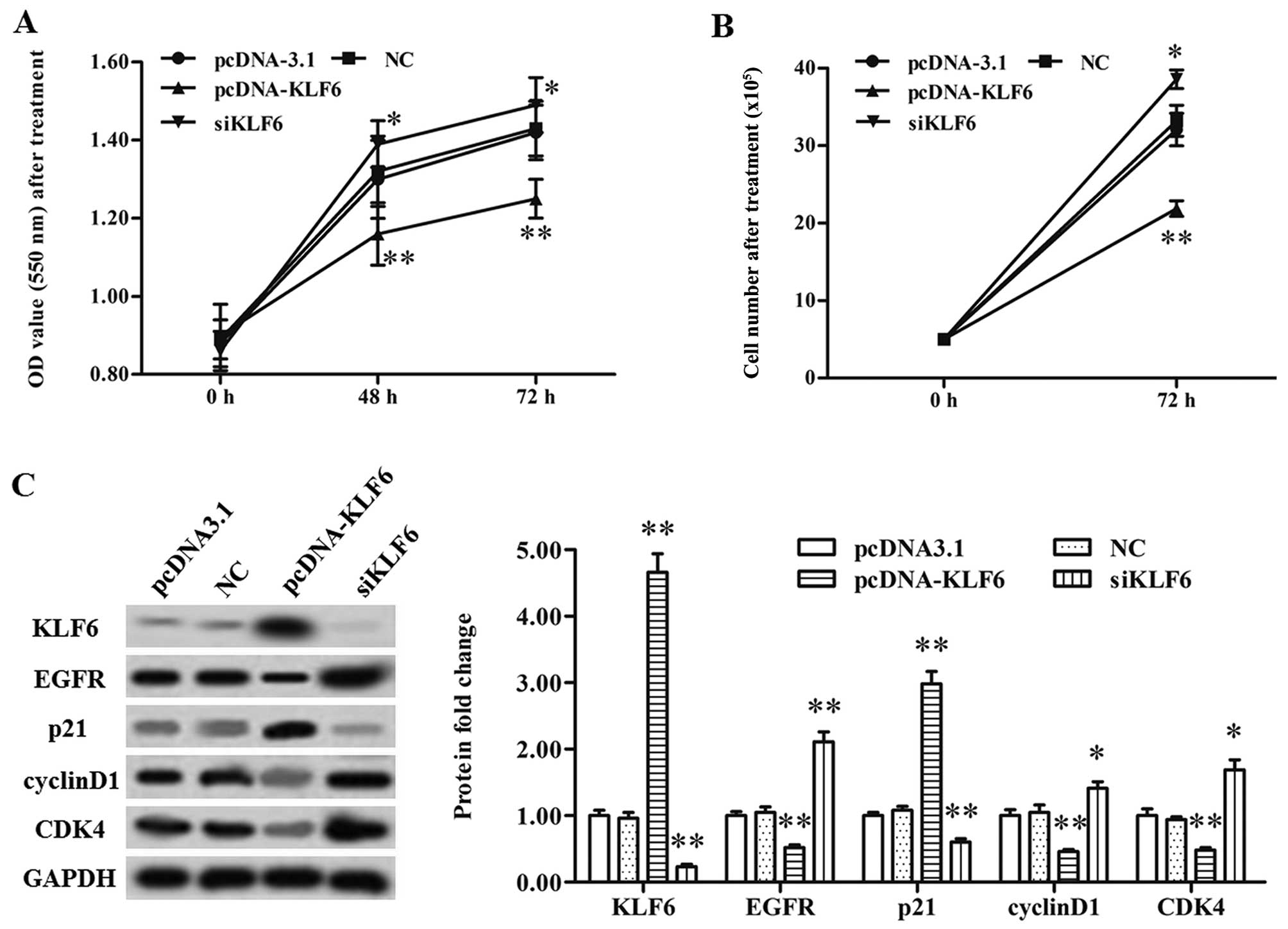 | Figure 4.KLF6 suppresses A375 cell
proliferation and the expression levels of EGFR and p21-mediated
CDK4. (A) A375 cell proliferation was suppressed by KLF6
overexpression and increased by KLF6 knockdown as detected by MTT
assay. (B) A375 cell proliferation was suppressed by KLF6
overexpression and increased by KLF6 knockdown as detected with
CellTiter-Blue H assay. (C) KLF6 suppressed the protein levels of
EGFR and p21-mediated CDK4. The pcDNA-KLF6, KLF6 siRNA, or relative
negative control vector or siRNA were transfected into the CMM A375
cells at 80% confluency. After incubation for 72 h, the protein
expression levels of KLF6, EGFR, p21, cyclin D1 and CDK4 were
detected using western blotting. *P<0.05, **P<0.01. KLF6,
Kruppel-like 6; EGFR, epidermal growth factor receptor; CDK4,
cyclin-dependent kinase 4. |
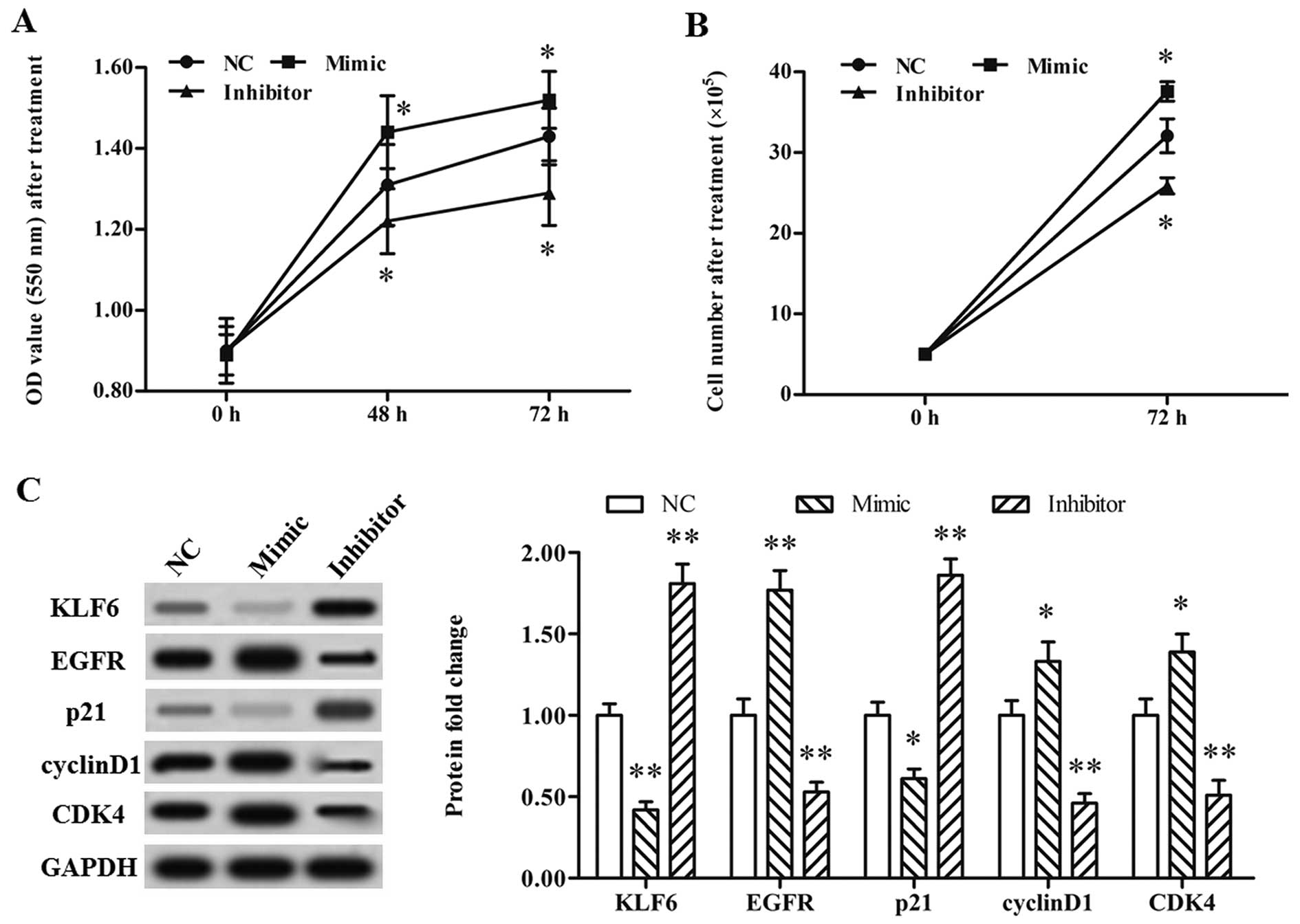
miR-4262 suppresses KLF6 expression
and promotes the proliferation of A375 cells
Finally, the role of miR-4262 in the proliferation
of CMM cells was explored. A single-stranded oligo miR-4262 mimic
and inhibitor were transfected individually into the A375 cells.
Our data from the MTT and CellTiter-Blue H assays showed that A375
cell proliferation was markedly promoted by the miR-4262 mimic and
reduced by the miR-4262 inhibitor (Fig.
5A and B). Moreover, the miR-4262 mimic significantly increased
the level of EGFR, reduced that of p21, and caused an increase in
the protein levels of cyclin D1 and CDK4 (Fig. 5C). The miR-4262 inhibitor had an
opposite effect on protein expression (Fig. 5C). These results demonstrated that
miR-4262 promoted the proliferation of CMM cells.
Discussion
miR-4262 was found in SOLiD ultra-deep small RNA
sequencing of the Ago2 complex in human embryonic stem cells and
neural precursors in 2009 (12),
suggesting a potential regulatory role of miR-4262 in cell
differentiation and proliferation. From then on, several studies
have revealed that miR-4262 is dysregulated in several pathological
processes, including tumor drug resistance, cancer metastasis and
tissue fibrosis (14–16). In this study, we assessed the
expression of miR-4262 in human CMM tissues and cell lines, as well
as in matched normal epithelial tissues and cell lines. Our results
showed that both pre- and mature miR-4262 were upregulated in the
CMM tissues and cell lines, suggesting that miR-4262 may promote
the progression of CMM.
As a newly discovered miRNA, the role of miR-4262
has been poorly studied. Here, we explored the effect of miR-4262
on the proliferation of CMM cell lines. Our data on miR-4262
overexpression and silencing both indicated that miR-4262 had a
positive effect on the proliferation of CMM A375 cells. However,
existing research results indicate that miR-4262 mainly plays a
suppressive role in cancer progression. Song et al reported
that the levels of miR-4262 were significantly decreased in
osteosarcoma specimens and the 5-year survival of osteosarcoma
patients with lower miR-4262 was reduced (17). Moreover, miR-4262 was found to
target the 3′-UTR of osteopontin (OPN) mRNA and inhibit
OPN-mediated cell invasion (17). A
study on sequence matching between zinc finger E-box-binding
homeobox 2 (ZEB2) and its targeting miRNA showed that miR-4262
potentially targets ZEB2, which was proven to be an E-cadherin
repressor, and may play a negative role in larynx carcinoma
progression (16). Another study on
acute lung injury also indicated that miR-4262 targets the
anti-apoptotic gene Bcl-2 to induce apoptosis in pulmonary
endothelial cells (18). We
speculated that miR-4262 may play multiple roles in different
biological processes. In fact, it is not a unique situation that
miRNAs play versatile roles in different types of cells, or even in
the same type of cells under different conditions. A typical
example is miR-210, an extensively investigated miRNA in cancer
biology that is regarded as a signature of hypoxia, that functions
either as a tumor promoter or a tumor suppressor, and can be a
positive or negative prognostic biomarker (19–22).
It has been proven that the function of KLF6 is
determined by the length of its splice variants (SVs).
KLF6-full-length (KLF6-FL or KLF6) protein is downregulated and
functions as a tumor suppressor in cancers, while truncated
KLF6-SVs are usually expressed at higher levels in tumors and drive
cancer growth and metastasis (5,23,24).
The length of KLF6 transcripts has been associated with the DNA
methylation level in the KLF6 promoter region (25). Treatment with the demethylating
agent 5-aza-2′-deoxycytidine caused upregulation of KLF6 expression
and a relative decrease in KLF6 SVs (26). As a tumor growth suppressor, KLF6
promotes the transactivation of the anti-tumorigenic gene p21 to
suppress the cyclin D1/CDK-mediated cell cycle (27). KLF6 was also found to negatively
regulate EGFR signaling in both cell culture and in vivo
models and was proposed as an anti-EGFR-based therapy for the
treatment of metastatic carcinomas (28). However, the exact role of KLF6 in
human CMM progression has not been well studied. In this study, we
reported that KLF6 was significantly downregulated in CMM cell
lines and played a negative regulatory role in the proliferation of
CMM cells. Moreover, KLF6 overexpression in CMM cells caused
downregulation of EGFR and a robust increase in p21 expression. In
contrast, knockdown of KLF6 increased EGFR and reduced p21
expression. KLF6 had a negative effect on CMM cell proliferation
through inactivation of the EGFR/PI3K/Akt and p21-mediated cyclin
D1/CDK pathways.
In conclusion, miR-4262 was found to be upregulated
in CMM tissues and cell lines compared with these levels in matched
normal epithelial tissues and normal cell lines. miR-4262 promoted
the proliferation of CMM cells through targeting of the tumor
suppressor KLF6.
Acknowledgements
This study was supported by the Fundamental Research
Funds for the Central Universities.
References
|
1
|
Moan JE, Baturaite Z, Dahlback A and
Porojnicu AC: Ultraviolet radiation and cutaneous malignant
melanoma. Adv Exp Med Biol. 810:359–374. 2014.PubMed/NCBI
|
|
2
|
Law MH, Bishop DT, Lee JE, Brossard M,
Martin NG, Moses EK, Song F, Barrett JH, Kumar R, Easton DF, et al:
GenoMEL Consortium; Essen-Heidelberg Investigators; SDH Study
Group; Q-MEGA and QTWIN Investigators; AMFS Investigators; ATHENS
Melanoma Study Group: Genome-wide meta-analysis identifies five new
susceptibility loci for cutaneous malignant melanoma. Nat Genet.
47:987–995. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Emri G, Emri E, Boros G, Hegedűs C, Janka
E, Gellén E and Remenyik E: Skin carcinogenesis: the pathogenetic
and therapeutic role of zinc. J Metallomics Nanotechnol. 2:19–26.
2015.
|
|
4
|
Atkins GB and Jain MK: Role of
Krüppel-like transcription factors in endothelial biology. Circ
Res. 100:1686–1695. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
DiFeo A, Martignetti JA and Narla G: The
role of KLF6 and its splice variants in cancer therapy. Drug Resist
Updat. 12:1–7. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Narla G, DiFeo A, Yao S, Banno A, Hod E,
Reeves HL, Qiao RF, Camacho-Vanegas O, Levine A, Kirschenbaum A, et
al: Targeted inhibition of the KLF6 splice variant, KLF6 SV1,
suppresses prostate cancer cell growth and spread. Cancer Res.
65:5761–5768. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Reeves HL, Narla G, Ogunbiyi O, Haq AI,
Katz A, Benzeno S, Hod E, Harpaz N, Goldberg S and Tal-Kremer S:
Kruppel-like factor 6 (KLF6) is a tumor-suppressor gene frequently
inactivated in colorectal cancer. Gastroenterology. 126:1090–1103.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kremer-Tal S, Reeves HL, Narla G, Thung
SN, Schwartz M, Difeo A, Katz A, Bruix J, Bioulac-Sage P,
Martignetti JA, et al: Frequent inactivation of the tumor
suppressor Kruppel-like factor 6 (KLF6) in hepatocellular
carcinoma. Hepatology. 40:1047–1052. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ebrahimi A, Nodushan SMHT, Mousavian A,
Mokarizadeh A, Abbasi M, Yahaghi E and Rasaei SM: Diagnostic and
prognostic potentials of KLF6 and HER3 expression alterations in
cutaneous malignant melanoma. Tumour Biol. Oct 16–2015.(Epub ahead
of print). View Article : Google Scholar
|
|
10
|
Cai D, Zhao J and Sun Q: Kruppel-like
factor 6 in the progression and prognosis of malignant melanoma. J
Int Med Res. 42:184–190. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lin WM, Baker AC, Beroukhim R, Winckler W,
Feng W, Marmion JM, Laine E, Greulich H, Tseng H, Gates C, et al:
Modeling genomic diversity and tumor dependency in malignant
melanoma. Cancer Res. 68:664–673. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Goff LA, Davila J, Swerdel MR, Moore JC,
Cohen RI, Wu H, Sun YE and Hart RP: Ago2 immunoprecipitation
identifies predicted microRNAs in human embryonic stem cells and
neural precursors. PLoS One. 4:e71922009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Rönnau CG, Verhaegh GW, Luna-Velez MV and
Schalken JA: Noncoding RNAs as novel biomarkers in prostate cancer.
Biomed Res Int. 2014:5917032014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Pouliot LM, Shen D-W, Suzuki T, Hall MD
and Gottesman MM: Contributions of microRNA dysregulation to
cisplatin resistance in adenocarcinoma cells. Exp Cell Res.
319:566–574. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Jing L, Jin C, Lu Y, Huo P, Zhou L, Wang Y
and Tian Y: Investigation of microRNA expression profiles
associated with human alcoholic cardiomyopathy. Cardiology.
130:223–233. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Gao S, Wang J, Xie J, Zhang T and Dong P:
Role of miR-138 in the regulation of larynx carcinoma cell
metastases. Tumour Biol. Oct 24–2015.(Epub ahead of print).
|
|
17
|
Song K, Liu N, Yang Y and Qiu X:
Regulation of osteosarcoma cell invasion through osteopontin
modification by miR-4262. Tumour Biol. 37:6493–6499. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Bao H, Gao F, Xie G and Liu Z:
Angiotensin-converting enzyme 2 inhibits apoptosis of pulmonary
endothelial cells during acute lung injury through suppressing
miR-4262. Cell Physiol Biochem. 37:759–767. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Huang X, Le Q-T and Giaccia AJ: MiR-210 -
micromanager of the hypoxia pathway. Trends Mol Med. 16:230–237.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chan YC, Banerjee J, Choi SY and Sen CK:
miR-210: The master hypoxamir. Microcirculation. 19:215–223. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wang H, Flach H, Onizawa M, Wei L, McManus
MT and Weiss A: Negative regulation of Hif1a expression and TH17
differentiation by the hypoxia-regulated microRNA miR-210. Nat
Immunol. 15:393–401. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
22
|
Qin Q, Furong W and Baosheng L: Multiple
functions of hypoxia-regulated miR-210 in cancer. J Exp Clin Cancer
Res. 33:502014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hatami R, Sieuwerts AM, Izadmehr S, Yao Z,
Qiao RF, Papa L, Look MP, Smid M, Ohlssen J, Levine AC, et al:
KLF6-SV1 drives breast cancer metastasis and is associated with
poor survival. Sci Transl Med. 5:169ra12. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhenzhen Z, De'an T, Limin X, Wei Y and
Min L: New candidate tumor-suppressor gene KLF6 and its splice
variant KLF6 SV2 counterbalancing expression in primary
hepatocarcinoma. Hepatogastroenterology. 59:473–476. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Andreoli V, Gehrau RC and Bocco JL:
Biology of Krüppel-like factor 6 transcriptional regulator in cell
life and death. IUBMB Life. 62:896–905. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chen CH, Huang PH, Chu PC, Chen MC, Chou
CC, Wang D, Kulp SK, Teng CM, Wang Q and Chen CS: Energy
restriction-mimetic agents induce apoptosis in prostate cancer
cells in part through epigenetic activation of KLF6 tumor
suppressor gene expression. J Biol Chem. 286:9968–9976. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lang UE, Kocabayoglu P, Cheng GZ,
Ghiassi-Nejad Z, Muñoz U, Vetter D, Eckstein DA, Hannivoort RA,
Walsh MJ and Friedman SL: GSK3β phosphorylation of the KLF6 tumor
suppressor promotes its transactivation of p21. Oncogene.
32:4557–4564. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Sangodkar J, Dhawan NS, Melville H, Singh
VJ, Yuan E, Rana H, Izadmehr S, Farrington C, Mazhar S, Katz S, et
al: Targeting the FOXO1/KLF6 axis regulates EGFR signaling and
treatment response. J Clin Invest. 122:2637–2651. 2012. View Article : Google Scholar : PubMed/NCBI
|