Introduction
Drugs enter into hepatocytes through influx
transporters in addition to the diffusion into blood flow, and will
be metabolized by hepatic enzymes and then be transported from
hepatocytes into blood flow or into bile to complete the process of
drug clearance (1). The absorption,
distribution, excretion and transportation of drugs by cell
membranes are inseparable from drug transporters (2). Drug transporters are divided into
influx transporters and efflux transporters, and influx
transporters include organic anion transporters, organic cation
transporters, organic anion transporting polypeptides and
Na+/taurocholate co-transporting polypeptides; and
efflux transporters consist of multidrug resistance proteins,
multidrug resistance-associated proteins, breast cancer resistance
proteins and bile salt efflux pumps (3). Organic anion transporting polypeptides
(OATPs) of influx transporter family plays an important role in the
transport of endogenous and exogenous substances, especially in the
transport of drugs (4).
Organic anion transporting polypeptides 1B1
(OATP1B1), one of the important proteins of OATPs family, is
specially distributed in the basolateral membrane of the liver
(5). OATP1B1 gene chromosome is
located in 12p12, and the encoding gene of its cDNA contains 2073
bases and encodes 691 amino acids, composed of 15 exons and 14
introns. Endogenous substances responsible for transport include
bile acids, thyroid hormones, free bilirubin, sulfate and
glucuronic acid complexes as well as estradiol-17β-glucuronide
(5). Drugs transported by OATP1B1
include statins (pitavastatin, pravastatin and atorvastatin),
methotrexate, rifampicin, olmesartan, nateglinide and repaglinide
(5). OATP1B1 has genetic
polymorphism with high mutation frequency in alleles and racial
differences. At present, more than forty mutation sites have been
found in people, of which 388A>G, 2000A>G and 1463G>C most
frequently occur in the United States blacks, with the frequencies
of 74%, 34% and 9%; 388A>G, 463C>A and 521T>C most
commonly occur in the United States whites with mutation
frequencies of 30%, 16% and 14%; and two kinds of single nucleotide
polymorphisms most commonly occurring in Chinese population are
388A>G and 521T>C with mutation frequencies of 73.4 and
14.0%, and the four main haplotypes of OATP1B1 include SLCO1B1*1a
(388A, 521T), SLCO1B1*1b (388G, 52lT), SLCO1B1*5 (388A, 521C) and
SLCO1B1*15 (388G, 52lC), among which SLCO1B1*1a belongs to
wild-type. Most studies showed that the mutation of 521 T>C
decreased the transport function of OATP1B1 and the efficiency of
drugs entering into hepatocytes, therefore improving the
concentration of drugs in blood and therapeutic effects (6). There has been no agreement on the role
of the mutation of bases located at the mutation of 388A>G in
the transport of OATP1B1, some studies suggested that mutation
occurring at the site did not affect the transport of proteins, but
some hold that it enhanced the transport function of proteins
(7).
National Comprehensive Cancer Network (NCCN)
recommends that patients with breast cancer whose expression of
estrogen receptor are positive, regardless of menstruation, age,
tumor size and lymph node status, should be treated with adjuvant
endocrine therapy after surgery (8). Tamoxifen, the basic drug of endocrine
therapy for breast cancer, is taken up into hepatocytes by OATP1B1
and then turns into active metabolite-endoxifen
(4-hydroxy-N-desmethyl tamoxifen) through the metabolism by liver
enzyme in vivo. Tamoxifen, an estrogen receptor antagonist,
has two isoforms such as Z-type which antagonizes the effect of
estrogen and E-type which has weak estrogen activity (5). Clinically, tamoxifen inhibits the
effect of estrogen by competing for estrogen binding receptors, and
thereby inhibiting the growth of breast cancer cells (9). However, tamoxifen also showed
significantly different effects in the treatment of breast cancer,
which could not be completely explained by the differences in liver
and kidney function, age and lifestyles, so we reveal the effects
of gene polymorphisms of drug transporters and drug metabolizing
enzymes on pharmacokinetics and pharmacodynamics of drugs from the
perspective of pharmacogenetics (10).
Tamoxifen is metabolized mainly by two pathways
in vivo, one is to metabolize tamoxifen into primary
metabolite N-desmethyl-tamoxifen (NDT) by CYP3A enzyme, which will
be further turned into endoxifen by CYP2D6, accounting for near 90%
of all metabolic pathways; the other is to metabolize tamoxifen
into 4-hydroxy-tamoxifen (4-OH-TAM) by CYP2D6, which will be turned
into endoxifen eventually through CYP3A, so CYP2D6 and CYP3A are
key enzymes to the metabolism of tamoxifen (10). FDA and FDA Advisory Committee also
proposed to use the genotyping results of CYP2D6 as the reference
of tamoxifen treatment, as patients with breast cancer who carry
the mutation of CYP2D6 genes speed up the metabolism of tamoxifen
into active metabolite-endoxifen and thereby decrease blood
concentration in the drug, and the effect of treatment. On the
contrary, patients carrying the mutation of CYP2D6 have low
metabolism and lead to the accumulation of drugs, thus causing
toxicity (11). The latest in
vivo experimental results showed that the concentration of
endoxifen was the main standard to predict whether tamoxifen could
achieve desired treatment effect in standard tamoxifen treatment.
Similar to the gene polymorphisms of drug metabolizing enzymes
affect the metabolism of drugs in vivo, the gene
polymorphisms of drug transporters influence the metabolism of
drugs in vivo (12).
The studies on the effect of the gene polymorphism
of organic anion transporting polypeptides 1B1 on the metabolism of
tamoxifen in vivo are very few, and the influence of the
gene polymorphism of OATP1B1 on the metabolism of tamoxifen into
endoxifen in vivo has not been reported. In China, up to
200,000 people are newly diagnosed of breast cancer each year, and
4–5 million people died of the disease (13). Tamoxifen, the gold standard drug for
the treatment of breast cancer, has more than 100 years of history,
but the age distribution and financial situations of patients with
breast cancer are varied. As OATP1B1 has polymorphism with racial
differences, it is necessary to combine the endocrine therapy of
patients with breast cancer in China based on racial differences of
OATP1B1, with OATP1B1*1a, OATP1B1*1b and OATP1B1*5 as research
subjects of OATP1B1 polymorphism, to analyze OATP1B1 gene
polymorphism on the concentration of tamoxifen and endoxifen, the
major metabolite of tamoxifen, thus to provide pharmacogenetic
basis for the prediction of the drug effect, and screen out
patients who benefit from the therapy, so as to avoid excessive
treatment and ensure reasonable and individualized administration
with drugs (5,6). Therefore, our objection is to
investigate whether or not OATP1B1 polymorphism affect the
metabolism of tamoxifen and its metabolite endoxifen.
Materials and methods
Overexpression of lentivirus cloning
by PCR and carrier enzyme
GV358 vector was used in this study, we designed PCR
primers with AgeI and AgeI, OATP1B1-F,
5′-GAGGATCCCCGGGTACCGGTCGCCACCATGGACCAAAATCAACATTTG-3′; OATP1B1-R,
5′-TCCTTGTAGTCCATACCACAATGTGTTTCACTATCTGC-3′. Total RNA was
extracted from liver tissue using TRIzol method (Invitrogen,
Carlsbad, CA, USA) according to the manufacturer's protocol. Total
RNA (1 µg) was reverse-transcribed using First Strand cDNA
Synthesis kit (Promega Biotech Co., Ltd., Beijing, China). PCR was
performed using GeneAmp PCR (2720 Thermal Cycler; Applied
Biosystems) by Taq polymerase (SinoBio, E001-02B) according to the
manufacturer's instructions. The qPCR steps were as follows:
initial denaturation at 98°C for 5 min, 30 cycles (98°C for 10 sec,
55°C for 10 sec, and 72°C for 90 sec), final extension at 72°C 8
min and hold at 4°C. The outcome of PCR and GV358 vector were
performed with dual-enzyme digestion and T4 ligase at 37°C
overnight.
Preparation of competent cells and
transformation
DH-5α Escherichia coli (1:100-1:50) was
inoculated with 1 ml of LB nutrient solution and cultivated at 37°C
for 2–3 h. Then, bacteria solution was placed in ice for 20 min and
centrifuged at 2,000 g for 10 min. After discarding the
supernatant, bacteria solution was incubated with 0.1 ml of
pre-cooling CaCl2 (100 mM) in ice for 20 min. After
centrifugation at 2,000 g for 10 min, bacteria solution was
resuspended with 0.1 ml of pre-cooling CaCl2 (100 mM) in
ice. Synthetic plasmid (10 µl) was added into 0.1 ml of competence
bacteria solution and incubated in ice for 30 min. Then, bacteria
solution was activated at 42°C for 90 sec and incubated in ice for
2 min. Next, bacteria solution was added into 0.5 ml of LB and
incubated at 37°C for 1 h. Lastly, bacteria solution was seeded at
solid state LB with antibiotic and incubated at 37°C for 12 h.
Bacterial colony was seeded into 3–5 ml of LB at 37°C for 12 h.
HEK293T cell culture and grouping
HEK293T cells were divided into six groups: Control
group (A group); HEK293T cells + tamoxifen (6, 30 and 150 ng/ml) or
endoxifen (3, 15 and 75 ng/ml) (B group); HEK293T cells + negative
plasmid + tamoxifen (6, 30 and 150 ng/ml) or endoxifen (3, 15 and
75 ng/ml) (C group); HEK293T cells + OATP1B1*1a plasmid + tamoxifen
(6, 30 and 150 ng/ml) or endoxifen (3, 15 and 75 ng/ml) (D group);
HEK293T cells + OATP1B1*1b (388 GG) plasmid + tamoxifen (6, 30 and
150 ng/ml) or endoxifen (3, 15 and 75 ng/ml) (E group); HEK293T
cells + OATP1B1*5 (521 CC) plasmid + tamoxifen (6, 30 and 150
ng/ml) or endoxifen (3, 15 and 75 ng/ml) (F group).
Real-time quantitative RT-PCR
Total RNA from cells using TRIzol method
(Invitrogen) according to the manufacturer's protocol. Total RNA (1
µg) was reverse-transcribed using First Strand cDNA Synthesis kit
(Promega). PCR was performed using GeneAmp PCR (2720 Thermal
Cycler; Applied Biosystems) by Taq polymerase (SinoBio, E001-02B)
according to the manufacturer's instructions. Real-time PCR assay
was performed by SYBR Premix Ex Taq™ (Takara, Japan). Real-time
PCR, denaturation at 95°C for 2 min; 40 cycles at 95°C for 15 sec
and 60°C for 30 sec; and a final dissociation stage (95°C for 5
min).
Western blotting
HEK293T cells were split with lysis buffer (RIPA,
Beyotime) and the supernatant was discarded after centrifugation at
12,000 g for 10 min at 4°C. Protein concentration was determined by
BCA assay (Beyotime). Equivalent protein (50 µg) was loaded for
separation into 10% gradient SDS-PAGE and transferred onto
nitrocellulose membranes (Santa Cruz Biotechnology, Inc.).
Membranes were blocked with 5% non-fat milk in TBST for 1 h at 37°C
and then incubated with primary antibodies: anti-OATP1B1 (1:1,000,
Abcam) and β-actin (1:2,000, Beyotime) overnight at 4°C followed by
horseradish peroxidase-conjugated secondary antibodies (1:5,000,
Beyotime) for 1 h at 37°C. The immunoreactive bands were visualized
by an ECL Western blotting detection kit (Beyotime) and analyzed
semi-quantitatively using Quantity One software (Bio-Rad
Laboratories, Milan, Italy).
MTT assay
HEK293T cells (1,000 cell/well) were seeded at
96-well plate and treated with 1–100 µM of tamoxifen or 0.1–1 µM of
endoxifen for 48 h. MTT(3-(4,5)-dimethylthiahiazo(−z-y1)-3,5-di-phenytetrazolium
bromide, 5 mg/ml, Sigma-Aldrich Co. LLC) was added into cells for 4
h at 37°C. DMSO (150 µl) was added into cell after removing medium
and dissolved for 10 min at 37°C. Absorbance was measured using
microplate reader at 492 nm, the inhibitory rate = (1 - absorbance
of experimental group/control of experimental group) × 100%.
HPLC-MS/MS
Mass spectrometry conditions are shown in Table I.
 | Table I.Mass spectrometry conditions. |
Table I.
Mass spectrometry conditions.
| Compound | Pair ion | DP | EP | CE | CXP |
|---|
| Tamoxifen | 372.1/72.2 | 65 | 6 | 40 | 4 |
| Endoxifen | 374.3/58.1 | 65 | 5 | 45 | 3 |
Liquid condition
A liquid, double distilled water (5 mM of ammonium
acetate and 1% formic acid); B liquid, methyl alcohol (1% formic
acid), column temperature was 20°C, and flow velocity was 0.3
ml/min. Injecting time was 8 min, the gradient was eluted.
Sample treatment methods
10 µl of interior label (Tegretol, 300 ng/ml) was
added into 100 µl of samples, then and samples were centrifuged at
1,000 g for 1 min. Methyl alcohol (300 µl) was added into samples
and shocked at 1,700 rpm for 2 min. After centrifugation with
1,0000 g for 5 min, clear liquid was absorbed and 5 µl clear liquid
was used to analyze tamoxifen and endoxifen levels.
Statistical analysis
All data are expressed as the mean ± standard
deviation. Differences between groups were tested by the Student's
t-test using the SPSS 18.10 program (Chicago, IL, USA). A p-value
of <0.05 was considered statistically significant.
Results
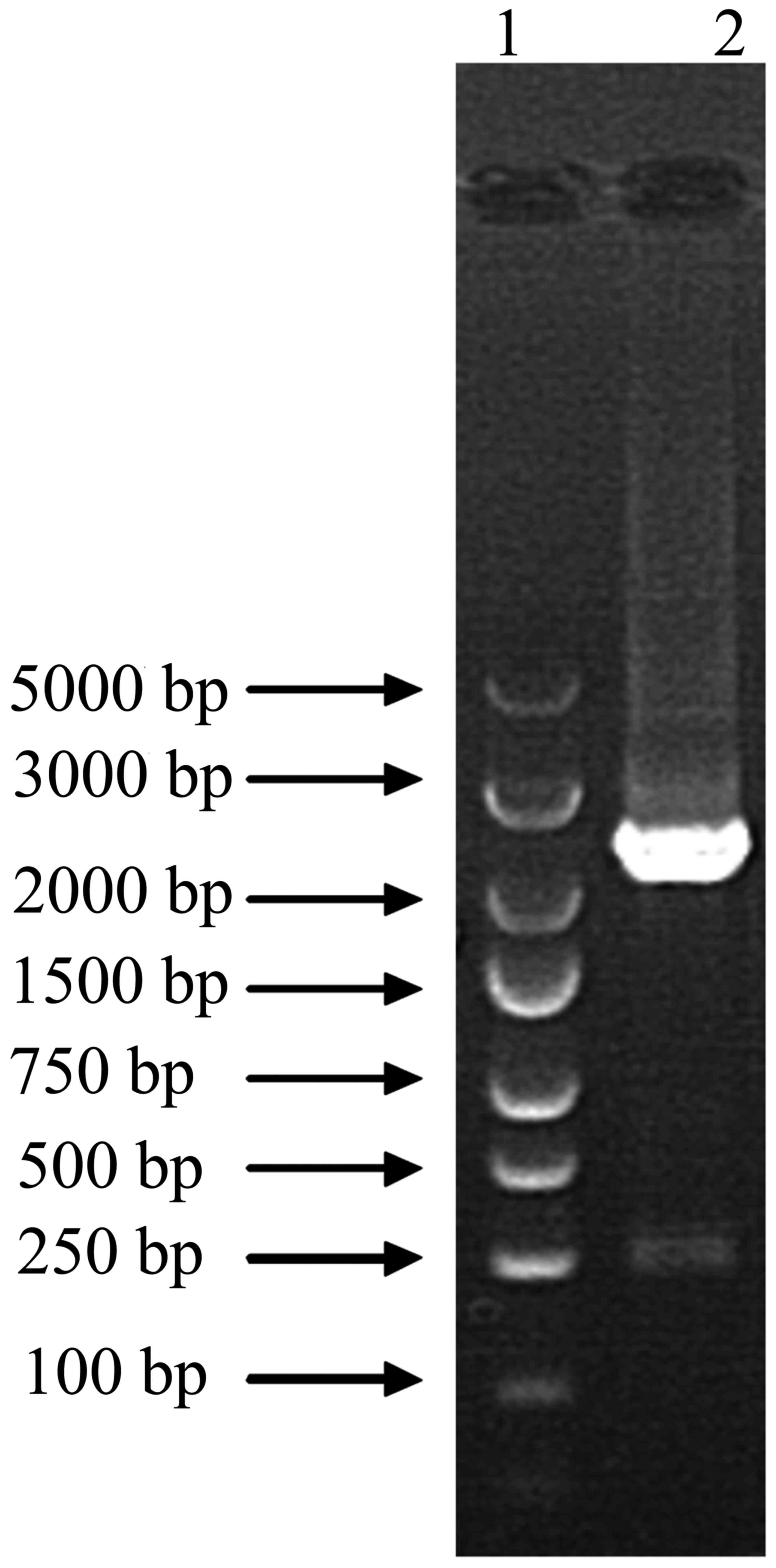
Target gene fragments
Firstly, target fragment of human OATP1B1 is 2,074
bp, and cDNA of this study is 2,117 bp with AgeI and
AgeI. We used PCR to compound target gene fragments, which
is 2,117 bp as shown in Fig. 1.
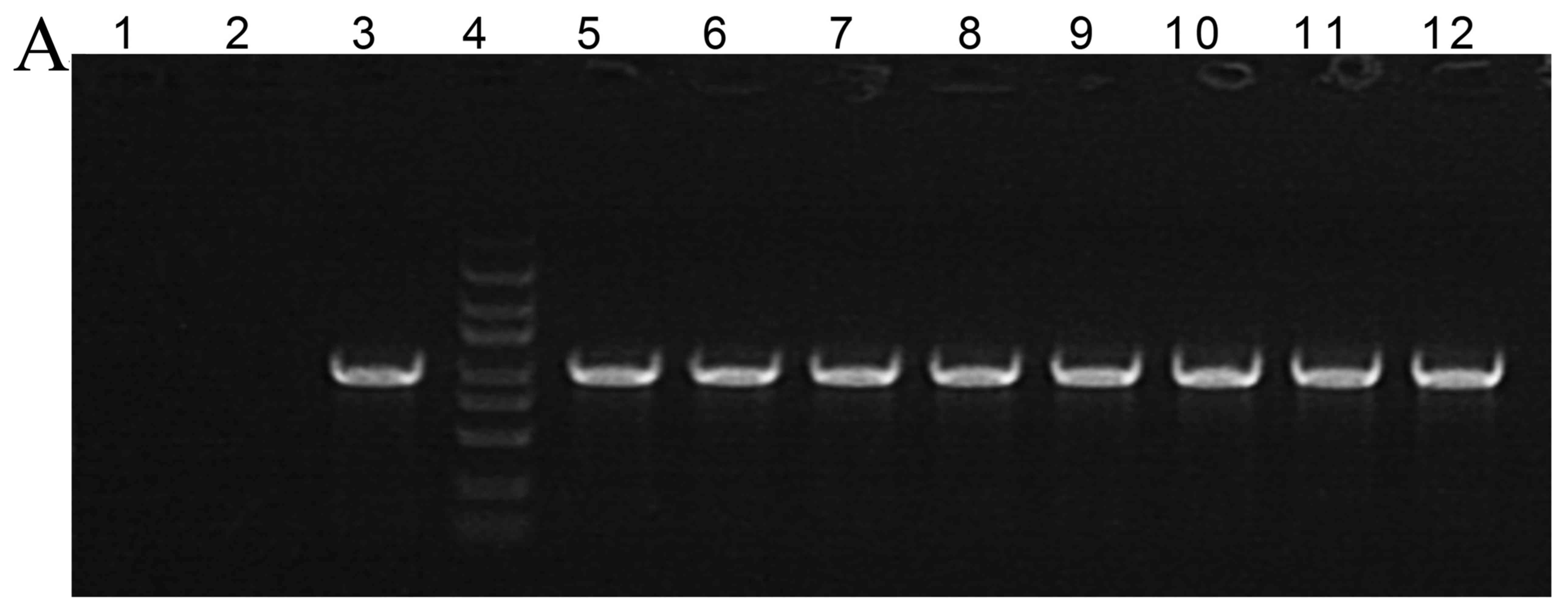
The construction of a recombinant
plasmid
Next, we used PCR to analyze construction of a
recombinant plasmid, 1,004-bp fragment is shown at Fig. 2A. As showed in Fig. 2B and C, 388 site is adenine (A), and
521 site is thymine (T).
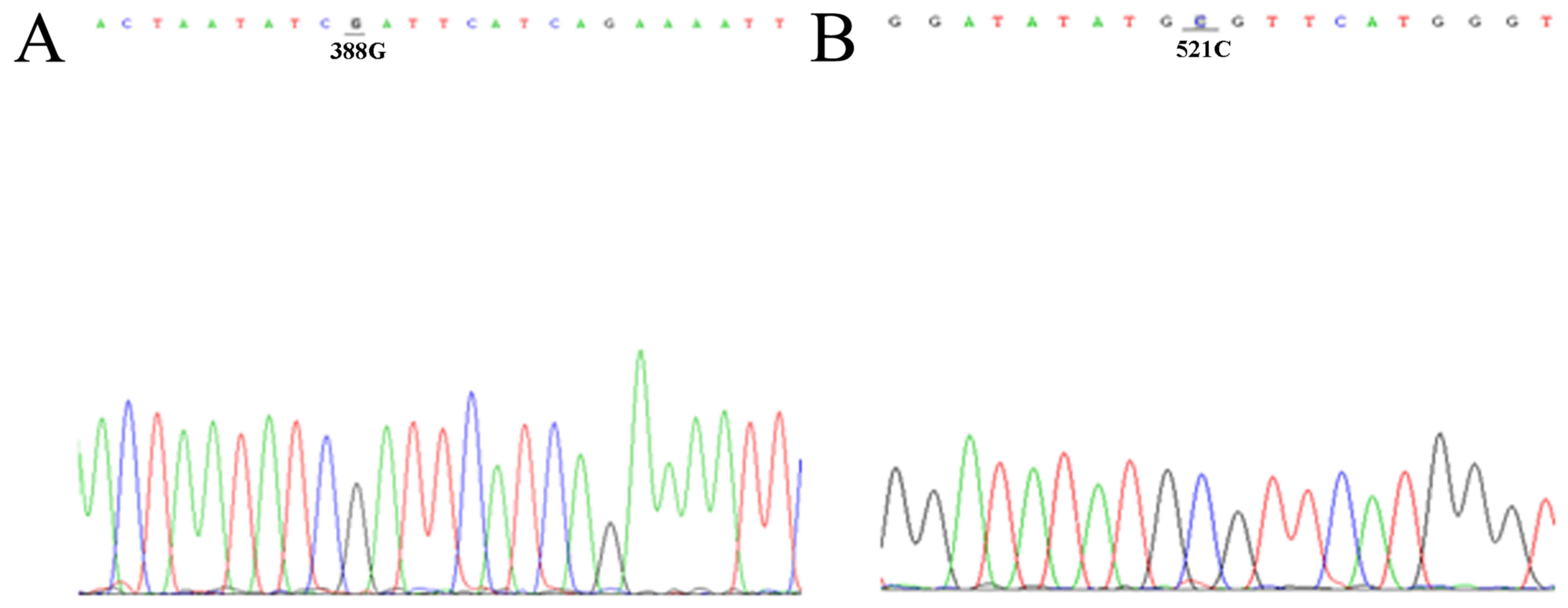
Site-specific mutagenesis of
recombinant plasmids
We used site-specific mutagenesis to induce mutation
at 388 and 521 sites, respectively; site-specific mutagenesis
production was DNA sequenced. As showed in Fig. 3, 388 site is guanine (G), and 521
site is cytosine (C).
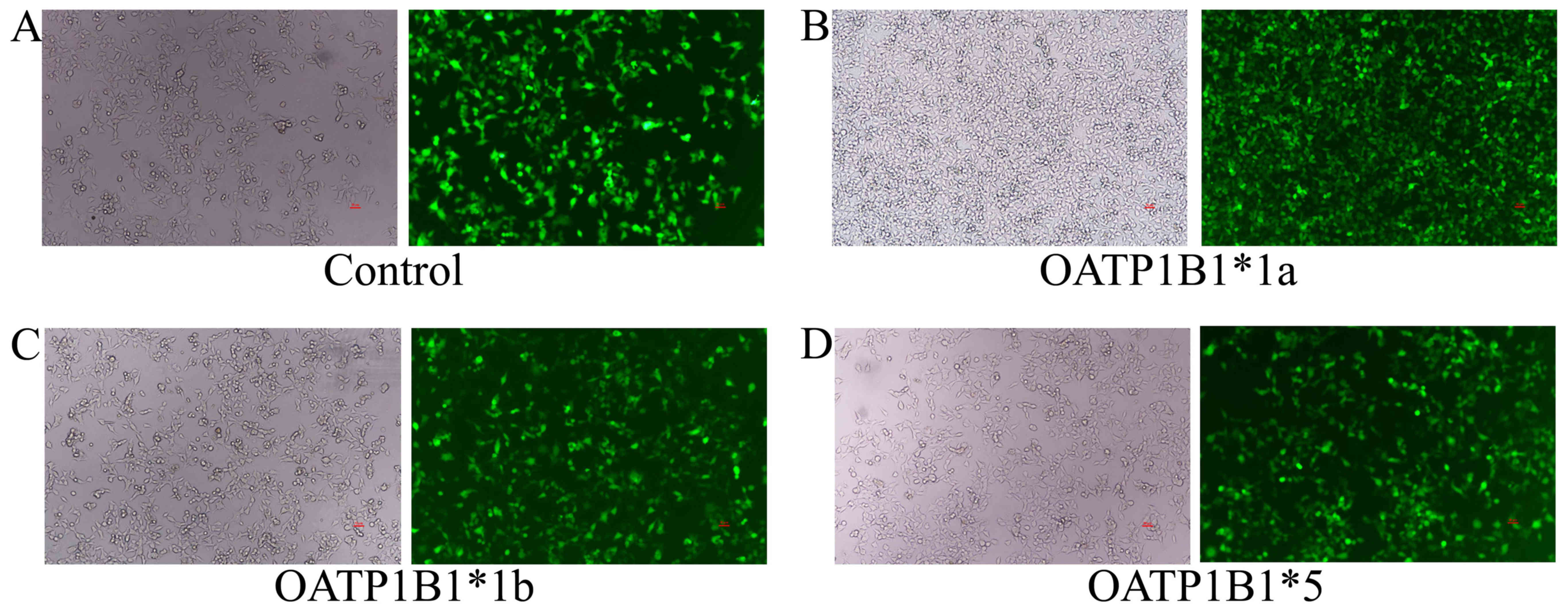
HEK293T cell infection
OATP1B1*1a, OATP1B1*1b (388 GG) and OATP1B1*5 (521
CC) plasmids were infected into HEK293T cells with Polybrene. After
infection for 72 h, fluorescent expression was observed using a
fluorescence microscope. There was an outstanding increase of
fluorescent expression in control, OATP1B1*1a, OATP1B1*1b (388 GG)
and OATP1B1*5 (521 CC) plasmids (Fig.
4).
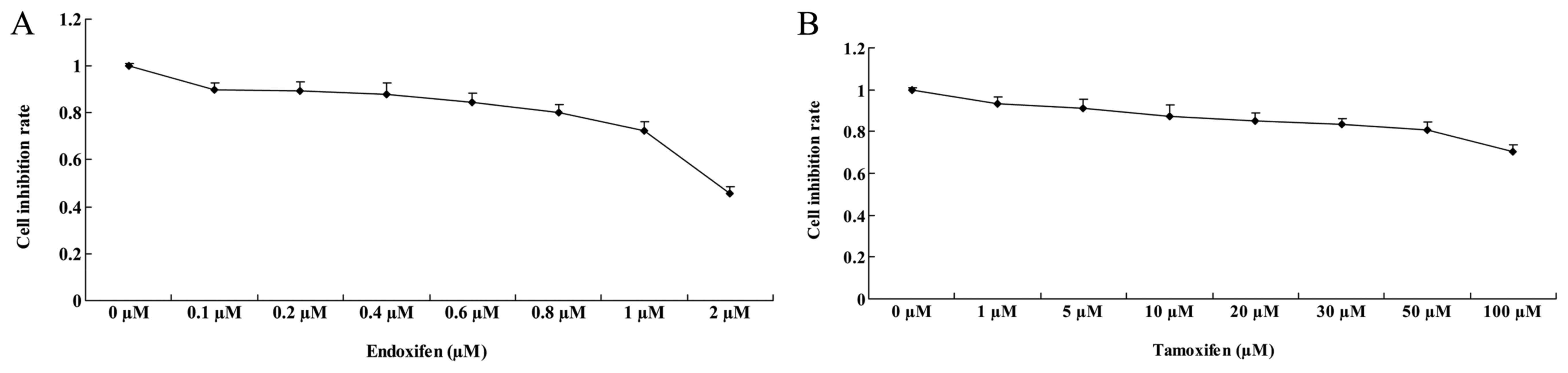
Cytotoxicity of HEK293T cells
Tamoxifen (1–100 µM) and 0.1–1 µM of endoxifen
treated in HEK293T cells for 48 h. Fig.
5A shows the inhibition rate of >80% treated with 0.1, 0.3
and 1 µM of endoxifen for 48 h in HEK293T cells. The inhibition
rate of HEK293T cell treated with 2 µM of endoxifen was 0.423±0.02%
(Fig. 5A). Inhibition of the rate
was >80% treated with 10, 25 and 50 µM of tamoxifen for 48 h
(Fig. 5B). The inhibition rate of
HEK293T cells treated with 100 µM of tamoxifen was 0.785±0.04%
(Fig. 5B).
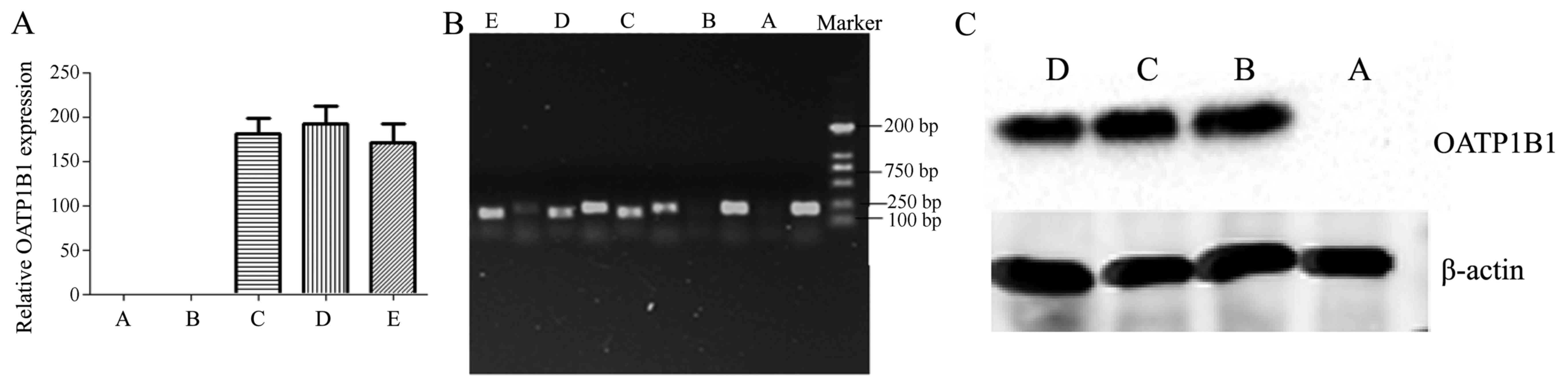
mRNA and protein expression in HEK293T
cells
Control, negative, OATP1B1*1a, OATP1B1*1b (388 GG)
and OATP1B1*5 (521 CC) plasmids were infected into HEK293T for 72
h. As showed in Fig. 6A and B,
there was marked increase of OATP1B1 mRNA expression in OATP1B1*1a,
OATP1B1*1b (388 GG) and OATP1B1*5 (521 CC) plasmids, compared with
negative group. The protein expression of OATP1B1 was significantly
induced in OATP1B1*1a, OATP1B1*1b (388 GG) and OATP1B1*5 (521 CC)
plasmid groups, compared with negative group (Fig. 6C).
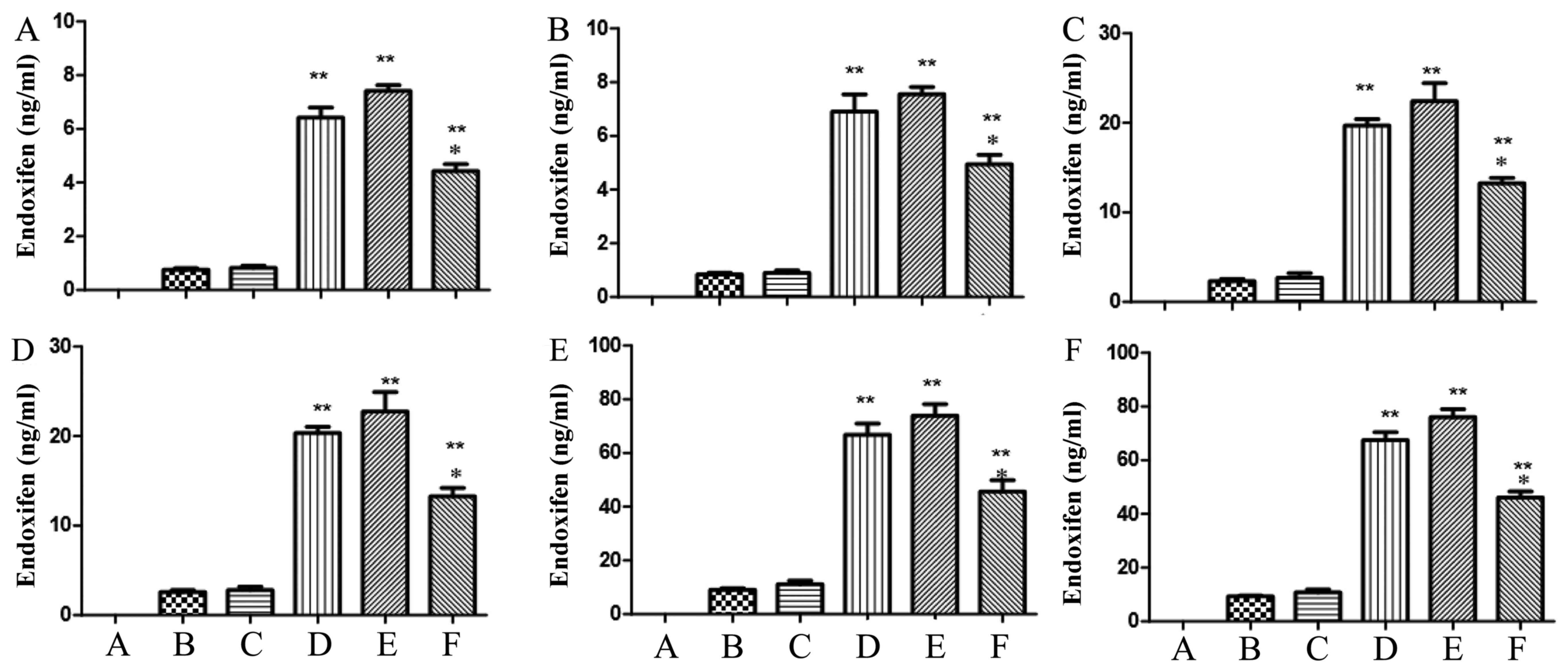
The methodology of tamoxifen level
using HPLC-MS/MS
Tamoxifen at 6, 30 and 150 ng/ml was added into the
lysate of HEK293T cells. In interblock, HPLC-MS/MS was used to
analyze tamoxifen levels, and the RSD (relative standard deviation)
of tamoxifen was 4.01, 1.17 and 4.27% in 6, 30 and 150 ng/ml of
tamoxifen group, respectively (Table
II). RE (relative error) of tamoxifen was 98.83, 99.04 and
100.38% in 6, 30 and 150 ng/ml of tamoxifen group, respectively
(Table II). Next, in interclass,
HPLC-MS/MS was used to analyze tamoxifen levels, and the RSD
(relative standard deviation) of tamoxifen was 0.90, 1.38 and 1.65%
in 6, 30 and 150 ng/ml of tamoxifen group, respectively (Table III). RE of tamoxifen was 99.50,
96.05 and 99.15% in 6, 30 and 150 ng/ml of tamoxifen group,
respectively (Table III).
 | Table II.Precision and accuracy of tamoxifen
in interblock. |
Table II.
Precision and accuracy of tamoxifen
in interblock.
| Concentration
(ng/ml) | 6 | 30 | 150 |
|---|
| Measured
concentrations | 6.11 | 29.4 | 146 |
|
| 5.98 | 30.2 | 147 |
|
| 6 | 29.9 | 145 |
|
| 6.03 | 29.4 | 159 |
|
| 5.51 | 29.6 | 156 |
| Average value (mean
± SD) | 5.93±0.24 | 29.70±0.35 | 150.6±6.43 |
| RSD (%) | 4.01 | 1.17 | 4.27 |
| RE (%) | 98.83 | 99.04 | 100.38 |
 | Table III.Precision and accuracy of tamoxifen
in interclass. |
Table III.
Precision and accuracy of tamoxifen
in interclass.
| Concentration
(ng/ml) | 6 | 30 | 150 |
|---|
| Measured
concentrations (mean ± SD) | 5.97±0.05 | 28.83±0.40 | 148.73±2.45 |
| RSD (%) | 0.90 | 1.38 | 1.65 |
| RE (%) | 99.50 | 96.05 | 99.15 |
The methodology of endoxifen level
using HPLC-MS/MS
Endoxifen (3, 15 and 75 ng/ml) was added into the
lysate of HEK293T cell. In interblock, HPLC-MS/MS was used to
analyze endoxifen levels, and the RSD (relative standard deviation)
of endoxifen was 7.66, 3.25 and 2.89% in 3, 15 and 75 ng/ml of
endoxifen group, respectively (Table
IV). RE of endoxifen was 98.33, 99.04 and 96.28% in 3, 15 and
75 ng/ml of endoxifen group, respectively (Table IV). Next, in interclass, HPLC-MS/MS
was used to analyze endoxifen levels, and the RSD of endoxifen was
2.38, 0.71 and 1.26% in 3, 15 and 75 ng/ml of endoxifen group,
respectively (Table V). RE of
endoxifen was 95.96, 92.4 and 93.87% in 6, 30 and 150 ng/ml of
endoxifen group, respectively (Table
V).
 | Table IV.Precision and accuracy of endoxifen
in interblock. |
Table IV.
Precision and accuracy of endoxifen
in interblock.
| Concentration
(ng/ml) | 3 | 15 | 75 |
|---|
| Measured
concentrations | 2.85 | 14.3 | 72.6 |
|
| 2.82 | 15 | 70.3 |
|
| 3.12 | 14.4 | 75.2 |
|
| 2.72 | 15.3 | 70.2 |
|
| 3.26 | 15.3 | 73 |
| Average value (mean
± SD) | 2.95±0.23 | 14.86±0.48 | 72.26±2.09 |
| RSD (%) | 7.66 | 3.25 | 2.89 |
| RE (%) | 98.33 | 99.04 | 96.28 |
 | Table V.Precision and accuracy of endoxifen
in interclass. |
Table V.
Precision and accuracy of endoxifen
in interclass.
| Concentration
(ng/ml) | 3 | 15 | 75 |
|---|
| Measured
concentrations (mean ± SD) | 2.88±0.07 | 13.86±0.10 | 70.40±0.89 |
| RSD (%) | 2.38 | 0.71 | 1.26 |
| RE (%) | 95.96 | 92.4 | 93.87 |
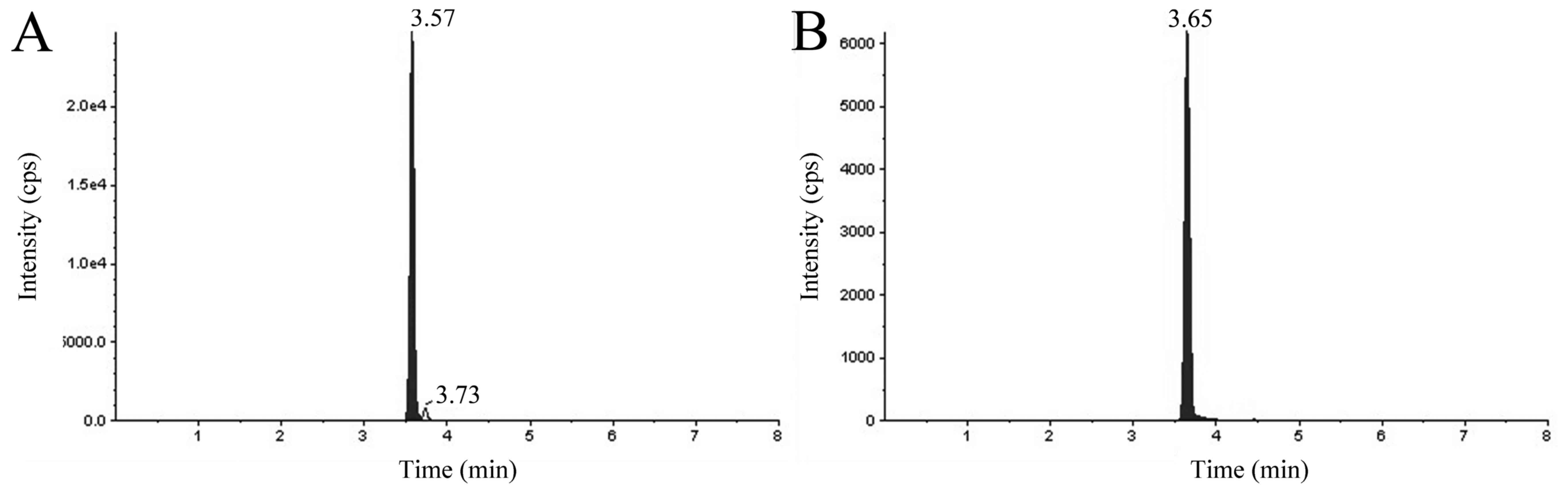
Special properties of the methods
Under the condition of the above experiments,
chromatographic peak of tamoxifen, endoxifen and Tegretol (interior
label) showed sharp and good separation, and this methods had good
specificity (Fig. 7). The retention
time of tamoxifen and Tegretol were 3.66 and 3.65 min, respectively
(Fig. 7A and B). Fig. 7C and D show that the retention time
of endoxifen and Tegretol are 3.57 and 3.65 min, respectively.
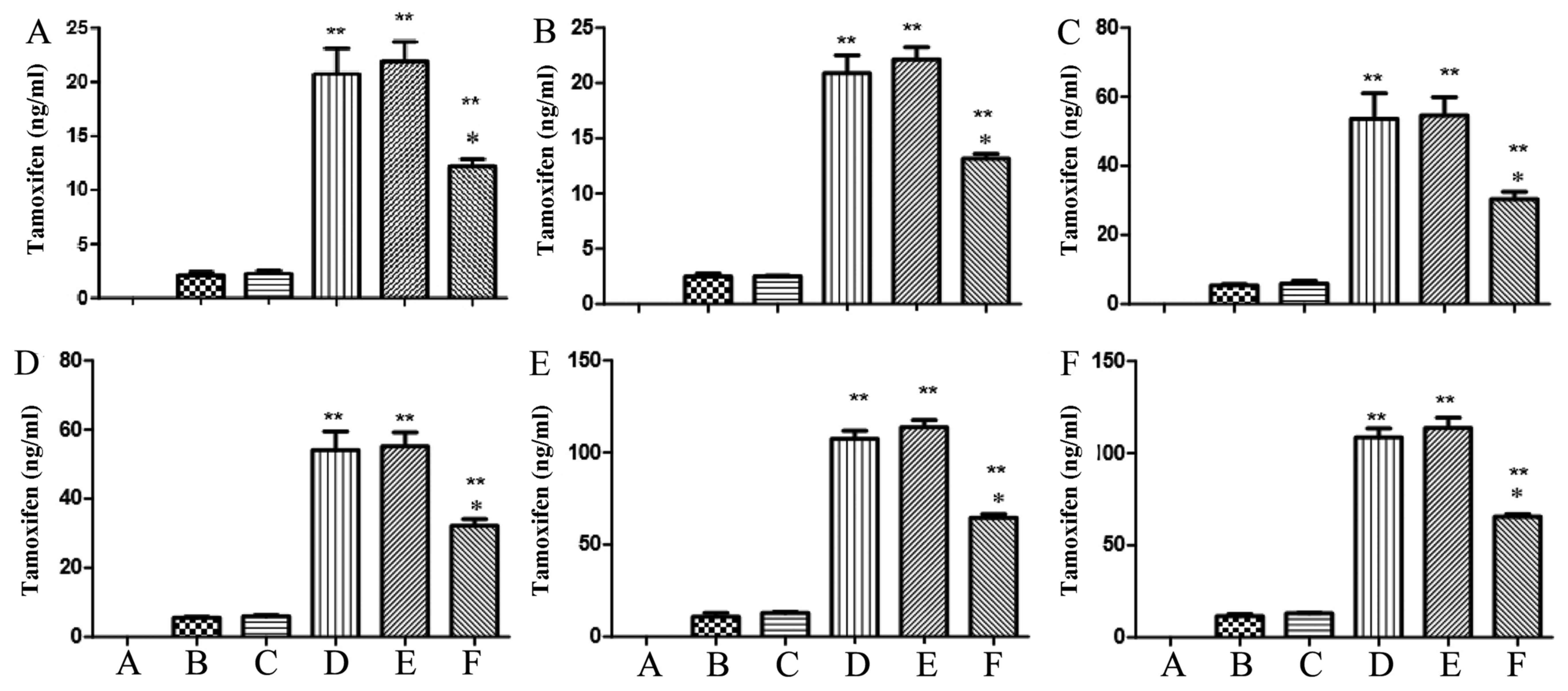
The content of tamoxifen using
HPLC-MS/MS
We explored the function of OATP1B1*1a, OATP1B1*1b
(388 GG) and OATP1B1*5 (521 CC) in the metabolism of tamoxifen
using HPLC-MS/MS. After infection at 24 and 48 h, there were
significant increases of tamoxifen levels in OATP1B1*1a, OATP1B1*1b
(388 GG) and OATP1B1*5 (521 CC) groups, compared with negative
group. However, the tamoxifen level of OATP1B1*5 (521 CC) group was
remarkably lower than that of OATP1B1*1a or OATP1B1*1b (388 GG)
(Fig. 8). However, the tamoxifen
level of OATP1B1*1a group was similar to that of OATP1B1*1b (388
GG) group (Fig. 8).
The content of endoxifen using
HPLC-MS/MS
We further explored the function of OATP1B1*1a,
OATP1B1*1b (388 GG) and OATP1B1*5 (521 CC) in the metabolism of
endoxifen using HPLC-MS/MS. After infection at 24 and 48 h,
significant increases of tamoxifen levels in OATP1B1*1a, OATP1B1*1b
(388 GG) and OATP1B1*5 (521 CC) groups were observed, compared with
negative group. However, the tamoxifen level of OATP1B1*5 (521 CC)
group was prominently lower than that of OATP1B1*1a or OATP1B1*1b
(388 GG) (Fig. 9). However, the
tamoxifen level of OATP1B1*1a group was also similar to that of
OATP1B1*1b (388 GG) group (Fig.
9).
Discussion
Endocrine therapy plays an important role in the
treatment of breast cancer, and tamoxifen is used as the major drug
of endocrine inhibitor therapy in the treatment of patients with
breast cancer with positive premenopausal estrogen receptor (ER)
and those with postmenopausal breast cancer (14). An in vitro experiment showed
that tamoxifen inhibited the expression of progesterone receptor in
MCF-7 cells (12). However, the
individual differences and adverse reactions of tamoxifen is of
increasing concern. Tamoxifens metabolites composed of 4-OH-TAM and
N-desmethyl-TAM (NDT) and endoxifen. Lin et al compared
pharmacokinetic parameters of endoxifen and tamoxifen, finding that
endoxifen was more easily absorbed and the concentration of drugs
in the blood peaked faster compared with tamoxifen (15). In the first in vivo
experiment of oral administration with different single doses of
endoxifen and single dose of tamoxifen, direct administration with
endoxifen reached Cmax faster, so orally taking smaller
dose of endoxifen can achieve the same effect as tamoxifen, so as
to avoid the adverse reaction of tamoxifen (16).
Clinical drugs transported by OATP1B1 transporters
include angiotensin converting enzyme inhibitors, statins,
angiotensin II receptor antagonists, antitumor drugs and
antibiotics (17). As OATP1B1 has
highly genetic polymorphism with racial differences in specific
mutations, its gene polymorphism affects the transport function of
transporters. A study of oral administration of 18 cases of Han
ethnicity people with 40 mg pravastatin showed that the drug
concentration of subjects with genotype OATP1B1*15 (388G, 521C),
especially the homozygous subjects increased significantly compared
with non-mutation subjects (18). A
study showed that the relationships between the gene polymorphism
of OATP1B1 transporters and rosuvastatin, finding that the
transport of rosuvastatin by mutant OATP1B1*5 was significantly
lower than that by wild-type OATP1B1*1a transporter, the difference
being statistically significant (p<0.05), and the result was
consistent with that of the in vivo experiment (19). When subjects with genotype OATP1B1*5
and genotype OATP1B1*1a took the same dose of rosuvastatin, the
concentration of drug in the blood of the former increased more
significantly, which was probably because the function of
transporter was weakened due to 521T>C mutations and thereby
resulting in the slowing down of drug metabolism (20). In addition, the association between
atorvastatin and atorvastatin with OATP1B1521T>C were also
reported (21).
There is no agreement to the impact of the mutation
of OATP1B1388A>G on drug transporting proteins, but some
experiments recently proved that the mutations of the 388 site was
related to the expression of proteins (22). Some studies found that OATP1B1*1b
could enhance the transport of drugs by transporters, and an in
vivo experiment reached the same conclusion (22,23).
Experimental results showed that area under the curve (AUC) of
pravastatin in people carrying genotype SLCO1B1*1B/*1B gene
decreased by 35% compared with those carrying wild-type SLCO1B1
gene (18). Some experiments held
that OATP1B1*1B inhibited the transport of drugs by
transporters.
At present, the studies on the correlation between
the gene polymorphism of OATP1B1 and breast cancer are few. The
research on the correlation between the gene polymorphism of
OATP1B1 and breast cancer conducted showed that the frequency of
mutant C allele of OATP1B1521T>C was higher in breast cancer
patients with positive estrogen receptor (ER) than those with
negative ER (5,6). However, breast cancer patients with
positive ER were more sensitive to endocrine therapy, suggesting
that the gene polymorphism of OATP1B1 may affect the prognosis of
patients (24). Previous studies on
the correlation between the gene polymorphism of OATP1B1 with
tamoxifen found that the mutation of OATP1B1388A>G and 521T>C
resulted in the decreased intake of tamoxifen by OATP1B1, while not
showing statistically significant differences compared with
wild-type group, mainly because the gene platform was previously
constructed by liposome transfection, which had low efficiency with
short period of gene expression (5,6,25). The
overexpression lentivirus cell platforms have been successfully
constructed, including OATP1B1*1a-HEK293T and OATP1B1*1b-HEK293T
and OATP1B1*5-HEK293T cell model, the infection efficiency is not
less than 80%. The gene expressions are high at mRNA and protein
level. The tamoxifen and endoxifen can be taken up into cells
through organic anion transporter polypeptide 1B1, and
OATP1B1521T>C inhibited the function of the transport protein,
resulting in the content of drug in cell lysis liquid in
OATP1B1*5-HEK293T group is lower than in OATP1B1*1a-HEK293T group
with statistical significance. OATP1B1388A>G makes the content
of drug in cell lysis liquid in OATP1B1*1b-HEK293T group and the
wild group+drug similarly, but there is no statistical
significance.
In conclusion, our results have demonstrated
tamoxifen and endoxifen can be transported by OATP1B1. However,
OATP1B1 521T>C can inhibit the function effects of OATP1B1 on
tamoxifen and endoxifen into cell. This study determined OATP1B1
participation in transportation of tamoxifen and endoxifen into
cells, which may represent a potential therapeutic strategy for
treatment of breast cancer.
Acknowledgements
This study was supported by Open Project of State
Key Laboratory of Natural Medicines, Key Laboratory of Drug
Metabolism and Pharmacokinetics, China Pharmaceutical University
(K-201401) and the Project for Jiangsu Province Key Laboratory of
Drug Metabolism and Pharmacokinetics (BM2012012) and Anhui
Provincial Excellent Young Talents Supporting Plan Key Projects
(gxyqZD2016178), Special Foundation for Talents of Yijishan
Hospital (YR201407, YR201501) and the Natural Science Foundation of
China (grant no. 81173134).
References
|
1
|
Oxenoid K and Chou JJ: A functional NMR
for membrane proteins: Dynamics, ligand binding, and allosteric
modulation. Protein Sci. 25:959–973. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Amaral MD, Farinha CM, Matos P and Botelho
HM: Investigating alternative transport of integral plasma membrane
proteins from the ER to the Golgi: Lessons from the cystic fibrosis
transmembrane conductance regulator (CFTR). Methods Mol Biol.
1459:105–126. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ramboer E, Rogiers V, Vanhaecke T and
Vinken M: Effects of Trichostatin A on drug uptake transporters in
primary rat hepatocyte cultures. EXCLI J. 14:567–576.
2015.PubMed/NCBI
|
|
4
|
Hubeny A, Keiser M, Oswald S, Jedlitschky
G, Kroemer HK, Siegmund W and Grube M: Expression of organic anion
transporting polypeptide 1A2 in red blood cells and its potential
impact on antimalarial therapy. Drug Metab Dispos. 44:1562–1568.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Pu Z, Zhang X, Chen Q, Yuan X and Xie H:
Establishment of an expression platform of OATP1B1 388GG and 521CC
genetic polymorphism and the therapeutic effect of tamoxifen in
MCF-7 cells. Oncol Rep. 33:2420–2428. 2015.PubMed/NCBI
|
|
6
|
Chae YJ, Lee KR, Lee JH, Lee W, Kim DD,
Chung SJ and Maeng HJ: Feasibility of the functional expression of
the human organic anion transporting polypeptide 1B1 (OATP1B1) and
its genetic variant 521T/C in the mouse liver. Eur J Pharm Sci.
96:28–36. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Chen M, Qu BX, Chen XL, Hu HH, Jiang HD,
Yu LS, Zhou Q and Zeng S: Construction of HEK293 cells stably
expressing wild-type organic anion transporting polypeptide 1B1
(OATP1B1*1a) and variant OATP1B1*1b and OATP1B1*15. Pharmazie.
71:337–339. 2016.PubMed/NCBI
|
|
8
|
Tryfonidis K, Basaran G, Bogaerts J,
Debled M, Dirix L, Thery JC, Tjan-Heijnen VC, Van den Weyngaert D,
Cufer T, Piccart M, et al: EORTC-Breast Cancer Group: A European
Organisation for Research and Treatment of Cancer randomized,
double-blind, placebo-controlled, multicentre phase II trial of
anastrozole in combination with gefitinib or placebo in hormone
receptor-positive advanced breast cancer (NCT00066378). Eur J
Cancer. 53:144–154. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Li K, Ying M, Feng D, Du J, Chen S, Dan B,
Wang C and Wang Y: Brachyury promotes tamoxifen resistance in
breast cancer by targeting SIRT1. Biomed Pharmacother. 84:28–33.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lei L, Wang X, Wu XD, Wang Z, Chen ZH,
Zheng YB and Wang XJ: Association of CYP2D6*10 (c.100C>T)
polymorphisms with clinical outcome of breast cancer after
tamoxifen adjuvant endocrine therapy in Chinese population. Am J
Transl Res. 8:3585–3592. 2016.PubMed/NCBI
|
|
11
|
Chang JH, Chen J, Liu L, Messick K and Ly
J: Rifampin-mediated induction of tamoxifen metabolism in a
humanized PXR-CAR-CYP3A4/3A7-CYP2D6 mouse model. Drug Metab Dispos.
44:1736–1741. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Charoenchokthavee W, Panomvana D,
Sriuranpong V and Areepium N: Prevalence of CYP2D6*2, CYP2D6*4,
CYP2D6*10, and CYP3A5*3 in Thai breast cancer patients undergoing
tamoxifen treatment. Breast Cancer (Dove Med Press). 8:149–155.
2016.PubMed/NCBI
|
|
13
|
Argalácsová S, Slanař O, Vítek P, Tesařová
P, Bakhouche H, Dražďáková M, Bartošová O, Zima T and Pertuželka L:
Contribution of ABCB1 and CYP2D6 genotypes to the outcome of
tamoxifen adjuvant treatment in premenopausal women with breast
cancer. Physiol Res. 64:(Suppl 4). S539–S547. 2015.PubMed/NCBI
|
|
14
|
Grzankowski KS, Szender JB,
Spring-Robinson CL, Lele SB, Odunsi KO and Frederick PJ: Evaluation
of metachronous breast and endometrial cancers: Preroutine and
postroutine adjuvant tamoxifen use. Int J Gynecol Cancer.
26:1440–1447. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lin G, Zhang K, Yi L, Han Y, Xie J and Li
J: National prociency testing result of CYP2D6*10 genotyping for
adjuvant tamoxifen therapy in China. PLoS One. 11:e01623612016.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Henderson SL, Teft WA and Kim RB: Profound
reduction in tamoxifen active metabolite endoxifen in a breast
cancer patient treated with rifampin prior to initiation of an
anti-TNFα biologic for ulcerative colitis: A case report. BMC
Cancer. 16:3042016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Durmus S, Lozano-Mena G, van Esch A,
Wagenaar E, van Tellingen O and Schinkel AH: Preclinical mouse
models to study human OATP1B1- and OATP1B3-mediated drug-drug
interactions in vivo. Mol Pharm. 12:4259–4269. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lu XF, Zhou Y, Bi KS and Chen XH: Mixed
effects of OATP1B1, BCRP and NTCP polymorphisms on the population
pharmacokinetics of pravastatin in healthy volunteers. Xenobiotica.
46:841–849. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Riedmaier Emami A, Burt H, Abduljalil K
and Neuhoff S: More power to OATP1B1: An evaluation of sample size
in pharmacogenetic studies using a rosuvastatin PBPK model for
intestinal, hepatic, and renal transporter-mediated clearances. J
Clin Pharmacol. 56:(Suppl 7). S132–S142. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Rose RH, Neuhoff S, Abduljalil K, Chetty
M, Rostami-Hodjegan A and Jamei M: Application of a physiologically
based pharmacokinetic model to predict OATP1B1-related variability
in pharmacodynamics of rosuvastatin. CPT Pharmacometrics Syst
Pharmacol. 3:e1242014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ulvestad M, Skottheim IB, Jakobsen GS,
Bremer S, Molden E, Asberg A, Hjelmesæth J, Andersson TB, Sandbu R
and Christensen H: Impact of OATP1B1, MDR1, and CYP3A4 expression
in liver and intestine on interpatient pharmacokinetic variability
of atorvastatin in obese subjects. Clin Pharmacol Ther. 93:275–282.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Higgins JW, Bao JQ, Ke AB, Manro JR,
Fallon JK, Smith PC and Zamek-Gliszczynski MJ: Utility of
Oatp1a/1b-knockout and OATP1B1/3-humanized mice in the study of
OATP-mediated pharmacokinetics and tissue distribution: Case
studies with pravastatin, atorvastatin, simvastatin, and
carboxydichlorofluorescein. Drug Metab Dispos. 42:182–192. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ide T, Sasaki T, Maeda K, Higuchi S,
Sugiyama Y and Ieiri I: Quantitative population pharmacokinetic
analysis of pravastatin using an enterohepatic circulation model
combined with pharmacogenomic Information on SLCO1B1 and ABCC2
polymorphisms. J Clin Pharmacol. 49:1309–1317. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Brenner S, Riha J, Giessrigl B, Thalhammer
T, Grusch M, Krupitza G, Stieger B and Jäger W: The effect of
organic anion-transporting polypeptides 1B1, 1B3 and 2B1 on the
antitumor activity of flavopiridol in breast cancer cells. Int J
Oncol. 46:324–332. 2015.PubMed/NCBI
|
|
25
|
Riha J, Brenner S, Böhmdorfer M, Giessrigl
B, Pignitter M, Schueller K, Thalhammer T, Stieger B, Somoza V,
Szekeres T, et al: Resveratrol and its major sulfated conjugates
are substrates of organic anion transporting polypeptides (OATPs):
Impact on growth of ZR-75-1 breast cancer cells. Mol Nutr Food Res.
58:1830–1842. 2014. View Article : Google Scholar : PubMed/NCBI
|