Introduction
Based on GIOBALCAN, an estimated 1.8 million new
lung cancer cases and 1.6 million deaths occurred in 2012. This
makes lung cancer the leading cause of cancer-related death among
males worldwide and females in developed countries (1). The poor prognosis is attributed to the
lack of efficient methods for early diagnosis and lack of
successful treatment for metastasis. Since non-small cell lung
cancer (NSCLC), which accounts for approximately 85% of all lung
cancer cases, is not very sensitive to chemotherapy and/or
radiation, surgery remains the treatment of choice. However, most
newly diagnosed NSCLC patients cannot undergo surgery due to local
invasion or distant metastasis. Therefore, it is particularly
important to study the molecular mechanisms underlying NSCLC, which
may provide novel molecular targets for the early diagnosis of lung
cancer.
MicroRNAs (miRNAs) are a group of non-coding RNAs
(~22 nucleotides) that can degrade target mRNA transcripts directly
or suppress their translation through complete or partial
complementarity recognizing the 3′UTR of target mRNAs (2). miRNAs have been proven to play an
important role in the post-transcriptional regulation of gene
expression and are involved in almost all aspects of cancer biology
such as tumor transformation, growth, angiogenesis and
epithelial-mesenchymal transition by inhibiting specific oncogenes
or tumor-suppressor genes. Accumulating data indicate that miRNAs
are present in body fluids including blood plasma and serum, urine,
saliva and semen (3,4) and circulating miRNA levels are more
accurate than the protein-coding gene profiles in tumor typing
(5). Therefore, miRNAs are more
likely to be novel molecular biomarkers in the screening and
monitoring of cancer patients (6).
In our previous study, we found that DAL-1
(differentially expressed in adenocarcinoma of the lung-1; also
known as EPB41L3, 4.1B) has an important role in the invasion and
metastasis of NSCLC (7). By using
microRNA.org, TargetScan and PicTar, we predicted four
miRNAs, miR-26a, miR-26b, miR-96 and miR-223, that regulate DAL-1.
Data from several studies previously showed that miR-223 does not
only promote the invasion of lung cancer cells but also the
metastasis of gastric cancer via targeting tumor suppressor DAL-1
(8,9). Our previous study demonstrated that
both miR-26a and DAL-1 gene expression are decreased in NSCLC, and
DAL-1 is not a real target gene of miR-26a (10). Both miR-26b and miR-26a belong to
the miR-26 family, and miR-26b has low expression levels in many
types of cancer, such as epithelial ovarian (EOC) (11), hepatocellular carcinoma (HCC)
(12), as well as colorectal cancer
(13). In this study, we chose
miR-96 as our research target.
MicroRNA-96 (hsa-miR-96, miR-96), located on
chromosome 7 (7q31~34), belongs to the miR-183 gene family, which
is the first gene cluster to be reported in the development and
function of ciliated ectodermal cells and organs and is essential
for the development and function of animal sensory organs (14,15).
With the growing interest in the miR-183 gene family, miR-96 has
been detected to be highly expressed in various human tumors and
involved in cancer development by regulating key genes in tumor
cell division and apoptosis (16–18).
Although studies have shown that miR-96 is overexpressed in lung
cancer (19,20), it still remains unclear whether
miR-96 could be used for early diagnosis and how miR-96 affects the
progression of lung cancer. Herein, we determined the expression of
miR-96 and the function of its target genes in lung cancer through
bioinformatic analysis, aiming to ascertain whether it is a
potential molecular biomarker for the early diagnosis of NSCLC and
to obtain clues for the pathogenesis of lung cancer.
Materials and methods
Affymetrix microarray
The microRNA expression profiles of lung cancer
(GSE51855, GSE48414, GSE63805, GSE68951) were downloaded from Gene
Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/), which are based on
the platform of Affymetrix Human Genome U133 Plus 2.0 Array.
Probe re-annotation
Four TEX texts (GPL7341, GPL16770, GPL18410,
GPL16770) were downloaded from GEO public data platform, to find
the probe number of the hsa-miR-96 gene in GSE51855, GSE48414,
GSE63805, GSE68951, respectively.
Cell culture
The following cell lines were cultured individually
in RPMI-1640 medium (Invitrogen; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA): human lung adenocarcinoma (A549, NCI-H1299 and
pAa), human lung large cell carcinoma (NCI-H460), human lung
squamous cell carcinoma (NCI-H520), human lung small cell carcinoma
(NCI-H446), human lung giant-cell carcinoma (95D) and human
bronchial epithelial (16HBE) cell lines. The medium was
supplemented with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher
Scientific, Inc.), 100 U/ml penicillin and 100 mg/ml streptomycin
(Hyclone; GE Healthcare Life Sciences, Logan, UT, USA). Cells were
maintained in 5% CO2 at 37°C.
Real-time quantitative PCR
Specific RT primers and TaqMan probe (American ABI
Company) were used for quantitative detection of hsa-miR-96 (cat
no. A25576) and reference gene U6 (cat no. 4426961). Total RNAs in
cells were isolated using TRIzol reagent (Invitrogen, Carlsbad, CA,
USA). The RNA yield and the ratio of absorbance at 260 to 280 nm
(A260/A280 ratio) were determined with the NanoDrop 2000
spectrophotometer (NanoDrop Technologies, Montchanin, DE, USA). The
cDNA synthesis and qRT-PCR were carried out using the TaqMan
MicroRNA Reverse Transcription kit and TaqMan MicroRNA assays and
TaqMan® Universal Master Mix, No AmpErase®
UNG (all from ABI, USA), respectively, according to the
manufacturers protocol. qRT-PCR was carried out using Applied
Biosystems® 7500 real-time PCR systems (Applied
Biosystems, Foster City, CA, USA). The experiment was repeated 3
times. The relative quantitative analysis was carried out using the
ΔΔCt method and the control was used for normalization of miRNA
expression.
Bioinformatic analysis of miR-96
target genes
The target genes of miR-96 were predicted using
miRecords. The intersection prediction results from at least 6
miRNA target gene prediction databases were analyzed to reduce the
false-positive rate. To explore the functional annotation and
pathway enrichment of those predicted genes, the Gene Ontology (GO)
and Kyoto Encyclopedia of Genes and Genomes (KEGG) database
analyses were conducted using a Database for Annotation,
Visualization and Integrated Discovery (DAVID) v6.7 online analysis
tool with P<0.05 as the significant threshold to obtain
significant gene sets.
Statistical analysis
All data are presented as mean ± SD and statistical
analyses were processed using SPSS 16.0 statistical software.
Wilcoxon's rank-sum test was used to compare the expression of
miR-96 between lung cancer and normal lung tissues/plasma in
GSE51855, GSE48414 and GSE68951. Wilcoxon matched-pairs signed
ranks sum test was used to analyzed the miR-96 expression in
GSE63805. Wilcoxon's rank-sum test and Kruskal-Wallis test were
conducted to analyze the correlation of miR-96 expression with the
clinicopathological features in GSE48414 and GSE51855.
Independent-sample t-test was conducted to evaluate the miR-96
expression in lung cancer cell lines and 16HBE cells. A P-value of
<0.05 was considered statistically significant.
Results
Expression of miR-96 in lung cancer
tissues, plasma and cell lines
We analyzed four microRNA expression profiling
datasets to explore the expression pattern of miR-96 in the tissues
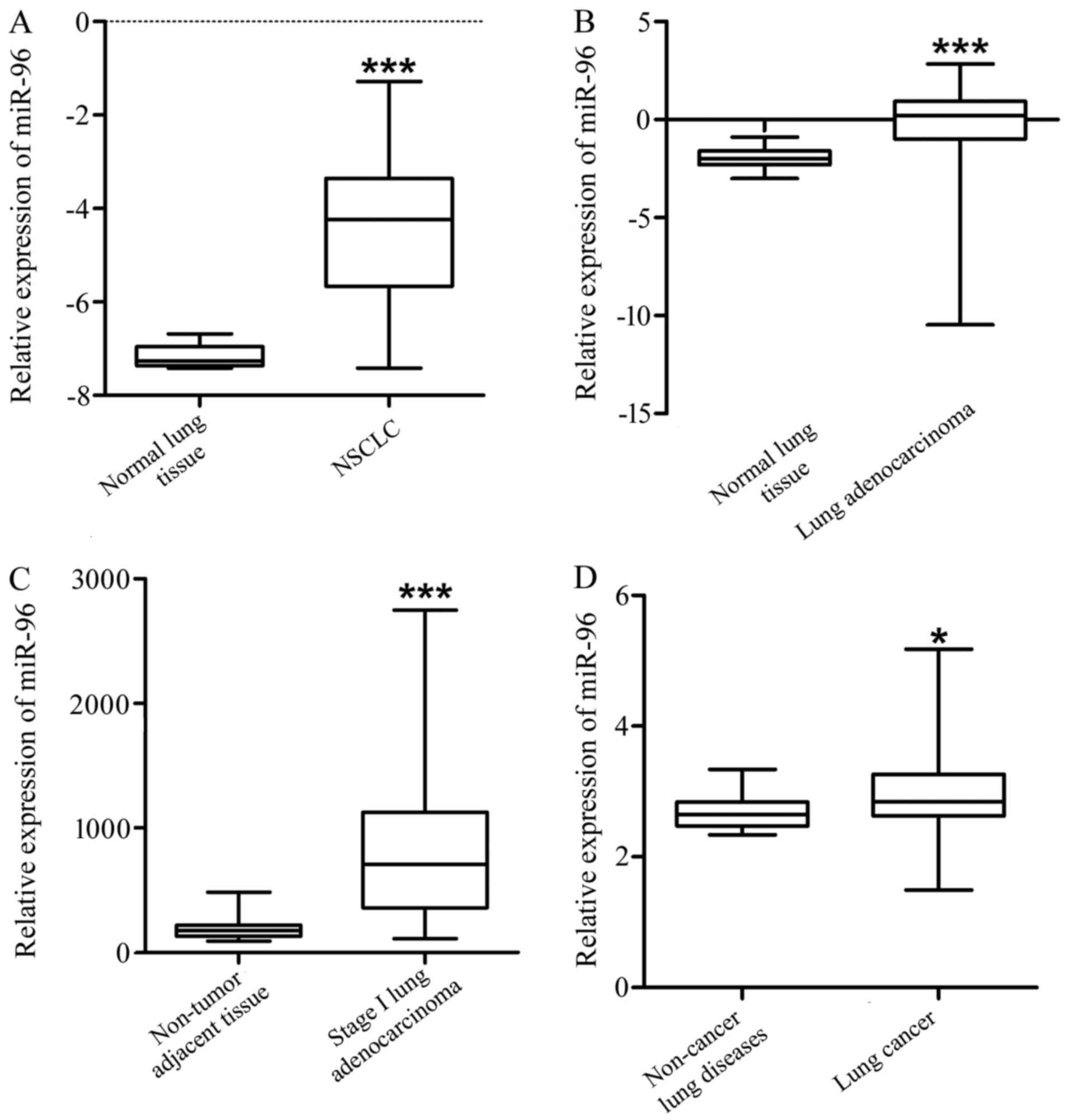
and plasma of lung cancer patients. The result indicate that,
compared with the normal lung tissues, miR-96 was significantly
increased in NSCLC (GSE51855, Fig.
1A and Table I, P<0.001),
lung adenocarcinoma (GSE48414, Fig.
1B and Table II, P<0.001)
and stage I adenocarcinoma tissues (GSE63805, Fig. 1C and Table III, P<0.001). In addition, the
expression level of miR-96 in the plasma (GSE68951) of the lung
cancer patients was significantly higher compared to that of the
non-cancer lung disease patients (Fig.
1D and Table IV,
P<0.05).
 | Table I.miR-96 expression in NSCLC and normal
lung tissues (GSE51855). |
Table I.
miR-96 expression in NSCLC and normal
lung tissues (GSE51855).
| Variables | N | Expression of
miR-96 | χ2/X | P-value |
|---|
| Total | 131 |
|
|
|
| NSCLC | 126 | −4.3623±0.12467 | −3.699 | 0.000 |
| NA | 5 |
−7.1807±0.12815 |
|
|
 | Table II.miR-96 expression in lung
adenocarcinoma and normal lung tissues (GSE48414). |
Table II.
miR-96 expression in lung
adenocarcinoma and normal lung tissues (GSE48414).
| Variables | N | Expression of
miR-96 |
χ2/Z | P-value |
|---|
| Total | 174 |
|
|
|
| ADC | 154 | −0.548±0.12819 | −5.804 | 0.000 |
| NA | 20 |
−1.9477±0.11705 |
|
|
 | Table III.miR-96 expression in adenocarcinoma
and adjacent non-tumor lung tissues (GSE63805). |
Table III.
miR-96 expression in adenocarcinoma
and adjacent non-tumor lung tissues (GSE63805).
| Variables | N | Expression of
miR-96 |
χ2/Z | P-value |
|---|
| Total | 62 |
|
|
|
| Stage I ADC | 32 | 849.71±114.63 | −4.638 | 0.000 |
| NA | 30 |
184.93±13.41056 |
|
|
 | Table IV.miR-96 expression in the serum of
NSCLC and non-cancerous pulmonary disease patients (GSE68951). |
Table IV.
miR-96 expression in the serum of
NSCLC and non-cancerous pulmonary disease patients (GSE68951).
| Variables | N | Expression of
miR-96 |
χ2/Z | P-value |
|---|
| Total | 38 |
|
|
|
| Lung cancer | 26 | 2.99409±0.0028 | −2.159 | 0.031 |
| Non-cancerous lung
diseases | 12 |
2.687812±0.0226 |
|
|
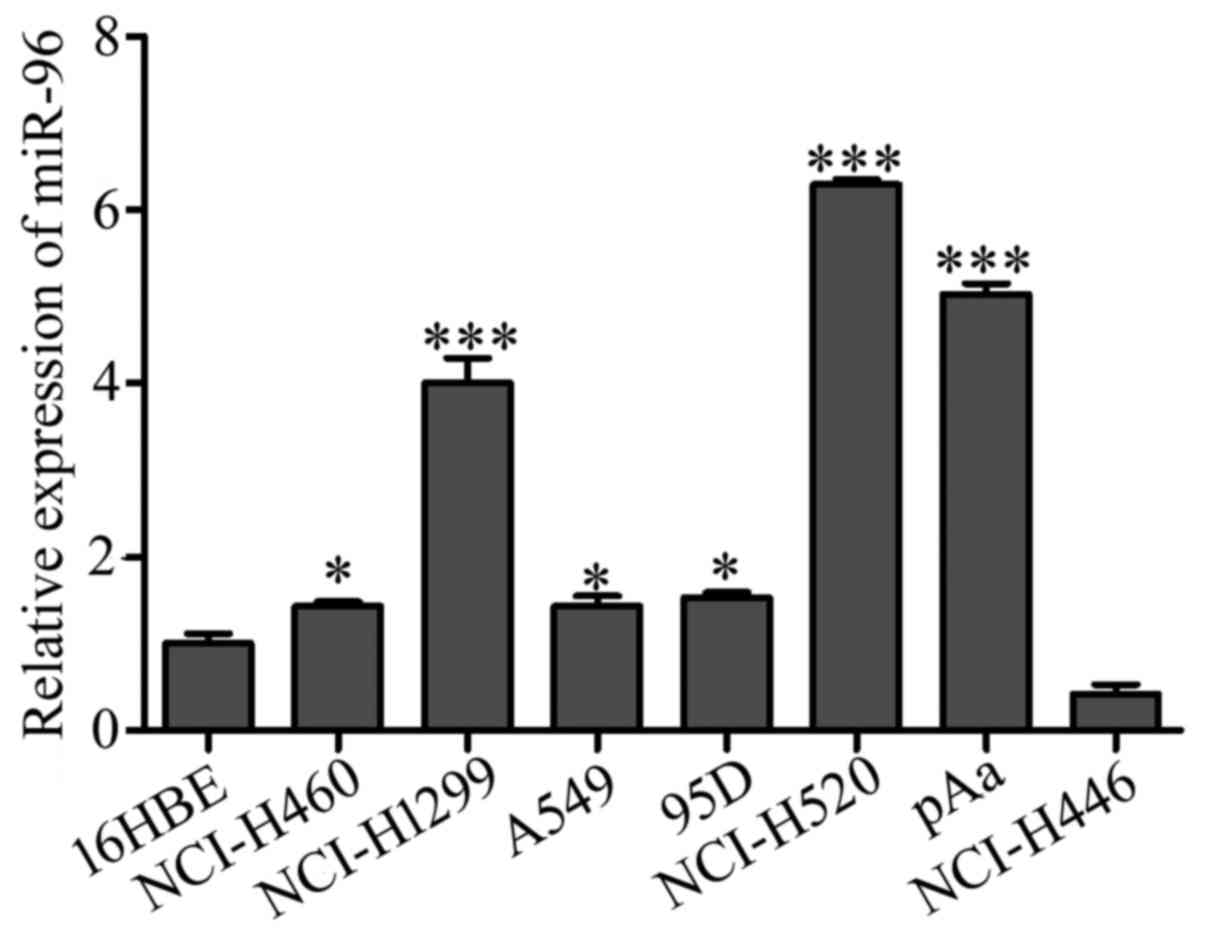
We subsequently examined the level of miR-96 in
different types of lung cancer cell lines and bronchial epithelial
16HBE cells using qRT-PCR. As shown in Fig. 2, the expression level of miR-96 was
elevated in all the 6 NSCLC cell lines but downregulated in the
small cell lung cancer NCI-H446 cells. The highest expression
levels for miR-96 were found in squamous cell carcinoma NCI-H520,
adenocarcinoma NCI-H1299 and pAa cells (P<0.001 for each).
Correlation between miR-96 expression
and clinicopathological features of NSCLC
We then analyzed the correlation between miR-96
expression and the clinicopathological features of NSCLC to further
explore the potential role of miR-96 in the development and
progression of lung cancer. Our results showed that the expression
level of miR-96 in the tumors was not related to the age (P=0.631),
sex (P=0.678), clinical stage (P=0.841) and histological subtype
(P=0.051) of the NSCLC patients (GSE48414 and GSE51855, Tables V and VI).
 | Table V.Correlation of miR-96 expression and
the clinicopathological characteristics of the lung adenocarcinoma
cases (GSE48414). |
Table V.
Correlation of miR-96 expression and
the clinicopathological characteristics of the lung adenocarcinoma
cases (GSE48414).
| Variables | N | Expression of
miR-96 |
χ2/Z | P-value |
|---|
| Total | 154 |
|
|
|
| Age (years) |
|
<65 | 68 |
−0.0922±0.15804 | −0.481 | 0.631 |
|
>65 | 86 |
−0.0244±0.14538 |
|
|
| Sex |
|
Male | 67 | 0.0161±0.18784 | −0.415 | 0.678 |
|
Female | 87 |
−0.1094±0.12377 |
|
|
| TNM
classification |
|
I+II | 126 | −0.069±0.14867 | 0.04 | 0.841 |
|
III+IV | 28 |
0.067259±0.04541 |
|
|
 | Table VI.Correlation of miR-96 expression and
the different histological types of NSCLC (GSE51855). |
Table VI.
Correlation of miR-96 expression and
the different histological types of NSCLC (GSE51855).
| Variables | N | Expression of
miR-96 |
χ2/X | P-value |
|---|
| Total | 126 |
|
|
|
| ADC | 76 |
−4.1497±0.15738 | 7.75 | 0.051 |
| SQC | 29 |
−4.8942±0.21236 |
|
|
| ASC | 4 |
−4.8262±0.74010 |
|
|
| LCC | 17 |
−4.3103±0.40235 |
|
|
Bioinformation analysis of miR-96
Prediction of miR-96 targets
Next, we used miRecords database to investigate
miR-96 targets. miRecords does not only provide the target gene
prediction of miRNAs but also the exact target genes regulated by
miRNAs, which have already been experimentally validated. As shown
in Table VII, a total of 71
target genes of miR-96 were predicted by at least six prediction
softwares involved in miRecords. Ten miR-96 target genes were found
and experimentally validated in the miRecords database, among which
ADCY6, IRS1 and MYRIP were also in the prediction list. Finally, 71
predicted and 7 validated miR-96 targets (Table VII) were involved in the GO and
KEGG analysis.
 | Table VII.The target genes of hsa-miR-96
investigated by miRecords database. |
Table VII.
The target genes of hsa-miR-96
investigated by miRecords database.
| No. | miRNA ID | Refseq | Symbol | Description | Note |
|---|
| 1 | hsa-miR-96 | NM_015270 | ADCY6 | Adenylate cyclase
6 | Predicted |
| 2 | hsa-miR-96 | NM_198715 | PTGER3 | Prostaglandin E
receptor 3 (subtype EP3) | Predicted |
| 3 | hsa-miR-96 | NM_016623 | FAM49B | Family with
sequence similarity 49, member B | Predicted |
| 4 | hsa-miR-96 | NM_016565 | CHCHD8 |
Coiled-coil-helix-coiled-coil-helix domain
containing 8 | Predicted |
| 5 | hsa-miR-96 | NM_015516 | TSKU | Tsukushin | Predicted |
| 6 | hsa-miR-96 | NM_015460 | MYRIP | Myosin VIIA and Rab
interacting protein | Predicted |
| 7 | hsa-miR-96 | NM_015215 | CAMTA1 | Calmodulin binding
transcription activator 1 | Predicted |
| 8 | hsa-miR-96 | NM_014946 | SPAST | Spastin | Predicted |
| 9 | hsa-miR-96 | NM_014943 | ZHX2 | Zinc fingers and
homeoboxes 2 | Predicted |
| 10 | hsa-miR-96 | NM_014839 | LPPR4 | Plasticity related
gene 1 | Predicted |
| 11 | hsa-miR-96 | NM_012300 | FBXW11 | F-box and WD repeat
domain containing 11 | Predicted |
| 12 | hsa-miR-96 | NM_012257 | HBP1 | HMG-box
transcription factor 1 | Predicted |
| 13 | hsa-miR-96 | NM_007198 | PROSC | Proline synthetase
co-transcribed homolog (bacterial) | Predicted |
| 14 | hsa-miR-96 | NM_006940 | SOX5 | SRY (sex
determining region Y)-box 5 | Predicted |
| 15 | hsa-miR-96 | NM_006861 | RAB35 | RAB35, member RAS
oncogene family | Predicted |
| 16 | hsa-miR-96 | NM_006791 | MORF4L1 | Mortality factor 4
like 1 | Predicted |
| 17 | hsa-miR-96 | NM_017974 | ATG16L1 | ATG16 autophagy
related 16-like 1 (S. cerevisiae) | Predicted |
| 18 | hsa-miR-96 | NM_018018 | SLC38A4 | Solute carrier
family 38, member 4 | Predicted |
| 19 | hsa-miR-96 | NM_018243 | 11-Sep | Septin 11 | Predicted |
| 20 | hsa-miR-96 | NM_198459 | DENND2C | DENN/MADD domain
containing 2C | Predicted |
| 21 | hsa-miR-96 | NM_194071 | CREB3L2 | cAMP responsive
element binding protein 3-like 2 | Predicted |
| 22 | hsa-miR-96 | NM_033505 | SELI | Selenoprotein
I | Predicted |
| 23 | hsa-miR-96 | NM_033260 | FOXQ1 | Forkhead box
Q1 | Predicted |
| 24 | hsa-miR-96 | NM_032560 | SMEK1 | SMEK homolog 1,
suppressor of mek1 (Dictyostelium) | Predicted |
| 25 | hsa-miR-96 | NM_032373 | PCGF5 | Polycomb group ring
finger 5 | Predicted |
| 26 | hsa-miR-96 | NM_032139 | ANKRD27 | Ankyrin repeat
domain 27 (VPS9 domain) | Predicted |
| 27 | hsa-miR-96 | NM_024915 | GRHL2 | Grainyhead-like 2
(Drosophila) | Predicted |
| 28 | hsa-miR-96 | NM_024811 | FLJ12529 | Pre-mRNA cleavage
factor I, 59 kDa subunit | Predicted |
| 29 | hsa-miR-96 | NM_022041 | GAN | Giant axonal
neuropathy (gigaxonin) | Predicted |
| 30 | hsa-miR-96 | NM_021229 | NTN4 | Netrin 4 | Predicted |
| 31 | hsa-miR-96 | NM_020871 | LRCH2 | Leucine-rich
repeats and calponin homology (CH) domain containing 2 | Predicted |
| 32 | hsa-miR-96 | NM_020795 | NLGN2 | Neuroligin 2 | Predicted |
| 33 | hsa-miR-96 | NM_020423 | SCYL3 | SCY1-like 3 (S.
cerevisiae) | Predicted |
| 34 | hsa-miR-96 | NM_020182 | PMEPA1 | Prostate
transmembrane protein, androgen induced 1 | Predicted |
| 35 | hsa-miR-96 | NM_006373 | VAT1 | Vesicle amine
transport protein 1 homolog (T. californica) | Predicted |
| 36 | hsa-miR-96 | NM_006283 | TACC1 | Transforming,
acidic coiled-coil containing protein 1 | Predicted |
| 37 | hsa-miR-96 | NM_006016 | CD164 | CD164 molecule,
sialomucin | Predicted |
| 38 | hsa-miR-96 | NM_002959 | SORT1 | Sortilin 1 | Predicted |
| 39 | hsa-miR-96 | NM_002923 | RGS2 | Regulator of
G-protein signaling 2, 24 kDa | Predicted |
| 40 | hsa-miR-96 | NM_002833 | PTPN9 | Protein tyrosine
phosphatase, non-receptor type 9 | Predicted |
| 41 | hsa-miR-96 | NM_002734 | PRKAR1A | Protein kinase,
cAMP-dependent, regulatory, type I, α (tissue specific extinguisher
1) | Predicted |
| 42 | hsa-miR-96 | NM_002515 | NOVA1 | Neuro-oncological
ventral antigen 1 | Predicted |
| 43 | hsa-miR-96 | NM_002265 | KPNB1 | Karyopherin
(importin) β 1 | Predicted |
| 44 | hsa-miR-96 | NM_002223 | ITPR2 | Inositol
1,4,5-triphosphate receptor, type 2 | Predicted |
| 45 | hsa-miR-96 | NM_002222 | ITPR1 | Inositol
1,4,5-triphosphate receptor, type 1 | Predicted |
| 46 | hsa-miR-96 | NM_002015 | FOXO1 | Forkhead box
O1 | Predicted |
| 47 | hsa-miR-96 | NM_001945 | HBEGF | Heparin-binding
EGF-like growth factor | Predicted |
| 47 | hsa-miR-96 | NM_001945 | HBEGF | Heparin-binding
EGF-like growth factor | Predicted |
| 48 | hsa-miR-96 | NM_001931 | DLAT | Dihydrolipoamide
S-acetyltransferase | Predicted |
| 49 | hsa-miR-96 | NM_001839 | CNN3 | Calponin 3,
acidic | Predicted |
| 50 | hsa-miR-96 | NM_000945 | PPP3R1 | Protein phosphatase
3 (formerly 2B), regulatory subunit B, α isoform | Predicted |
| 51 | hsa-miR-96 | NM_000935 | PLOD2 | Procollagen-lysine,
2-oxoglutarate 5-dioxygenase 2 | Predicted |
| 52 | hsa-miR-96 | NM_000663 | ABAT | 4-aminobutyrate
aminotransferase | Predicted |
| 53 | hsa-miR-96 | NM_002998 | SDC2 | Syndecan 2 | Predicted |
| 54 | hsa-miR-96 | NM_003060 | SLC22A5 | Solute carrier
family 22 (organic cation/carnitine transporter), member 5 | Predicted |
| 55 | hsa-miR-96 | NM_003182 | TAC1 | Tachykinin,
precursor 1 | Predicted |
| 56 | hsa-miR-96 | NM_006007 | ZFAND5 | Zinc finger,
AN1-type domain 5 | Predicted |
| 57 | hsa-miR-96 | NM_005766 | FARP1 | FERM, RhoGEF
(ARHGEF) and pleckstrin domain protein 1 (chondrocyte-derived) | Predicted |
| 58 | hsa-miR-96 | NM_005544 | IRS1 | Insulin receptor
substrate 1 | Predicted |
| 59 | hsa-miR-96 | NM_005502 | ABCA1 | ATP-binding
cassette, sub-family A (ABC1), member 1 | Predicted |
| 60 | hsa-miR-96 | NM_005400 | PRKCE | Protein kinase C,
ε | Predicted |
| 61 | hsa-miR-96 | NM_005277 | GPM6A | Glycoprotein
M6A | Predicted |
| 62 | hsa-miR-96 | NM_004985 | KRAS | v-Ki-ras2 Kirsten
rat sarcoma viral oncogene homolog | Predicted |
| 63 | hsa-miR-96 | NM_004958 | FRAP1 | FK506 binding
protein 12-rapamycin associated protein 1 | Predicted |
| 64 | hsa-miR-96 | NM_004926 | ZFP36L1 | Zinc finger protein
36, C3H type-like 1 | Predicted |
| 65 | hsa-miR-96 | NM_004731 | SLC16A7 | Solute carrier
family 16, member 7 (monocarboxylic acid transporter 2) | Predicted |
| 66 | hsa-miR-96 | NM_004514 | FOXK2 | Forkhead box
K2 | Predicted |
| 67 | hsa-miR-96 | NM_004481 | GALNT2 |
UDP-N-acetyl-α-D-galactosamine:polypeptide
N-acetylgalactosaminyltransferase 2 (GalNAc-T2) | Predicted |
| 68 | hsa-miR-96 | NM_003654 | CHST1 | Carbohydrate
(keratan sulfate Gal-6) sulfotransferase 1 | Predicted |
| 69 | hsa-miR-96 | NM_003379 | EZR | Ezrin | Predicted |
| 70 | hsa-miR-96 | NM_003342 | UBE2G1 |
Ubiquitin-conjugating enzyme E2G 1 (UBC7
homolog, yeast) | Predicted |
| 71 | hsa-miR-96 | NM_000332 | ATXN1 | Ataxin 1 | Predicted |
| 72 |
| NM_000863 | HTR1B |
| Validated |
| 73 |
| NM_198159 | MITF |
| Validated |
| 74 |
| NM_002015.3 | Foxo1 |
| Validated |
| 75 |
| NM_0016513 | AQp5 |
| Validated |
| 76 |
| NM_001408 | CELSR2 |
| Validated |
| 77 |
| NM_153437 | ODF2 |
| Validated |
| 78 |
| NM_001005861 | RYK |
| Validated |
Gene ontology and KEGG pathway enrichment
analysis of miR-96 target genes
Gene ontology enrichment analysis was performed to
analyze 78 miR-96 target genes (Table
VII). In total, 42 GO terms were obtained, which included 24
biological processes, 15 cellular components and 3 molecular
functions. These 42 GO terms were sorted by P-values for further
analysis and are listed in Tables
VIII and IX.
 | Table VIII.GO gene function (biological_process)
analysis of the miR-96 targets. |
Table VIII.
GO gene function (biological_process)
analysis of the miR-96 targets.
| GO ID | GO ontology | GO term | Counts | P-value |
|---|
| GO:0009725 |
Biological_process | Response to hormone
stimulus | 9 | 3.95E-04 |
| GO:0009719 |
Biological_process | Response to
endogenous stimulus | 9 | 7.57E-04 |
| GO:0010033 |
Biological_process | Response to organic
substance | 11 | 0.002442 |
| GO:0016197 |
Biological_process | Endosome
transport | 4 | 0.002826 |
| GO:0032228 |
Biological_process | Regulation of
synaptic transmission, GABAergic | 3 | 0.003041 |
| GO:0016055 |
Biological_process | Wnt receptor
signaling pathway | 5 | 0.004019 |
| GO:0032868 |
Biological_process | Response to insulin
stimulus | 4 | 0.012812 |
| GO:0044057 |
Biological_process | Regulation of
system process | 6 | 0.0173 |
| GO:0007169 |
Biological_process | Transmembrane
receptor protein tyrosine kinase signaling pathway | 5 | 0.023729 |
| GO:0042325 |
Biological_process | Regulation of
phosphorylation | 7 | 0.025784 |
| GO:0032870 |
Biological_process | Cellular response
to hormone stimulus | 4 | 0.027124 |
| GO:0001666 |
Biological_process | Response to
hypoxia | 4 | 0.027651 |
| GO:0043279 |
Biological_process | Response to
alkaloid | 3 | 0.028506 |
| GO:0050804 |
Biological_process | Regulation of
synaptic transmission | 4 | 0.028721 |
| GO:0051174 |
Biological_process | Regulation of
phosphorus metabolic process | 7 | 0.030557 |
| GO:0019220 |
Biological_process | Regulation of
phosphate metabolic process | 7 | 0.030557 |
| GO:0070482 |
Biological_process | Response to oxygen
levels | 4 | 0.03149 |
| GO:0010648 |
Biological_process | Negative regulation
of cell communication | 5 | 0.032803 |
| GO:0007612 |
Biological_process | Learning | 3 | 0.03461 |
| GO:0051969 |
Biological_process | Regulation of
transmission of nerve impulse | 4 | 0.034993 |
| GO:0031998 |
Biological_process | Regulation of fatty
acid beta-oxidation | 2 | 0.03838 |
| GO:0031644 |
Biological_process | Regulation of
neurological system process | 4 | 0.038689 |
| GO:0043434 |
Biological_process | Response to peptide
hormone stimulus | 4 | 0.039324 |
| GO:0046907 |
Biological_process | Intracellular
transport | 8 | 0.040338 |
 | Table IX.GO gene function (cellular_component)
analysis of the miR-96 targets. |
Table IX.
GO gene function (cellular_component)
analysis of the miR-96 targets.
| GO ID | GO ontology | Term | Count | P-value |
|---|
| GO:0042995 |
Cellular_component | Cell
projection | 11 | 9.15E-04 |
| GO:0005815 |
Cellular_component | Microtubule
organizing center | 6 | 0.005289516 |
| GO:0005624 |
Cellular_component | Membrane
fraction | 10 | 0.009055844 |
| GO:0005626 |
Cellular_component | Insoluble
fraction | 10 | 0.011347551 |
| GO:0031095 |
Cellular_component | Platelet dense
tubular network membrane | 2 | 0.013319656 |
| GO:0005813 |
Cellular_component | Centrosome | 5 | 0.017582227 |
| GO:0031094 |
Cellular_component | Platelet dense
tubular network | 2 | 0.017720687 |
| GO:0000267 |
Cellular_component | Cell fraction | 11 | 0.020267702 |
| GO:0044463 |
Cellular_component | Cell projection
part | 5 | 0.02029103 |
| GO:0045202 |
Cellular_component | Synapse | 6 | 0.020676761 |
| GO:0012505 |
Cellular_component | Endomembrane
system | 9 | 0.021888271 |
| GO:0005955 |
Cellular_component | Calcineurin
complex | 2 | 0.022102431 |
| GO:0044430 |
Cellular_component | Cytoskeletal
part | 10 | 0.024057867 |
| GO:0045121 |
Cellular_component | Membrane raft | 4 | 0.025855924 |
| GO:0005856 |
Cellular_component | Cytoskeleton | 12 | 0.039405823 |
Among the 24 biological process GO terms, the top 10
terms were: GO:0009725 (response to hormone stimulus), GO:0009719
(response to endogenous stimulus), GO:0010033 (response to organic
substance), GO:0016197 (endosome transport), GO:0032228 (regulation
of synaptic transmission, GABAergic), GO:0016055 (Wnt receptor
signaling pathway), GO:0032868 (response to insulin stimulus),
GO:0044057 (regulation of system process), GO:0007169
(transmembrane receptor protein tyrosine kinase signaling pathway)
and GO:0042325 (regulation of phosphorylation) (Table VIII).
The 15 cellular component GO terms were: GO:0042995
(cell projection), GO:0005815 (microtubule organizing center),
GO:0005624 (membrane fraction), GO:0005626 (insoluble fraction),
GO:0031095 (platelet dense tubular network membrane), GO:0005813
(centrosome), GO:0031094 (platelet dense tubular network),
GO:0000267 (cell fraction), GO:0044463 (cell projection part),
GO:0045202 (synapse), GO:0012505 (endomembrane system), GO:0005955
(calcineurin complex), GO: 0044430 (cytoskeletal part), GO:0045121
(membrane raft) and GO:0005856 (cytoskeleton) (Table IX).
In regards to the molecular function of the GO
terms, GO:0005220 (inositol 1,4,5-trisphosphate-sensitive
calcium-release channel activity), GO:0008095 (inositol-1,4,5-
trisphosphate receptor activity) and GO:0005516 (calmodulin
binding) were the highest presented terms (Table X).
 | Table X.GO gene function (molecular_function)
analysis of the miR-96 targets. |
Table X.
GO gene function (molecular_function)
analysis of the miR-96 targets.
| GO ID | GO ontology | Term | Count | P-value |
|---|
| GO:0005220 |
Molecular_function | Inositol
1,4,5-trisphosphate-sensitive calcium-release channel activity | 2 | 0.01449 |
| GO:0008095 |
Molecular_function |
Inositol-1,4,5-trisphosphate receptor
activity | 2 | 0.01927 |
| GO:0005516 |
Molecular_function | Calmodulin
binding | 4 | 0.03047 |
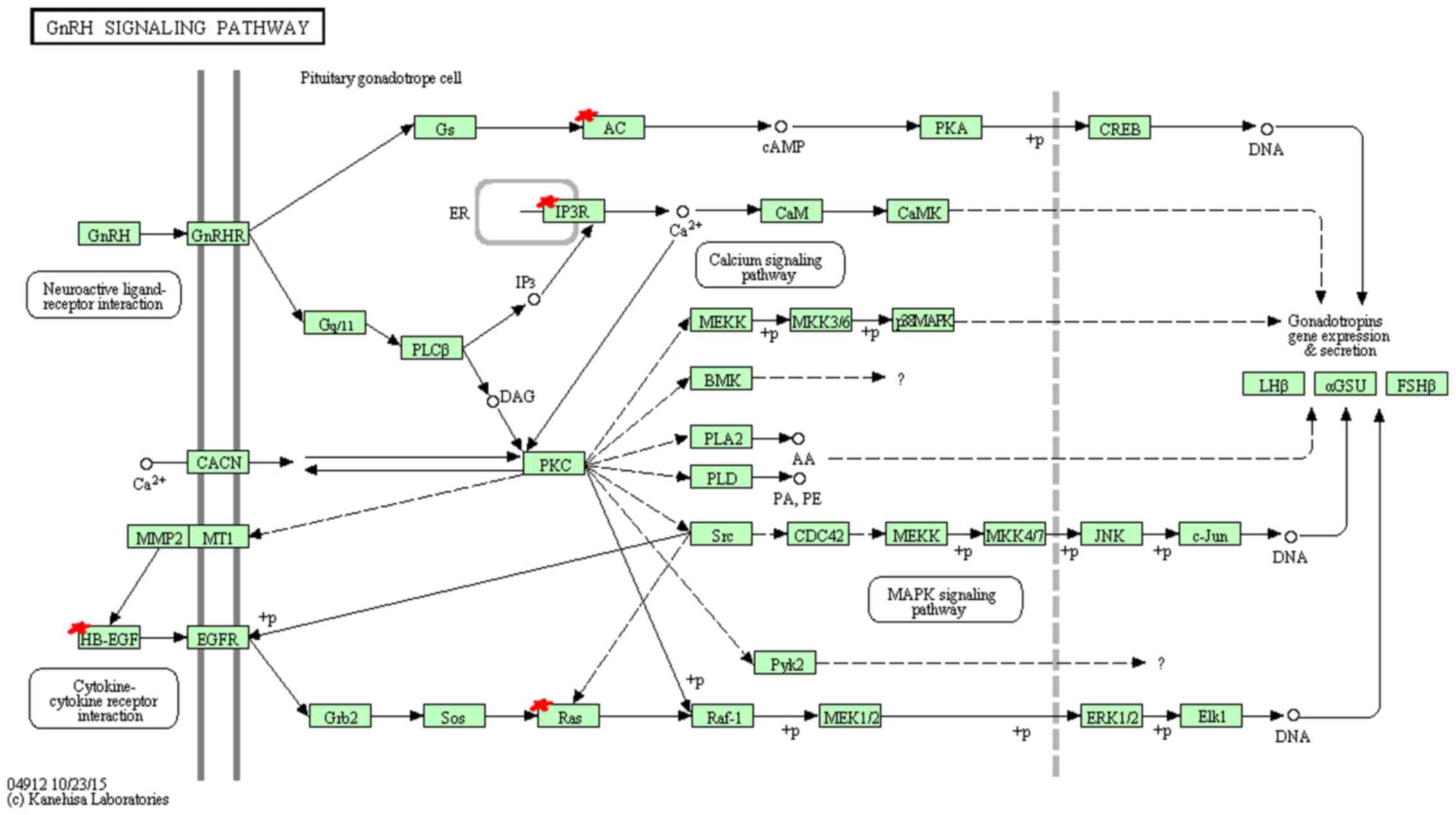
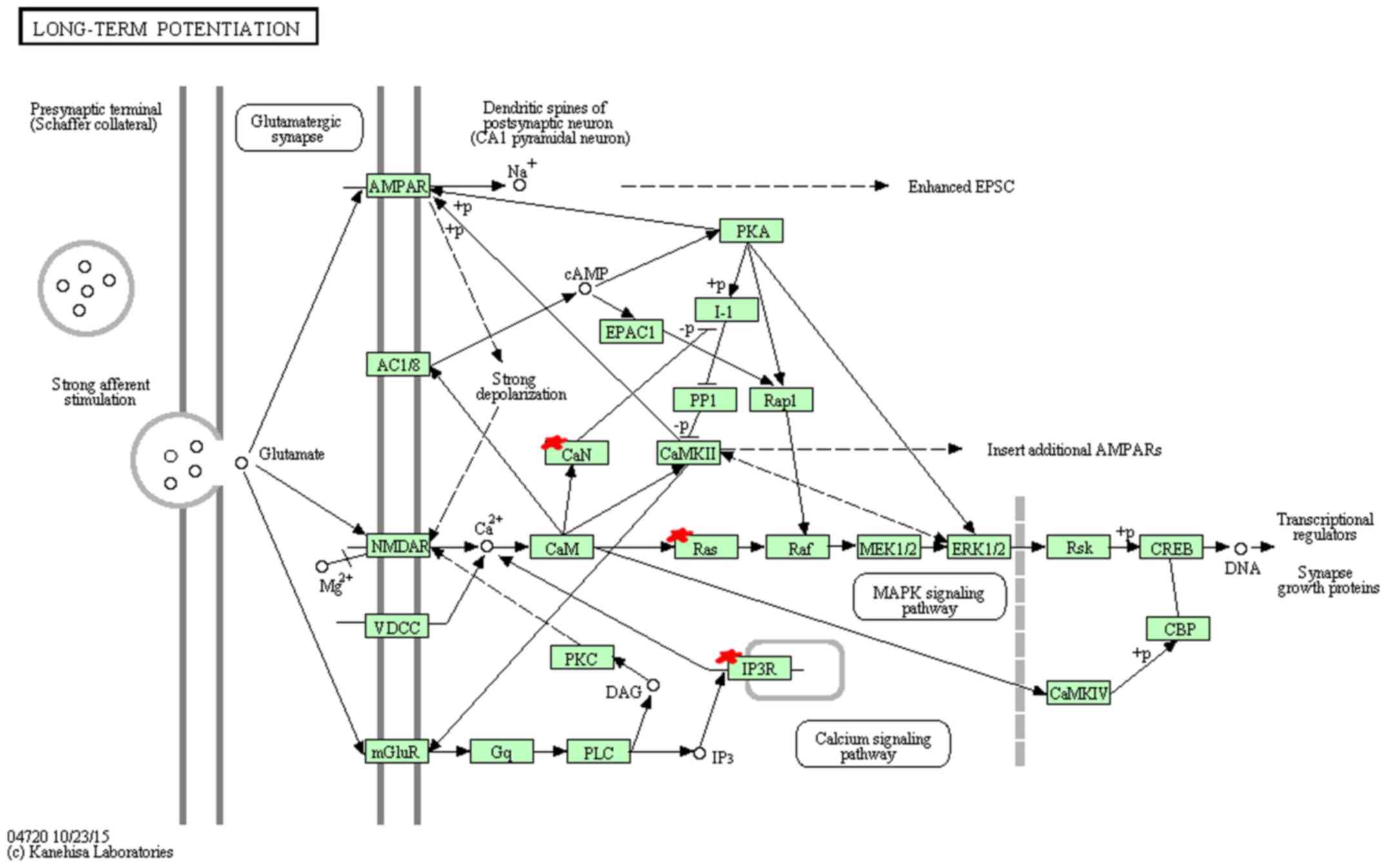
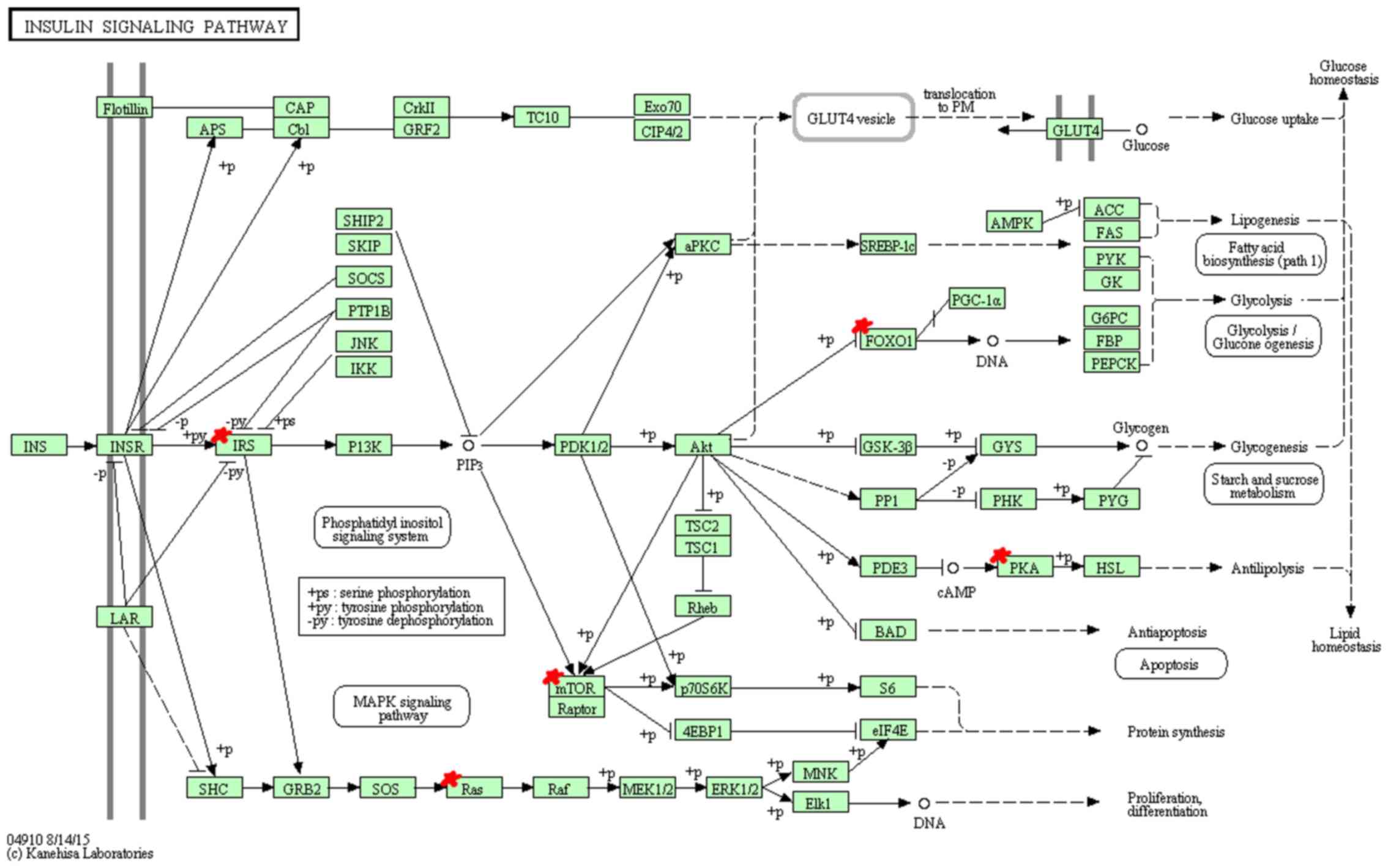
KEGG pathway analysis indicated that miR-96 target
genes are mainly enriched in 9 pathways (Table XI). Among these pathways, hsa04912
(GnRH signaling pathway) (Fig. 3),
hsa04114 (oocyte meiosis), hsa04720 (long-term potentiation)
(Fig. 4), hsa04910 (insulin
signaling pathway) (Fig. 5),
hsa05215 (prostate cancer) and hsa04540 (gap junction) showed
significantly higher enrichment, followed by hsa04916
(melanogenesis), hsa04270 (vascular smooth muscle contraction) and
hsa04930 (Type II diabetes mellitus).
 | Table XI.Pathways enrichment analysis of the
miR-96 tagets (KEGG). |
Table XI.
Pathways enrichment analysis of the
miR-96 tagets (KEGG).
| GO ID | Name of
pathway | Count | P-value | Genes |
|---|
| hsa04912 | GnRH signaling
pathway | 5 | 0.001862149 | NM_004985,
NM_002223, NM_002222, NM_015270, NM_001945 |
| hsa04114 | Oocyte meiosis | 5 | 0.002843987 | NM_012300,
NM_002223, NM_002222, NM_015270, NM_000945 |
| hsa04720 | Long-term
potentiation | 4 | 0.005899778 | NM_004985,
NM_002223, NM_002222, NM_000945 |
| hsa04910 | Insulin signaling
pathway | 5 | 0.005930538 | NM_004958,
NM_002015, NM_004985, NM_005544, NM_002734 |
| hsa05215 | Prostate
cancer | 4 | 0.01237797 | NM_004958,
NM_002015, NM_004985, NM_194071 |
| hsa04540 | Gap junction | 4 | 0.01237797 | NM_004985,
NM_002223, NM_002222, NM_015270 |
| hsa04916 | Melanogenesis | 4 | 0.016481296 | NM_004985,
NM_015270, NM_198159, NM_194071 |
| hsa04270 | Vascular smooth
muscle contraction | 4 | 0.02283774 | NM_002223,
NM_002222, NM_015270, NM_005400 |
| hsa04930 | Type II diabetes
mellitus | 3 | 0.027134231 | NM_004958,
NM_005544, NM_005400 |
Discussion
Owing to its elevated expression, much effort has
been dedicated to study the role of miR-96 in various types of
cancers (21–23). In the majority of the tumors, miR-96
acts as an oncogene to promote the proliferation and invasion of
cancer cells by inhibiting transcription factor FOXO1 (24), FOXO3a (25), tumor suppressor protein RECK
(26) and metastasis suppressor
protein MTSS1 (27). However, in
pancreatic cancer, miR-96 functions as a tumor suppressor by
targeting HERG1 and NUAK1 (28,29).
There is no explicit conclusion whether miR-96 could
affect the development and progression of lung cancer and serve as
a molecular biomarker for the clinical diagnosis of lung cancer. By
analyzing four microRNA expression profiles and qRT-PCR, we showed
that miR-96 was markedly increased in NSCLC, lung adenocarcinoma,
stage I adenocarcinoma tissues and NSCLC cell lines. Consistent
with our result, Ma et al reported that miR-96 was
significantly upregulated in six NSCLC tissues and its expression
was then validated in an independent set of 35 pairs of tumors and
their adjacent normal tissues as well as in the serum of patients
with NSCLC (19). To verify the
microRNA expression signatures of lung cancer, Vosa et al
performed a comprehensive meta-analysis of 20 published microRNA
expression studies in lung cancer and identified a statistically
significant microRNA meta-signature of seven upregulated microRNAs,
including miR-21, miR-210, miR-182, miR-183, miR-31, miR-200b and
miR-205. Since miR-182, miR-183 and miR-96 all belong to the
miR-183 family, in conjunction with our results, miR-96 may serve
as a novel molecular biomarker to distinguish early NSCLC patients
from healthy individuals.
miRNAs are present not only in tissues but also in
body fluids, such as blood, plasma, serum and sputum. Shen et
al conducted several studies to assess the function of miRNAs
in the sputum and plasma of lung cancer patients (30). They showed that the expression
profile of plasma miR-21, miR-126, miR-210 and miR-486-5p produce
high sensitivity and specificity in identifying stage I NSCLC
patients. Zhu et al examined 70 pairs of lung cancer and
non-cancerous tissues as well as serum samples. They found that
miR-96 expression in tumors was positively associated with its
expression in serum (31). Our data
revealed that miR-96 expression in the plasma of lung cancer was
significantly higher compared to that of non-cancer lung disease
patients, suggesting that miR-96 may serve as a potential
non-invasive marker for lung cancer diagnosis.
Although studies have shown that miR-96 is
associated with poor overall survival in patients with pancreatic
cancer (32), liver cancer
(33) and colorectal cancer
(34), our results did not
demonstrate any significant correlation between the expression
level of miR-96 and clinical stage as well as the histological
subtype of the NSCLC patients. These discrepancies may be due to
the different samples and databases that were used. Further studies
are needed to confirm whether miR-96 could serve as a prognostic
biomarker for lung cancer.
To date, computational methods have been widely used
for the prediction of miRNAs and their target genes. However, the
most commonly used miRNA target prediction websites, such as
TargetScan, microRNA.org and PicTar, could not yield
consistent results due to their different algorithm. miRecords is
an integrated microRNA target database which includes a total of 11
established prediction programs. In this study, we selected the
results predicted by at least six softwares in miRecords as the
putative miR-96 target gene set and a collection of 78 predicted
target genes were involved in GO/KEGG functional enrichment
analysis. Since the GO hierarchy contains an added complexity by
allowing terms to have multiple parents or ascendants, we used
Fishers exact 0.01 to reduce the redundancy in lists of enriched GO
terms. Our data showed that among the 24 biological process GO
terms obtained, the top 10 terms could be roughly grouped into
several different categories including response to the stimulus
(GO:0009725, GO:0009719, GO:0010033 and GO:0032868), signaling
pathway (GO:0016055, GO:0032868, GO:0007169) and neurotransmission
(GO:0032228). Tyrosine kinase signaling (GO:0007169) is currently
known as the most successful molecular-targeted therapeutic
approach for lung cancer (35). The
canonical Wnt signaling pathway (GO:0016055), is another important
regulator of proliferation (36)
and metastasis (37) of non-small
lung cancer cells. In addition, the 15 cellular component GO terms
were significantly enriched in various specific processes with high
frequency, such as cell division (GO:0005815, GO:0005813), cell
communication (GO:0042995, GO:0044463) and cell migration
(GO:0042995, GO:0044463, GO:0044430, GO:0005856), indicating that
miR-96 may function as a regulator for the motility, migration and
invasion of tumor cells. Moreover, three highly enriched molecular
function GO terms (GO: 0005220, GO:0008095, and GO:0005516) suggest
a potential new role of miR-96 in regulating calcium signaling
important for tumor cell proliferation, apoptosis and
migration.
In the KEGG annotation, GnRH signaling pathway
(hsa04912), oocyte meiosis (hsa04114), long-term potentiation
(hsa04720), insulin signaling pathway (hsa04910) and prostate
cancer (hsa05215) showed the highest enrichment. GnRH has been
reported to participate in the self-renewal of A549-derived lung
cancer stem-like cells by upregulating the JNK signaling pathway
(38). Insulin, bound to insulin
receptor, promotes cell proliferation through the RAS-RAF-MAP
kinase signaling pathway and regulates cell survival process
through (PI3K)-Akt-mammalian target of rapamycin (mTOR) pathway,
playing an important role in the clinical treatment of NSCLC
(39). Long-term potentiation and
prostate cancer pathway, related to transcription regulation,
cancer cell survival and proliferation respectively, suggest the
potential function for miR-96 in cancer growth.
Although DAL-1 was not in the list of the 78
targets, DAL-1 was predicted as the target gene of miR-96 by 5
predicted databases of miRecords: MirTarget2, PicTar, PITA,
RNAhybird, and TargetScan/TargetScanS (data not shown). For future
studies, comprehensive screening, confirmation experiments and
further bioinformatic analysis using available web tools such as
Ingenuity Pathway Analysis (IPA) and STRINGProtein-Protein
Interaction Networks need to be carried out on the predicted
targets to explore the novel regulatory mechanism of miR-96 in
cancer metastasis.
In conclusion, our results showed that miR-96,
functioning as an oncogene, may play an important role in the
development and progression of lung cancer. Both in tissue and
plasma, miR-96 may have the potential to serve as a molecular
biomarker for the early diagnosis of NSCLC.
Acknowledgements
This study was funded by the National Nature Science
Foundation of China (no. 81401391), Ph.D. Programs Foundation of
the Ministry of Education of China (no. 20134423110001); National
Nature Science Foundation of Guangdong Province
(no.S2012010010181); Science and Technology Project of Guangzhou
City (no. 2014Y2-00171) and Education System Innovative Academic
Team of Guangzhou City (no. 13C06); Guangzhou City-Belonged
Universities Scientific Research Program (no. 2012C130); National
Natural Science Foundation of Guangdong Province (no.
2015A030313452).
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Conte I, Banfi S and Bovolenta P:
Non-coding RNAs in the development of sensory organs and related
diseases. Cell Mol Life Sci. 70:4141–4155. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Mitchell PS, Parkin RK, Kroh EM, Fritz BR,
Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O'Briant
KC, Allen A, et al: Circulating microRNAs as stable blood-based
markers for cancer detection. Proc Natl Acad Sci USA.
105:10513–10518. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Weber JA, Baxter DH, Zhang S, Huang DY,
Huang KH, Lee MJ, Galas DJ and Wang K: The microRNA spectrum in 12
body fluids. Clin Chem. 56:1733–1741. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Boutros PC, Lau SK, Pintilie M, Liu N,
Shepherd FA, Der SD, Tsao MS, Penn LZ and Jurisica I: Prognostic
gene signatures for non-small-cell lung cancer. Proc Natl Acad Sci
USA. 106:2824–2828. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Roepman P, Jassem J, Smit EF, Muley T,
Niklinski J, van de Velde T, Witteveen AT, Rzyman W, Floore A,
Burgers S, et al: An immune response enriched 72-gene prognostic
profile for early-stage non-small-cell lung cancer. Clin Cancer
Res. 15:284–290. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Chen X, Guan X, Zhang H, Xie X, Wang H,
Long J, Cai T, Li S, Liu Z and Zhang Y: DAL-1 attenuates
epithelial-to mesenchymal transition in lung cancer. J Exp Clin
Cancer Res. 34:32015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Liang H, Yan X, Pan Y, Wang Y, Wang N, Li
L, Liu Y, Chen X, Zhang CY, Gu H, et al: MicroRNA-223 delivered by
platelet-derived microvesicles promotes lung cancer cell invasion
via targeting tumor suppressor EPB41L3. Mol Cancer. 14:582015.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Li X, Zhang Y, Zhang H, Liu X, Gong T, Li
M, Sun L, Ji G, Shi Y, Han Z, et al: miRNA-223 promotes gastric
cancer invasion and metastasis by targeting tumor suppressor
EPB41L3. Mol Cancer Res. 9:824–833. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Cai T, Guan X, Wang H, Fang Y, Long J, Xie
X, et al: miR-26a regulates ANXA1, rather than DAL-1, in the
development of lung cancer. Oncol Lett (accepted).
|
|
11
|
Lin J, Zhang L, Huang H, Huang Y, Huang L,
Wang J, Huang S, He L, Zhou Y, Jia W, et al: miR-26b/KPNA2 axis
inhibits epithelial ovarian carcinoma proliferation and metastasis
through downregulating OCT4. Oncotarget. 6:23793–23806. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Shen G, Lin Y, Yang X, Zhang J, Xu Z and
Jia H: MicroRNA-26b inhibits epithelial-mesenchymal transition in
hepatocellular carcinoma by targeting USP9X. BMC Cancer.
14:3932014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang C, Tong J and Huang G: Nicotinamide
phosphoribosyl transferase (Nampt) is a target of microRNA-26b in
colorectal cancer cells. PLoS One. 8:e699632013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Pierce ML, Weston MD, Fritzsch B, Gabel
HW, Ruvkun G and Soukup GA: MicroRNA-183 family conservation and
ciliated neurosensory organ expression. Evol Dev. 10:106–113. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kuhn S, Johnson SL, Furness DN, Chen J,
Ingham N, Hilton JM, Steffes G, Lewis MA, Zampini V, Hackney CM, et
al: miR-96 regulates the progression of differentiation in
mammalian cochlear inner and outer hair cells. Proc Natl Acad Sci
USA. 108:2355–2360. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Guo Y, Liu H, Zhang H, Shang C and Song Y:
miR-96 regulates FOXO1-mediated cell apoptosis in bladder cancer.
Oncol Lett. 4:561–565. 2012.PubMed/NCBI
|
|
17
|
Yu N, Fu S, Liu Y, Xu Z, Liu Y, Hao J,
Wang B and Zhang A: miR-96 suppresses renal cell carcinoma invasion
via downregulation of Ezrin expression. J Exp Clin Cancer Res.
34:1072015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li C, Du X, Tai S, Zhong X, Wang Z, Hu Z,
Zhang L, Kang P, Ji D, Jiang X, et al: GPC1 regulated by miR-96-5p,
rather than miR-182-5p, in inhibition of pancreatic carcinoma cell
proliferation. Int J Mol Sci. 15:6314–6327. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ma L, Huang Y, Zhu W, Zhou S, Zhou J, Zeng
F, Liu X, Zhang Y and Yu J: An integrated analysis of miRNA and
mRNA expressions in non-small cell lung cancers. PLoS One.
6:e265022011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Guo H, Li Q, Li W, Zheng T, Zhao S and Liu
Z: miR-96 downregulates RECK to promote growth and motility of
non-small cell lung cancer cells. Mol Cell Biochem. 390:155–160.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wu Z, Liu K, Wang Y, Xu Z, Meng J and Gu
S: Upregulation of microRNA-96 and its oncogenic functions by
targeting CDKN1A in bladder cancer. Cancer Cell Int. 15:1072015.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Fendler A, Jung M, Stephan C, Erbersdobler
A, Jung K and Yousef GM: The antiapoptotic function of miR-96 in
prostate cancer by inhibition of FOXO1. PLoS One. 8:e808072013.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lin H, Dai T, Xiong H, Zhao X, Chen X, Yu
C, Li J, Wang X and Song L: Unregulated miR-96 induces cell
proliferation in human breast cancer by downregulating
transcriptional factor FOXO3a. PLoS One. 5:e157972010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Song HM, Luo Y, Li D-F, Wei CK, Hua KY,
Song JL, Xu H, Maskey N and Fang L: MicroRNA-96 plays an oncogenic
role by targeting FOXO1 and regulating AKT/FOXO1/Bim pathway in
papillary thyroid carcinoma cells. Int J Clin Exp Pathol.
8:9889–9900. 2015.PubMed/NCBI
|
|
25
|
Gao F and Wang W: MicroRNA-96 promotes the
proliferation of colorectal cancer cells and targets tumor protein
p53 inducible nuclear protein 1, forkhead box protein O1 (FOXO1)
and FOXO3a. Mol Med Rep. 11:1200–1206. 2015.PubMed/NCBI
|
|
26
|
Zhang J, Kong X, Li J, Luo Q, Li X, Shen
L, Chen L and Fang L: miR-96 promotes tumor proliferation and
invasion by targeting RECK in breast cancer. Oncol Rep.
31:1357–1363. 2014.PubMed/NCBI
|
|
27
|
Xu L, Zhong J, Guo B, Zhu Q, Liang H, Wen
N, Yun W and Zhang L: miR-96 promotes the growth of prostate
carcinoma cells by suppressing MTSS1. Tumour Biol. 37:12023–12032.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Feng J, Yu J, Pan X, Li Z, Chen Z, Zhang
W, Wang B, Yang L, Xu H, Zhang G, et al: HERG1 functions as an
oncogene in pancreatic cancer and is downregulated by miR-96.
Oncotarget. 5:5832–5844. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Huang X, Lv W, Zhang JH and Lu DL: miR 96
functions as a tumor suppressor gene by targeting NUAK1 in
pancreatic cancer. Int J Mol Med. 34:1599–1605. 2014.PubMed/NCBI
|
|
30
|
Shen J, Liu Z, Todd NW, Zhang H, Liao J,
Yu L, Guarnera MA, Li R, Cai L, Zhan M, et al: Diagnosis of lung
cancer in individuals with solitary pulmonary nodules by plasma
microRNA biomarkers. BMC Cancer. 11:3742011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhu W, Liu X, He J, Chen D, Hunag Y and
Zhang YK: Overexpression of members of the microRNA-183 family is a
risk factor for lung cancer: A case control study. BMC Cancer.
11:3932011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Ebrahimi S, Hosseini M, Ghasemi F,
Shahidsales S, Maftouh M, Akbarzade H, Parizadeh SA, Hassanian SM
and Avan A: Circulating microRNAs as potential diagnostic,
prognostic and therapeutic targets in pancreatic cancer. Curr Pharm
Des. 22:6444–6450. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Leung WK, He M, Chan AW, Law PT and Wong
N: Wnt/β-catenin activates miR-183/96/182 expression in
hepatocellular carcinoma that promotes cell invasion. Cancer Lett.
362:97–105. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Sun Y, Liu Y, Cogdell D, Calin GA, Sun B,
Kopetz S, Hamilton SR and Zhang W: Examining plasma microRNA
markers for colorectal cancer at different stages. Oncotarget.
7:11434–11449. 2016.PubMed/NCBI
|
|
35
|
Kobayashi S, Boggon TJ, Dayaram T, Jänne
PA, Kocher O, Meyerson M, Johnson BE, Eck MJ, Tenen DG and Halmos
B: EGFR mutation and resistance of non-small-cell lung cancer to
gefitinib. N Engl J Med. 352:786–792. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Huang C, Ma R, Xu Y, Li N, Li Z, Yue J, Li
H, Guo Y and Qi D: Wnt2 promotes non-small cell lung cancer
progression by activating WNT/β-catenin pathway. Am J Cancer Res.
5:1032–1046. 2015.PubMed/NCBI
|
|
37
|
Chen X, Song X, Yue W, Chen D, Yu J, Yao Z
and Zhang L: Fibulin-5 inhibits Wnt/β-catenin signaling in lung
cancer. Oncotarget. 6:15022–15034. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Lu C, Huang T, Chen W and Lu H: GnRH
participates in the self-renewal of A549-derived lung cancer
stem-like cells through upregulation of the JNK signaling pathway.
Oncol Rep. 34:244–250. 2015.PubMed/NCBI
|
|
39
|
Scagliotti GV and Novello S: The role of
the insulin-like growth factor signaling pathway in non-small cell
lung cancer and other solid tumors. Cancer Treat Rev. 38:292–302.
2012. View Article : Google Scholar : PubMed/NCBI
|