Introduction
The p53 tumor suppressor mediates an adequate
reaction of cells to stress. It functions mainly as a transcription
factor responding to various stress stimuli by controlling
expression of its target genes. The p53 protein participates
in control of cell cycle, DNA repair, apoptosis, senescence, cell
metabolism and maintaining of genome integrity (1,2).
Mutations and deletions of the TP53 gene are the most
frequent genetic alterations detected in human tumors, though they
are rather less frequent in lymphomas (3,4).
Recently, the revision of the World Health
Organization (WHO) classification of lymphoid neoplasms was
published reflecting and summarizing new achievements in the field.
The increasing impact of cytogenetic and molecular approaches,
including next-generation sequencing (NGS), was emphasized as they
incessantly bring new insights and deepen understanding of the
lymphoma development, and thus improve diagnosis, prognosis and
treatment of this disease. The biologically relevant subgrouping of
heterogeneous diffuse large B-cell lymphomas (DLBCL) and
appropriate recognition and classification of double/triple-hit
lymphomas (DHL/THL) belong to the highlighted diagnostic topics
among mature B-cell lymphomas (5).
Concurrently, the European Expert Group on NGS-based Diagnostics in
Lymphomas (EGNL) summarized the gathered data and divided recurrent
mutations in specific lymphomas into five categories having: i)
immediate impact on therapy; ii) diagnostic impact; iii) prognostic
impact; iv) potential clinical impact in the near future; and v)
mutations to be considered for research purposes only. Tumor
suppressor TP53 ‘gets through’ a few categories depending on
the particular diagnosis. The assessment of its status is essential
for clinical decision-making in patients with chronic lymphocytic
leukemia (CLL). In addition, the prognostic value of the
TP53 status was recognized in splenic marginal zone lymphoma
(SMZL), mantle cell lymphoma (MCL) and the subgroup of DLBCL
concordantly bearing MYC translocation (6).
The role of p53 in lymphoma has been attributed to
its ability to induce apoptosis. Stressed lymphocytes are prone to
apoptosis and disruption of the p53-dependent apoptosis seems to be
essential for the development and progression of lymphoma. However,
direct loss of p53 function by mutation of the TP53 gene is
not the only way how to accomplish it. The p53 protein level and
function itself might be abrogated by several mechanisms. In
addition, inactivation of some other proapoptotic genes (e.g.
BIM or NOXA) or hyperactivation of antiapoptotic
factors (e.g. Bcl-2) can fulfill this role (3). Current genomic technologies allow deep
molecular characterization of particular types of lymphoma and
their genomic signatures. Recently, the data have been reviewed and
summarized for DLBCL, follicular lymphoma (FL), Burkitt lymphoma
(BL), mantle cell lymphoma (MCL), marginal zone B-cell lymphoma
(MZBL) and primary mediastinal B-cell lymphoma (PMBL). Acquisition
of TP53 mutation was demonstrated as one of characteristic
markers for DLBCL and MCL, reaching frequency 20 and 25%,
respectively, but not for other lymphomas (7). This supports the concept of
alternative ways abrogating the p53-dependent apoptosis. We have
been studying the TP53 aberrations in MCL and DLBCL for a
long time. In our previous study, we analyzed the TP53
status in 33 cases of MCL and 75 cases of DLBCL and assessed its
prognostic impact (8,9). Recently, we have enlarged our cohorts
and deepened our effort. In the present study, we present the
complex analysis of the TP53 status in 57 cases of MCL and
131 cases of DLBCL and reconsider its impact on outcome of the
diseases.
Materials and methods
Clinical material
Fifty-seven patients diagnosed with MCL (44 men and
13 women) and 131 patients diagnosed with DLBCL (76 men and 55
women) were included in the present study. All patients underwent
surgical biopsy of the tumor tissue at the University Hospital Brno
and were diagnosed by pathologist according to the WHO
classification. Median age at MCL diagnosis was 65 years (ranging
from 34 to 80 years). DLBCL cohort comprises 105 de novo
cases, 26 patients developed DLBCL as a secondary tumor. Median age
at DLBCL diagnosis was 57 years (ranging from 20 to 85 years). For
all patients, the fresh-frozen tissue samples as well as
formalin-fixed, paraffin-embedded (FFPE) tumor tissue blocks were
available. All patients were included in this study only after
signing the informed consent approved by the ethics committee of
the hospital.
FASAY and split assay
FASAY was performed as previously described
(10,11) with small modifications. Total RNA
was extracted using NucleoSpin RNA kit (New England BioLabs, Inc.,
Ipswich, MA, USA) and stored at −80°C until further processing.
cDNA was synthesized by ProtoScript® II (New England
BioLabs) using primer oligo(dT)12. PCR was performed
using primers P3 (5′-CCTTGCCGTCCCAAGCAATGGATGAT-3′), P4
(5′-ACCCTTTTTGGACTTCAGGTGGCTGGAACC0-3′) and Phusion DNA polymerase
(New England BioLabs). Yeast cells were co-transformed with the PCR
product, linearized pSS16 plasmid and salmon sperm DNA carrier
(Life Technologies, Inc., Carlsbad, CA, USA) by the lithium acetate
procedure (12). Transformed yeast
cells were plated on minimal medium lacking leucine and containing
adenine (5 µg/ml), followed by incubation at 35°C for 2–3 days and
then at room temperature for 2–3 days. For split assay, PCR of the
TP53 5′- part was performed with primers P3 and P17
(5′-GCCGCCCATGCAGGAACTGTTACACAT-3′), the 3′- part with primers P4
and P16 (5′-GCGATGGTCTGGCCCCTCCTCAGCATCGCG0-3′). Yeast cells were
transformed with linearized vectors pFW35 and pFW34 (13).
FASAY deduces the functional status of p53 from
color of colonies of transformed yeast cells. Expression of
functional p53 results in formation of large white colonies,
inactive p53 leads to smaller red ones. The background frequency of
red yeast colonies typically does not exceed 10%. Thus, samples
providing <10% of red colonies are considered to contain only
wild-type TP53 alleles, while samples providing >10% of
red colonies are suspicious to possess a TP53 mutation. The
ratio of red colonies scoring between 10 and 20% can result from a
presence of clonal TP53 mutation in rather small fraction of
cells or from increased rate of RNA degradation. To distinguish
these two possibilities, version of FASAY, called split assay, was
established. In the split assay, the 5′- and 3′- parts of the
TP53 cDNA are tested separately (13).
Purification of the plasmids from
transformed yeast cells and sequencing of the TP53 cDNA
Yeast cells from individual yeast colonies were
harvested, resuspended in TSN (2% Triton X-100, 1% SDS, 100 mM
NaCl, 10 mM Tris pH 8.0, 1 mM EDTA), and ground by vortexing with
glass beads; plasmid DNA was extracted by phenol/chloroform
procedure. The TP53 cDNA was amplified using the P3 and P4
primers and Taq polymerase (Life Technologies) and subjected
to agarose gel electrophoresis. The PCR product was purified by
MinElute PCR purification kit (Qiagen, Hilden, Germany) and
sequenced by BigDye Terminator v3.1 cycle sequencing kit (Applied
Biosystems, Darmstadt, Germany) using ABI PRISM 3130x Genetic
Analyzer (Applied Biosystems).
Isolation and sequencing of the TP53
gDNA
Genomic DNA was isolated from FFPE blocks or from
the frozen tumor samples using Puregene DNA Isolation kit (Gentra
Systems, Minneapolis, MN, USA) according to the manufacturer's
instructions. The TP53 exons were amplified by PCR and
automated fluorescent sequencing was performed by using BigDye
Terminator kit and ABI 3130 Sequencer (Applied Biosystems,
Carlsbad, CA, USA). Either a picked exon of interest or all exons
2–11 were analyzed. Primers and conditions were adopted from IARC
TP53 Database (14).
Immunoblotting
Tissue samples were lysed in 150 mM NaCl, 50 mM NaF,
50 mM Tris (pH 8.0), 5 mM EDTA, 1% NP-40 and 1 mM
phenylmethylsulfonyl fluoride in ice for 30 min, and the cell
extract was centrifuged at 17,000 × g for 30 min to remove cell
debris. Protein concentration was measured by the Bradford assay.
Solubilized proteins were resolved by 10% SDS-PAGE and transferred
onto a nitrocellulose membrane. Blots were blocked in 0.1% Tween-20
and 5% low-fat milk in phosphate-buffered saline (PBS) for 1 h and
probed with anti-p53 mouse monoclonal antibody DO-1 (Dako,
Glostrup, Denmark) and PC10 (Abcam, Cambridge, UK) at 4°C. Blots
were developed with Dako peroxidase-conjugated rabbit anti-mouse
immunoglobulin (Dako) using the ECL chemiluminescence detection kit
(GE Healthcare UK, Ltd., Little Chalfont, UK).
Fluorescence in situ hybridization
(FISH)
FISH was performed on tissue sections prepared from
FFPE blocks. For the TP53-specific locus analysis, the
ZytoLight® SPEC TP53/CEN17 Dual Color Probe were used
(ZytoVision GmbH, Bremerhaven, Germany). Hybridization was
performed according to the manufacturer's instructions. Images were
scanned by DM 5500 B microscope equipped with Leica DFC290 HD
camera. Fluorescence signals were analyzed using Leica LAS X
software (Leica Microsystems GmbH, Wetzlar, Germany). One hundred
cells per case were analyzed. The cut-off level was defined by the
mean value plus three times the SD of the frequency of control
cells exhibiting one TP53 and two CEN17 signals (9.6%).
Statistical analyses
Standard descriptive statistics was applied in the
analysis; absolute and relative frequencies for categorical
variables and median supplemented with minimum-maximum range for
continuous variables. The influence of monitored parameters on
survival and progress-free survival was assessed by hazard ratio
estimates from univariate Cox models. Graphic visualization of
patients survival according to monitored parameters was performed
using Kaplan-Meier survival curves. Statistical significance of
differences in survival among groups of patients was tested using
the log-rank test. Alpha = 0.05 was used as a level of statistical
significance. Analyses were performed in statistical software IBM
SPSS Statistics 22.0.0.1 for Windows (IBM Corp., Armonk, NY,
USA).
Results
Assessment of the TP53 status in MCL
cases
In our previous study we analyzed the TP53
status in tumor tissues of 33 MCL patients, detecting TP53
mutations in 9 of them. Recently, we extended the cohort and
enrolled 24 new patients. First, we isolated RNA from frozen tumor
tissue, performed FASAY and assessed frequency of red colonies.
Twenty cases scored under the 10% cut-off level, while 4 cases
scored clearly above it (ranged from 39.9 to 78.8%). For the
positive cases, the TP53 expression vector was recovered
from 3 to 6 red yeast colonies, and these cDNAs were used as
templates for DNA sequencing. In all cases, unambiguous clonal
TP53 mutation was identified (Table I). We found that: i) two missense
mutations (cases M42 and M55), including the p.R273H that appeared
in the cohort for the second time; ii) one nonsense mutation (M43;
p.Q331*); and iii) one mutation (M35) representing the frame shift
mutation due to one nucleotide insertion and causing formation of
premature termination codon (PTC). For the cases M35 and M43, gDNA
was isolated from frozen tumor tissue and the corresponding
TP53 exons (7 and 9, respectively) were amplified by PCR and
sequenced. Both mutations were confirmed.
 | Table I.Summary of the TP53 status
analyses in samples of mantle cell lymphoma. |
Table I.
Summary of the TP53 status
analyses in samples of mantle cell lymphoma.
| Case | FASAYa | cDNA/gDNA
sequencing | TP53
status | FISHb | IB | Comment |
|---|
| M6 | 88.3 | c.844C>T | p.R282W | 69.0 | +++ |
|
| M9 | 32.2 | c.712T>A | p.C238S | 28.0 | +++ |
|
| M14 | 89.6 | c.524G>A | p.R175H | 0 | +++ |
|
| M17 | 82.5 | c.488A>G | p.Y163C | 6.6 | +++ |
|
| M19 | 92.0 | c.818G>A | p.R273H | 0 | ++ |
|
| M20 | 90.6 | c.742C>T | p.R248W | 48.0 | +++ |
|
| M22 | 75.4 | c.824G>A | p.C275Y | 0 | ++ |
|
| M27 | 33.1 | c.329G>T | p.R110L | 0 | ++ | tsc |
| M31 | 17.6 | c.425delC | p.P142Lfs*27 | 3.4 | − | RNA decay |
| M35 | 60.3 | c.681insT | p.D228* | 0 | ++ | Truncated
protein |
| M42 | 75.6 | c.838A>G | p.R280G | 9.6 | +++ |
|
| M43 | 39.9 | c.991C>T | p.Q331* | 8.3 | +++ | Truncated
protein |
| M55 | 78.8 | c.818G>A | p.R273H | 61.0 |
|
|
The p53 protein accumulation in tumor tissue of all
newly enrolled cases was assessed by immunoblotting using the
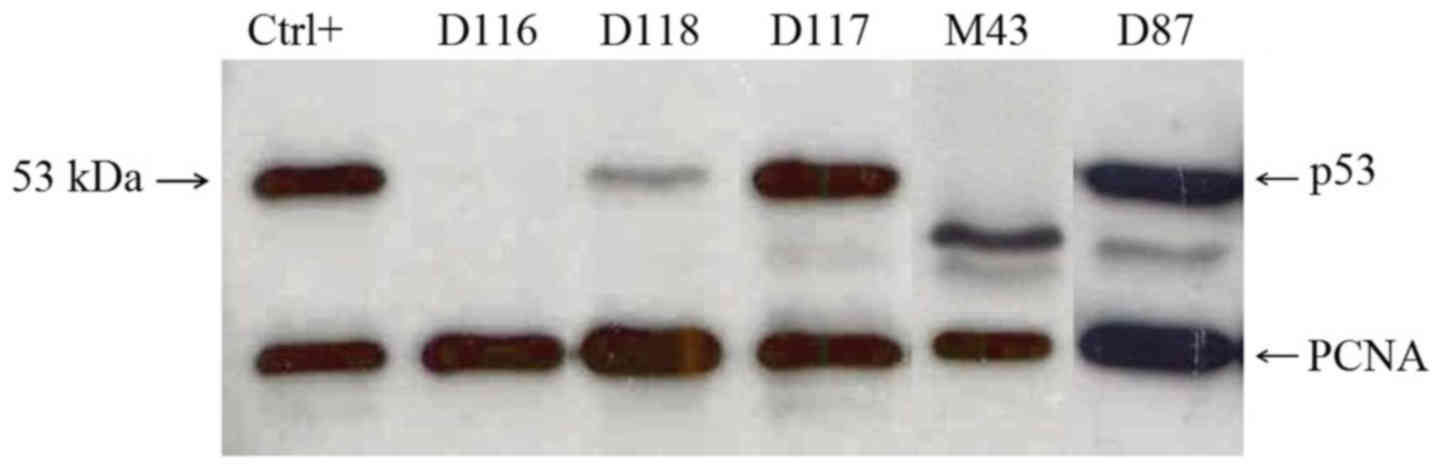
p53-specific DO-1 monoclonal antibody (Fig. 1). Among 24 cases, 9 exhibited either
strong (+++) or intermediate (++) p53 level, including all 4 cases
bearing a clonal TP53 mutation. In cases M35 and M43, the
truncated p53 proteins were detected corresponding to the shortened
coding sequence terminated by the PTC causing TP53 mutation.
The remaining 15 cases exhibited low (+) or none level (−) of the
p53 protein.
We performed FISH analyses of the TP53
alleles using the locus-specific and centromeric probes. We
analyzed 22 cases, including all cases bearing the TP53
clonal mutation. Two cases, including M55, scored positive. The
other case with the loss of the TP53-specific locus, M47, did not
evince a TP53 mutation by FASAY. Therefore, we isolated DNA
from the frozen tumor tissue and performed gDNA sequencing of exons
2–11. No TP53 mutation was detected.
Characteristics of TP53 mutations
detected in MCL
Altogether, we detected 13 TP53 mutations
(Table I) among 57 MCL cases
(22.8%). All 13 mutations were localized in the region coding for
DNA-binding domain. Ten mutations (76.9%) were the missense ones,
including the one that was temperature sensitive. Only one
mutation, p.R273H, appeared repeatedly in two cases. There were
three mutations causing PTC formation (in positions 169, 228 and
331, respectively); one of them was the nonsense one, one insertion
and one deletion. Only the termination codon in position 169 (case
M31) induced the mRNA degradation via the PTC-mediated mRNA decay
mechanism as we previously showed (8). It was also the only mutation which was
not associated with high level of the p53 protein in nuclei of
tumor cells. Concerning the TP53 wild-type cases, the p53
protein accumulation was found in 5 of them, 2 cases were not
analyzed. Altogether, the p53 protein was accumulated in 17 cases
(30.9%) and the concordance between the TP53 mutation and
the p53 protein accumulation was 89.1% (49/55). The loss of
TP53-specific locus was analyzed altogether in 54 MCL cases.
Five positive cases were detected (9.3%); 4 of them bore
concurrently missense mutation on the other TP53 allele.
Nine TP53 mutation-positive cases were negative for the
TP53-specific locus loss (Table
I).
Correlation between the TP53 status
and MCL outcome
In our previous study, 33 MCL patients were
enrolled, female represented 24.2%, and the average age at the time
of diagnosis was 63.4 years. Seventy percent of patients were in
stage IV. Their median survival time reached 51.7 months and we
showed that TP53 mutation (present in 27.3% of cases) was
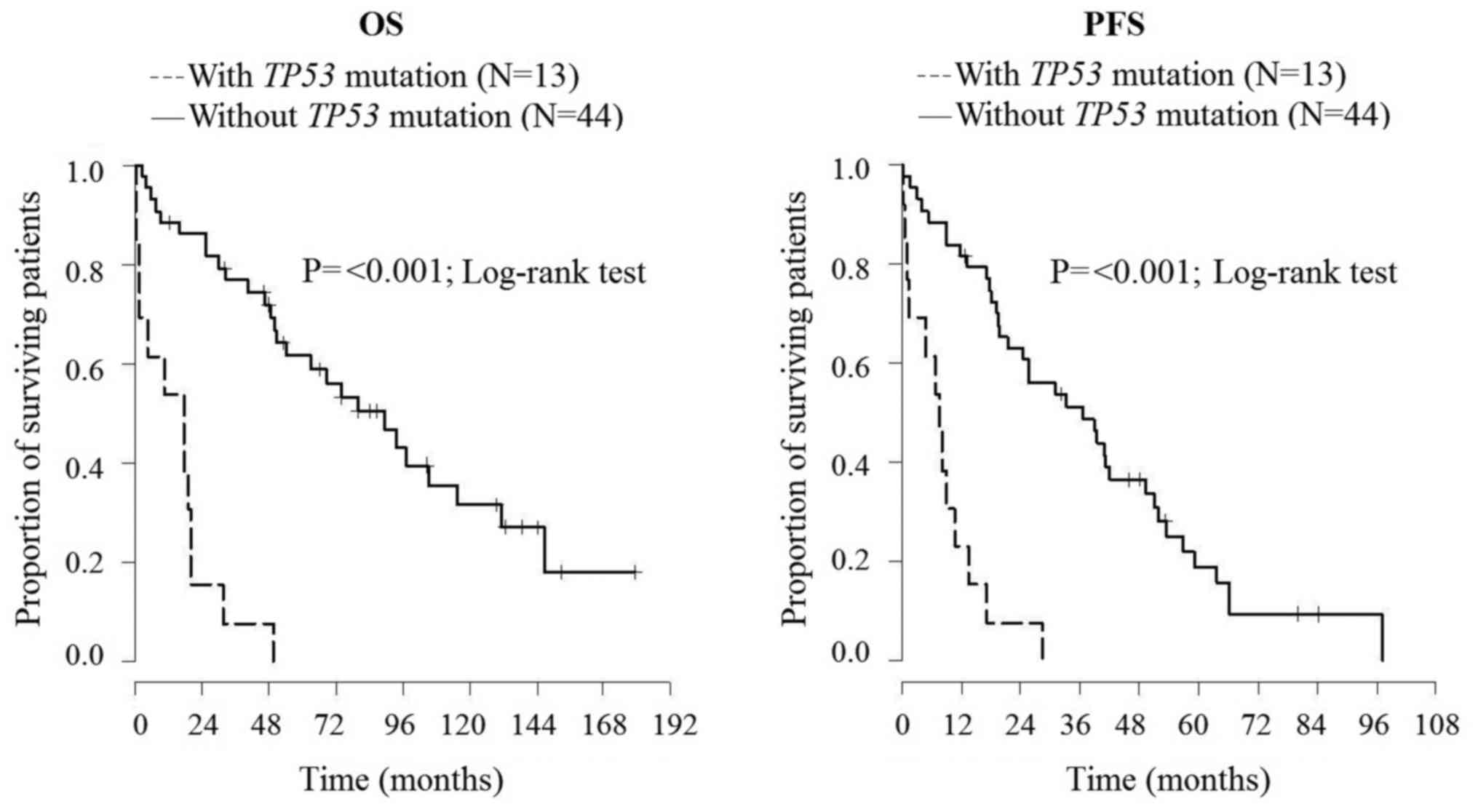
associated with shorter overall survival (P=0.045) (8). The enlarged cohort comprised 57 MCL
patients (average age, 65 years; 22.8% of female; 86% in stage IV)
with median survival time 48.4 months. In this enlarged cohort, we
confirmed that the presence of TP53 mutation (22.8% cases)
was associated not only with shorter overall survival but also with
shorter progression-free survival (P<0.001) (Fig. 2).
Assessment of the TP53 status in DLBCL
cases
In our previous study we analyzed the TP53
status in tumor tissue of 75 DLBCL patients including 54 de
novo cases, detecting 16 TP53 mutations (21.3%). The
recently enlarged cohort comprises 131 patients including 105 de
novo cases. First, we isolated RNA from frozen tumor tissue of
all 56 new cases, performed FASAY/split assay and assessed the
frequency of red colonies. Forty-six cases scored under the 10%
cut-off level and were considered to lack the TP53 mutation.
The other 10 cases scored above the cut-off level (ranged from 22.7
to 93.8%). The cDNA isolated from the positive yeast colonies was
sequenced and 13 TP53 mutations were detected (Table II). Three cases (D86, D87 and D88)
bore two mutations, one in each allele. The missense mutations were
found in both D97 and D99 cases: one fully inactivating and the
second one temperature-sensitive (p. V157A and p. S127T), based on
the specific phenotypes of transformed yeast colonies (15). Yet, another temperature-sensitive
mutation (p.Y234H) was detected in case D78. In case D87,
combination of nonsense mutation in one allele and short
insertion/splicing mutation in the second allele was detected. In
the 3 cases with two independent mutations, the gDNA was isolated
from frozen tumor tissues, the corresponding TP53 exons were
amplified by PCR and sequenced; all 6 mutations detected on the
cDNA level were clearly confirmed (Table II).
 | Table II.Summary of the TP53 status
analyses in samples of diffuse large B-cell lymphoma. |
Table II.
Summary of the TP53 status
analyses in samples of diffuse large B-cell lymphoma.
| Case | FASAYa | cDNA/gDNA
sequencing | TP53
status | FISHb | IB | Comment |
|---|
| D3 | 70.0 | c.403T>C | p.C135R | 63.7 | +++ | tsc |
| D4 | 94.0 | c.177_182del | p.P60_D61del | 49.1 | +++c | Truncated
protein |
| D5 | 21.7 | c.730G>A | p.G244S | 0.0 |
|
|
| D7 | 96.8 | c.734G>A | p.G245D | 0.0 | +++ |
|
| D8 | 12.1 | c.586C>T | p.R196* | 0.0 | − | RNA decay |
| D18 | 28.4 | c.818G>A | p.R273H | 0.0 | + |
|
| D28 | 78.0 | C403T>C | p.C135R | 27.5 | + |
|
| D30 | 73.6 | c.761T>A | p.I254N | 22.4 | + | tsc |
| D35 | 32.0 | c.770T>G | p.L257R | 18.8 | − |
|
| D40 | 60.0 | c.713G>T | p.C238F | 0.0 | +++ |
|
| D44 | 83.0 | c.815T>G | p.V272G | 46.0 | +++ |
|
| D46 | 46.3 | c.817C>T | p.R273C | 0.0 | +++ |
|
| D49 | 74.5 | c.524G>A | p.R175H | 29.6 | +++ |
|
| D50 | 80.3 | c.742C>G | p.R248G | 0.0 | − |
|
| D66 | 62.4 | c.826-835del10 | p.A276Gfs*66 | 1.9 | +c | Truncated
protein |
| D72 | 74.1 | c.818G>A | p.R273H | 1.8 | +++ |
|
| D78 | 53.0 | c.700T>C | p.Y234H | 0.4 | ++ | tsc |
| D86A | 80.8 | c.406C>G | p.Q136E | 0.0 | +++ |
|
| D86B |
| c.470T>C | p.V157A |
|
| tsc |
| D87A | 47.9 | c.783-2A>T
(r.783_784insTGT) |
p.S261_G262insC | 0.0 | +++ |
|
| D87B |
| c.958A>T | p.K320* |
|
|
|
| D88A | 93.8 | c.376T>G | p.Y126D | 53.0 | +++ |
|
| D88B |
| c.379T>A | p.S127T |
|
| tsc |
| D94 | 22.7 | c.406C>G | p.Q136E | 0.0 | ++ |
|
| D105 | 37.8 | c.743G>A | p.R248Q | 8.7 | + |
|
| D109 | 63.6 | c.485T>A | p.I162N | 26.0 | +++ |
|
| D117 | 78.2 | c.839G>T | p.R280I | 4.0 | +++ |
|
| D124 | 78.8 | c.794T>G | p.L265R | 44.0 | +++ |
|
| D131 | 61.0 | c.818G>A | p.R273H | 16.0 | ++ |
|
The p53 protein accumulation was assessed by
immunoblotting in tumor tissue of all 56 newly enrolled cases
(Fig. 1). Ten cases exhibited
strong (+++) p53 protein level, including 6 cases bearing a clonal
TP53 mutation. No p53 protein was detected in 4 cases with a
clonal missense TP53 mutation. Four cases with no
TP53 mutation found by functional analysis scored positive
by immunoblotting. Forty-three cases without a TP53 mutation
scored negative by immunoblotting.
We performed FISH analysis of the TP53
alleles using the locus-specific and centromeric probes in all 57
newly enrolled cases. Ten cases scored positive, including 4 cases
bearing the TP53 clonal mutation. Six cases with the loss of
the TP53-specific locus did not evince a TP53
mutation by FASAY. We randomly chose 2 of them (D113 and D130),
isolated gDNA from frozen tumor tissues, amplified and sequenced
exons 4–10. No TP53 mutation was detected.
Characteristics of TP53 mutations
detected in DLBCL
Altogether, we detected 29 TP53 mutations in
26 tumor samples (Table II) among
131 DLBCL cases (19.8%). All mutations were localized in the
DNA-binding domain-coding region. Twenty-four mutations (82.8%)
were the missense ones; including 5 temperature sensitive mutations
(20.8%). The mutation p.R273H appeared repeatedly in 3 cases (D18,
D72 and D131), the mutation p.Q136E was detected twice (D86A and
D94). Two mutations were the nonsense ones (D8 and D87B) causing
PTC formation in positions 196 and 320. Two mutations were
recognized as deletions, one in frame (D4), another causing the
reading frame shift and PTC formation in position 345 (D66; 9). The
TP53 mutation detected in case D87A was the splicing site
mutation (c.783-2A>T) resulting in one amino acid insertion at
the protein level (p.S261_G262insC), as documented on mRNA from
yeast colonies (r.783_784insTGT). Only the termination codon in
position 196 (case D8) induced the mRNA degradation via the
PTC-mediated mRNA decay mechanism as we previously shown (9). This mutation, as well as other 11
cases with the TP53 mutation, was not associated with high
level of the p53 protein in nuclei of tumor cells. The case D66
bearing short deletion and PTC in position 345 was also classified
as the p53 protein-negative, although slight (+) band of truncated
p53 protein was detected by immunoblotting. The proteins detected
in D4 and D87 cases were also truncated (Fig. 1). Concerning the TP53
wild-type cases, the p53 protein accumulation was found in 6 of
them. Altogether, the p53 protein was accumulated in 20 cases
(15.3%) and the concordance between the TP53 mutation and
the p53 protein accumulation was 86.3% (113/131). The loss of
TP53-specific locus was analyzed in all 131 DLBCL cases.
Twenty positive cases were detected (15.3%); 10 of them bore
concurrently a TP53 mutation. Ten TP53
mutation-positive cases were negative for the TP53-specific
locus loss (Table II).
Correlation between the TP53 status
and DLBCL outcome
In our previous study, 75 DLBCL patients were
enrolled. Twenty-one cases developed into DLBCL by transformation
from less aggressive disease, 54 patients were de novo
cases. Among the de novo DLBCL patients, 46 were treated
with R-CHOP therapy. The only statistically significant impact of
the TP53 status on clinical parameters was found for the
R-CHOP-treated de novo DLBCL patients: the TP53
mutation and/or deletion decreased their progression-free survival
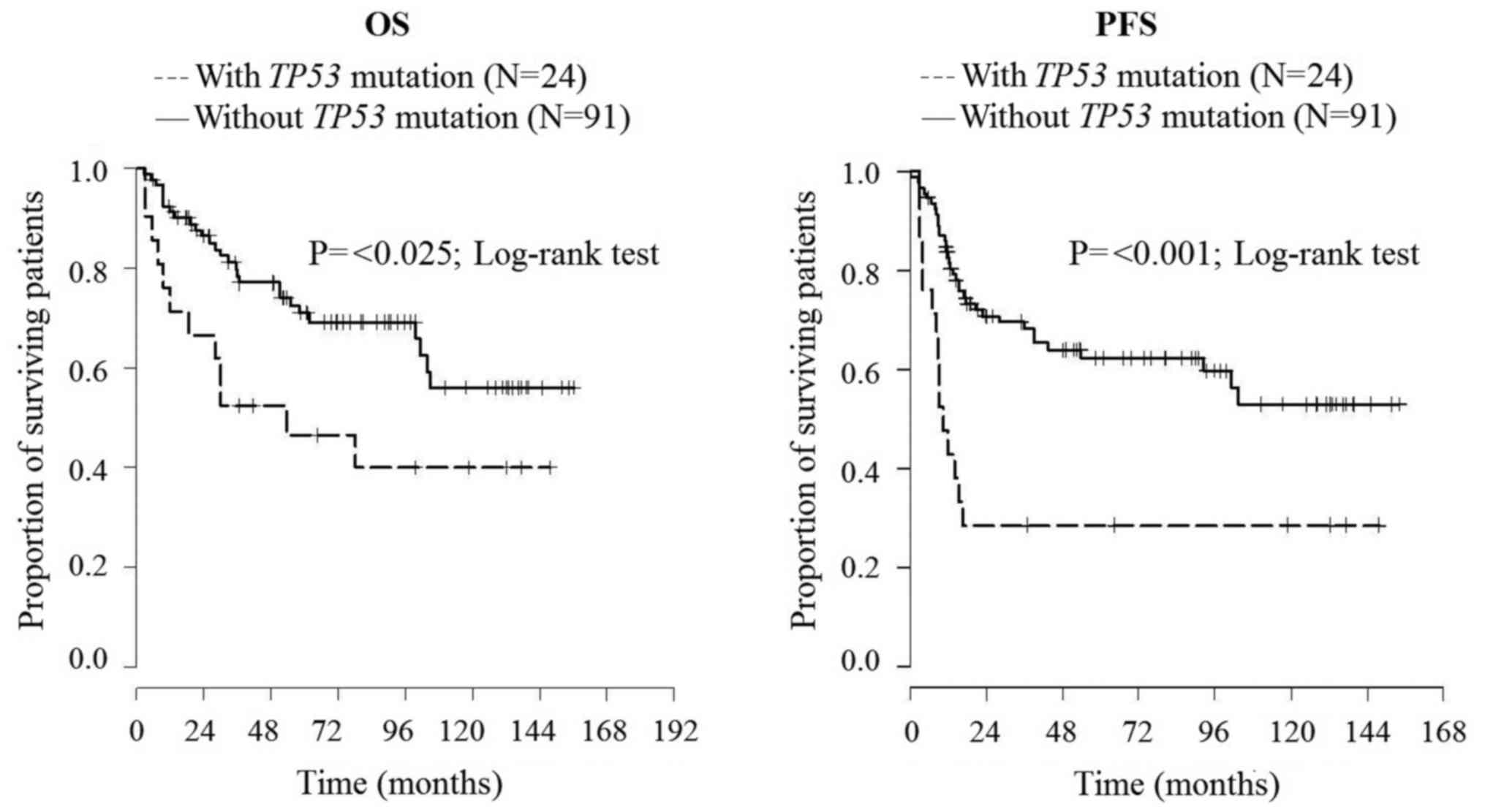
(P=0.021) (9). The enlarged cohort
comprised 131 DLBCL patients: 26 secondary and 105 de novo
cases. Altogether 115 patients were treated with rituximab. In this
extended study, we proved that TP53 mutation shorten both OS
and PFS (P=0.029, P=0.001, respectively) of R-CHOP-treated DLBCL
patients and decreases both OS and PFS of patients with secondary
DLBCL disease (both P=0.021) (Fig.
3).
Discussion
In the present study, we performed detailed analyses
of the TP53 status in 24 new cases of MCL and 56 new cases
of DLBCL. Extending our previous studies (8,9), we
altogether assembled cohorts of 57 MCL and 131 DLBCL cases. This
enabled us not only to verify and deepen the previous
accomplishments but also to compare the TP53 status of two
different types of lymphoma.
We studied the cohort of MCL patients that was
significantly extended, almost doubled, and the results concerning
TP53 confirmed the originally reached findings. In addition,
the results were in very good agreement with recently published
data of other authors. The frequency of the TP53 mutations
in the present cohort was 22.8%, compared to 27.3% in the original
one. The frequencies of the TP53 aberrations recently
reported by others ranged from 13.7 to 28.0% depending on cohort
size and used methodical approach (16–22).
In our cohort, the TP53 mutations occurred more frequently
in the aggressive MCL variants (blastoid and pleomorphic) reaching
41.2 and 54.5%, respectively, compared to 14.6 and 13.6%,
respectively, in the classical cases. Similar results were reported
by Slotta-Huspenina et al (17) who detected the TP53 mutations
in 10% of classical cases and in 91% of blastoid cases. On the
other hand, comparable frequency of the TP53 mutations among
classical and blastoid variants (13 vs. 18%) was also reported
(16). The ratio of missense
mutations was 69.2% in our presented cohort, compared to 88.9% in
the original one. It is comparable to 71.4% (14/15) and 81.8%
(9/11) reached by others (16,19,20).
This study showed that the presence of TP53 mutation is
associated with both shorter overall survival and progression-free
survival, while in the original cohort the association between
TP53 mutation and PFS did not reach the statistical
significance. In some recent studies, the adverse impact of the
TP53 mutations and aberrations on OS of MCL patients was
confirmed (16,17). We found rather low frequency of the
TP53 locus deletion among the MCL cases reaching 9.3 and
9.1%, respectively. This aberration occurs almost exclusively in
the cases with the other allele being inactivated by missense
mutation. Altogether, only one third (30.8 and 33.3%, respectively)
of our TP53-mutated cases had lost the second TP53
allele. The 17p loss was found in 50% of TP53 mutated cases
and altogether in 32% of MCL cases by Halldórsdóttir et al
(16). Beà et al (18) even found the 17p alterations in 6 of
8 TP53-mutated cases (75%) while the one of the remaining
cases bore two independent TP53 mutations. Delfau-Larue
et al (22) found deletion
of TP53-specific locus in 22% of their MCL cases. The
proportion of MCL cases with the TP53 locus deletion thus
remains rather unclear.
We were unable to find, even in the extended cohort,
statistically significant association between the disease course
and the p53 protein accumulation or the TP53 locus deletion
independently on the presence of TP53 mutation, while the
former was documented by Nordström et al (21) and the latter by Delfau-Larue et
al (22). In both studies, the
size of respective cohorts was substantially larger comprising 127
and 134 cases, respectively. On the other hand, in the large cohort
of 119 MCL cases described by Halldórsdóttir et al (16), the presence or absence of
TP53 locus, though found in high proportion of cases (31.9%)
did not influence patient survival, thus confirming similar
conclusions made by Stocklein et al (23).
The TP53 status found in our extended DLBCL
cohort also confirmed and deepened our original findings and
corresponded very well with the data published recently by
International DLBCL Rituximab-CHOP Consortium. This crucial,
extensive, multicentric study comprises data obtained by analysis
of 506 DLBCL patients treated with R-CHOP where 133 TP53
mutations were collected in total (24). In our present cohort, 29 TP53
mutations in 26 tumor samples among 131 DLBCL cases were detected.
The total frequency of the TP53 mutations reached 19.8%,
compared to 22.1% documented by the Consortium cohort. The ratio of
missense mutations reached 82.8% in our cohort compared with 81%
described by the Consortium collection. In the Consortium study,
the IARC TP53 database (http://www.iarc.fr) was used to retrieve functional
level of the detected TP53 mutations. Sixteen mutations were
recognized as partially functional, representing 18.2% (24). Using the functional analysis FASAY
for detection of the TP53 mutations allowed us to directly
recognize five partially functional, temperature-dependent
mutations representing 17.2% of our TP53 mutations.
Collection of TP53 mutations detected by the Consortium was
large enough to assemble mutational spectrum. The mutation in codon
273 appearing repeatedly in four our cases was the second most
frequent mutation position found by the Consortium (detected in 6
cases). In the most often mutated codon 248 according to the
Consortium (mutated in 11 cases), we detected two mutations. In the
Consortium study, 17 patients representing 15.2% of all patients
with the mutated TP53 status exhibited multiple mutations.
Three of our patients with two independent mutations in separated
alleles represented comparable 11.5% of our cohort of 26 patients
with mutated the TP53 gene. From seven Consortium patients
carrying a mutation in a splicing site, three bore another
TP53 mutation, the missense one. Notably, our only patient
bearing the splicing site mutation (c.783-2A>T; p.262insC; D87A)
also exhibited the second TP53 mutation (c.958A>T; p.K320*;
D87B). Both these mutations are rather rare as the D87A mutation
was reported eight times only and D87B twice only according to IARC
database (http://www.iarc.fr, version R18; 14). In
our cohort, both these mutations, as well as other multiple
mutations, were confirmed by gDNA sequencing.
We found 20 cases with the loss of the TP53
locus, only half of them bearing concurrent TP53 mutation.
This rather weak association between TP53 allelic deletion
and TP53 mutation was reported also by the Consortium.
Similarly to the study, we did not find a significant impact of
TP53 allelic deletion on either OS or PFS, thus, supporting
conclusion that TP53 mutations and not TP53 locus deletion
drive DLBCL progression (24).
In the present study, we showed that the TP53
mutations shorten OS and PFS of DLBCL patients treated with
rituximab and decreases both OS and PFS of patients with secondary
DLBCL disease. These results significantly extended our previous
study (9), where the only
statistically significant association was found between the
TP53 mutation status and PFS of the R-CHOP-treated de
novo DLBCL patients. Nevertheless, the TP53 allelic
loss, and the p53 protein accumulation without concerning the
presence of the TP53 mutation were not shown to have any
clinical impact. It is in good agreement with the Consortium study
detecting the TP53 allelic loss in comparable low proportion
of cases (12.3%) and without significant association with decreased
OS or PFS (24). In the same study,
the p53 protein overexpression analysis by immunohistochemistry was
found to have only limited prognostic impact strongly dependent on
used cut-offs (24).
The present study allowed us to compare results of
the TP53 status analyses of two different lymphoma types,
MCL and DLBCL. In many regards, the results were similar. For
example, the frequency of TP53 mutations was comparable,
reaching 22.8% in MCL and 19.8% in DLBCL. These results match well
with the recently published comprehensive data presenting 25% for
MCL and 20 % for DLBCL (7).
Similarly, the ratio of missense TP53 mutations was
comparable for MCL and DLBCL reaching 76.9 and 82.8%, respectively.
The detected frequency of TP53 locus deletion was similarly
low in both diseases reaching 9.3% in MCL and 15.3% in DLBCL. Its
low association with TP53 mutations was found also in both
diseases. All these results strongly support involvement of the
TP53 tumor suppressor mutations, rather than deletion or
overexpression, in development and progression of both MCL and
DLBCL. Although the frequency of TP53 mutations was very
similar in both types of lymphoma, the indispensable sizes of the
cohorts allowing conclusive assessment of their clinical impacts
are very different due to strong molecular difference between MCL a
DLBCL. While MCL represents rather clearly defined type of lymphoma
that is easy distinguishable from other lymphomas, DLBCL as
diagnostic unit is characterized by its heterogeneity (5,7,25). Any
conclusive results would always be conditioned by assembling
extensive cohorts of patients.
Acknowledgements
The present study was supported by the grant
NT/13784-4/2012 of the Internal Grant Agency of the Ministry of
Health of the Czech Republic, MH CZ-DRO (FNBr, 65269705), and MEYS
CR CEITEC 2020 (LQ1601).
References
|
1
|
Harris SL and Levine AJ: The p53 pathway:
Positive and negative feedback loops. Oncogene. 24:2899–2908. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Sherr CJ: Principles of tumor suppression.
Cell. 116:235–246. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Xu-Monette ZY, Medeiros LJ, Li Y, Orlowski
RZ, Andreeff M, Bueso-Ramos CE, Greiner TC, McDonnell TJ and Young
KH: Dysfunction of the TP53 tumor suppressor gene in lymphoid
malignancies. Blood. 119:3668–3683. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Leroy B, Anderson M and Soussi T: TP53
mutations in human cancer: Database reassessment and prospects for
the next decade. Hum Mutat. 35:672–688. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Swerdlow SH, Campo E, Pileri SA, Harris
NL, Stein H, Siebert R, Advani R, Ghielmini M, Salles GA, Zelenetz
AD, et al: The 2016 revision of the World Health Organization
classification of lymphoid neoplasms. Blood. 127:2375–2390. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Rosenquist R, Rosenwald A, Du MQ, Gaidano
G, Groenen P, Wotherspoon A, Ghia P, Gaulard P, Campo E and
Stamatopoulos K: European Research Initiative on CLL (ERIC) and the
European Association for Haematopathology (EAHP): Clinical impact
of recurrently mutated genes on lymphoma diagnostics:
State-of-the-art and beyond. Haematologica. 101:1002–1009. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Iqbal J, Naushad H, Bi C, Yu J, Bouska A,
Rohr J, Chao W, Fu K, Chan WC and Vose JM: Genomic signatures in
B-cell lymphoma: How can these improve precision in diagnosis and
inform prognosis? Blood Rev. 30:73–88. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Stefancikova L, Moulis M, Fabian P,
Ravcukova B, Vasova I, Muzik J, Malcikova J, Falkova I, Slovackova
J and Smardova J: Loss of the p53 tumor suppressor activity is
associated with negative prognosis of mantle cell lymphoma. Int J
Oncol. 36:699–706. 2010.PubMed/NCBI
|
|
9
|
Stefancikova L, Moulis M, Fabian P, Vasova
I, Zedek F, Ravcukova B, Muzik J, Kuglik P, Vranova V, Falkova I,
et al: Prognostic impact of p53 aberrations for R-CHOP-treated
patients with diffuse large B-cell lymphoma. Int J Oncol.
39:1413–1420. 2011.PubMed/NCBI
|
|
10
|
Flaman JM, Frebourg T, Moreau V,
Charbonnier F, Martin C, Chappuis P, Sappino AP, Limacher IM, Bron
L and Benhattar J: A simple p53 functional assay for screening cell
lines, blood, and tumors. Proc Natl Acad Sci USA. 92:3963–3967.
1995. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Smardová J, Nemajerová A, Trbusek M,
Vagunda V and Kovarík J: Rare somatic p53 mutation identified in
breast cancer: A case report. Tumour Biol. 22:59–66. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ishioka C, Frebourg T, Yan YX, Vidal M,
Friend SH, Schmidt S and Iggo R: Screening patients for
heterozygous p53 mutations using a functional assay in yeast. Nat
Genet. 5:124–129. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Waridel F, Estreicher A, Bron L, Flaman
JM, Fontolliet C, Monnier P, Frebourg T and Iggo R: Field
cancerisation and polyclonal p53 mutation in the upper
aero-digestive tract. Oncogene. 14:163–169. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Bouaoun L, Sonkin D, Ardin M, Hollstein M,
Byrnes G, Zavadil J and Olivier M: TP53 variations in human
cancers: New lessons from the IARC TP53 database and genomics Data.
Hum Mutat. 37:865–876. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Grochova D, Vankova J, Damborsky J,
Ravcukova B, Smarda J, Vojtesek B and Smardova J: Analysis of
transactivation capability and conformation of p53
temperature-dependent mutants and their reactivation by amifostine
in yeast. Oncogene. 27:1243–1252. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Halldórsdóttir AM, Lundin A, Murray F,
Mansouri L, Knuutila S, Sundström C, Laurell A, Ehrencrona H,
Sander B and Rosenquist R: Impact of TP53 mutation and 17p deletion
in mantle cell lymphoma. Leukemia. 25:1904–1908. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Slotta-Huspenina J, Koch I, de Leval L,
Keller G, Klier M, Bink K, Kremer M, Raffeld M, Fend F and
Quintanilla-Martinez L: The impact of cyclin D1 mRNA isoforms,
morphology and p53 in mantle cell lymphoma: p53 alterations and
blastoid morphology are strong predictors of a high proliferation
index. Haematologica. 97:1422–1430. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Beà S, Valdés-Mas R, Navarro A, Salaverria
I, Martín-Garcia D, Jares P, Giné E, Pinyol M, Royo C, Nadeu F, et
al: Landscape of somatic mutations and clonal evolution in mantle
cell lymphoma. Proc Natl Acad Sci USA. 110:18250–18255. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Meissner B, Kridel R, Lim RS, Rogic S, Tse
K, Scott DW, Moore R, Mungall AJ, Marra MA, Connors JM, et al: The
E3 ubiquitin ligase UBR5 is recurrently mutated in mantle cell
lymphoma. Blood. 121:3161–3164. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhang J, Jima D, Moffitt AB, Liu Q, Czader
M, Hsi ED, Fedoriw Y, Dunphy CH, Richards KL, Gill JI, et al: The
genomic landscape of mantle cell lymphoma is related to the
epigenetically determined chromatin state of normal B cells. Blood.
123:2988–2996. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Nordström L, Sernbo S, Eden P, Grønbaek K,
Kolstad A, Räty R, Karjalainen ML, Geisler C, Ralfkiaer E,
Sundström C, et al: SOX11 and TP53 add prognostic information to
MIPI in a homogenously treated cohort of mantle cell lymphoma: a
Nordic Lymphoma Group study. Br J Haematol. 166:98–108. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Delfau-Larue MH, Klapper W, Berger F,
Jardin F, Briere J, Salles G, Casasnovas O, Feugier P, Haioun C,
Ribrag V, et al: European Mantle Cell Lymphoma Network: High-dose
cytarabine does not overcome the adverse prognostic value of CDKN2A
and TP53 deletions in mantle cell lymphoma. Blood. 126:604–611.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Stöcklein H, Hutter G, Kalla J, Hartmann
E, Zimmermann Y, Katzenberger T, Adam P, Leich E, Höller S,
Müller-Hermelink HK, et al: Genomic deletion and promoter
methylation status of Hypermethylated in Cancer 1 (HIC1) in mantle
cell lymphoma. J Hematop. 1:85–95. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Xu-Monette ZY, Wu L, Visco C, Tai YC,
Tzankov A, Liu WM, Montes-Moreno S, Dybkaer K, Chiu A, Orazi A, et
al: Mutational profile and prognostic significance of TP53 in
diffuse large B-cell lymphoma patients treated with R-CHOP: Report
from an International DLBCL Rituximab-CHOP Consortium Program
Study. Blood. 120:3986–3996. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lu TX, Young KH, Xu W and Li JY: TP53
dysfunction in diffuse large B-cell lymphoma. Crit Rev Oncol
Hematol. 97:47–55. 2016. View Article : Google Scholar : PubMed/NCBI
|