Introduction
Triple-negative breast cancer (TNBC) accounts for
~15% of all breast cancers and is characterized by the absence of
estrogen receptor (ER), progesterone receptor (PR) and human
epidermal growth factor receptor 2 (HER2) expression (1). Without effective therapeutic targets,
chemotherapy is the only systemic treatment strategy for patients
with TNBC. Neoadjuvant chemotherapy (NAC) for breast cancer is an
important therapeutic modality that can improve the clinical
outcome of individuals with breast cancer, particularly those with
TNBC (2). It is widely accepted
that pathological complete response (pCR) is a reliable predictor
of good prognosis in patients with TNBC and a number of studies
have investigated potential biomarkers for the pCR of TNBC patients
to NAC (3–6). However, these investigations mainly
focused on clinicopathological parameters, as well as imaging data
and lymphocyte infiltration. Since its advent and development,
high-throughput sequencing has been widely used to investigate the
underlying mechanisms of numerous disorders. In particular,
microarray analysis has dual significance, as it may be used to
identify individuals with a higher chance of pCR to NAC and to
explore potential therapeutic targets in TNBC patients, by using
high-throughput sequencing data to identify reliable
biomarkers.
At present, anthracycline-taxane-based NAC is among
the most commonly administered regimens for patients with TNBC.
Thus, there is an urgent necessity to identify reliable biomarkers
for predicting the chemotherapeutic response and clinical outcome
of TNBC patients following taxane-anthracycline-based NAC, in order
to avoid the administration of toxic agents and to help doctors
tailor treatment strategies for individual patients. Therefore, in
the present study, we extracted gene expression data on 64 TNBC
patients (with pCR and non-pCR to NAC) from the GSE41998 dataset of
Gene Expression Omnibus (GEO), and identified five core genes that
were closely associated with the pCR of TNBC patients, namely NOL7,
GFER, COMMD4, sirtuin 5 (SIRT5) and SRC, via a genetic
algorithm-support vector machine (GA-SVM)-based method.
SIRT5 is a member of the sirtuins (SIRTs), a family
of nicotinamide adenine dinucleotide (NAD+)-dependent
deacetylases that participate in various biological processes,
including the regulation of metabolism, cell division, aging and
oxidative stress (7). In recent
years, studies have evaluated the potential roles of SIRTs in
different cancer types (8–10). In particular, the expression of
SIRT5 has been assessed in various types of cancer, including
endometrial carcinoma, head and neck squamous cell carcinoma, basal
cell carcinoma, lung and breast cancer (11–14).
However, to the best of our knowledge, no study has investigated
the value of SIRT5 in predicting the pCR of TNBC patients to NAC,
or the potential association of SIRT5 expression with the
clinicopathological characteristics and prognosis of breast cancer
patients.
In the present study, we aimed to clarify the
relevance of SIRT5 expression to the pCR to NAC via online database
analyses. Using bioinformatic methods, public microarray data were
downloaded to determine whether the expression of SIRT5 was
associated with the pCR to anthracycline-taxane-based NAC in TNBC
patients. The biological functions of SIRT5 were also assessed by
Gene Ontology (GO) analysis. Additionally, Oncomine database
analysis was used to demonstrate whether the differential
expression of SIRT5 in breast cancer patients was associated with
the clinicopathological features, clinical outcomes and response to
anthracycline-taxane-based chemotherapy in breast cancer
patients.
Materials and methods
GEO dataset
The gene expression profile microarray dataset
GSE41998, which was originated from a randomized, multicenter,
open-label, phase II trial (NCT00455533), was obtained from the GEO
database (www.ncbi.nlm.nih.gov/geo). This dataset was deposited
by Horak et al (15), who
enrolled previously untreated women with histologically-confirmed
primary invasive breast adenocarcinoma and treated them with four
cycles of AC (doxorubicin plus cyclophosphamide), followed by 1:1
randomization to ixabepilone (n=148) or paclitaxel (n=147). The
gene expression data from 64 TNBC patients who received the
AC-Taxol (paclitaxel) regimen were analyzed in this study. The
dataset was originally produced using an Affymetrix Human Genome
U133A 2.0 Array. The gene expression data of all samples were
pre-processed through background correction, quantile
normalization, probe summarization and probe ID to gene symbol
conversion using the MAS5 algorithm in the Affymetrix package
(version 1.54.0) of Bioconductor.
Differentially expressed gene (DEG)
analysis, data preprocessing and GA-SVM-based classification
The limma package of Bioconductor was used for
identification of DEGs. Genes with P<0.01 and a minimum absolute
log2 (FC) >log2 (1.5) were considered to be differentially
expressed between pCR and non-pCR patients. Visualization of the
identified DEGs on a heat map was achieved using the ComplexHeatmap
version 1.16.0 package (Bioconductor). The GA-SVM method was used
as previously described (16) with
a search iteration number of 100 and a population size of 20.
Annotations for the probe arrays were obtained using the Affymetrix
Human Genome U133A 2.0 Array annotation data (hgu133a2.db version
3.2.3 package; Bioconductor). For cases of multiple probe sets
mapping to the same gene, the averages of the probe set values were
taken as the expression values.
Functional enrichment analysis of
SIRT5
The Pearson's correlation analysis was employed to
evaluate statistically significant associations between
‘221010_s_at’ (SIRT5) and other gene probes in the 64 TNBC patients
who underwent neoadjuvant doxorubicin/cyclophosphamide therapy
followed by paclitaxel treatment in the GSE41998 set. The
significance threshold was set at P<0.001 and the resulting
correlation networks were visualized with Cytoscape (version 3.5.1;
http://cytoscape.org/). Subsequently, to identify
the potential underlying mechanisms of SIRT5 in the
anthracycline-taxane-based NAC response, we searched genes that
were significantly associated with SIRT5 in the Database for
Annotation, Visualization and Integrated Discovery (DAVID; version
6.8, david.ncifcrf.gov) (17). In GO analysis, the categories
screened were ‘cellular component’, ‘biological process’ and
‘molecular function’ terms. P<0.05 was considered to indicate
statistically significant differences.
Oncomine database analysis
The expression levels of SIRT5 in breast cancers
were analyzed in Oncomine gene expression array datasets
(www.oncomine.org) (18). We searched SIRT5 in the database,
using thresholds of P<0.05 and fold change >1.5. A total of 6
independent datasets were extracted for the differential expression
analysis of SIRT5, through which normal breast tissues were
compared with breast cancer tissues. In addition, SIRT5 expression
levels were evaluated in breast cancers with differences in
clinopathological characteristics, including histological,
molecular and pathological subtype and clinical outcome.
Additionally, to assess the predictive value of SIRT5 on the
response of TNBC patients to neoadjuvant anthracycline-taxane-based
chemotherapy, the expression levels of SIRT5 were also examined in
epirubicin/cyclophosphamide-docetaxel chemotherapy responders and
non-responders.
Survival analysis
The prognostic value of SIRT5 was determined by
Kaplan-Meier analysis using the KM plotter online software
(http://kmplot.com/analysis/) on 5,143
breast cancer patients (2017 version). SIRT5 gene (probe set,
221010_s_at) was entered into the database (http://kmplot.com/breast/) to obtain Kaplan-Meier
survival plot where the number at risk was indicated below the main
plot (17). Hazard ratio (HR) with
95% confidence intervals (CIs) and log-rank P-value were calculated
and displayed on the webpage. The primary endpoint of interest was
distant metastasis-free survival (DMFS) and the secondary was
overall survival (OS).
Statistical analysis
The differentiated expression analysis and Pearson's
correlation test were applied for two-class differential expression
analyses (e.g., pCR vs. non-pCR) and multiclass ordinal analyses
(e.g., grade I vs. II breast cancer) during the Oncomine database
analysis (www.oncomine.org). Unless otherwise
indicated, in all analyses P<0.05 was considered to indicate a
statistically significant difference.
Results
Identification of DEGs between pCR and
non-pCR patients
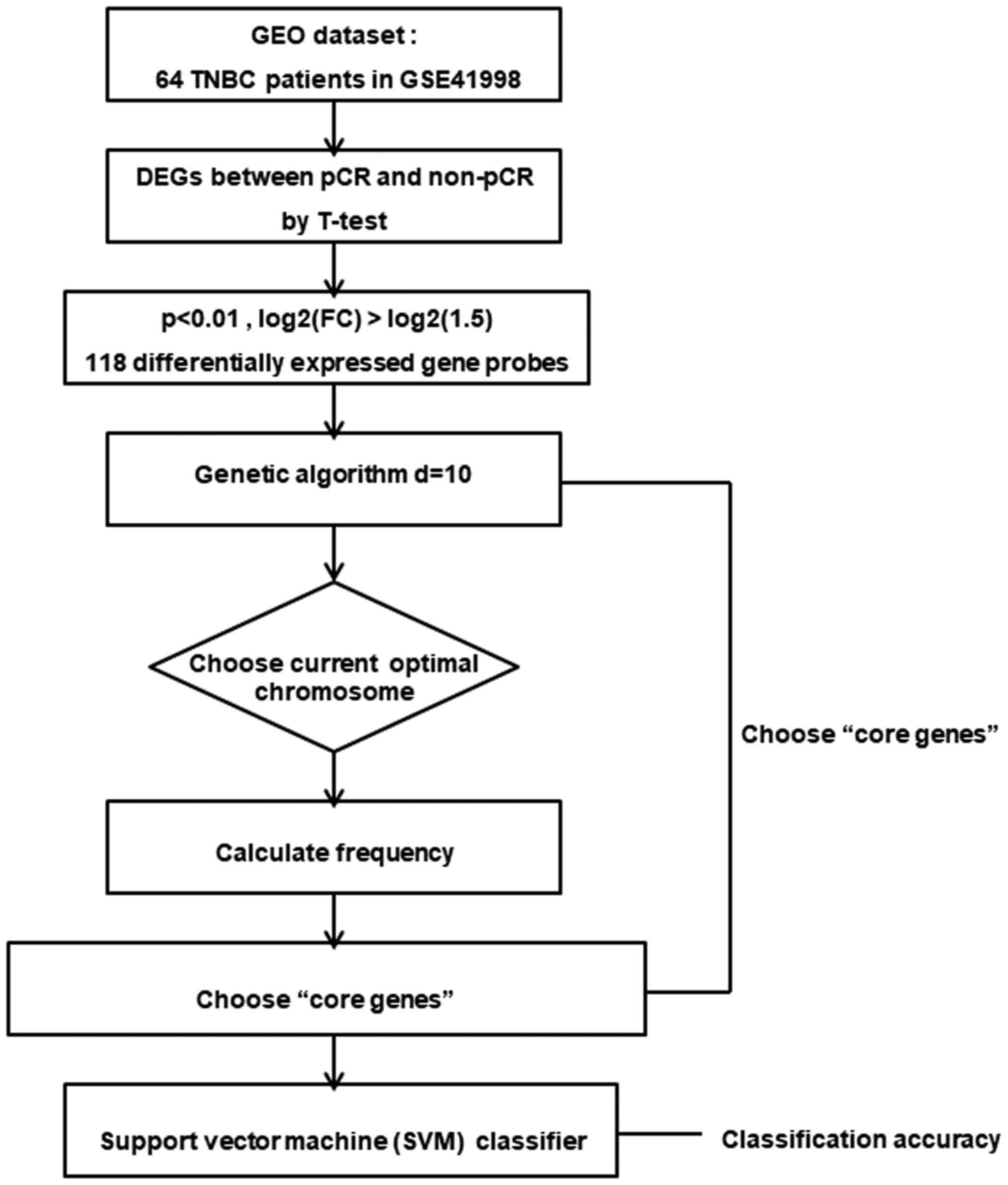
The overall procedure of analysis in the GEO
database is displayed in Fig. 1.
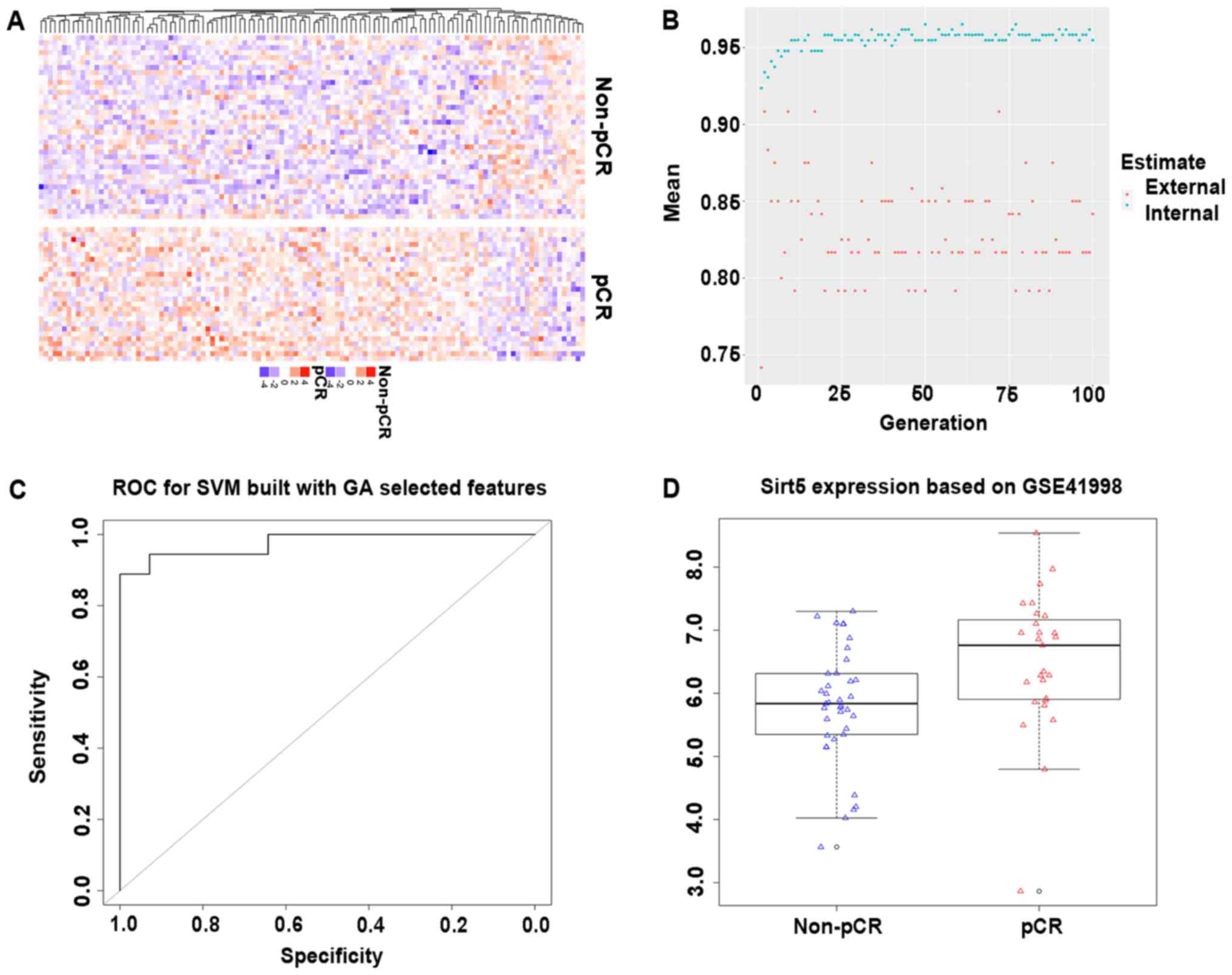
Based on the public microarray dataset GSE41998, we first compared
gene expression profiles among patients with pCR and non-pCR, and
identified 118 DEG probes, including 99 upregulated and 19
downregulated DEGs (Fig. 2A).
GA-SVM-based pCR classifiers
In order to identify optimal biomarkers for
predicting pCR, the GA of the GA-SVM method was used to narrow down
the number of genes, using the DEGs as features and their
expression levels as feature values. A total of 61 gene probes
mostly related to the predictive classification were selected
(Fig. 2B). Subsequently, these 61
candidate biomarkers were integrated into risk classifiers with
regard to the identification of TNBC patients with pCR. SVM
analysis with a 10-fold cross-validation procedure was performed to
evaluate the predictive performance of the SVM-based classifiers.
The area under the receiver operating characteristic curve was
0.9762 (Fig. 2C). Five core genes
were identified to have better predictive performance regarding pCR
to NAC. The five core genes were 202881_x_at, 204660_at,
209133_s_at, 221010_s_at, 221281_at (NOL7, GFER, COMMD4, SIRT5 and
SRC, respectively; Table I). To the
best of our knowledge, SIRT5 is closely related to cell metabolism
and had a significant positive association with pCR to NAC in TNBC
patients (Fig. 2D). These data
indicated the value of SIRT5 in predicting the pCR of TNBC patients
to taxane-anthracycline-based NAC.
 | Table I.Five core genes identified by using
GA-SVM based classifier to discriminate TNBC patients with pCR
after anthracycline-taxane based NAC from those without pCR
(GSE41998). |
Table I.
Five core genes identified by using
GA-SVM based classifier to discriminate TNBC patients with pCR
after anthracycline-taxane based NAC from those without pCR
(GSE41998).
| Probe ID | Gene symbol | P-value | Adjusted
P-value | B-statistic |
|---|
| 202881_x_at | NOL7 | 0.004161662 | 0.795911511 | −2.86518995 |
| 204660_at | GFER | 0.001336072 | 0.762088046 | −2.35709256 |
| 209133_s_at | COMMD4 | 0.007963704 | 0.795911511 | −3.15483672 |
| 221010_s_at | SIRT5 | 0.006766523 | 0.795911511 | −3.08223489 |
| 221281_at | SRC | 0.007015234 | 0.795911511 | −3.0983295 |
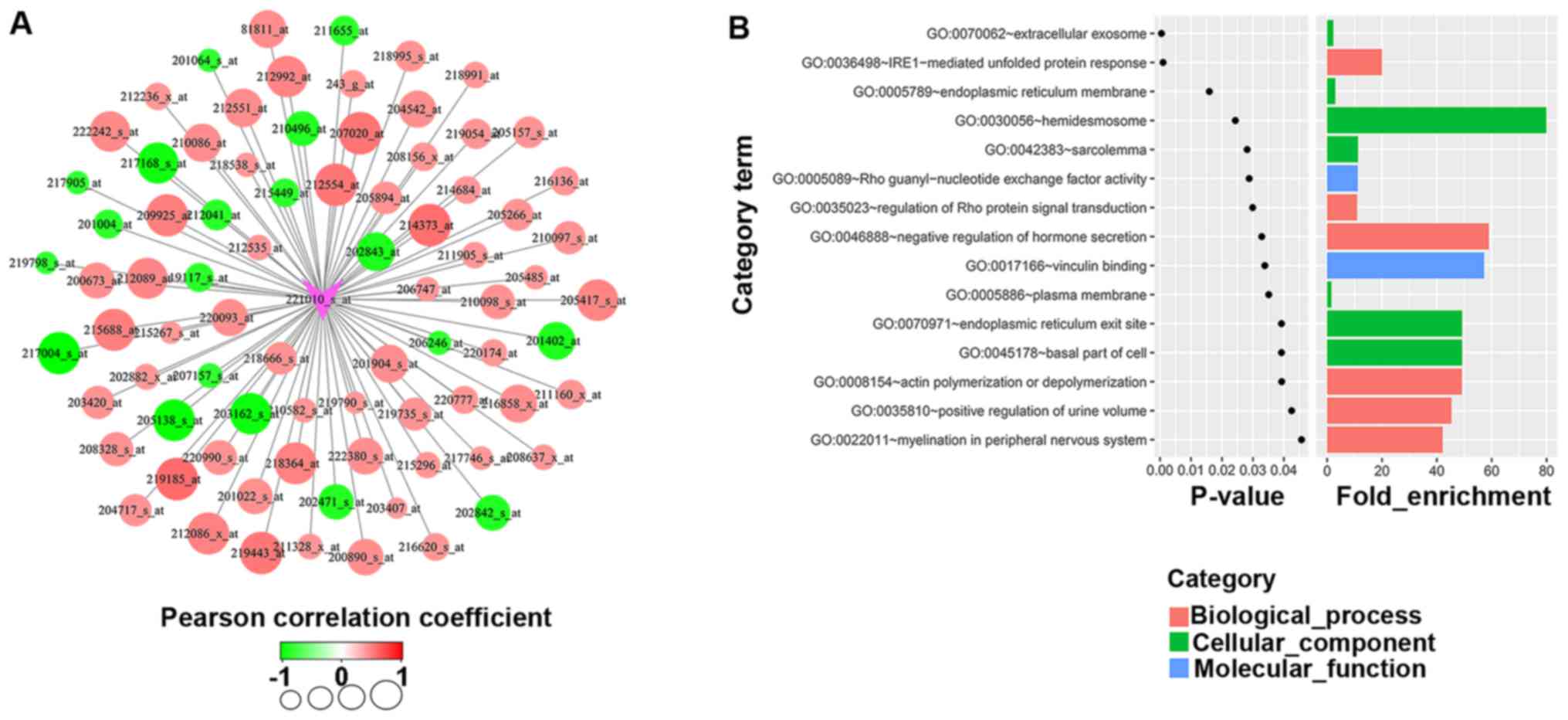
Functional enrichment analysis of
SIRT5
To investigate the potential altered biological
functions of SIRT5 associated with the pCR of TNBC patients to NAC,
we first screened 81 probes that were significantly associated with
probe ‘221010_s_at’ (SIRT5) in the TNBC samples in GSE41998
(Fig. 3A). In silico GO
analysis of these 81 differentially probes was subsequently
performed with a threshold of P<0.05. As displayed in Fig. 3B, SIRT5 was associated with all
three GO functions (cellular component, biological process and
molecular function), among which the molecular function involved
Rho guanyl-nucleotide exchange factor activity (3 genes, P=0.029)
and vinculin binding (2 genes, P=0.034). The cellular component
involved extracellular exosomes (20 genes, P=0.0047), the
endoplasmic reticulum membrane (ER) (8 genes, P=0.016),
hemidesmosomes (2 genes, P=0.024), the sarcolemma (3 genes,
P=0.028), the plasma membrane (20 genes, P=0.035), endoplasmic
reticulum exit sites (2 genes, P=0.039) and the basal part of the
cell (2 genes, P=0.039). The biological processes involved the
IRE1-mediated unfolded protein response (4 genes, P=0.001),
regulation of Rho protein signal transduction (3 genes, P=0.03),
negative regulation of hormone secretion (2 genes, P=0.033), actin
polymerization or depolymerization (2 genes, P=0.039), positive
regulation of urine volume (2 genes, P=0.043) and myelination in
peripheral nervous system (2 genes, P=0.046). It is interesting to
note that the Rho pathway was reflected in the enriched GO cellular
component terms, indicating that aberrations in the Rho pathway may
play an important role in the putative SIRT5-mediated response to
anthracycline-taxane based NAC.
SIRT5 expression analysis in the
Oncomine database
By querying ‘SIRT5’ in the Oncomine database, the
expression of this gene in various cancer types and cancer subtypes
was evaluated. In the present study, we further analyzed the
expression levels of SIRT5 in breast cancers, through which the
log-transformed and normalized expression values of SIRT5 were
extracted, analyzed and interpreted from bar charts. On analysis of
SIRT5 expression in cancer versus normal tissues, higher expression
levels of SIRT5 were observed in breast cancer tissues in three
datasets (data not shown), while lower levels were observed in
breast cancer tissues in another three datasets (data not shown).
These results indicated that SIRT5 may not be a tumorigenic gene in
breast cancer.
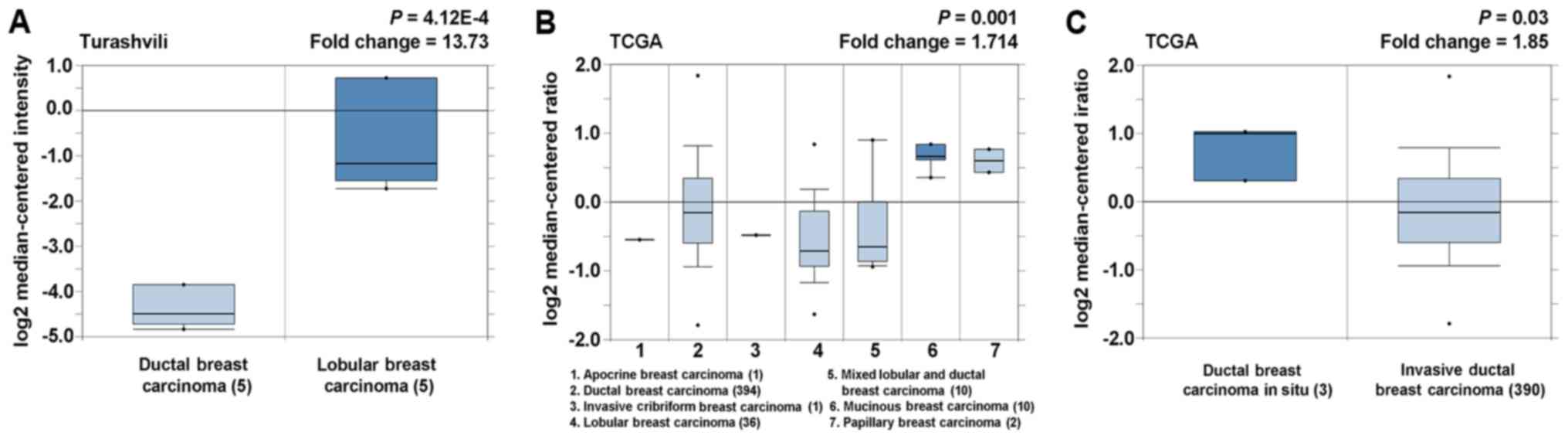
The results of cancer histological subtype analysis
are displayed in Fig. 4. Compared
with ductal breast carcinoma, lobular breast carcinoma was found to
express higher levels of SIRT5 (Fig.
4A). Overexpression of SIRT5 was observed in mucinous breast
carcinoma, relative to the other histological subtypes (Fig. 4B). Meanwhile, ductal breast
carcinoma in situ expressed higher levels of SIRT5 than
invasive breast carcinoma (Fig.
4C). These results indicated that varying degrees of SIRT5
upregulation may be specific to certain histological subtypes.
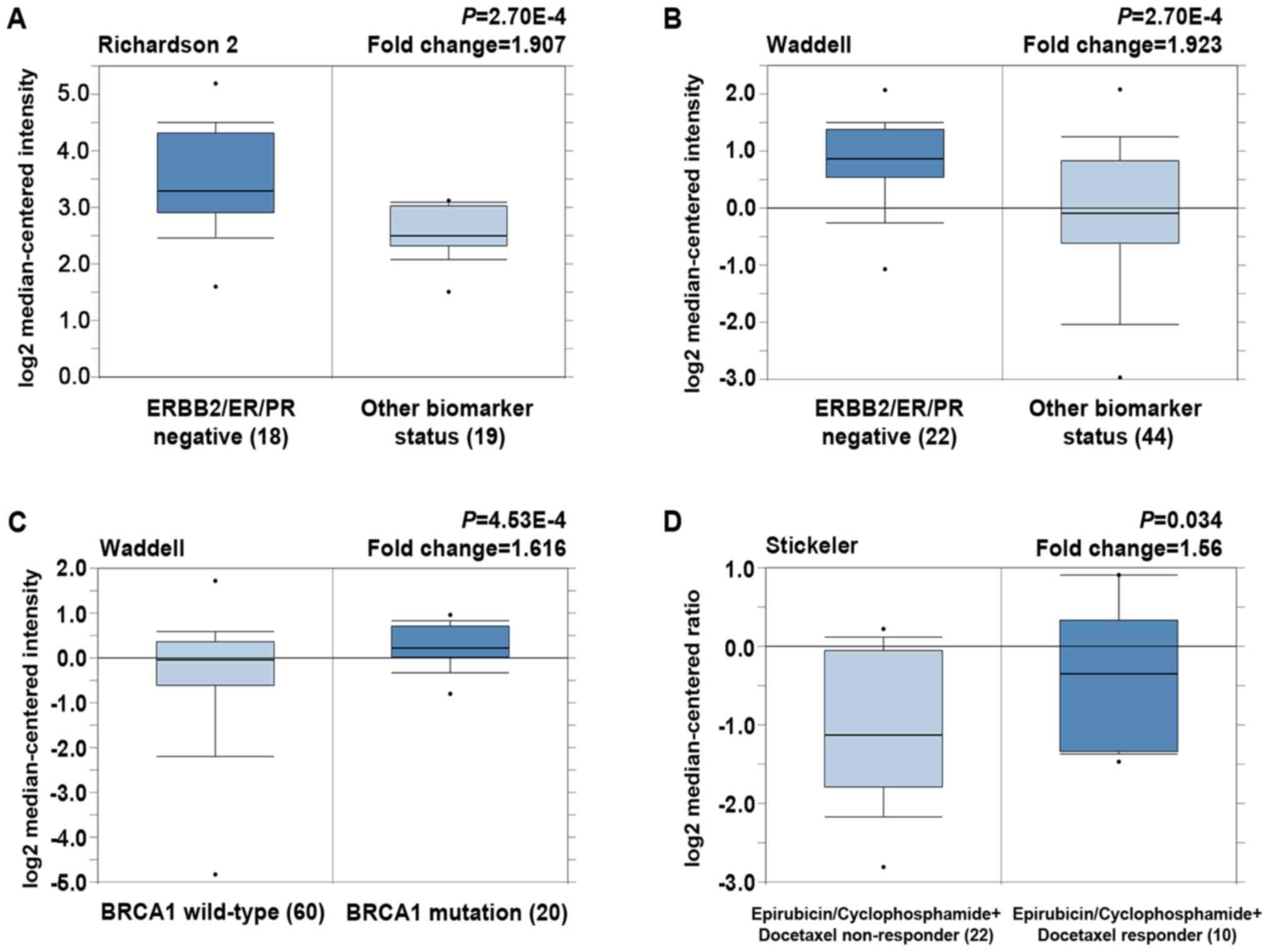
With regard to the expression of SIRT5 in different
molecular subtypes, SIRT5 was overexpressed in TNBC (Fig. 5A and B). Higher levels of SIRT5 were
also expressed in ER-negative, PR-negative and HER2-negative breast
carcinoma, compared with ER-positive, PR-positive and HER2-positive
carcinomas, respectively (Table
II). Furthermore, patients with BRCA1 mutations exhibited
elevated SIRT5 levels compared with those expressing wild-type
BRCA1 (Fig. 5C). On analysis of
SIRT5 expression with regard to patient treatment response, a
significant 1.560-fold elevation in SIRT5 expression level was
observed in epirubicin/cyclophosphamide-docetaxel responders
relative to non-responders (Fig.
5D). Collectively, these results demonstrated that high-level
expression of SIRT5 was positively associated with the TNBC and
BRCA1 mutant subtypes and the response to anthracycline-taxane
based treatment.
 | Table II.Elevated SIRT5 expression in hormone
receptor negative or HER2-negative breast cancer tissues compared
to hormone receptor positive or HER2-positive breast cancer tissues
(Oncomine database). |
Table II.
Elevated SIRT5 expression in hormone
receptor negative or HER2-negative breast cancer tissues compared
to hormone receptor positive or HER2-positive breast cancer tissues
(Oncomine database).
| Study name | Cancer type | Negative (n) | Positive (n) | Total (n) | Fold change | P-value |
|---|
| Waddell breast | ER-negative vs.
ER-positive | 27 | 42 | 69 | −1.625 | 9.64E-4 |
| Richardson breast
2 | ER-negative vs.
ER-positive | 24 | 15 | 39 | −1.632 | 7.72E-4 |
| Waddell breast | PR-negative vs.
PR-positive | 28 | 38 | 66 | −1.570 | 0.003 |
| Richardson breast
2 | PR-negative vs.
PR-positive | 26 | 13 | 39 | −1.626 | 5.34E-4 |
| Waddell breast | HER2-negative vs.
HER2-positive | 43 | 12 | 55 | −1.816 | 0.013 |
| Richardson breast
2 | HER2-negative vs.
HER2-positive | 29 | 8 | 37 | −1.501 | 0.016 |
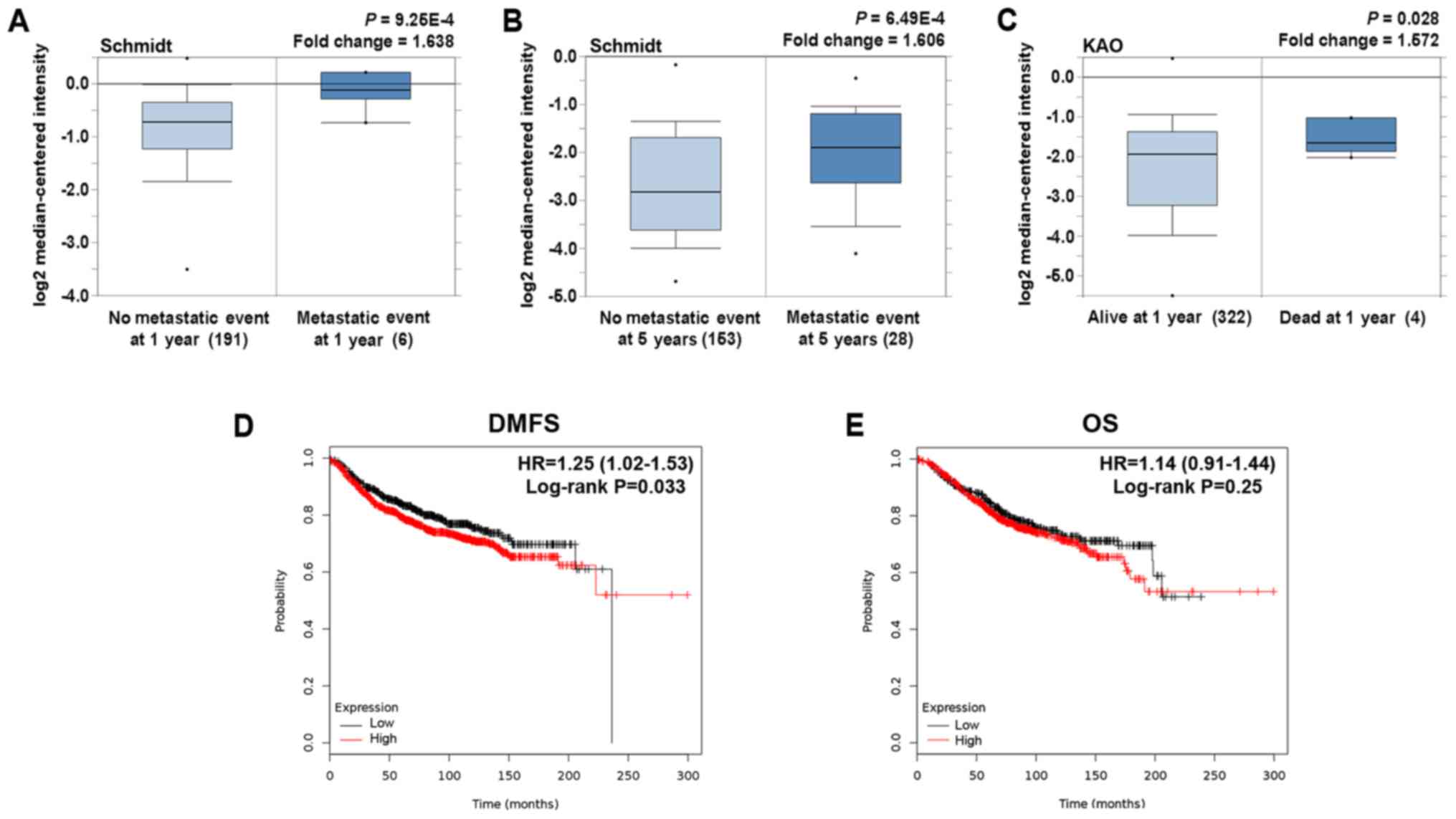
On clinical outcome analysis, higher SIRT5
expression was prone to occur in patients presenting with
metastasis at 1 and 5 years (Fig. 6A
and B). Additionally, in comparison with patients who were
still alive at 1 year, those who died within 1 year exhibited SIRT5
overexpression (Fig. 6C).
Meanwhile, differential expression of SIRT5 was observed between
the primary tumor site and metastatic site, as well as between
breast carcinomas with other variable clinicopathological
characteristics (grade, stage and distant metastatic status;
Table III). Collectively, these
results indicated that breast patients with elevated SIRT5 mRNA
levels have relatively low 1- and 5-year distant metastasis-free
survival rates and 1-year survival rates.
 | Table III.SIRT5 expression analysis in primary
sites relative to metastasis and in different pathological subtypes
of breast cancer (Oncomine database). |
Table III.
SIRT5 expression analysis in primary
sites relative to metastasis and in different pathological subtypes
of breast cancer (Oncomine database).
| Study name | Cancer type | Total (n) | Fold change | P-value |
|---|
| Radvanyi
breast | Primary site vs.
metastasis (33 vs. 4) | 37 |
2.398 | 9.53E-6 |
| Bittner breast
2 | Primary site vs.
metastasis (327 vs. 9) | 336 | −1.850 | 0.035 |
| Ma breast 4 | Grade 1 vs. grade 2
(4 vs. 3) |
7 |
1.534 | 0.014 |
| Desmadt breast
2 | Grade 2 vs. grade 3
(5 vs. 4) |
9 | −2.127 | 0.028 |
| Bittner breast | Grade 1 vs. grade 2
(5 vs. 23) | 28 | −1.522 | 0.034 |
| Miyake breast
2 | Stage II vs. grade
III (97 vs. 18) | 115 |
1.701 | 3.52E-5 |
| Bittner breast | M0 vs. M1 (176 vs.
5) | 181 | −1.523 | 0.026 |
Survival analysis of SIRT5 in breast
cancer patients
Subsequently, to evaluate the prognostic effect of
SIRT5, we constructed Kaplan-Meier plots and log-rank analyses in
an online database. We performed this analysis using the ‘best
cutoff’ value of mRNA of SIRT5 (all percentiles are computed and
the best performing threshold is automatically chosen as the
cut-off). We observed that high mRNA expression of SIRT5 was an
indicator of increased risk for distant metastasis (P=0.033;
Fig. 6D). Although it was suggested
that SIRT5 may not be a predictor of OS (Fig. 6E), there was predictive value of
SIRT5 for early death in breast cancer patients, therefore further
study is needed.
Discussion
NAC is one of the well-established treatments for
breast cancer and is particularly relevant in the treatment of
TNBCs, as it has been demonstrated that TNBCs have a better
response to NAC than other molecular subtypes of breast cancer
(19). However, it should be noted
that only a minority of patients with TNBC have been reported to
achieve a pCR, which has recently been proposed as a surrogate for
relapse-free survival (RFS) in TNBC patients after NAC and is
well-accepted as the strongest predictive factor of good prognosis
in patients with TNBC (20,21). Therefore, it is necessary to
identify reliable predictors of pCR to NAC in TNBC patients. In the
present study, we used high-throughput sequencing data and reported
the role of SIRT5 in predicting pCR to anthracycline-taxane-based
NAC in TNBC patients for the first time. Using the gene expression
and treatment response data of 64 TNBC patients (GSE41998 dataset
of the GEO database) who underwent neoadjuvant
doxorubicin/cyclophosphamide therapy followed by paclitaxel
treatment, SIRT5 was screened as one of the five core genes
associated with the pCR of TNBC patients by generating GA-SVM-based
pCR classifiers. A positive association was also observed between
SIRT5 expression and the pCR to NAC. Additionally, in the Oncomine
database analysis, epirubicin/cyclophosphamide-docetaxel responders
were found to express elevated levels of SIRT5 when compared with
epirubicin/cyclophosphamide-docetaxel non-responders. Collectively,
these findings indicated the promising value of SIRT5 in predicting
the pCR of TNBC patients to anthracycline-taxane-based NAC.
SIRT5 is one of three mitochondrial SIRTs and has
recently been demonstrated to affect cellular metabolism by
regulating the post-translational modifications (PTMs) of multiple
mitochondrial proteins (22).
Previous studies have indicated that SIRT5 can affect ammonia
detoxification through the PTMs of carbamoyl phosphate synthetase 1
(23). Results of in vitro
experiments in the MDA-MB-231 human breast carcinoma cell line
indicated that silenced SIRT5 resulted in the accumulation of
ammonia; ammonia-induced autophagy was subsequently activated and
enabled tumor cells to survive in stressful conditions, including
those induced by chemotherapy (24). Additionally, it was found that
autophagy inhibition by proteasome inhibitors increased
anthracycline-induced apoptosis in breast cancer cells (25). Thus, it is plausible that elevated
SIRT5 expression may increase the sensitivity of cancer cells to
anthracycline treatment by promoting ammonia metabolism and
inhibiting autophagy.
In addition to cellular metabolism regulation,
multiple potential mechanisms through which SIRT5 may affect the
response of TNBC patients to NAC were identified by searching genes
that were positively correlated with SIRT5 expression in the DAVID
database to obtain the GO annotations. First, in the cellular
component ontology, the genes were primarily enriched in cell
membrane-related structures. The most significant item was
extracellular exosomes. Secretion of exosomes containing breast
cancer resistance proteins has been suggested to heighten the
response of breast cancer cells to doxorubicin (26). In the GO category of molecular
function, the enriched items were Rho guanyl-nucleotide exchange
factor activity and vinculin binding. Subsequently, it was found
that these genes were enriched in a variety of biological processes
involved in the endocrine, urinary and nervous systems.
Furthermore, it should be noted that the regulation of Rho proteins
was also enriched. It has been reported that the Rho signaling
pathway exerts its effects on cellular activities primarily through
its central role in actin cytoskeleton dynamics (27). A reciprocal relationship has also
been explored between Rho proteins and microtubules (28,29)
and it has been demonstrated that the activation of the Rho pathway
altered the activity of microtubule associated protein Tau (MAPT)
via phosphorylation to regulate microtubule stabilization (30). MAPT was indicated as a potential
predictor of pCR to taxane-containing NAC in advanced breast cancer
patients (31). Therefore, these
results collectively indicate that SIRT5 may increase the pCR to
anthracycline-taxane-based NAC by inhibiting ammonia-induced
autophagy and regulating the Rho signaling pathway.
With regard to SIRT5 and tumorigenesis, findings
from previous studies have demonstrated that SIRT5 may have
different effects on different cancer types by acting as a
tumor-suppressor or tumor-promoter. In comparison with normal
tissues, SIRT5 was significantly under expressed in head and neck
squamous cell carcinoma samples, indicating the role of SIRT5 as a
tumor-suppressor (13). Similarly,
the expression levels of SIRT5 were substantially downregulated in
endometrial carcinoma tissues compared with non-neoplastic
endometrium (11). However, no
significant change in SIRT5 expression was observed in basal cell
carcinoma tissues relative to non-cancerous tissues (12). For breast cancer, a previous study
reported significantly elevated expression levels of SIRT5 in
breast cancer tissues (14).
Nevertheless, in the Oncomine database analysis conducted in the
present study, the findings were inconsistent, with either higher
or lower expression levels of SIRT5 observed in the breast tumor
tissues of different datasets when compared with normal breast
tissues. Thus, we speculated that the effects of SIRT5 may even
vary in a specific cancer with different clinicopathological
characteristics.
To some extent, the results of the present study
indeed revealed an association of SIRT5 expression with the
clinicopathological characteristics of breast cancer. It was
observed that lobular and mucinous breast carcinoma expressed
higher levels of SIRT5 in comparison with ductal breast carcinoma.
SIRT5 was also overexpressed in breast carcinoma in situ
relative to invasive ductal breast carcinoma. These results
indicated that SIRT5 upregulation may be specific to certain
histopathological type and early-stage breast cancers. However, the
reason for this remains unclear at present. Furthermore, compared
with other molecular subtypes, the TNBC and BRCA1 mutation subtypes
expressed substantially elevated levels of SIRT5. In addition it
was observed that elevated expression levels of SIRT5 were
significantly associated with 1- and 5-year distant metastasis and
with patient death at 1 year. These correlations are consistent
with BRCA1 mutations generally being associated with metastatic and
aggressive disease, and with TNBC patients having the worst overall
and disease-free survival and poor distant metastasis-free
survival. Furthermore, the results of online KM plotter analysis
also indicated that SIRT5 was a reliable biomarker for DMFS and
short OS in breast cancer patients. Collectively, these findings
indicated that elevated SIRT5 expression may be involved in the
formation of specific histological subtypes of breast cancer and
confer a stronger invasive ability on breast cancer cells.
In conclusion, the findings of the present study
implicated SIRT5 as a predictor of pCR to
anthracycline-taxane-based NAC in TNBC patients. GO analysis
indicated that the Rho pathway may be the mechanism through which
SIRT5 impacted the response of TNBC to NAC. Additionally, it was
found that the expression of SIRT5 was positively correlated with
the subtypes of TNBC and BRCA1 mutation, as well as closely
associated with poor clinical outcomes and poor prognostic
clinicopathological parameters. The results of the present study
may provide valuable indications for the basic research and
clinical treatment of SIRT5-expressing responders with TNBC to NAC.
However, further investigations are required to confirm our
hypothesis.
Acknowledgements
We would like to thank all the participants
recruited in the present study.
Glossary
Abbreviations
Abbreviations:
|
SIRT5
|
sirtuin 5
|
|
NAC
|
neoadjuvant chemotherapy
|
|
TNBC
|
triple-negative breast cancer
|
|
ER
|
estrogen receptor
|
|
PR
|
progesterone receptor
|
|
HER2
|
human epidermal growth factor receptor
2
|
|
pCR
|
pathological complete response
|
|
DEGs
|
differentially expressed genes
|
|
GA-SVM
|
genetic algorithm-support vector
machine
|
|
RFS
|
relapse-free survival
|
|
PTMs
|
post-translational modifications
|
References
|
1
|
Yardley DA, Brufsky A, Coleman RE, Conte
PF, Cortes J, Glück S, Nabholtz JM, O'Shaughnessy J, Beck RM, Ko A,
et al: Phase II/III weekly nab-paclitaxel plus gemcitabine or
carboplatin versus gemcitabine/carboplatin as first-line treatment
of patients with metastatic triple-negative breast cancer (the
tnAcity study): Study protocol for a randomized controlled trial.
Trials. 17:632016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
von Minckwitz G and Martin M: Neoadjuvant
treatments for triple-negative breast cancer (TNBC). Ann Oncol. 23
Suppl 6:vi35–vi39. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hatzis C, Symmans WF, Zhang Y, Gould RE,
Moulder SL, Hunt KK, Abu-Khalaf M, Hofstatter EW, Lannin D, Chagpar
AB, et al: Relationship between complete pathologic response to
neoadjuvant chemotherapy and survival in triple-negative breast
cancer. Clin Cancer Res. 22:26–33. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Song IH, Heo SH, Bang WS, Park HS, Park
IA, Kim YA, Park SY, Roh J, Gong G and Lee HJ: Predictive value of
tertiary lymphoid structures assessed by high endothelial venule
counts in the neoadjuvant setting of triple-negative breast cancer.
Cancer Res Treat. 49:399–407. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Vici P, Di Benedetto A, Ercolani C,
Pizzuti L, Di Lauro L, Sergi D, Sperati F, Terrenato I, Dattilo R,
Botti C, et al: Predictive significance of DNA damage and repair
biomarkers in triple-negative breast cancer patients treated with
neoadjuvant chemotherapy: An exploratory analysis. Oncotarget.
6:42773–42780. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Humbert O, Riedinger JM, Charon-Barra C,
Berriolo-Riedinger A, Desmoulins I, Lorgis V, Kanoun S, Coutant C,
Fumoleau P, Cochet A, et al: Identification of biomarkers including
18FDG-PET/CT for early prediction of response to
neoadjuvant chemotherapy in triple-negative breast cancer. Clin
Cancer Res. 21:5460–5468. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Yang L, Ma X, He Y, Yuan C, Chen Q, Li G
and Chen X: Sirtuin 5: A review of structure, known inhibitors and
clues for developing new inhibitors. Sci China Life Sci.
60:249–256. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Vellinga TT, Borovski T, de Boer VC,
Fatrai S, van Schelven S, Trumpi K, Verheem A, Snoeren N, Emmink
BL, Koster J, et al: SIRT1/PGC1α-dependent increase in oxidative
phosphorylation supports chemotherapy resistance of colon cancer.
Clin Cancer Res. 21:2870–9. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bai L, Lin G, Sun L, Liu Y, Huang X, Cao
C, Guo Y and Xie C: Upregulation of SIRT6 predicts poor prognosis
and promotes metastasis of non-small cell lung cancer via the
ERK1/2/MMP9 pathway. Oncotarget. 7:40377–40386. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kugel S, Sebastián C, Fitamant J, Ross KN,
Saha SK, Jain E, Gladden A, Arora KS, Kato Y, Rivera MN, et al:
SIRT6 suppresses pancreatic cancer through control of Lin28b. Cell.
165:1401–1415. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bartosch C, Monteiro-Reis S, Almeida-Rios
D, Vieira R, Castro A, Moutinho M, Rodrigues M, Graça I, Lopes JM
and Jerónimo C: Assessing sirtuin expression in endometrial
carcinoma and non-neoplastic endometrium. Oncotarget. 7:1144–1154.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Temel M, Koç MN, Ulutaş S and Göğebakan B:
The expression levels of the sirtuins in patients with BCC. Tumour
Biol. 37:6429–6435. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lai CC, Lin PM, Lin SF, Hsu CH, Lin HC, Hu
ML, Hsu CM and Yang MY: Altered expression of SIRT gene family in
head and neck squamous cell carcinoma. Tumour Biol. 34:1847–1854.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Igci M, Kalender ME, Borazan E, Bozgeyik
I, Bayraktar R, Bozgeyik E, Camci C and Arslan A: High-throughput
screening of Sirtuin family of genes in breast cancer. Gene.
586:123–128. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Horak CE, Pusztai L, Xing G, Trifan OC,
Saura C, Tseng LM, Chan S, Welcher R and Liu D: Biomarker analysis
of neoadjuvant doxorubicin/cyclophosphamide followed by ixabepilone
or Paclitaxel in early-stage breast cancer. Clin Cancer Res.
19:1587–1595. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Peng S, Xu Q, Ling XB, Peng X, Du W and
Chen L: Molecular classification of cancer types from microarray
data using the combination of genetic algorithms and support vector
machines. FEBS Lett. 555:358–362. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Györffy B, Lanczky A, Eklund AC, Denkert
C, Budczies J, Li Q and Szallasi Z: An online survival analysis
tool to rapidly assess the effect of 22,277 genes on breast cancer
prognosis using microarray data of 1,809 patients. Breast Cancer
Res Treat. 123:725–731. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Rhodes DR, Kalyana-Sundaram S, Mahavisno
V, Varambally R, Yu J, Briggs BB, Barrette TR, Anstet MJ,
Kincead-Beal C, Kulkarni P, et al: Oncomine 3.0: Genes, pathways,
and networks in a collection of 18,000 cancer gene expression
profiles. Neoplasia. 9:166–180. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Liedtke C, Mazouni C, Hess KR, André F,
Tordai A, Mejia JA, Symmans WF, Gonzalez-Angulo AM, Hennessy B,
Green M, et al: Response to neoadjuvant therapy and long-term
survival in patients with triple-negative breast cancer. J Clin
Oncol. 26:1275–1281. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Caudle AS, Yu TK, Tucker SL, Bedrosian I,
Litton JK, Gonzalez-Angulo AM, Hoffman K, Meric-Bernstam F, Hunt
KK, Buchholz TA, et al: Local-regional control according to
surrogate markers of breast cancer subtypes and response to
neoadjuvant chemotherapy in breast cancer patients undergoing
breast conserving therapy. Breast Cancer Res. 14:R832012.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Li J, Chen S, Chen C, Di G, Liu G, Wu J
and Shao Z: Pathological complete response as a surrogate for
relapse-free survival in patients with triple negative breast
cancer after neoadjuvant chemotherapy. Oncotarget. 8:18399–18408.
2017.PubMed/NCBI
|
|
22
|
Kumar S and Lombard DB: Mitochondrial
sirtuins and their relationships with metabolic disease and cancer.
Antioxid Redox Signal. 22:1060–1077. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Nakagawa T, Lomb DJ, Haigis MC and
Guarente L: SIRT5 Deacetylates carbamoyl phosphate synthetase 1 and
regulates the urea cycle. Cell. 137:560–570. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Polletta L, Vernucci E, Carnevale I,
Arcangeli T, Rotili D, Palmerio S, Steegborn C, Nowak T,
Schutkowski M, Pellegrini L, et al: SIRT5 regulation of
ammonia-induced autophagy and mitophagy. Autophagy. 11:253–270.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Guo W, Wang Y, Wang Z, Wang YP and Zheng
H: Inhibiting autophagy increases epirubicin's cytotoxicity in
breast cancer cells. Cancer Sci. 107:1610–1621. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kong JN, He Q, Wang G, Dasgupta S, Dinkins
MB, Zhu G, Kim A, Spassieva S and Bieberich E: Guggulsterone and
bexarotene induce secretion of exosome-associated breast cancer
resistance protein and reduce doxorubicin resistance in MDA-MB-231
cells. Int J Cancer. 137:1610–1620. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Hanna S and El-Sibai M: Signaling networks
of Rho GTPases in cell motility. Cell Signal. 25:1955–1961. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wojnacki J, Quassollo G, Marzolo MP and
Cáceres A: Rho GTPases at the crossroad of signaling networks in
mammals: Impact of Rho-GTPases on microtubule organization and
dynamics. Small GTPases. 5:e284302014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Hervé JC and Bourmeyster N: Rho GTPases at
the crossroad of signaling networks in mammals. Small GTPases.
6:43–48. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Amano M, Kaneko T, Maeda A, Nakayama M,
Ito M, Yamauchi T, Goto H, Fukata Y, Oshiro N, Shinohara A, et al:
Identification of Tau and MAP2 as novel substrates of Rho-kinase
and myosin phosphatase. J Neurochem. 87:780–790. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Li ZH, Xiong QY, Tu JH, Gong Y, Qiu W,
Zhang HQ, Wei WS, Hou YF and Cui WQ: Tau proteins expressions in
advanced breast cancer and its significance in taxane-containing
neoadjuvant chemotherapy. Med Oncol. 30:5912013. View Article : Google Scholar : PubMed/NCBI
|