Introduction
Gastric cancer (GC) is one of the most common
cancers and the second leading cause of cancer-associated mortality
worldwide (1). There are
approximately 750,000 newly diagnosed cases each year (2). However, patients with GC have a poor
prognosis with a 5-year overall survival (OS) rate less than 25%
(2). The OS of GC patients is
considered to be associated with age, tumor node metastasis stage,
histological grade and surgical approach (3). The unsatisfactory clinical outcome is
greatly due to a lack of knowledge regarding the molecular
pathogenesis of GC. Therefore, the identification of potential
biomarkers and effective targeted therapies for GC to predict
prognosis are urgently required.
Non-coding RNAs (ncRNAs) are RNA molecules that
broadly exist in high eukaryotic organisms but have no ability to
code proteins (4). Sana et
al (5) reported that ncRNAs may
play important biological roles in cellular development, chromosome
formation, transcriptional regulation and RNA modification.
Long non-coding RNAs (lncRNAs), once considered
transcriptional ‘noise’, are one subtype of ncRNA that are >200
nucleotides in length (6). To date,
over 12,000 lncRNAs encoded in the human genome have been
identified. Increasing evidence has revealed that these lncRNAs
regulate proliferation, invasion and metastasis in various cancers
(5,7–10).
Furthermore, several studies have revealed that lncRNAs have
potential in the prediction of the diagnosis and prognosis of
malignant tumors including GC (11–13).
LncRNAs play a role in crucial biological functions
in multiple ways. Dysregulated lncRNAs can function as oncogenes or
tumor suppressors to alter cellular pathways. LncRNAs have also
been demonstrated to promote tumor cell migration and metastasis by
inducing the epithelial-mesenchymal transition. Furthermore,
several lncRNAs have significant effects on multidrug resistance,
which is responsible for chemotherapy failure. In 2011, Salmena
et al (14) proposed the
competitive endogenous RNA (ceRNA) hypothesis that RNA transcripts
communicate with and regulate each other by using shared miRNA
response elements (MREs). This competition between mRNAs, lncRNAs
and pseudogene transcripts regulates their expression using MREs to
compete for binding with microRNAs (miRNAs), which is important for
tumor initiation and progression (15,16).
Later, this hypothesis was validated experimentally by other
researchers (17,18).
The functions of various lncRNAs in GC have already
been verified (18–21). However, there is still a lack of
studies with large sample sizes and cancer-specific lncRNA
biomarkers in GC. Furthermore, fewer studies have been designed to
identify the potential ceRNA network.
In the present study, the authors used data from 372
tumor tissues and 32 adjacent non-tumor tissues from The Cancer
Genome Atlas (TCGA), which provides information on RNA sequencing
including mRNA, miRNA and lncRNA data. Then, the ceRNA network in
GC was constructed. To verify the reliability of these results,
four lncRNAs from the ceRNA network were randomly selected and
their expression levels and functions in the GC cell line SGC-7901
were examined.
Materials and methods
Data
GC patients with RNA sequence data were enrolled in
a comprehensive integrated analysis from TCGA database (www.portal.gdc.cancer.gov). The enrollment
criteria were as follows: i) Gastric adenocarcinoma; ii) enough
data for analysis; iii) without another type of malignant tumor and
iv) naive to preoperative radiotherapy and chemotherapy. Finally,
404 samples including 372 tumor tissues and 32 adjacent non-tumor
tissues were included. The study was performed in accordance with
the publication guidelines provided by TCGA (www.cancergenome.nih.gov/publications/publicationguidelines).
RNA sequence data and differentially
expressed analysis
The stomach adenocarcinoma (STAD) RNA expression
profile data (level 3) of the corresponding patients were obtained
from TCGA-STAD database (retrospect to Sep 1, 2017), which provides
normalized transcriptome profiling data from high-throughput
sequencing, including lncRNA, mRNA and miRNA sequencing profiles.
Each sample consisted of the corresponding RNAseq, miRNAseq and
clinical data. Next, the differential expression levels of lncRNAs,
mRNAs and miRNAs were compared between tumor tissues and adjacent
non-tumor gastric tissues. This was conducted using ‘edgeR’
(22), a bioconductor package based
on R language (Version 3.4.3, 2017, http://www.r-project.org), to identify the
differentially expressed lncRNAs (DElncRNAs) and differentially
expressed mRNAs (DEmRNAs) with thresholds of log fold change (FC)
>2.0 and the false discovery rate (FDR)-adjusted P-values
<0.01, as well as differentially expressed miRNAs (DEmiRNAs)
with thresholds of log FC >1.5 and FDR-adjusted P-values
<0.01. The ‘ggplot2’ package in R was used to build the volcano
plots of DElncRNAs, DEmRNAs and DEmiRNAs (23). The DEmRNAs were submitted to the
Database for Annotation, Visualization, and Integrated Discovery
(DAVID) (www.david.abcc.ncifcrf.gov) to be classified into
different Gene Ontology (GO) and Kyoto Encyclopedia of Genes and
Genomes (KEGG) annotation groups. Upregulated and downregulated
genes were analyzed. The criteria were set as P<0.05 and fold
enrichment score >1.5.
Construction of the ceRNA network
In the present study, lncRNA-miRNA-mRNA interactions
were predicted by the overlapping of the miRNA seed sequence
binding site both on the chosen dysregulated lncRNAs and the
significantly dysregulated mRNAs. The miRcode (www.mircode.org) was used to predict the lncRNA-miRNA
interactions (24). MiRNA-targeted
mRNAs were determined by using miRDB (www.mirdb.org), Targetscan (www.targetscan.org) and miRTarBase (mirtarbase.mbc.nctu.edu.tw). To further enhance
the ceRNA network reliability, the intersective lncRNAs and mRNAs
were selected on the miRNA prediction and the differentially
expressed data form TCGA-STAD. Those intersection lncRNAs not
included in the GENCODE lncRNA annotation (www.gencodegenes.org, V27) were discarded. Cytoscape
v3.5.0 software was used to construct the interactive and visual
ceRNA network (25). The DEmRNA in
the ceRNA network were enriched in the KEGG pathway by KOBAS 3.0
(http://kobas.cbi.pku.edu.cn) to analyze
the potential functions.
Cell culture and transfection
Human GC cell line SGC-7901 and normal gastric
epithelial cell line GES-1 were purchased from Shanghai Cell Bank,
Chinese Academy of Sciences (Shanghai, China). Cells was cultured
in Dulbecco's modified Eagles medium (DMEM; Gibco; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal
bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.), 100
IU/ml penicillin, and 100 mg/ml streptomycin in a humidified
atmosphere at 37°C in an environment containing 5% CO2.
For transfection, cells were cultured up to 70% confluency and
transfected with 50 nM specific lncRNAs small interfering RNA
(siRNA, si-ERVMER61, 5′-GGGUACUGUGUGUGAUAUC-3′; si-DSCR4-IT1,
5′-GAGCCAUCCAAGGAUACAA-3′; si-HULC, 5′-GGAAUUGGAGCCUUUACAA-3′;
si-LINC00200, 5′-UCGCACGCUUUGCGUAGAU-3′) or nontargeting siRNA
(si-NC, 5′-GCAAGUAUAGCCGUAAGCA-3′) (Guangzhou RiboBio Co., Ltd,
Guangzhou, China), using Lipofectamine® 3000
(Invitrogen; Thermo Fisher Scientific, Inc.) by incubating with
OptiMem-I (Gibco; Thermo Fisher Scientific, Inc.) media for 4 h.
The cells were then cultured in fresh DMEM with 10% FBS. After 12 h
of transfection, the relative levels of lncRNAs in transfected
cells were examined using reverse transcription-quantitative
polymerase chain reaction (RT-qPCR).
RNA extraction and RT-qPCR
validation
Total RNA was extracted from cultured cells using
TRIzol® LS reagent (Thermo Fisher Scientific, Inc.)
according to the manufacturer's protocol. For lncRNA reverse
transcription, cDNA was synthesized at 37°C for 1 h using 5×RT
Master Mix (Toyobo Life Science, Osaka, Japan). LncRNA expression
levels were quantified using qPCR with the SYBR Green I (Takara
Bio, Inc., Otsu, Japan) dye detection method on ABI vii7 PCR
instrument (Applied Biosystems; Thermo Fisher Scientific, Inc). The
sequences of the primers used for the PCR are presented in Table I. GAPDH was used as an internal
control for the expression analysis of lncRNAs. The PCR cycling
conditions were as follows: 95°C for 5 min, 40 cycles of 95°C for
10 sec, 60°C for 20 sec, dissociation at 95°C for 10 sec, 60°C for
10 sec. Relative quantification of lncRNA expression was calculated
using the 2−∆∆Cq method (26). The RT-qPCR reactions were all
repeated three times.
 | Table I.Primer sequences for reverse
transcription-quantitative polymerase chain reaction. |
Table I.
Primer sequences for reverse
transcription-quantitative polymerase chain reaction.
| Gene name | Primer sequences
(5′-3′) |
|---|
| ERVMER61 |
|
| F |
CAACCCACAGCAATTACACTTC |
| R |
CCCAAGACTAGCCCTACAAATC |
| DSCR4-IT1 |
|
| F |
GAGCCATCCAAGGATACAATCA |
| R |
AGTGAGCAAACACACAGAGG |
| HULC |
|
| F |
CATGATGGAATTGGAGCCTTTAC |
| R |
CCGGCCTTTACTTCAGAGTT |
| LINC00200 |
|
| F |
TTCCACACACAGGACCAAAG |
| R |
GCCCGATACATCAAAGCTACA |
| GAPDH |
|
| F |
TGACTTCAACAGCGACACCCA |
| R |
CACCCTGTTGCTGTAGCCAAA |
Cell proliferation assay
The cell proliferation ability was assessed using a
Cell Counting Kit-8 (CCK-8) assay (Beyotime Institute of
Biotechnology, Haimen, China), and was conducted according to the
manufacturer's protocol. In brief, the siRNA transfected groups and
the si-NC group of SGC-7901 cells (5×104) were seeded
into each well of a 96-well plate and cultured in 100 µl DMEM
supplemented with 10% FBS for 24 h. At 0, 24, 48 and 72 h, medium
was exchanged for 10 µl CCK-8 reagent, and the cells were incubated
for 1 h. The absorbance was measured for each well at a wavelength
of 450 nm. Cell growth was monitored every 24 h over 3 days. Each
experiment was performed at least 3 times.
Transwell invasion assay
The co-culture system was used to evaluate the
regulation of invasiveness in SGC-7901 cells. Cell invasion assays
were performed using transwell chambers (8 µm pore size; BD
Biosciences, Franklin Lakes, NJ, USA) precoated with Matrigel
(Corning Life Sciences, Tewksbury, MA, USA). Approximately
1×106 GC cells in serum-free DMEM media were seeded into
the upper chambers following siRNA or si-NC transfection. The lower
chamber contained medium with 10% FBS as a chemoattractant. After
incubation for 24 h, the non-invading cells and gel were removed
from the upper chamber with cotton tipped swabs, whereas cells
attached to the lower surface of the chamber were fixed and stained
with crystal violet solution for 10 min at room temperature, after
which the positive cell number was counted under an inverted
microscope (IX71; Olympus Corporation, Tokyo, Japan) in five random
fields. At least three chambers from three different experiments
were analyzed.
Wound healing assay
Cell migration was evaluated by a wound healing
assay to determine whether these lncRNAs could be involved in the
regulation of the migration of GC cells. In brief, the siRNA
transfected groups and the si-NC group of SGC-7901 cells were
incubated in 6-well plates, respectively. A small wound area was
made in the 90% confluent monolayer by using a 200 µl pipette tip
in a lengthwise stripe. Cells were then incubated in serum-free
DMEM media at 37°C in a 5% CO2 incubator for 24 h.
Photographs were taken at the indicated time points using a Bx50
microscope (Olympus Corporation). A total of ten measurements were
made at random intervals along the wound length. Data were averaged
and expressed as a percentage of the original width. The wound
healing assay was conducted in triplicate.
Statistical analysis
To identify the DERNAs associated with prognosis in
the ceRNA network, the survival curves of differentially expressed
lncRNAs, miRNAs and mRNAs were plotted using the ‘survival’ package
in R (27). The log-rank test was
used to compare significant differences between subgroups.
Student's t-test was used to compare the differences between two
groups. All analyses were performed using SPSS software version
24.0 for Windows (IBM SPSS, Armonk, NY, USA). P<0.05 was
considered to indicate a statistically significant difference.
Results
Patient characteristics
The detailed clinicopathological characteristics of
enrolled patients are listed in Table
II. All 372 patients were pathologically diagnosed with gastric
adenocarcinoma. The median age of the patients was 67 (ranging from
35 to 90). There were 239 (64.2%) males and 133 (35.8%) females.
The majority of patients were Caucasian (63.7%) and without distant
metastasis (87.9%).
 | Table II.Clinicopathological characteristics
of 372 patients with gastric cancer. |
Table II.
Clinicopathological characteristics
of 372 patients with gastric cancer.
| Parameter | Patients | (%) |
|---|
| Age (years) |
|
≤60 | 123 | 33.1 |
|
>60 | 249 | 66.9 |
| Gender |
|
Male | 239 | 64.2 |
|
Female | 133 | 35.8 |
| Race |
|
Asian | 73 | 19.6 |
|
White | 237 | 63.7 |
| Black
or African American | 11 | 3.0 |
| Not
available | 51 | 13.7 |
| Histologic
grade |
| High
(G1-2) | 145 | 39.0 |
| Low
(G3) | 218 | 58.6 |
|
GXa | 9 | 2.4 |
| Histologic
subtype |
| Signet
ring type | 10 | 2.7 |
| Diffuse
type | 63 | 16.9 |
| Not
otherwise specified | 206 | 55.4 |
|
Mucinous type | 19 | 5.1 |
|
Papillary type | 5 | 1.3 |
| Tubular
type | 68 | 18.3 |
| Not
available | 1 | 0.3 |
| Pathologic
stage |
| Stage
I | 52 | 14.0 |
| Stage
II | 110 | 29.6 |
| Stage
III | 152 | 40.9 |
| Stage
IV | 37 | 9.9 |
| Not
available | 21 | 5.6 |
| Pathologic T |
| T1 | 19 | 5.1 |
| T2 | 79 | 21.2 |
| T3 | 166 | 44.6 |
| T4 | 100 | 26.9 |
|
TXb | 8 | 2.2 |
| Pathologic N |
| N0 | 110 | 29.6 |
| N1 | 96 | 25.8 |
| N2 | 76 | 20.4 |
| N3 | 73 | 19.6 |
|
NXc | 17 | 4.6 |
| Pathologic M |
| M0 | 327 | 87.9 |
| M1 | 25 | 6.7 |
|
MXd | 20 | 5.4 |
| Status |
|
Death | 149 | 40.1 |
|
Alive | 222 | 59.7 |
| Not
available | 1 | 0.3 |
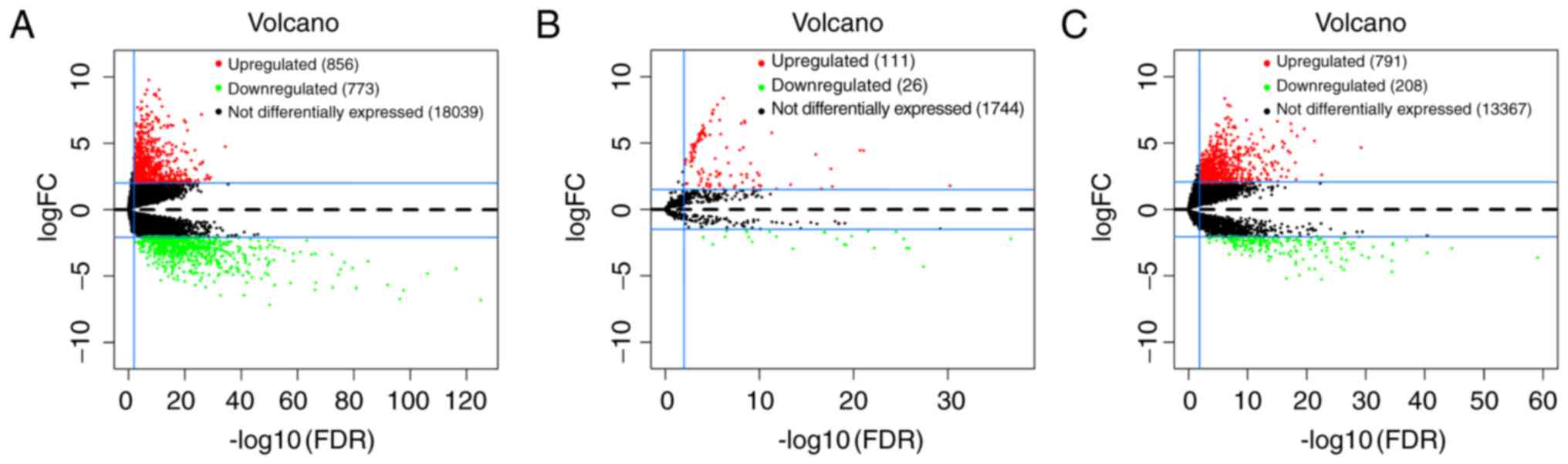
DEmRNAs and DEmiRNAs in GC
The present study identified the significant DEmRNAs
and DEmiRNAs in 372 tumor samples compared with the 32 adjacent
non-tumor samples from the TCGA database. With logFC >2 and
adjusted P-value <0.01, a total of 1629 DEmRNAs were identified
by the ‘edgeR’ package in R, including 856 (52.5%) upregulated and
773 (47.5%) downregulated genes. Furthermore, 137 miRNAs that were
differentially expressed from the TCGA database were identified
with logFC >1.5, adjusted P-value <0.01, of which 111 (81.0%)
were upregulated and 26 (23.4%) were downregulated. The volcano
plots of DEmRNAs and DEmiRNAs were built using the ‘ggplot2’
package in R (Fig. 1A and B). The
volcano plot of DElncRNAs was built using the same method (Fig. 1C).
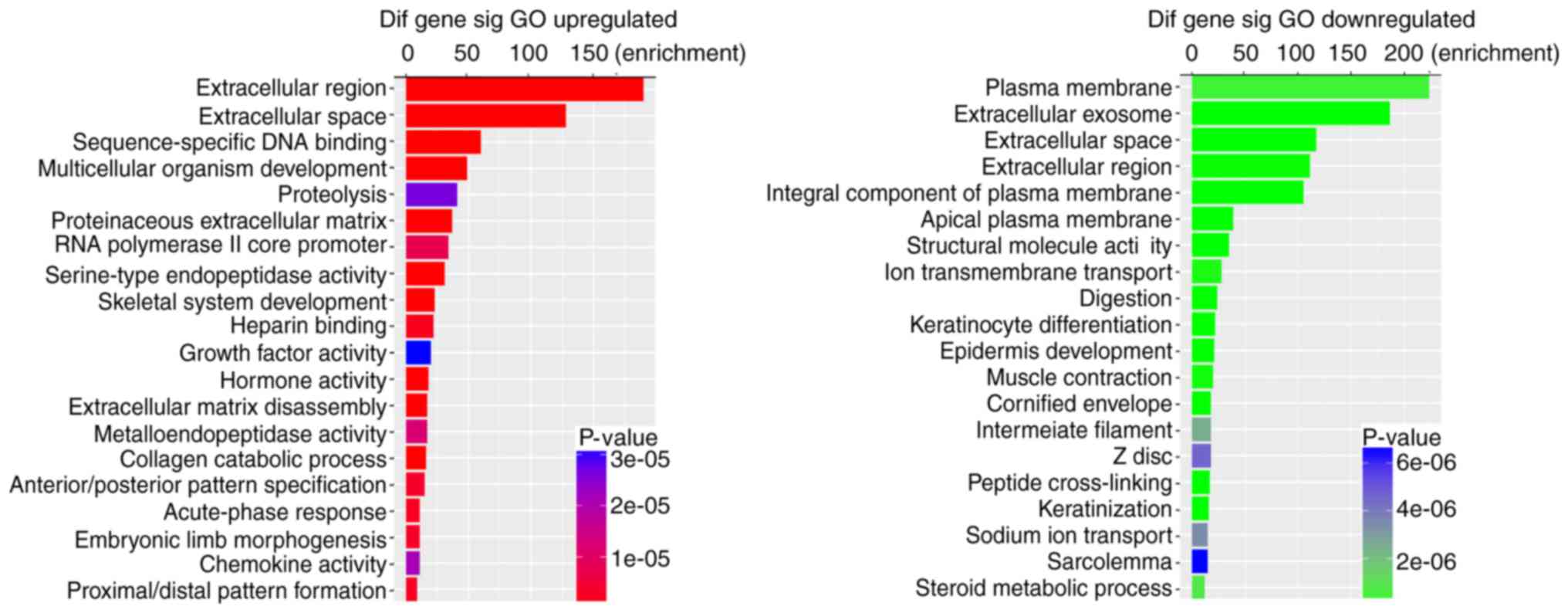
To further validate the potential functional
implication of 1629 DEmRNAs, functional enrichment analysis of
DEmRNA was performed based on the GO and KEGG pathways (P-value
<0.05 and fold enrichment score >1.5). It was demonstrated
that upregulated DEmRNAs were significantly enriched in 165 GO
terms, with 115 in biological process (BP), 17 in cellular
component (CC) and 33 in molecular function (MF). The downregulated
DEmRNAs were significantly enriched in 220 GO terms, with 123 in
BP, 38 in CC and 59 in MF. Fig. 2
shows the top 20 enriched GO terms for DEmRNAs based on the
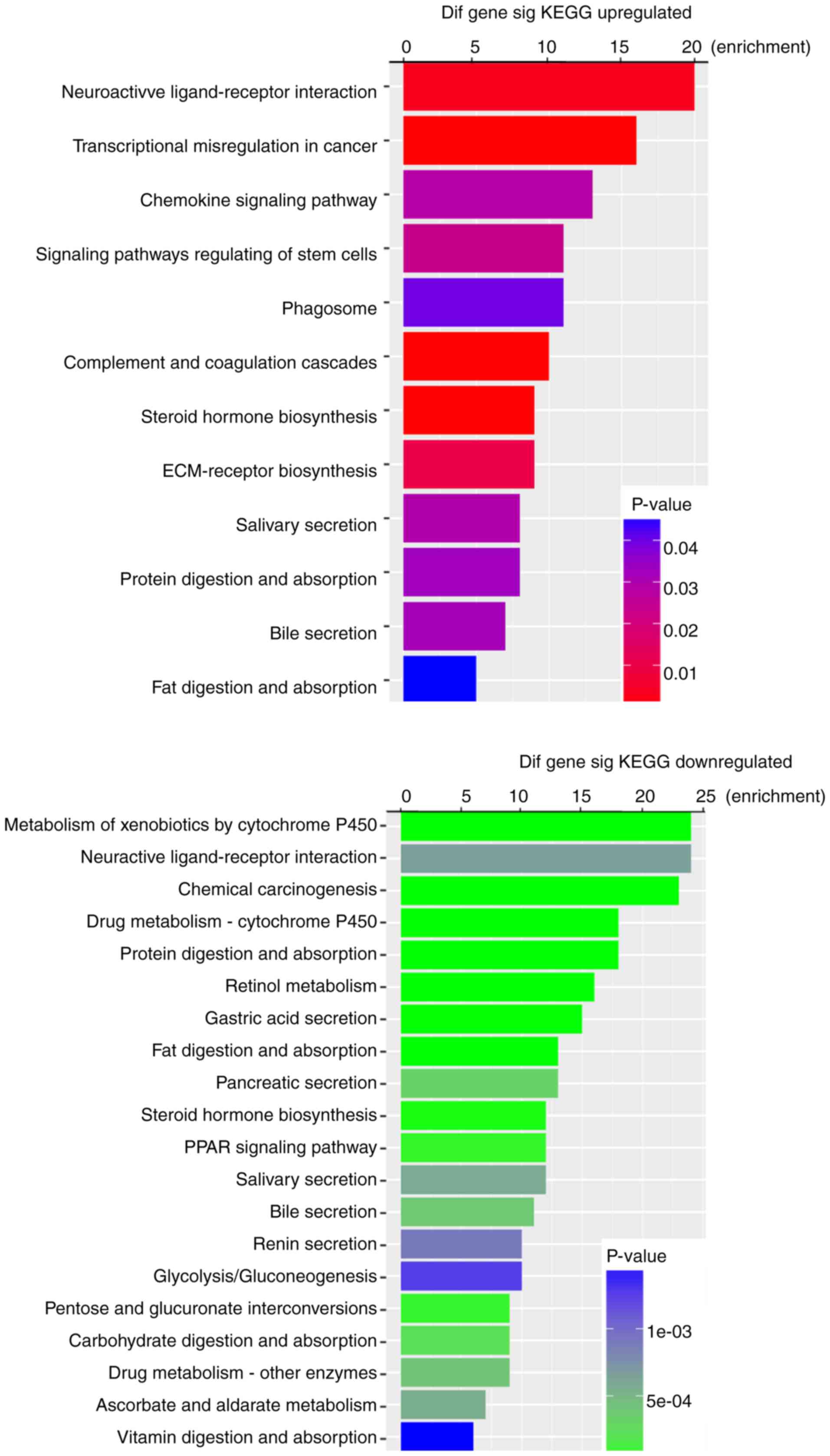
P-values. KEGG pathway analysis indicated that 12 pathways
corresponded to upregulated DEmRNAs and 43 pathways corresponded to
downregulated DEmRNAs. ‘Neuroactive ligand-receptor interaction’
and ‘Metabolism of xenobiotics by cytochrome P450’ were the most
enriched pathways in the upregulation and downregulation groups,
respectively (Fig. 3). Furthermore,
among these pathways, chemical carcinogenesis, transcriptional
misregulation in cancer, the chemokine signaling pathway and
signaling pathway regulation of stem cells were considered
cancer-associated pathways.
Next, 137 DEmiRNAs that targeted mRNAs were
predicted using miRDB, Targetscan and miRTarBase, and a total of
925 mRNAs were included in all the three databases. Finally, 24
mRNAs were selected that interacted with 9 DEmiRNAs (Table III) by intersecting 925 targeted
mRNAs and 1629 DEmRNAs. These 24 mRNAs were further used to
construct the ceRNA network.
 | Table III.miRNAs that may target gastric cancer
specific mRNAs. |
Table III.
miRNAs that may target gastric cancer
specific mRNAs.
| miRNAs | mRNAs |
|---|
| hsa-mir-96 | TRIB3 |
| hsa-mir-143 | COL1A1,
SERPINE1 |
| hsa-mir-145 | MEST, SERPINE1 |
| hsa-mir-195 | ALOX12, BTG2, CBX2,
CCNE1, CEP55, CLSPN, E2F7, HOXA10, KIF23, TMEM100, TPM2 |
| hsa-mir-204 | CHRDL1, HOXC8,
IL11, NPTX1 |
| hsa-mir-205 | ESRRG |
| hsa-mir-372 | ATAD2, CADM2,
LEFTY1, TMEM100 |
| hsa-mir-373 | ATAD2, CADM2,
LEFTY1, TMEM100 |
| hsa-mir-519d | ATAD2, FAM129A,
KIF23 |
DElncRNAs in GC
According to the cut-off criteria of logFC >2 and
adjusted P-value <0.01, the present study then identified 999
DElncRNAs between GC tumor tissues and adjacent non-tumor tissues
from TCGA database, of which 791 were upregulated and 208 were
downregulated. The volcano plot of DElncRNAs is presented in
Fig. 1C. The 9 key DEmiRNAs
mentioned in Table III were then
used to predict the corresponding lncRNAs using the miRcode
database. It was revealed that 9 DEmiRNAs interacted with the 63
DElncRNAs retrieved in the miRcode database (Table IV). Moreover, the selected 63
DElncRNAs (42 upregulated, 21 downregulated) were all identified in
the GENCODE lncRNA annotation (V27), and all of them were used to
build the ceRNA network.
 | Table IV.miRNAs that may target gastric cancer
specific lncRNAs. |
Table IV.
miRNAs that may target gastric cancer
specific lncRNAs.
| miRNAs | lncRNAs |
|---|
| hsa-mir-96 | ADAMTS9-AS1,
ADAMTS9-AS2, C8orf31, DLEU7-AS1, ERVMER61-1, FRMD6-AS2, HCG22,
LINC00114, LINC00221, LINC00534, LRRC3-AS1, MYB-AS1, NKX2-1-AS1,
RBMS3-AS3, UCA1 |
| hsa-mir-143 | AC074035.1,
AC110491.1, ADAMTS9-AS2, AL139002.1, AL391152.1, ARHGEF26-AS1,
C15orf54, C17orf77, C2orf48, CECR3, FRMD6-AS2, HOTAIR, HOTTIP,
LINC00114, LINC00163, LINC00221, LINC00200, LINC00284, LINC00460,
LINC00524, MIR205HG, PART1, UCA1 |
| hsa-mir-145 | ADAMTS9-AS1,
ADAMTS9-AS2, AP003027.1, DLX6-AS1, HCG22, LINC00052, LINC00184,
LINC00330, MIR205HG, NKX2-1-AS1, PART1 |
| hsa-mir-195 | ABCA9-AS1,
AC034229.1, AC087269.1, AC092422.1, AP002478.1, AP003027.1,
C15orf54, C2orf48, CECR3, DLEU7-AS1, DLX6-AS1, HCG22, HOTTIP,
IL20RB-AS1, LINC00200, LINC00284, LINC00326, LINC00355, LINC00330,
PART1 |
| hsa-mir-204 | AC006449.1,
AC034229.1, AC110491.1, ADAMTS9-AS2, ANO1-AS2, ARHGEF26-AS1,
C17orf77, C2orf48, C8orf31, DLX6-AS1, DSCR4-IT1, ERVMER61-1,
FRMD6-AS2, HOTAIR, HOTTIP, HULC, LINC00114, LINC00221, LINC00200,
LINC00330, LINC00332, LINC00410, LINC00501, LINC00524, LRRC3-AS1,
NKX2-1-AS1, MIR205HG, PART1, RBMS3-AS3, VCAN-AS1 |
| hsa-mir-205 | AC006449.1,
AC110491.1, ADAMTS9-AS2, AP002478.1, ARHGEF26-AS1, C20orf166-AS1,
ERVMER61-1, HOTTIP, IL20RB-AS1, LINC00184, LINC00284, LINC00326,
LINC00330, LINC00410, LINC00524, LINC00534, MIR205HG, PART1 |
| hsa-mir-372 | AC011374.1,
AC061975.6, ADAMTS9-AS2, ANO1-AS2, AP002478.1, ARHGEF26-AS1,
C15orf54, C17orf77, C20orf166-AS1, C2orf48, C8orf31, CECR3,
DLX6-AS1, HOTTIP, LINC00184, LINC00221, LINC00330, LINC00393,
LINC00534, VCAN-AS1 |
| hsa-mir-373 | AC011374.1,
AC061975.6, ADAMTS9-AS2, ANO1-AS2, AP002478.1, ARHGEF26-AS1,
C15orf54, C17orf77, C20orf166-AS1, C2orf48, C8orf31, CECR3,
DLX6-AS1, HOTTIP, LINC00184, LINC00221, LINC00330, LINC00393,
LINC00534, VCAN-AS1 |
| hsa-mir-519d | AC061975.6,
AC092422.1, AL391152.1, ANO1-AS2, AP002478.1, AP003027.1,
ARHGEF26-AS1, C17orf77, C20orf166-AS1, C2orf48, DLX6-AS1, H19,
HOTAIR, HOTTIP, IGF2-AS, LINC00052, LINC00184, LINC00200,
LINC00221, LINC00284, LINC00330, LINC00365, LINC00410, LINC00454,
NKX2-1-AS1, TDRG1, VCAN-AS1 |
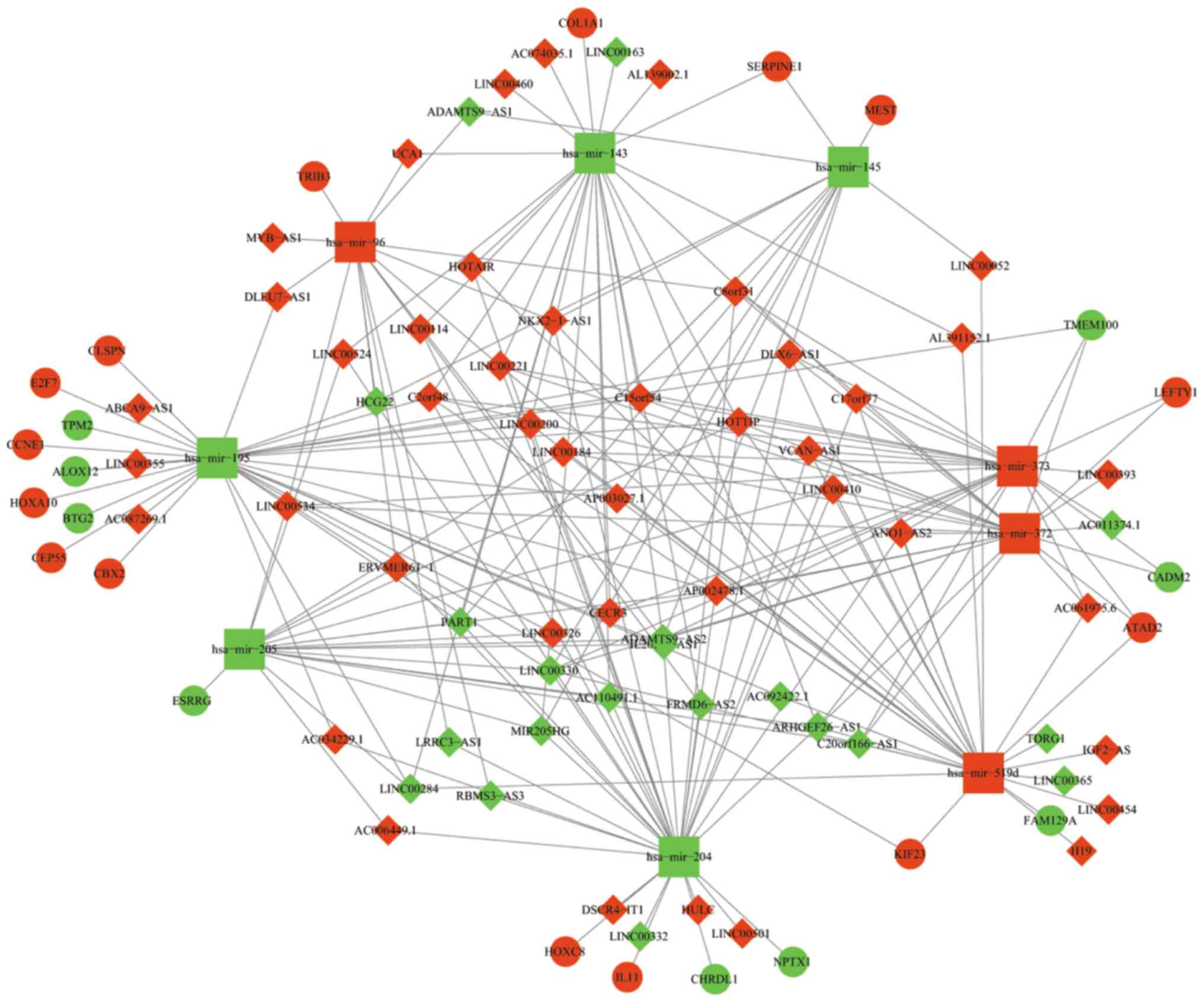
Construction of a ceRNA network
To better understand the mechanism by which lncRNA
mediates mRNA through combining with miRNA in GC, the present study
constructed a lncRNA-miRNA-mRNA ceRNA network using Cytoscape 3.5.0
according to the information provided in Tables III and IV. The network is presented in Fig. 4. In total, 63 DElncRNAs, 9 DEmiRNAs
and 24 DEmRNAs were involved in the network. Certain mRNAs involved
in the ceRNA network have been reported to be cancer-associated
genes, including Tribbles Pseudokinase 3, Serpin Family E Member 1
(SERPINE1), Mesoderm Specific Transcript, BTG Anti-Proliferation
Factor 2, Chromobox 2, Cyclin E1 (CCNE1), Centrosomal Protein 55
(CEP55), E2F Transcription Factor 7, Homeobox (HOX)A10, Kinesin
Family Member 23, Transmembrane Protein 100, HOXC8, interleukin 11
and ATPase Family, AAA Domain Containing 2 (ATAD2). Subsequently, a
Kaplan-Meier curve was used to analyze the association between the
differentially expressed genes in the ceRNA network and the OS of
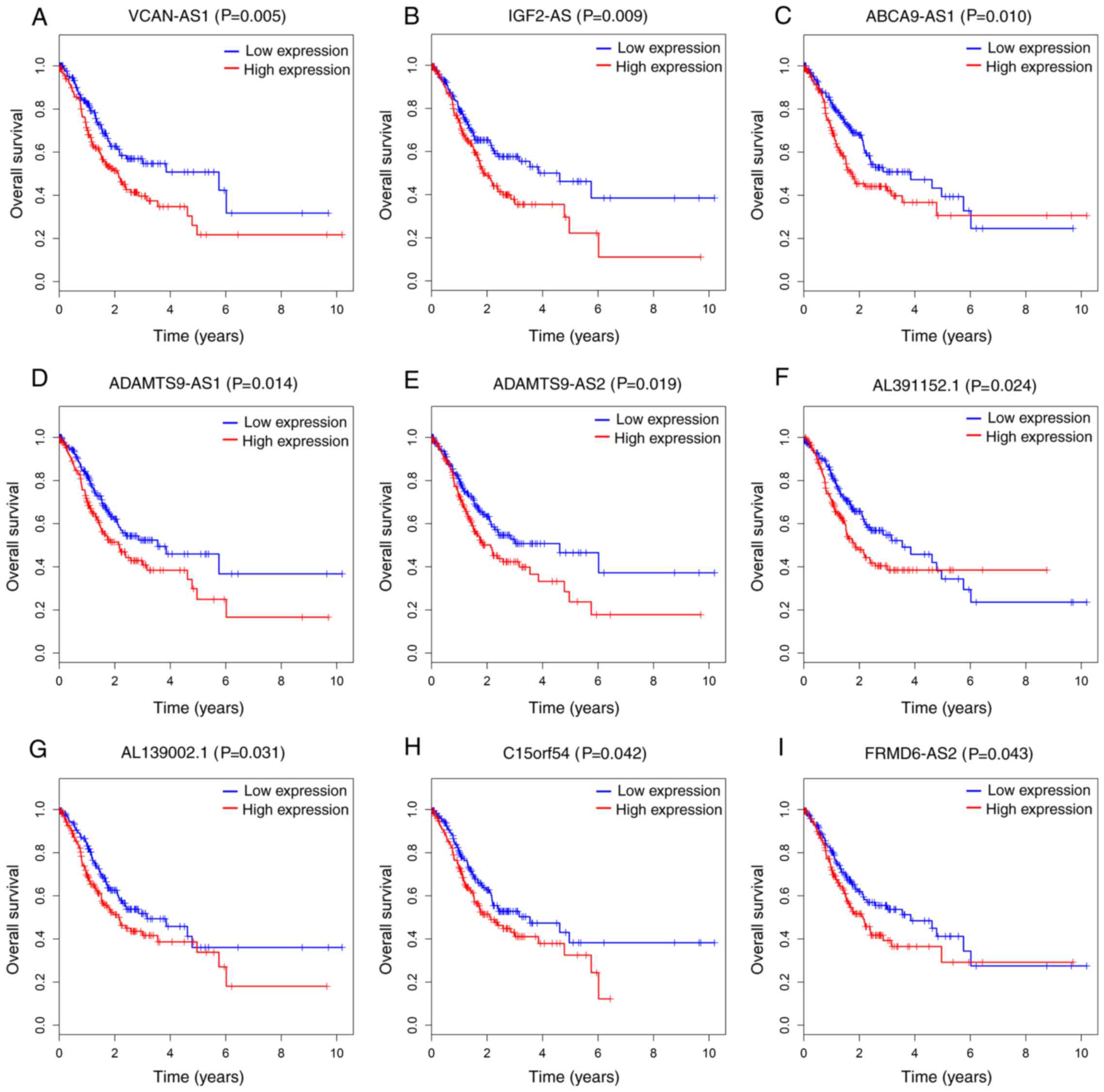
patients with GC. As a result, 9 out of 63 DElncRNAs were all
significantly negatively associated with OS (P<0.05; Fig. 5). Additionally, 6 out of 24 DEmRNAs
were considered to be significantly associated with OS. Increased
levels of CEP55, Claspin (CLSPN) and HOXA10 were associated with
longer survival time, while higher levels of Chordin Like 1
(CHRDL1), Neuronal Pentraxin 1 and SERPINE1 were associated with
poorer prognosis (P<0.05; Fig.
6). However, no association was observed between the key 9
DEmiRNAs and OS. Finally, the present study also analyzed the 24
DEmRNAs involved in the ceRNA network to understand the signaling
pathways indirectly regulated by lncRNAs by KOBAS 3.0 (http://kobas.cbi.pku.edu.cn). As presented in Table V, 4 KEGG pathways with statistical
significance were identified. A total of three of the pathways,
including the p53 signaling pathway, microRNAs in cancer and the
Phosphatidylinositol 3-kinase (PI3K)- RAC-a
serine/threonine-protein kinase (Akt) signaling pathway, were
involved in the pathogenesis of GC. One gene (CCNE1) was involved
in more than one KEGG pathway in accordance with GC
development.
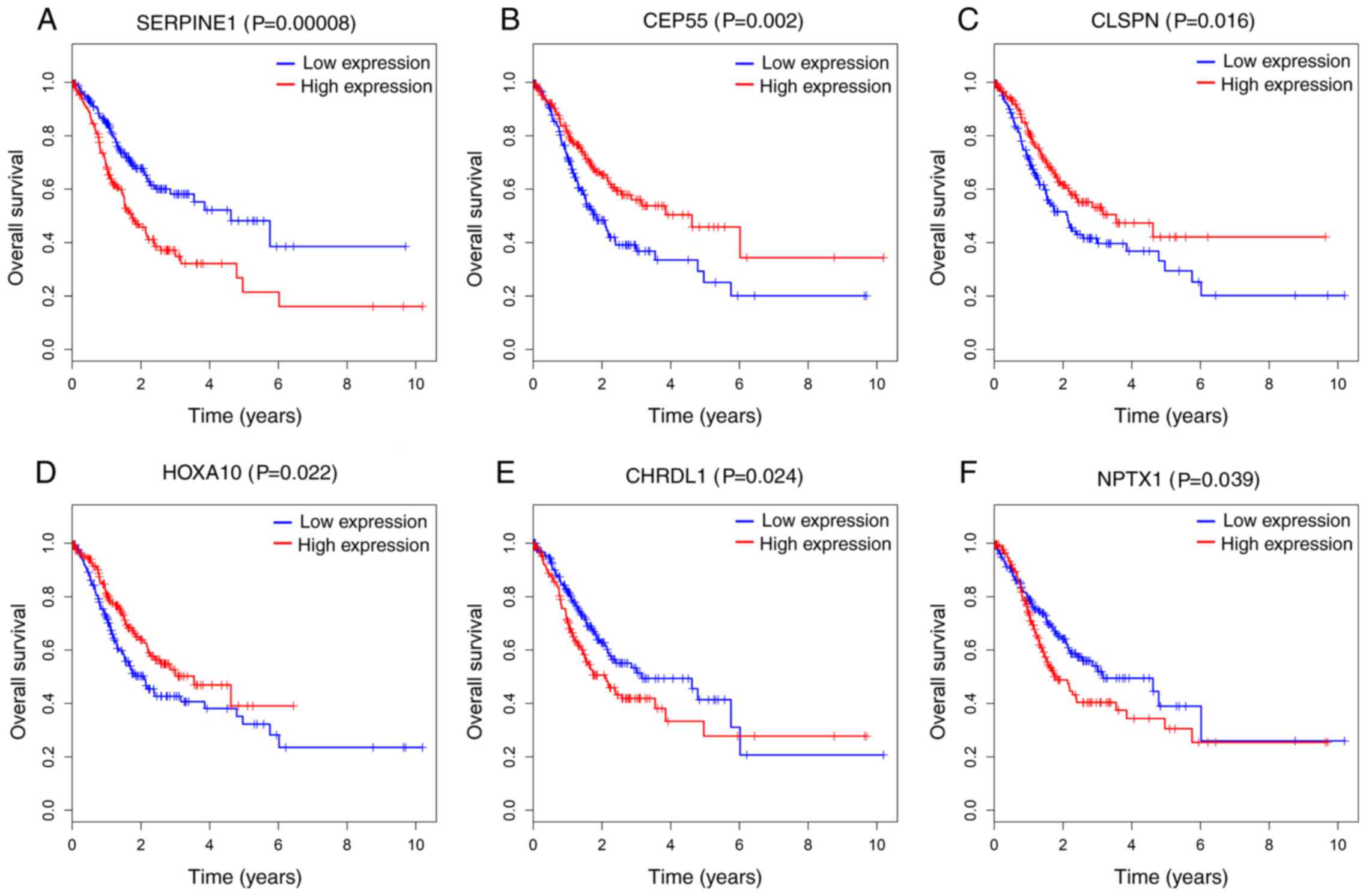 | Figure 6.Kaplan-Meier survival curves for 6
protein-coding genes associated with overall survival. Horizontal
axis: overall survival time (years); vertical axis: Survival
function. Six DEmRNAs are presented (P<0.05), including (A)
SERPINE1, (B) CEP55, (C) CLSPN, (D) HOXA10, (E) CHRDL1 and (F)
NPTX1. SERPINE 1, Serpin Family E Member 1; CEP55, Centrosomal
Protein 55; CLSPN, Claspin; HOXA10, Homeobox A10; CHRDL1, Chordin
Like 1; NPTX1, Neuronal Pentraxin 1. |
 | Table V.Kyoto Encyclopedia of Genes and
Genomes pathways enriched by the coding genes involved in competing
endogenous RNA network. |
Table V.
Kyoto Encyclopedia of Genes and
Genomes pathways enriched by the coding genes involved in competing
endogenous RNA network.
| Pathways ID | Description | P-value | Genes |
|---|
| hsa04115 | p53 signaling
pathway | 0.002088 | CCNE1,
SERPINE1 |
| hsa04933 | AGE-RAGE signaling
pathway in diabetic complications | 0.004497 | COL1A1,
SERPINE1 |
| hsa05206 | MicroRNAs in
cancer | 0.028122 | CCNE1, KIF23 |
| hsa04151 | PI3K-Akt signaling
pathway | 0.042544 | CCNE1, COL1A1 |
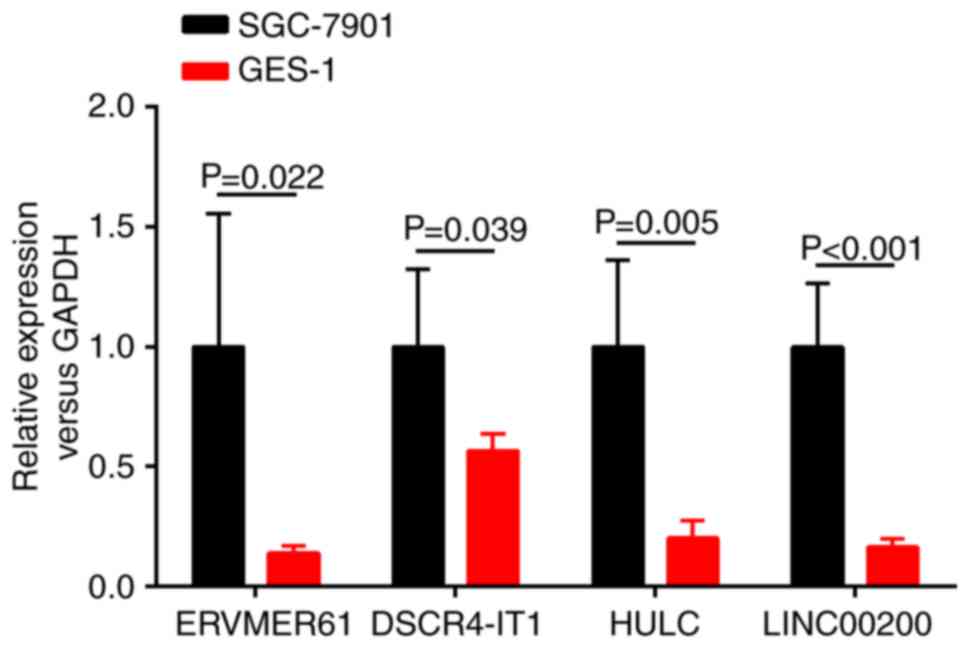
RT-qPCR validation of key lncRNAs
To confirm the credibility of the bioinformatics
results and the validity of the aforementioned analyzed results,
the present study randomly selected 4 upregulated key lncRNAs
(ERVMER61, DSCR4-IT1, HULC and LINC00200) from the ceRNA network
and examined the expression levels of these 4 lncRNAs between the
SGC-7901 cell line and the human gastric epithelial mucosa cell
line GES-1 by RT-qPCR. As presented in Fig. 7, the results revealed that the
expression levels of these 4 lncRNAs were all significantly
increased in the SGC-7901 cell line relative to the human gastric
epithelial mucosa cell line GES-1 (P<0.05). The conclusions were
consistent with TCGA database and bioinformatics predicted
results.
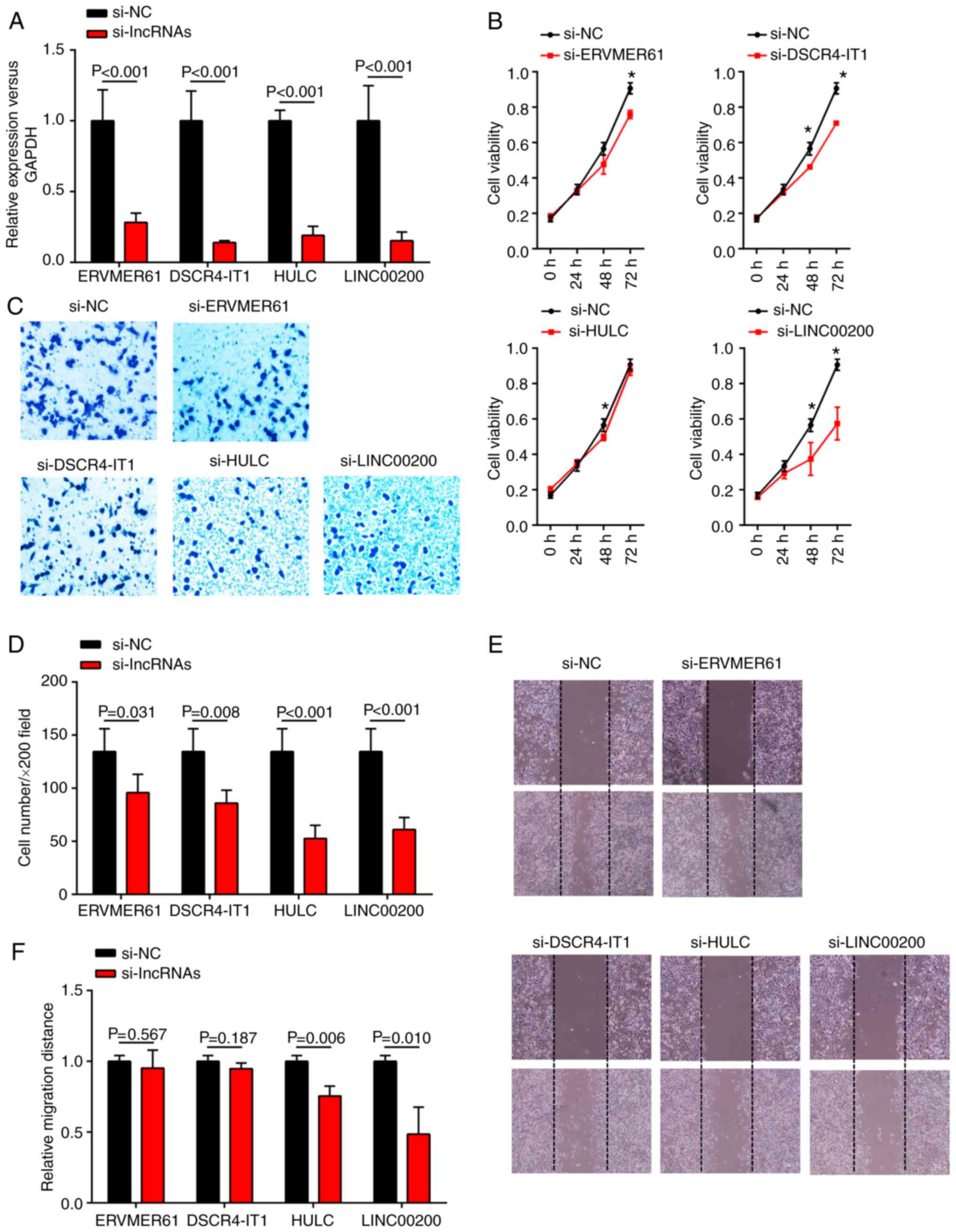
Exploration of the biological
functions of key lncRNAs
To better understand the biological functions of the
four key lncRNAs, the present study further investigated their
impact on tumor proliferation, invasion and migration in the
SGC7901 cell line. The 4 key lncRNAs (ERVMER61, DSCR4-IT1, HULC and
LINC00200) were knocked down in SGC-7901 cells via transfection
with corresponding siRNAs. The expression levels of the four
lncRNAs in SGC-7901 cells were evidently downregulated following
siRNA transfection (Fig. 8A).
Growth curves generated from CCK8 proliferation assays demonstrated
that knockdown of DSCR4-IT1 and LINC00200 significantly inhibited
cell proliferation in SGC-7901 cells at both 48 and 72 h
(P<0.05), whereas knockdown of ERVMER61 and HULC only resulted
in significant inhibition of the growth ability of SGC-7901 cells
at 72 and 48 h, respectively (Fig.
8B). These findings indicated that DSCR4-IT1 and LINC00200 may
behave as oncogenes to promote GC cell proliferation. In addition,
reduced cell invasion in SGC-7901 cells was observed after
si-ERVMER61, si-DSCR4-IT1, si-HULC and si-LINC00200 transfection
(Fig. 8C and D). Furthermore, a
decrease in cell migration was also observed when HULC or LINC00200
were significantly downregulated in a wound healing assay (Fig. 8E and F), suggesting that HULC and
LINC00200 can promote GC cell migration in vitro.
Discussion
Previously, multiple studies have suggested that
lncRNAs play a crucial role in the modulation of tumor behavior
through various complex mechanisms such as epigenetic regulation,
transcriptional regulation, and post-transcriptional regulation
(28–30). Until now, however, there have been
few studies of the expression profiles of lncRNAs in GC, and a few
of them were reported through a microarray or sequencing with a
small sample size (31). There is a
complex regulatory network association between lncRNAs and miRNAs
or lncRNAs and mRNAs in GC, and these networks play important roles
in the pathogenesis and progression of GC (32,33).
However, limited data are available. In the present study, the
authors aimed to investigate the interactions among lncRNAs, miRNAs
and mRNAs by constructing a ceRNA network and searched for lncRNAs
that may be promising biomarkers for GC.
In the present study, 1629 mRNAs were differentially
expressed from 372 GC tumor tissues and 32 non-tumor gastric
tissues based on the RNA sequence data from TCGA. With
bioinformatic technologies, 24 DEmRNAs were selected to construct
the ceRNA network. Then, the enrichment of functions and signaling
pathways of the DEmRNAs were analyzed using GO and KEGG,
respectively. The GO results indicated the functions of significant
differences mostly in terms of the biological process, cellular
component and molecular function. Among the KEGG pathway analysis
results, some were considered to be cancer associated, such as
transcriptional misregulation in cancer, signaling pathway
regulation of stem cells, p53 signaling pathway, PI3K-Akt signaling
pathways, and microRNAs in cancer. Various previous studies have
revealed that the PI3K-Akt and p53 signaling pathways play an
important role in cell proliferation and cell apoptosis reduction
(34–36). It has also been reported that the
lncRNAs AK023391 and MEG3 impact GC cell function via the PI3K-Akt
or p53 signaling pathway (37,38).
Furthermore, three DEmRNAs (SERPINE1, CCNE1 and COL1A1) from the
ceRNA network involved in the PI3K-Akt or p53 signaling pathway
were revealed to play a crucial role in the pathogenesis, prognosis
and molecular therapy of GC (39–41).
In addition, other cancer-associated pathways identified in the
study, including miRNAs in cancer, transcriptional misregulation in
cancer and chemical carcinogenesis, were also reported in previous
studies (21,42), indicating that the present results
are highly reliable.
The ceRNA hypothesis has been proposed as a novel
regulatory mechanism between non-coding RNA and coding RNA
(14). LncRNA can regulate gene
expression by interacting with the miRNAs by MREs. For example, Jin
et al (43) revealed that
the lncRNA SNHG15 contributes to non-small cell lung cancer (NSCLC)
tumorigenesis by regulating the CDK14 protein via sponging miR-486,
providing novel insight into NSCLC pathogenesis and a potential
therapeutic strategy for NSCLC patients. Yu et al (44) demonstrated that lncRNA PVT1 can
promote the metastasis and proliferation of colon cancer by
suppressing the miR-30d-5p/Runt Related Transcription Factor 2
axis. To understand the internal contact between lncRNA-miRNA-mRNA
in the development and progression of GC, the present study
constructed the ceRNA network by bioinformatics prediction based on
differences in lncRNA, miRNA and mRNA expression from TCGA. Certain
lncRNAs that exist in the network have been proven to interact with
miRNA and mRNA and may be potential biomarkers in the diagnosis,
therapy and prognosis of GC, including UCA1, HOTTIP, HOTAIR and H19
(45–48). Additionally, a few genes from the
ceRNA network, including CHRDL1, COL1A1, HOXC8 and ATAD2, have been
reported to act as tumor oncogenes or suppressor genes,
participating in tumor growth, invasion and metastasis (41,49–51).
Then, the present study analyzed the association
between the 63 key lncRNAs and OS. The results indicated that 9 of
them were significantly associated with survival and could be
considered as potential prognostic markers for GC. Among the 9
lncRNAs, high IGF2-AS expression has been reported to be associated
with poor clinical outcomes in ovarian cancer (52), and ADAMTS9-AS2 has also been
identified as a poor prognostic biomarker of colorectal cancer
(53) and glioma (54) with DNA methyltransferase-1. However,
the roles of remaining 7 lncRNAs (ABCA9-AS1, ADAMTS9-AS1,
AL139002.1, AL391152.1, C15orf54, FRMD6-AS2 and VCAN-AS1) have not
yet been reported.
Finally, four key lncRNAs (ERVMER61, DSCR4-IT1,
HULC, LINC00200) were randomly selected from the network and their
expression levels analyzed in the SGC-7901 and GEC-1 cell lines.
The expression data from TCGA and the verification results of the
GC cell line were in accordance. The present study also explored
the functions of the 4 lncRNAs in the SGC-7901 cell line and tried
to search for biomarkers that may affect the biological behaviors
of GC cells. The results demonstrated that all 4 lncRNAs were
associated with tumor invasion, and parts of them were associated
with tumor proliferation and/or migration. It was worth noting that
LINC00200 played an important role in the tumor proliferation and
progression of GC, which provides us with a novel biomarker and
perhaps a potential target for GC.
There are still some limitations to the present
study. First, the present study validated the results of TCGA in a
GC cell line in vitro. Thus, an in vivo experiment
using patient GC samples is required to further confirm the
results. Second, other GC cell lines with different
differentiations are needed to explore and verify the functions of
key lncRNAs.
In conclusion, the results identified the
cancer-specific mRNAs and noncoding RNAs in GC by bioinformatics
analysis of large-scale samples from TCGA database. The study
therefore provides insights into the ceRNA regulatory network in GC
and describes lncRNAs that are associated with GC as diagnostic,
therapeutic and prognostic biomarkers.
Acknowledgements
Not applicable.
Funding
The present study was supported by grants from the
National Natural Science Foundation of China (grant nos. 31570877
and 31570908), Changzhou Health and Family Planning Commission
Youth Talent Science and Technology Project (grant no. QN201709),
Changzhou High-Level Medical Talents Training Project (grant no.
2016CZBJ054), Changzhou Health and Family Planning Commission
Program (grant no. ZD201608) and Wujin Sci and Tech Program (grant
no. WS201606).
Availability of data and materials
The datasets used during the present study are
available from the corresponding author upon reasonable
request.
Authors' contributions
WH conceived, designed and performed the study. DZ,
XL, JW, XY, QW, WL, JJ and CW were also involved in the conception
of the study and gave their advice in the process of the research.
DZ assisted WH with the data analysis. WH wrote the paper. WH and
DZ reviewed and edited the manuscript. All authors have read and
approved the manuscript and agree to be accountable for all aspects
of the research in ensuring that the accuracy or integrity of any
part of the work are appropriately investigated and resolved.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2016. CA Cancer J Clin. 66:7–30. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Cheng Y, Jin Z, Agarwal R, Ma K, Yang J,
Ibrahim S, Olaru AV, David S, Ashktorab H, Smoot DT, et al: LARP7
is a potential tumor suppressor gene in gastric cancer. Lab Invest.
92:1013–1019. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wang J, Guo W, Wu Q, Zhang R and Fang J:
Impact of combination epidural and general anesthesia on the
long-term survival of gastric cancer patients: A retrospective
study. Med Sci Monit. 22:2379–2385. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Yamamura S, Imai-Sumida M, Tanaka Y and
Dahiya R: Interaction and cross-talk between non-coding RNAs. Cell
Mol Life Sci. 75:467–484. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sana J, Faltejskova P, Svoboda M and Slaby
O: Novel classes of non-coding RNAs and cancer. J Transl Med.
10:1032012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ponting CP, Oliver PL and Reik W:
Evolution and functions of long noncoding RNAs. Cell. 136:629–641.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Pan W, Liu L, Wei J, Ge Y, Zhang J, Chen
H, Zhou L, Yuan Q, Zhou C and Yang M: A functional lncRNA HOTAIR
genetic variant contributes to gastric cancer susceptibility. Mol
Carcinog. 55:90–96. 2016. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Huang C, Cao L, Qiu L, Dai X, Ma L, Zhou
Y, Li H, Gao M, Li W, Zhang Q, et al: Upregulation of H19 promotes
invasion and induces epithelial-to-mesenchymal transition in
esophageal cancer. Oncol Lett. 10:291–296. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wang F, Xie C, Zhao W, Deng Z, Yang H and
Fang Q: Long non-coding RNA CARLo-5 expression is associated with
disease progression and predicts outcome in hepatocellular
carcinoma patients. Clin Exp Med. 17:33–43. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wang Y, Liu X, Zhang H, Sun L, Zhou Y, Jin
H, Zhang H, Zhang H, Liu J, Guo H, et al: Hypoxia-inducible
lncRNA-AK058003 promotes gastric cancer metastasis by targeting
gamma-synuclein. Neoplasia. 16:1094–1106. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Li L, Zhang L, Zhang Y and Zhou F:
Increased expression of LncRNA BANCR is associated with clinical
progression and poor prognosis in gastric cancer. Biomed
Pharmacother. 72:109–112. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gong Z, Zhang S, Zeng Z, Wu H, Yang Q,
Xiong F, Shi L, Yang J, Zhang W, Zhou Y, et al: LOC401317, a
p53-regulated long non-coding RNA, inhibits cell proliferation and
induces apoptosis in the nasopharyngeal carcinoma cell line HNE2.
PLoS One. 9:e1106742014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Svoboda M, Slyskova J, Schneiderova M,
Makovicky P, Bielik L, Levy M, Lipska L, Hemmelova B, Kala Z,
Protivankova M, et al: HOTAIR long non-coding RNA is a negative
prognostic factor not only in primary tumors, but also in the blood
of colorectal cancer patients. Carcinogenesis. 35:1510–1515. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Salmena L, Poliseno L, Tay Y, Kats L and
Pandolfi PP: A ceRNA hypothesis: The rosetta stone of a hidden RNA
language? Cell. 146:353–358. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Tay Y, Rinn J and Pandolfi PP: The
multilayered complexity of ceRNA crosstalk and competition. Nature.
505:344–352. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Bassett AR, Azzam G, Wheatley L, Tibbit C,
Rajakumar T, McGowan S, Stanger N, Ewels PA, Taylor S, Ponting CP,
et al: Understanding functional miRNA-target interactions in vivo
by site-specific genome engineering. Nat Commun. 5:46402014.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang J, Liu X, Wu H, Ni P, Gu Z, Qiao Y,
Chen N, Sun F and Fan Q: CREB up-regulates long non-coding RNA,
HULC expression through interaction with microRNA-372 in liver
cancer. Nucleic Acids Res. 38:5366–5383. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chen X, Chen Z, Yu S, Nie F, Yan S, Ma P,
Chen Q, Wei C, Fu H, Xu T, et al: Long noncoding RNA LINC01234
functions as a competing endogenous RNA to regulate CBFB expression
by sponging miR-204-5p in gastric cancer. Clin Cancer Res.
24:2002–2014. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Gu W, Gao T, Sun Y, Zheng X, Wang J, Ma J,
Hu X, Li J and Hu M: LncRNA expression profile reveals the
potential role of lncRNAs in gastric carcinogenesis. Cancer
Biomark. 15:249–258. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ye H, Liu K and Qian K: Overexpression of
long noncoding RNA HOTTIP promotes tumor invasion and predicts poor
prognosis in gastric cancer. OncoTargets Ther. 9:2081–2088.
2016.
|
|
21
|
Li F, Huang C, Li Q and Wu X: Construction
and comprehensive analysis for dysregulated long non-coding RNA
(lncRNA)-associated competing endogenous RNA (ceRNA) network in
gastric cancer. Med Sci Monit. 24:37–49. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Robinson MD, McCarthy DJ and Smyth GK:
edgeR: A Bioconductor package for differential expression analysis
of digital gene expression data. Bioinformatics. 26:139–140. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wickham H: ggplot2: Elegant Graphics for
Data Analysis. 1st edition. Springer-Verlag; New York, NY: 2009,
View Article : Google Scholar
|
|
24
|
Jeggari A, Marks DS and Larsson E:
miRcode: A map of putative microRNA target sites in the long
non-coding transcriptome. Bioinformatics. 28:2062–2063. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2−ΔΔCT method. Methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Therneau T: A Package for Survival
Analysis in S. version 2.38. 2015.https://CRAN.R-project.org/package=survival
|
|
28
|
Muers M: RNA: Genome-wide views of long
non-coding RNAs. Nat Rev Genet. 12:7422011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kornienko AE, Guenzl PM, Barlow DP and
Pauler FM: Gene regulation by the act of long non-coding RNA
transcription. BMC Biol. 11:592013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wahlestedt C: Targeting long non-coding
RNA to therapeutically upregulate gene expression. Nat Rev Drug
Discov. 12:433–446. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Song H, Sun W, Ye G, Ding X, Liu Z, Zhang
S, Xia T, Xiao B, Xi Y and Guo J: Long non-coding RNA expression
profile in human gastric cancer and its clinical significances. J
Transl Med. 11:2252013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Chen X: Predicting lncRNA-disease
associations and constructing lncRNA functional similarity network
based on the information of miRNA. Sci Rep. 5:131862015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Kong R, Zhang EB, Yin DD, You LH, Xu TP,
Chen WM, Xia R, Wan L, Sun M, Wang ZX, et al: Long noncoding RNA
PVT1 indicates a poor prognosis of gastric cancer and promotes cell
proliferation through epigenetically regulating p15 and p16. Mol
Cancer. 14:822015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Fu H, Wang C, Yang D, Wei Z, Xu J, Hu Z,
Zhang Y, Wang W, Yan R and Cai Q: Curcumin regulates proliferation,
autophagy, and apoptosis in gastric cancer cells by affecting PI3K
and P53 signaling. J Cell Physiol. 233:4634–4642. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Qiu YS, Liao GJ and Jiang NN: REG3A
overexpression suppresses gastric cancer cell invasion,
proliferation and promotes apoptosis through PI3K/Akt signaling
pathway. Int J Mol Med. 41:3167–3174. 2018.PubMed/NCBI
|
|
36
|
Zhang J, Wei W, Jin HC, Ying RC, Zhu AK
and Zhang FJ: Programmed cell death 2 protein induces gastric
cancer cell growth arrest at the early S phase of the cell cycle
and apoptosis in a p53-dependent manner. Oncol Rep. 33:103–110.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Huang Y, Zhang J, Hou L, Wang G, Liu H,
Zhang R, Chen X and Zhu J: LncRNA AK023391 promotes tumorigenesis
and invasion of gastric cancer through activation of the PI3K/Akt
signaling pathway. J Exp Clin Cancer Res. 36:1942017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Wei GH and Wang X: lncRNA MEG3 inhibit
proliferation and metastasis of gastric cancer via p53 signaling
pathway. Eur Rev Med Pharmacol Sci. 21:3850–3856. 2017.PubMed/NCBI
|
|
39
|
Ooi A, Oyama T, Nakamura R, Tajiri R,
Ikeda H, Fushida S and Dobashi Y: Gene amplification of CCNE1,
CCND1, and CDK6 in gastric cancers detected by multiplex
ligation-dependent probe amplification and fluorescence in situ
hybridization. Hum Pathol. 61:58–67. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Ju H, Lim B, Kim M, Noh SM, Kim WH, Ihm C,
Choi BY, Kim YS and Kang C: SERPINE1 intron polymorphisms affecting
gene expression are associated with diffuse-type gastric cancer
susceptibility. Cancer. 116:4248–4255. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Li J, Ding Y and Li A: Identification of
COL1A1 and COL1A2 as candidate prognostic factors in gastric
cancer. World J Surg Oncol. 14:2972016. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Li CY, Liang GY, Yao WZ, Sui J, Shen X,
Zhang YQ, Peng H, Hong WW, Ye YC, Zhang ZY, et al: Integrated
analysis of long non-coding RNA competing interactions reveals the
potential role in progression of human gastric cancer. Int J Oncol.
48:1965–1976. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Jin B, Jin H, Wu HB, Xu JJ and Li B: Long
non-coding RNA SNHG15 promotes CDK14 expression via miR-486 to
accelerate non-small cell lung cancer cells progression and
metastasis. J Cell Physiol. 233:7164–7172. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Yu X, Zhao J and He Y: Long non-coding RNA
PVT1 functions as an oncogene in human colon cancer through
miR-30d-5p/RUNX2 axis. J BUON. 23:48–54. 2018.PubMed/NCBI
|
|
45
|
Gu L, Lu LS, Zhou DL and Liu ZC: UCA1
promotes cell proliferation and invasion of gastric cancer by
targeting CREB1 sponging to miR-590-3p. Cancer Med. 7:1253–1263.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Zhao R and Zhang Y, Zhang X, Yang Y, Zheng
X, Li X, Liu Y and Zhang Y: Exosomal long noncoding RNA HOTTIP as
potential novel diagnostic and prognostic biomarker test for
gastric cancer. Mol Cancer. 17:682018. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Xue M, Chen LY, Wang WJ, Su TT, Shi LH,
Wang L, Zhang W, Si JM, Wang LJ and Chen SJ: HOTAIR induces the
ubiquitination of Runx3 by interacting with Mex3b and enhances the
invasion of gastric cancer cells. Gastric Cancer. 21:756–764. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Yan J, Zhang Y, She Q, Li X, Peng L, Wang
X, Liu S, Shen X, Zhang W, Dong Y, et al: Long noncoding RNA
H19/miR-675 axis promotes gastric cancer via FADD/caspase 8/caspase
3 signaling pathway. Cell Physiol Biochem. 42:2364–2376. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Pei YF, Zhang YJ, Lei Y, Wu DW, Ma TH and
Liu XQ: Hypermethylation of the CHRDL1 promoter induces
proliferation and metastasis by activating Akt and Erk in gastric
cancer. Oncotarget. 8:23155–23166. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Liu H, Zhang M, Xu S, Zhang J, Zou J, Yang
C, Zhang Y, Gong C, Kai Y and Li Y: HOXC8 promotes proliferation
and migration through transcriptional up-regulation of TGFβ1 in
non-small cell lung cancer. Oncogenesis. 7:12018. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Chen D, Maruschke M, Hakenberg O,
Zimmermann W, Stief CG and Buchner A: TOP2A, HELLS, ATAD2, and TET3
are novel prognostic markers in renal cell carcinoma. Urology.
102:265 e261–265 e267. 2017. View Article : Google Scholar
|
|
52
|
Dong Y, Li J, Han F, Chen H, Zhao X, Qin
Q, Shi R and Liu J: High IGF2 expression is associated with poor
clinical outcome in human ovarian cancer. Oncol Rep. 34:936–942.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Li Q, Dai Y, Wang F and Hou S:
Differentially expressed long non-coding RNAs and the prognostic
potential in colorectal cancer. Neoplasma. 63:977–983. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Yao J, Zhou B, Zhang J, Geng P, Liu K, Zhu
Y and Zhu W: A new tumor suppressor LncRNA ADAMTS9-AS2 is regulated
by DNMT1 and inhibits migration of glioma cells. Tumour Biol.
35:7935–7944. 2014. View Article : Google Scholar : PubMed/NCBI
|