Oncogenesis transforms a normal cell into a tumor
cell through the acquisition of basic tumor characteristics denoted
by the hallmarks of cancer, including sustained proliferative
signaling, growth suppressor avoidance, cell death resistance,
replicative immortality, angiogenesis, invasion and metastasis
activation (1,2). Cancer research has focused on the
search for genetic biomarkers capable of regulating the acquisition
of oncogenic characteristics, but less than half of the reported
effects were related to a cancer-specific endpoint (3). However, epigenetic factors have shown
their crucial importance in gene expression regulation through
posttranslational modifications without altering DNA sequences
(4). Additionally, transcription
factors have been identified as a group of proteins with the
ability to regulate expression by binding to a large number of gene
promoters (5,6). Transcription factors have been
consistently deregulated in human cancer due to the presence of
translocations, deletions, amplifications and point mutations;
additionally, they serve as terminal regulators and convergence
points of important oncogenic signaling pathways, becoming novel
and promising cancer therapy targets (5,7).
Runt-related transcription factor (RUNX) proteins
belong to a transcription factor family of embryonic development
master regulators that are involved in essential cellular
processes, including proliferation, differentiation, cell lineage
specification and even apoptosis (8). Mammals have 3 RUNX genes with
very dynamic expression patterns, depending on the differentiation
and developmental stages, and microenvironmental signals of cancer
(9). Functionally, RUNX1 is
important for hematopoietic cell differentiation (10,11),
RUNX2 is essential for osteogenesis (12–14)
and RUNX3 regulates gastric epithelium growth (15). In cancer, RUNX1 has been
associated with leukemia (16–18),
and solid tumor development on the skin, lung, intestine and breast
(19,20), while RUNX2 has been
associated with osteosarcoma (21–23),
papillary carcinoma, thyroid carcinoma (24,25),
and breast and prostate cancer (26–28),
and RUNX3 with gastric cancer (29).
RUNX proteins belong to a family of transcription
factors conserved in evolution that regulate proliferation,
differentiation and cell growth in different tissues and specific
contexts (33,34). RUNX genes can be identified
in C. elegans (35).
Bilateria organisms only have one RUNX gene with at least
two introns, suggesting that the multiple RUNX genes in
vertebrates and insects come from independent duplication events
within every lineage (36).
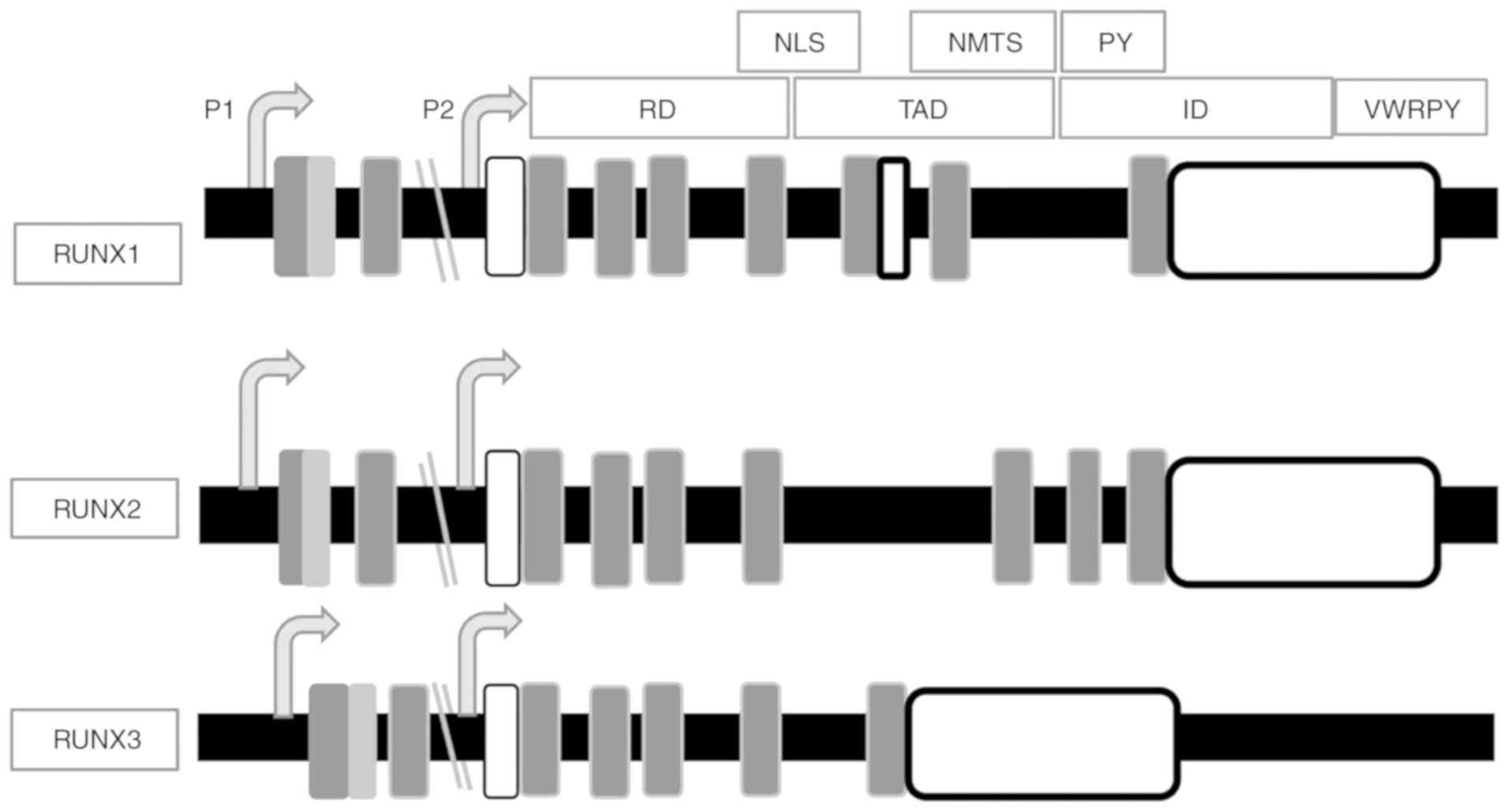
The Runt homology domain (RD; exons 2, 3 and 4)
mediates DNA binding and the transactivation domain (TAD; exon 6)
mediates protein-protein interactions (46,47).
The RD has a highly conserved motif of 128 amino acids near the
N-terminus, with a homology degree close to 90%, which binds to a
TGt/cGGT element present in its target gene promoter (38,39).
The RD three dimensional conformation in its DNA-binding state is
an S-type immunoglobulin (Ig) domain (48). The Ig domain is involved in
molecular recognition and DNA binding of other transcription
factors, including cellular tumor antigen p53 (p53), nuclear
factor-κB (NF-κB), nuclear factor of activated T-cells and signal
transducer and activator of transcription (49). RD has only been identified in
Bilateria organisms, suggesting that it may be a creation of
metazoans (8).
At the C-terminus, there is an inhibitory domain
(ID), which negatively regulates protein expression (50). There is also a highly conserved
valine-tryptophan-arginine-proline-tyrosine (VWRPY) motif for the
interaction with the Groucho/transducin-like enhancer protein (TLE)
family of corepressors (51). A 40
amino acid sequence acts as a RUNX activated protein target
(52–54). There is also a sequence of nine
amino acid located following the RD, called the nuclear
localization signal (NLS) (55).
The proline-tyrosine (PY) sequence has a proline-rich motif
important for protein interaction with a WW domain (56). Comparing amino acid sequences from
different species revealed highly conserved RD, VWRPY and PY motifs
at the C-terminus (57). However,
RUNX proteins have less homology in regulatory elements and protein
binding sequence regions, which functionally characterize every
RUNX protein (42).
RUNX complex function is determined by a diverse and
highly dynamic range of posttranslational modifications
(specifically, methylation, phosphorylation, acetylation and
ubiquitination) that affect its gene expression, protein activity,
subcellular location and stability (39). The DNA and multiple
posttranslational modifications of the RUNX family determine how
the activities of the transcription factors regulate cell cycle
progression or the response to external stimuli (8).
RUNX proteins are the substrates of several kinases
such as serine/threonine-protein kinase pim-1 (Pim-1),
mitogen-activated protein kinase (ERK, also known as MAPK) and
cyclins-cdk (75). Pim-1 is
a proto-oncogene that phosphorylates all RUNX proteins (75). Pim-1 phosphorylates RUNX3 to enhance
its stability and cytoplasm location (76). Increased expression of RUNX2
and Pim-1 leads to T-cell lymphoma synergistic development
(77). RUNX1 is phosphorylated by
the serine/threonine kinase ERK2, increasing its transactivation
ability (78), but reducing protein
stability due to Sin3A corepressor dissociation (79). Homeodomain-interacting protein
kinase 2, a protein kinase, phosphorylates RUNX1 to promote
cooperation between RUNX1, histone acetyltransferase KAT6A (MOZ)
and p300 to activate transcription (80,81).
RUNX proteins are weak transcriptional regulatory
factors when acting independently; therefore, they require
interaction with other proteins to increase or decrease their
activity (9). Additionally, RUNX
proteins form functional complexes with other proteins to activate
and repress the transcription of key regulators associated with
cell growth and differentiation, demonstrating a dual function of
this family (39).
The RUNX family recruits' corepressors to repress
the transcription of multiple target genes through its VWRPY motif
interaction with the Groucho/TLE family of corepressor proteins
(82). Corepressor mSin3A
association allows the recruitment of histone deacetylases (HDACs)1
and 2, for cyclin-dependent kinase inhibitor 1
(p21Wafl/Cip) expression in NIH3T3 cells, a fibroblast
cell line sensitive to the foci formation of leukemia and sarcoma
viruses (83). RUNX1 is clearly
associated with HDACs 1, 3 and 9, and weakly associated with HDACs
2, 5 and 6; RUNX2 recruits HDAC6, whereas RUNX1 and RUNX3 recruit
SUV39H1 to suppress transcription (84).
The RUNX family recruits coactivators to activate
the transcription of multiple target genes (9). RUNX1 binds to ETS1 through its ID,
eliminating its requirement for CBFβ and leading to a better DNA
binding ability, which encourages transactivation and synergistic
promoter activation (85). RUNX1
binds ETS-related transcription factor Elf (NERF)-2 and NERF-1 to
activate and repress tyrosine-protein kinase Blk, a B cell-specific
gene, respectively (71). RUNX1 and
RUNX2 bind to proto-oncogene c-Fos and transcription factor AP-1
through the RD to activate the collagenase-3 gene promoter
(72). TAD acts through the
recruitment of histone acetyl transferases, including MOZ and
mortality factor 4-like protein, which physically interact with
RUNX1 and RUNX2, clearly stimulating transactivation activity
(73).
RUNX1 forms heterodimers with CBFβ through an RD
consensus sequence, enhancing gene transcription when interacting
with coactivators, including p300 and CREB-binding protein, and
suppressing gene transcription when interacting with
transcriptional corepressors, including Sin3A, TLE and histone
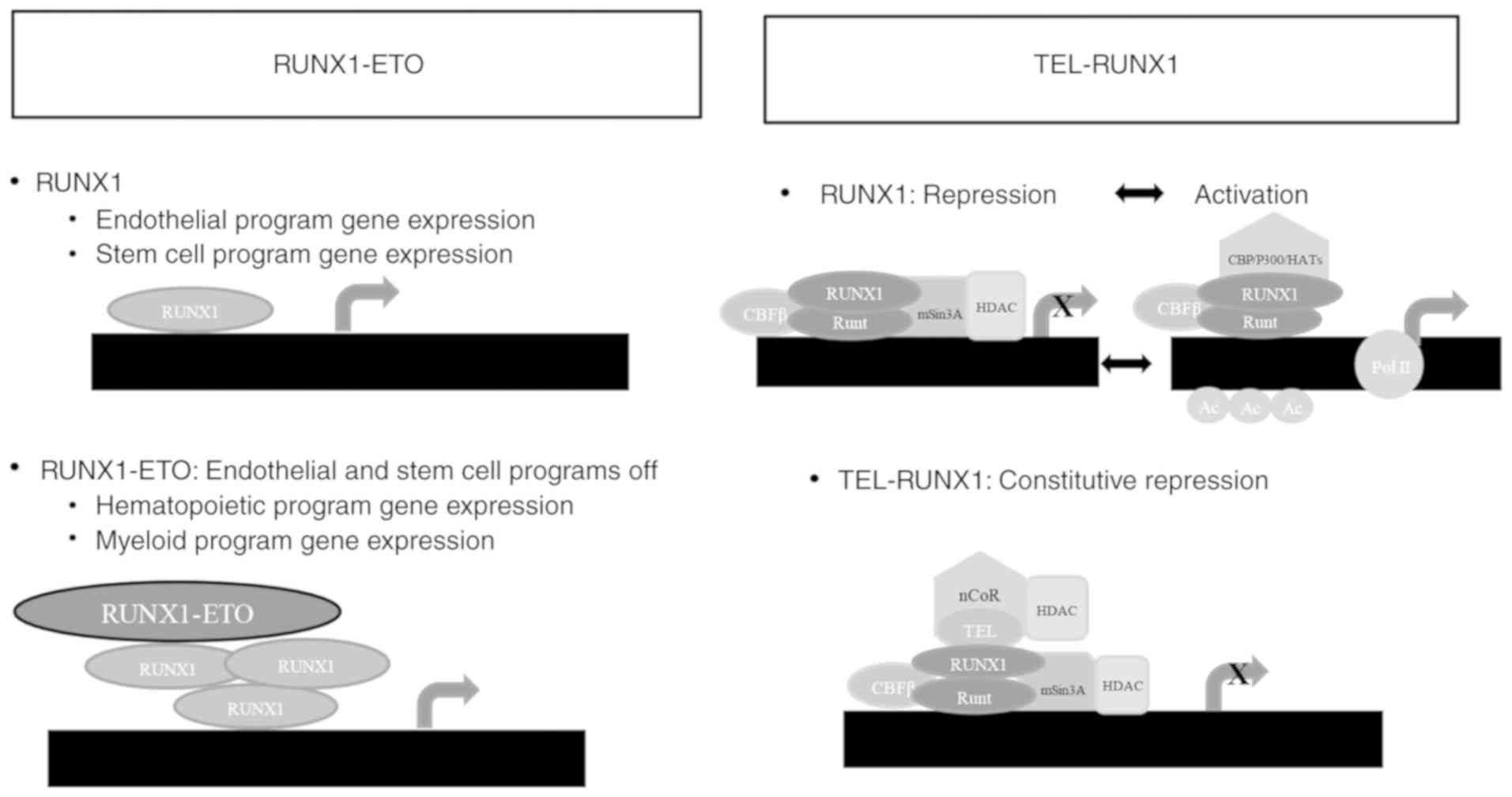
deacetylases (11). The RUNX1-ETO
and TEL-RUNX1 association with CBFβ and mSin3A represses
transcription through indirect HDAC recruitment, which removes
acetyl groups from histones H3 and H4's lysine residues, allowing
compacted or repressed chromatin formation, which reduces the
accessibility of transcriptional machinery promoters (86–88).
RUNX1 dissociates from mSin3A/HDAC and associates with p300,
reversing the process following properly stimulation (89).
The RUNX family collaborates with the SWItch/Sucrose
Non-Fermentable (SWI/SNF) chromatin remodeling complex for
transcriptional activation (90).
RUNX1-SWI/SNF association controls gene expression during
hematopoiesis, a process associated with chromatin-activating
modifications, including histone H4 acetylation and histone H3
lysine 4 demethylation (90).
Decreases in RUNX1 expression reduce the co-occupation of SWI/SNF,
transcription activator BRG1 and SWI/SNF-related matrix-associated
actin-dependent regulator of chromatin subfamily B member 1
components in RUNX1 target gene promoters; therefore, RUNX1 is
important in the regulation of hematopoietic functions (90). RUNX2 also associates with SWI/SNF
but through CCAAT/enhancer-binding protein β to favor the specific
transcriptional activation of osteoblastic
differentiation-associated genes (91).
RUNX1 interactions with multiple proteins through
its terminal C domain allows it to control its target gene
expression, which is mainly involved in hematopoietic
differentiation, ribosomal biogenesis, cell cycle regulation, and
TGF-β and p53 signaling pathways (62,96).
RUNX1 is essential for the definitive establishment of
hematopoiesis during embryogenic development, and is required for
hematopoietic stem and progenitor cell regulation (97). In adults, RUNX1 serves a role in
lymphocyte and megakaryocyte maturation. The polycomb
group-polycomb repressive complex 1 core complex and polycomb group
RING finger protein 1 (Pcgf1) inhibit progenitor cell self-renewal
by negatively regulating homeobox protein Hox genes, whereas RUNX1
drives cell differentiation where self-renewal has been limited by
Pcgf (97). RUNX1 and Pcgf1 joint
action demonstrates a required epigenetic and transcriptional
regulation association for hematopoietic differentiation (97). The cell differentiation of myeloid
progenitors into granulocytes requires RUNX1, meanwhile the absence
or reduction of RUNX1 expression activates cell proliferation
(98,99).
Specific RUNX2 levels contribute to cell cycle
entry, exit and progression in osteoblasts and endothelial cells
(100). RUNX2 suppresses
pre-osteoblast proliferation, affecting cell cycle progression in
the G1 phase (100,101). RUNX2 acts as a master regulator
for osteoblastic lineage formation, either directly or indirectly
controlling key gene expression (collagen 1, osteocalcin,
osteopontin, alkaline phosphatase and bone sialoprotein) for early
differentiation of osteoblasts (12). Osteoblastic lineage progression from
pluripotent mesenchymal cells to mature osteocytes is regulated by
multiple physiological signals, including transforming growth
factor (TGF)β, bone morphogenetic protein, vitamin D and
glucocorticoids (102). RUNX2
expression is very high in hematopoietic stem cells, even higher
than RUNX1, but decreases during myeloid differentiation (103). RUNX2 also regulates lymphoid
lineage in the early stages and B cell differentiation (104,105).
RUNX protein aberrant expression and mutations have
been associated with different types of cancer, where they may act
as tumor suppressors and oncogenes depending on the biological
context (9). Additionally, in
fibroblasts with overexpressed RUNX proteins, their ability to
regulate multiple targets associated with specific functions during
oncogenesis and development was demonstrated (108–110).
RUNX1 haploinsufficiency causes a predisposition to
leukemia, but its overexpression is necessary for solid tumor
formation in the skin, lungs, intestines and breasts (19,20,111).
Leukemia development has been associated with RUNX1 point
mutations, amplifications and translocations (16–18).
RUNX1 frequent chromosomal translocations in leukemia
generate unique fusion proteins with great oncogenic potential that
affect the TAD, but not the RD, making RUNX1 a dominant
negative inhibitor (17).
In the majority of cases, chimeric genes that
involve the RUNX1 locus inhibit its function, but it's function is
increased in other cases (18,119).
If RUNX1 chimeric genes inactivate the function of RUNX1 to cause
leukemia, then functional loss by mutations must also cause acute
myeloid leukemia; in fact, patients with different types of
leukemia possess heterozygous or homozygous sporadic and familial
mutations (120,121). However, an extra RUNX1 copy in
megakaryoblastic leukemia associated with Down syndrome in newborns
and children generates RUNX1 overexpression, which can also lead to
leukemia development (122).
RUNX2 protein is the only one in the family that
has a polyglutamine-polyalanine motif at the N-terminal prior to
the RD, which has been associated with the formation of spiral
structures, and aggregation and toxicity during the establishment
of human genetic diseases (127).
RUNX2 oncogenic activity was first demonstrated in transgenic mice,
where it was associated with T-cell lymphoma induction when there
was Myc proto-oncogene protein (c-Myc) ectopic expression (128).
In breast and prostate cancer, RUNX2 is
overexpressed and associated with an increase in metastatic
capacity (26–28). RUNX2 expression increases markedly
in neoplastic breast cells, especially in metastatic cells
(135). RUNX2 increases breast
tumor cell metastatic capacity by increasing the expression of
several factors, including vascular endothelial growth factor,
matrix metalloproteases (MMP2, MMP9 and MMP13) and bone
sialoprotein (BSP), facilitating the process (136). CADD522 was a identified from a
computer-assisted drug design screen as cholecalciferol (a
prohormone and precursor of 25-OH Vitamin D3 prohormone) (137) and is a small molecule capable of
inhibiting RUNX2 expression through the blockade of its protein
domain or binding pocket interactions, leading to growth
inhibition, clonogenic survival, tumorsphere formation and the
invasion of breast cancer cells (138).
Bone morphogenetic protein-3b (BMP-3b/GDF10) is a
tumor growth inhibitor and a member of TGFβ family (139). RUNX2 is highly expressed in
lung tumor cells that negatively regulate BMP-3b (139). The molecular mechanism that
mediates BMP-3b suppression by RUNX2 is based on the recruitment of
histone-lysine N-methyltransferase SUV39H1 to the BMP-3b proximal
promoter of the specific methyltransferase for histone H3 lysine 9
(H3K9), which increases methylation levels (139). In RUNX2 knockout H1299
cells, a significant decrease in H3K9 methylation levels at the
BMP-3b promoter was observed, thereby increasing BMP-3b expression
levels (139). Meanwhile, RUNX2
overexpression increased the wound healing process in response to
TGF-β. One study suggests that RUNX2 is a potential therapeutic
target to block tumor suppressor gene silencing in lung tumor cells
(139). However, it is necessary
to include clinical studies to prove this hypothesis in
patients.
RUNX3 is located in a chromosomal region identified
as a tumor suppression center, where there are a large number of
genes that are inhibited during different tumor processes, due to
its ability to positively regulate other tumor suppressor genes
(140). RUNX3 nonspecific
localization in the cytoplasm has been reported as the major form
of RUNX3 inactivation (141) due
to Src tyrosine kinase activation, as has been observed in cancer
cell lines (142) in addition to
gastric (141) and breast cancer
cells from patients of the University Hospital Tissue Repository
and the Pathology Department, National University of Singapore
(143). Therefore, RUNX3
expression in tumor stroma has been associated with a good clinical
prognosis (144).
Pancreatic ductal adenocarcinoma (PDAC) studies may
have helped solve the inconsistences in RUNX3 tumor suppressor
function, as they suggest that it is instead a switch for
metastatic control (160,161). Studies demonstrating RUNX3 as a
tumor suppressor (145) and as an
oncogene (157) demonstrate its
dual role in cancer, as in PDAC where it acts as a tumor suppressor
slowing proliferation and as an oncogene promoting metastasis and
invasion, controlling the balance of local growth and metastasis in
primary and metastatic tumors (160). RUNX3 expression has been
associated with mothers against decapentaplegic homolog 4 (SMAD4,
also known as DPC4) copy number variants, and level patterns have
been directly associated with relapse and the response to therapies
(160). RUNX3 expression has also
been associated with combined epigenetic programs and metabolic
processes when it is part of the retinoic acid receptor
β/RUNX3/collagen α-1(VI) chain signal axis, linking hypoglycemia
with local invasion and angiogenesis, and hyperglycemia with
metastatic colonization (162).
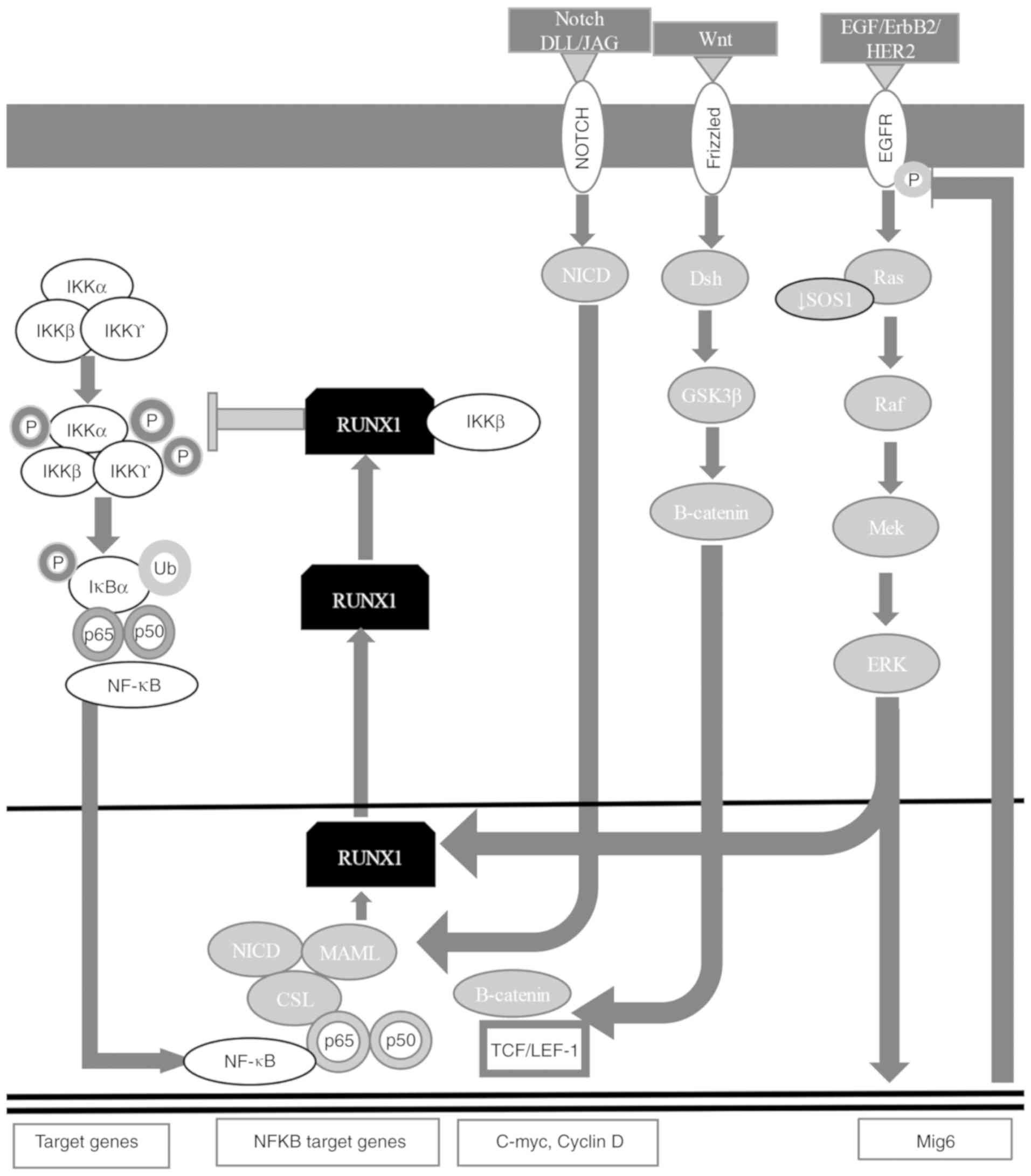
RUNX1 is highly expressed in the mesenchymal and
epithelial compartments of embryonic and postnatal lungs with
lipopolysaccharide-induced lung inflammation, regulating the NF-κB
signaling pathway through the interaction with the inhibitor of
nuclear factor-κB kinase (IKK) complex or the NF-κB subunit p50 in
the cytoplasm (Fig. 3) (7,60,167,168). RUNX1 is targeted in mesenchymal
and epithelial compartments of the skin during embryogenesis,
deregulating lymphoid enhancer-binding factor (Lef)1 and protein
Wnt (Wnt) signaling in opposite directions, decreasing Lef1 and
activating canonical Wnt signaling (169). RUNX1 controls the epidermal growth
factor receptor (EGFR) signaling pathway in non-small-cell lung
cancer cells, regulating ERBB receptor feedback inhibitor 1
expression, which is a negative feedback regulator of the EGFR
phosphorylated form (Fig. 3)
(7,60,167,168,170). RUNX1 translocates to the cytoplasm
to form a complex with IKKβ that inhibits the NF-κB signaling
pathway (Fig. 3) (7,60,167,168).
The neurogenic locus notch homolog protein (Notch)
signaling pathway is important for cell fate (170). Notch is associated with the
increase in NF-κB expression, activating zinc finger protein SNAI1
and EMT, and stabilizing β-catenin (171). The Notch1-4 receptor has an
extracellular and an intracellular (NICD) domain that binds to the
Notch ligand [delta-like protein (DLL)/jagged (JAG)] from a
different cell, releasing its NICD, which translocates to the
nucleus and interacts with CSL [acronym for recombining binding
protein suppressor of hairless (also known as CBF1)/suppressor of
hairless protein/DNA-binding protein LAG-1 (Figs. 3 and 4) (7,60,167,168,172–175). The NICD-CSL complex displaces
corepressors and recruits mastermind-like protein (MAML) to form
the Notch-CSL-MAML complex, which recruits members of the Notch
transcriptional complex to activate gene expression (176). RUNX1 is regulated by Notch1 in
NIH3T3 cells (177), in
hematopoietic stem cell development (178) and in mesodermal cells (179). RUNX2 inhibits the Notch signaling
pathway during normal osteoblast differentiation (180) and during bone remodeling, and
regulates osteopontin in osteoblastic cells (181). RUNX3 is a direct target of Notch
in endothelial cells (182).
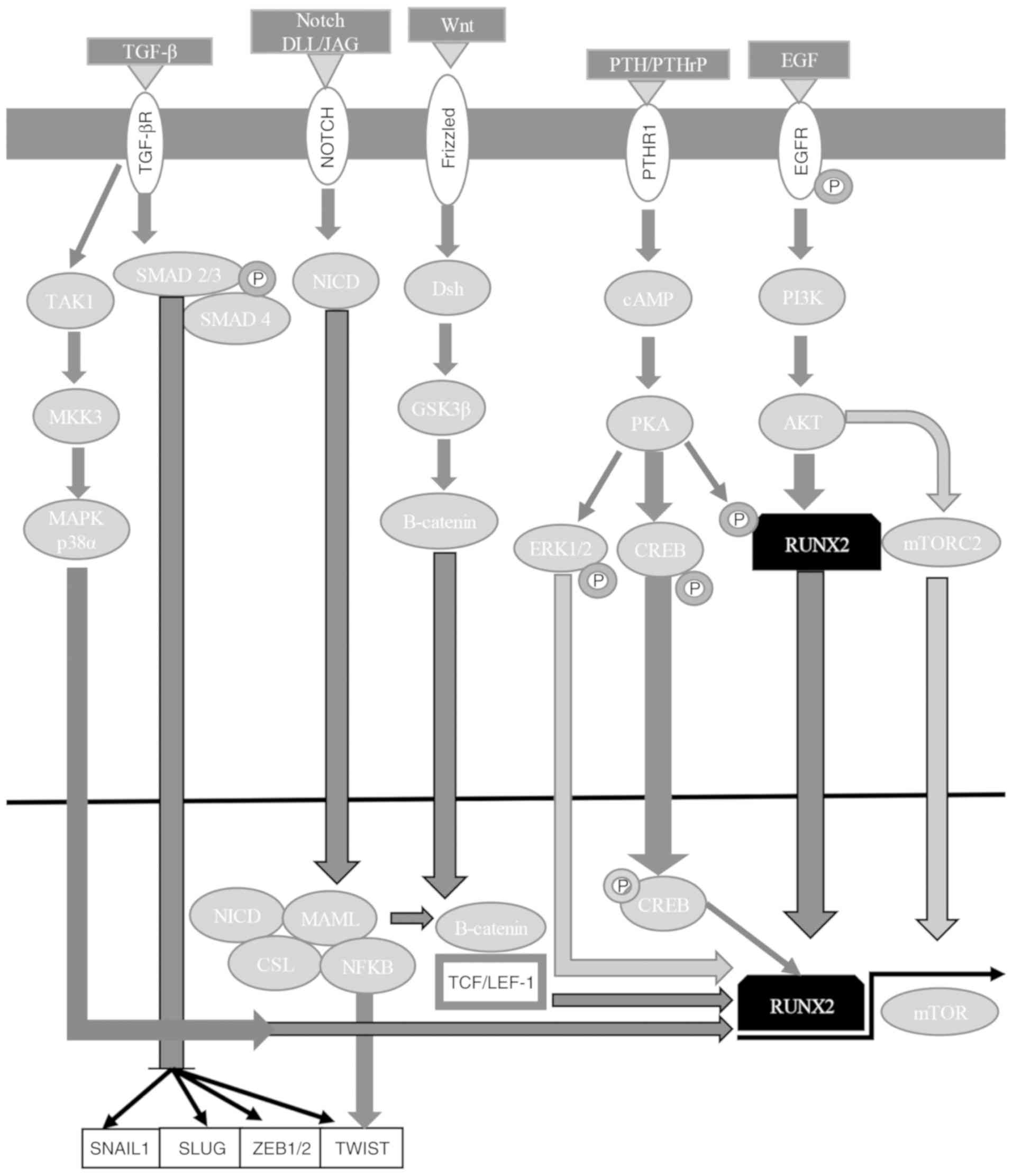
In osteoblasts and chondrocytes, Wnt signaling
induces differentiation and chondrocytic hypertrophy through
RUNX2 positive regulation (187), whereas during osteogenesis,
RUNX2 is a direct target of β-catenin/Lef1 to stimulate bone
formation (188). Wnt signaling is
associated with TGF-β signaling (189). β-catenin and the SMAD2/3-SMAD4
complex can activate Lef1, behaving as a molecular node that links
the Wnt signaling pathway with other signaling pathways associated
with EMT (7). RUNX3 can activate
the Wnt signaling pathway to control TCF-4/β-catenin complex
stabilization on the Wnt target gene promoter, suppressing
tumorigenesis in KatoIII cells; however, RUNX3-TCF-4/β-catenin
complex binding can also repress the Wnt signaling pathway
depending on cell context mechanisms (190).
RUNX1 and RUNX3 tumor suppressor activities are
mediated in part by estrogen signaling antagonism, as previously
described regarding RUNX2 activity (191). RUNX1 interacts with estrogen
receptor (ER)α to attenuate estrogen signaling (Fig. 3) (7,60,167–169). RUNX1 positively regulates the
receptor tyrosine-protein kinase erbB-2 (ErbB2/HER2) signaling
pathway in gastric cancer by binding to the son of sevenless
homolog 1 (SOS1) promoter. Therefore, RUNX1 knockdown is associated
with decreased SOS1 expression and ErbB2/HER2 dephosphorylation,
which suppresses gastric cancer cell proliferation (192). RUNX2 has been demonstrated to
reduce ERα (also known as ESR1) activity, binding to the ESR1 gene
promoter (193). Furthermore,
RUNX2 is inhibited by estrogens, which may help to explain their
context-dependent non-osseous anti-metastatic roles, as ERα is only
associated with the increased skeletal dissemination of breast
cancer cells (194). RUNX2
regulates cAMP-associated G-protein-coupled receptor signaling,
activating the G-protein coupled estrogen receptor 1 gene and
repressing the expression of the regulator of G-protein signaling 2
gene in osteoblasts to respectively increase and reduce mitogenic
signal sensitivity, allowing cell cycle progression and
osteoblastic lineage commitment (195). RUNX3 mediates ERα ubiquitination
and degradation (144), possibly
because the binding of RUNX3-ERα alters its posttranslational
modifications, changing its stability (196,197) or facilitating E3 ligase (E3
ubiquitin-protein ligase Mdm2 and Smurfs) recruitment (Fig. 5) (144,184,198,199). RUNX3 inhibits the
estrogen-dependent proliferation and transformation potential of
ERα-positive MCF-7 breast cancer cells, reducing ERα stability
(200).
RUNX2 is phosphorylated/activated by cAMP-dependent
protein kinase (PK)A and MAPK signaling pathways; it is also
enhanced by factors that stimulate signal transduction pathways,
including parathyroid hormone/parathyroid hormone-related protein
from the PKA and PKC signaling pathways, and BMPs of Smad proteins,
suggesting a fundamental role in directing osteoblast
differentiation (173). RUNX2 has
been associated with metastasis in breast cancer, activating SNAI2
expression in the TGF-β and Wnt signaling pathways (194). RUNX2's interaction with the
phosphatidylinositol 4,5-bisphosphate 3-kinase/RAC-α
serine/threonine-protein kinase (PI3K/AKT) signaling pathway is
essential to control cancer growth and metastasis, where AKT
phosphorylates/activates RUNX2 or phosphorylates/inactivates RUNX2
regulators (Fig. 4) (7,167,172–175). RUNX2 also activates the PI3K/AKT
signaling pathway, regulating its different components in
non-transformed and transformed cells (175). Therefore, AKT activation and high
levels of RUNX2 may induce tumor progression and aggressiveness
(Fig. 4) (7,167,172–175,199).
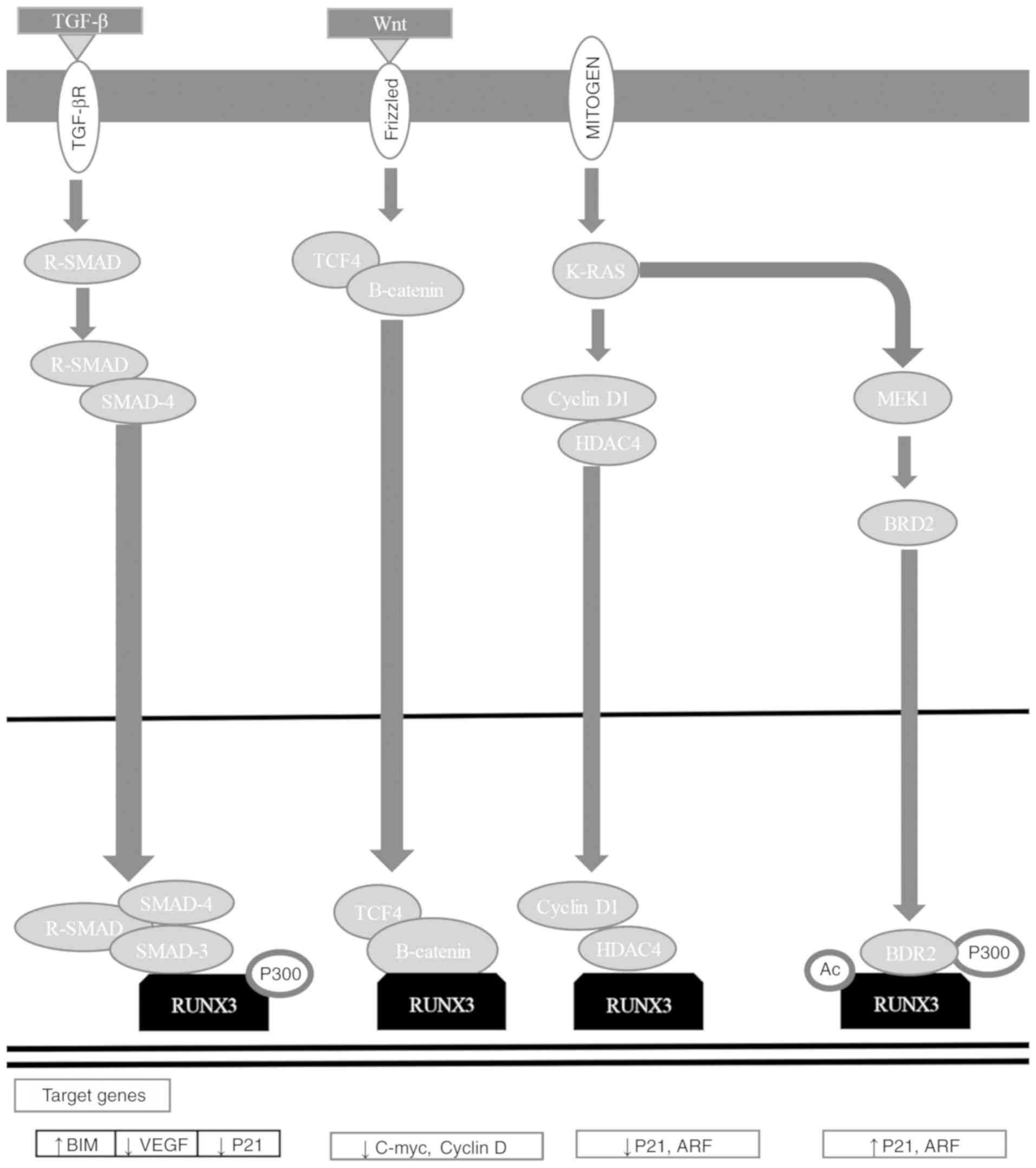
RUNX3 is a downstream effector of the TGF-β
signaling pathway, and has critical functions in apoptosis
regulation, angiogenesis, EMT, and cell migration and invasion
processes (184). RUNX3 functions
as an initiator of tumorigenesis, participating in the Wnt
oncogenic signaling pathway and the TGF-β tumor suppressor
signaling pathway (Fig. 5)
(184,196). RUNX3 associates with SMAD3/SMAD4
to activate growth inhibition reliant on TGF-β and apoptosis by p21
and Bcl-2-like protein 11 induction (Fig. 4) (7,167,172–175,201–203). RUNX3 inhibits the oncogenic
signaling pathway by forming a complex with TCF-4/β-catenin, which
avoids binding to its target gene promoters [c-Myc and
G1/S-specific cyclin D1 (cyclin D1)], regulating apoptosis and the
cell cycle (Fig. 5) (184,204). RUNX3 inhibits EMT, avoiding the
Wnt signaling pathway (184,205,206). Mitogenic stimulation induces
RUNX3-bromodomain-containing protein 2 (BRD2) complex formation,
and p21 and ADP-ribosylation factor (ARF) expression, while a
decrease in GTPase KRas (K-Ras) signaling pathway activation and an
increase in cyclin D1 converts the RUNX3-BRD2 complex into a
RUNX3-HDAC4 complex, shutting down ARF and p21 expression (184,207). When K-Ras is constitutively
activated, the oncogenic Ras-activated dual specificity
mitogen-activated protein kinase kinase 1 signaling pathway
inhibits conversion between complexes, keeping ARF1 and p21
expression active (Fig. 5)
(184,207).
Transcription factor coding genes are deregulated
in cancer since they can be amplified, deleted, chromosomally
translocated and affected by point mutations (5,7).
Transcription factors deregulatory mechanisms in cancer suggest its
importance in aberrant gene expression during cell transformation
and justify considering them as therapeutic targets.
RUNX genes have proved to be essential regulators
of cell fate in development but have opposite effects in cancer,
acting as dominant oncogenes or tumor suppressors (30–32,39,208,209). RUNX protein complexes control the
expression of multiple genes by binding to their promoters or
enhancers, which are relevant for cell fate, a feature that may
also be involved in tumor cell gene regulation (77). RUNX complex regulation is lineage
and stage specific, and includes crucial decisions between stopping
the cell cycle and continuing with proliferation, and between
differentiation and self-renewal (9).
RUNX can act as an expression activator or
repressor of a specific target gene, depending on the interacting
coactivators or corepressors, since RUNX proteins can join and
recruit a large group of them and regulate target promoters
(11). RUNX proteins have some
common characteristics in the transactivation/inhibition domains
and in some specific conserved motifs, including the nuclear-matrix
binding signal and VWRPY motif that interacts with corepressors
(210). In general, the conserved
RD and the divergent C-terminal domains (Fig. 1) (44) suggest that RUNX proteins have a
redundant function in some cellular contexts and that they exert
unique effects in others (9,211).
The majority of the genes involved in cancer
etiology are classified as oncogenes or tumor suppressors; however,
RUNX genes have not been classified within either of these groups,
as there is experimental evidence of a dual function in different
types of cancer (9,31,32).
The suppressed expression of RUNX genes in some types of cancer has
been associated with the presence of inactivating mutations, gene
deletions and hypermethylation, whereas retroviral insertion in
murine models has been associated with gene activation (119).
RUNX family oncogenic potential can be based on the
fact that the family has diverged evolutionarily in function, or
that its functions arose from the develop of specific controls in
its gene expression (9). Trials
with transcriptional reporters demonstrated essentially identical
effects of the three genes in a series of target promoters, and
hematopoietic development rescue by the knock-in of coding exons at
RUNX2 and RUNX3 3′end in RUNX1, which reveal
at least a partial functional overlap (212,213).
The contrasting roles of RUNX proteins can be
explained by generating specific biological contexts for lineage
and cancer or the developmental stage at which these abnormalities
have been detected. For example, myeloid leukemia cases are
associated with chromosome 21 polysomies and with RUNX1
amplification (216). In addition,
lymphoid neoplasms have been demonstrated to be activated by the
proviral insertion murine RUNX genes and RUNX1 amplification in
humans (32).
Scientific research on cancer has revealed that the
oncogenic potential of RUNX proteins depends on specific gene
expression patterns at different types and stages of cancer
(217). RUNX family oncogenic
potential can be based in principle on its gene structure, which
allows them to use different promoters and perform alternative
splicing for the formation of multiple isoforms (42). RUNX protein isoforms can provide
specific characteristics to act as transcription factors with the
ability to regulate a certain number of genes involved in oncogenic
signaling pathways.
The ability of RUNX proteins to form functional
complexes with other proteins can enable them to activate and
repress the transcription of key process regulators associated with
oncogenic development, including cell growth and cell
differentiation. Likewise, posttranscriptional modifications in
RUNX protein expression regulation, which is associated with their
overexpression and functional loss, may partially demonstrate the
dual function of these transcription factors (9).
Experimental evidence on the dual function of RUNX
in cancer suggests that the therapeutic control of their expression
can change their oncogenic function and turn them into tumor
suppressor genes, leading them to positively regulate tumor
suppressor genes and negatively regulate oncogenes, reversing the
tumorigenic processes in patients (39,125).
Likewise, RUNX proteins could be identified as a group of relevant
biomarkers that could be used to develop early detection techniques
(39). The experimental
determination of the molecular context in which RUNX proteins
change their oncogenic function into tumor suppressors is the key
to their use as biomarkers and therapeutic targets in cancer
treatment.
Not applicable.
The current review was funded by the grant number
8740 from Pontificia Universidad Javeriana. The work of the author
BAOO is supported by the doctoral fellowships given by The
Administrative Department of Science, Technology and Innovation of
Colombia.
Not applicable.
AR conceptualized the design of the present review.
BAOO did the literature search and contributed to the manuscript
writing. BH and LLK made several revisions of the text, making
crucial contributions to the scientific analysis and discussion of
the thesis presented in the review. All authors approved the final
manuscript.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
|
1
|
Hanahan D and Weinberg R: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hanahan D and Weinberg R: The hallmarks of
cancer. Cell. 100:57–70. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Green G, Carmona R, Zakeri K, Lee CH,
Borgan S, Marhoon Z, Sharabi A and Mell LK: Specificity of genetic
biomarker studies in cancer research: A systematic review. PLoS
One. 11:e01564892016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Sharma S, Kelly TK and Jones PA:
Epigenetics in cancer. Carcinogenesis. 31:27–36. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bhagwat AS and Vakoc CR: Targeting
transcription factors in cancer. Trends Cancer. 1:53–65. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Yeh JE, Toniolo PA and Frank DA: Targeting
transcription factors: Promising new strategies for cancer therapy.
Curr Opin Oncol. 25:652–658. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Darnell JE: Transcription factors as
targets for cancer therapy. Nat Rev Cancer. 2:740–749. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chuang LS, Ito K and Ito Y: RUNX family:
Regulation and diversification of roles through interacting
proteins. Int J Cancer. 132:1260–1271. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Blyth K, Cameron ER and Neil JC: The RUNX
genes: Gain or loss of function in cancer. Nat Rev Cancer.
5:376–387. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Dowdy CR, Xie R, Frederick D, Hussain S,
Zaidi SK, Vradii D, Javed A, Li X, Jones SN, Lian JB, et al:
Definitive hematopoiesis requires Runx1 C-terminal-mediated
subnuclear targeting and transactivation. Hum Mol Genet.
19:1048–1057. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yamagata T, Maki K and Mitani K:
Runx1/AML1 in normal and abnormal hematopoiesis. Int J Hematol.
82:1–8. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lian JB, Javed A, Zaidi SK, Lengner C,
Montecino M, van Wijnen AJ, Stein JL and Stein GS: Regulatory
controls for osteoblast growth and differentiation: Role of
Runx/Cbfa/AML factors. Crit Rev Eukaryot Gene Expr. 14:1–41. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lian JB and Stein GS: Runx2/Cbfa1: A
multifunctional regulator of bone formation. Curr Pharm Des.
9:2677–2685. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Komori T: Requisite roles of Runx2 and
Cbfb in skeletal development. J Bone Miner Metab. 21:193–197.
2003.PubMed/NCBI
|
|
15
|
Fukamachi H: Runx3 controls growth and
differentiation of gastric epithelial cells in mammals. Dev Growth
Differ. 48:1–13. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Osato M: Point mutations in the RUNX1/AML1
gene: Another actor in RUNX leukemia. Oncogene. 23:4284–4296. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
De Braekeleer E, Férec C and De Braekeleer
M: RUNX1 translocations in malignant hemopathies. Anticancer Res.
29:1031–1037. 2009.PubMed/NCBI
|
|
18
|
Niini T, Kanerva J, Vettenranta K,
Saarinen-Pihkala UM and Knuutila S: AML1 gene amplification: A
novel finding in childhood acute lymphoblastic leukemia.
Haematologica. 85:362–366. 2000.PubMed/NCBI
|
|
19
|
Chuang LS and Ito Y: RUNX3 is
multifunctional in carcinogenesis of multiple solid tumors.
Oncogene. 29:2605–2615. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Taniuchi I, Osato M and Ito Y: Runx1: No
longer just for leukemia. EMBO J. 31:4098–4099. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Sadikovic B, Thorner P, Chilton-MacNeill
S, Martin JW, Cervigne NK, Squire J and Zielenska M: Expression
analysis of genes associated with human osteosarcoma tumors shows
correlation of RUNX2 overexpression with poor response to
chemotherapy. BMC Cancer. 10:2022010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kurek KC, Del Mare S, Salah Z, Abdeen S,
Sadiq H, Lee SH, Gaudio E, Zanesi N, Jones KB, DeYoung B, et al:
Frequent attenuation of the WWOX tumor suppressor in osteosarcoma
is associated with increased tumorigenicity and aberrant RUNX2
expression. Cancer Res. 70:5577–5586. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lau CC, Harris CP, Lu XY, Perlaky L,
Gogineni S, Chintagumpala M, Hicks J, Johnson ME, Davino NA, Huvos
AG, et al: Frequent amplification and rearrangement of chromosomal
bands 6p12-p21 and 17p11.2 in osteosarcoma. Genes Chromosomes
Cancer. 39:11–21. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Endo T, Ohta K and Kobayashi T: Expression
and function of Cbfa-1/Runx2 in thyroid papillary carcinoma cells.
J Clin Endocrinol Metab. 93:2409–2412. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Dalle Carbonare L, Frigo A, Francia G,
Davì MV, Donatelli L, Stranieri C, Brazzarola P, Zatelli MC,
Menestrina F and Valenti MT: Runx2 mRNA expression in the tissue,
serum, and circulating non-hematopoietic cells of patients with
thyroid cancer. J Clin Endocrinol Metab. 97:E1249–E1256. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Barnes GL, Javed A, Waller SM, Kamal MH,
Hebert KE, Hassan MQ, Bellahcene A, Van Wijnen AJ, Young MF, Lian
JB, et al: Osteoblast-related transcription factors Runx2
(Cbfa1/AML3) and MSX2 mediate the expression of bone sialoprotein
in human metastatic breast cancer cells. Cancer Res. 63:2631–2637.
2003.PubMed/NCBI
|
|
27
|
Barnes GL, Hebert KE, Kamal M, Javed A,
Einhorn TA, Lian JB, Stein GS and Gerstenfeld LC: Fidelity of Runx2
activity in breast cancer cells is required for the generation of
metastases-associated osteolytic disease. Cancer Res. 64:4506–4513.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Leong DT, Lim J, Goh X, Pratap J, Pereira
BP, Kwok HS, Nathan SS, Dobson JR, Lian JB, Ito Y, et al:
Cancer-related ectopic expression of the bone-related transcription
factor RUNX2 in non-osseous metastatic tumor cells is linked to
cell proliferation and motility. Breast Cancer Res. 12:R892010.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Friedrich MJ, Rad R, Langer R, Voland P,
Hoefler H, Schmid RM, Prinz C and Gerhard M: Lack of RUNX3
regulation in human gastric cancer. J Pathol. 210:141–146. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Kilbey A, Terry A, Cameron ER and Neil JC:
Oncogene-induced senescence: An essential role for Runx. Cell
Cycle. 7:2333–2340. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Blyth K, Vaillant F, Jenkins A, McDonald
L, Pringle MA, Huser C, Stein T, Neil J and Cameron ER: Runx2 in
normal tissues and cancer cells: A developing story. Blood Cells
Mol Dis. 45:117–123. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Cameron ER and Neil JC: The Runx genes:
Lineage-specific oncogenes and tumor suppressors. Oncogene.
23:4308–4314. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Coffman JA: Runx transcription factors and
the developmental balance between cell proliferation and
differentiation. Cell Biol Int. 27:315–324. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Sullivan JC, Sher D, Eisenstein M,
Shigesada K, Reitzel AM, Marlow H, Levanon D, Groner Y, Finnerty JR
and Gat U: The evolutionary origin of the Runx/CBFbeta
transcription factors-studies of the most basal metazoans. BMC Evol
Biol. 8:2282008. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Nam S, Jin YH, Li QL, Lee KY, Jeong GB,
Ito Y, Lee J and Bae SC: Expression pattern, regulation, and
biological role of runt domain transcription factor, run, in
Caenorhabditis elegans. Mol Cell Biol. 22:547–554. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Rennert J, Coffman JA, Mushegian AR and
Robertson AJ: The evolution of Runx genes I. A comparative study of
sequences from phylogenetically diverse model organisms. BMC Evol
Biol. 3:42003. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Robertson AJ, Dickey-Sims C, Ransick A,
Rupp DE, McCarthy JJ and Coffman JA: CBFbeta is a facultative Runx
partner in the sea urchin embryo. BMC Biol. 4:42006. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Warren AJ, Bravo J, Warren AJ and Rabbitts
TH: Structural basis for the heterodimeric interaction between the
acute leukaemia-associated transcription factors AML1 and CBFβ.
EMBO J. 19:3004–3015. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Ito Y, Bae SC and Chuang LS: The RUNX
family: Developmental regulators in cancer. Nat Rev Cancer.
15:81–95. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Levanon D, Negreanu V, Bernstein Y, Bar-Am
I, Avivi L and Groner Y: AML1, AML2, and AML3, the human members of
the runt domain gene-family: CDNA structure, expression, and
chromosomal localization. Genomics. 23:425–432. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Bae SC, Takahashi E, Zhang YW, Ogawa E,
Shigesada K, Namba Y, Satake M and Ito Y: Cloning, mapping and
expression of PEBP2 alpha C, a third gene encoding the mammalian
Runt domain. Gene. 159:245–248. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Levanon D and Groner Y: Structure and
regulated expression of mammalian RUNX genes. Oncogene.
23:4211–4219. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Bangsow C, Rubins N, Glusman G, Bernstein
Y, Negreanu V, Goldenberg D, Lotem J, Ben-Asher E, Lancet D,
Levanon D and Groner Y: The RUNX3 gene-sequence, structure and
regulated expression. Gene. 279:221–232. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Marshall LJ, Moore AC, Ohki M, Kitabayashi
I, Patterson D and Ornelles DA: RUNX1 permits E4orf6-directed
nuclear localization of the adenovirus E1B-55K protein and
associates with centers of viral DNA and RNA synthesis. J Virol.
82:6395–408. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Stock M and Otto F: Control of RUNX2
isoform expression: The role of promoters and enhancers. J Cell
Biochem. 95:506–517. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Miyoshi H, Ohira M, Shimizu K, Mitani K,
Hirai H, Imai T, Yokoyama K, Soeda E and Ohki M: Alternative
splicing and genomic structure of the AML1 gene involved in acute
myeloid leukemia. Nucleic Acids Res. 23:2762–2769. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Kagoshima H, Shigesada K, Satake M, Ito Y,
Miyoshi H, Ohki M, Pepling M and Gergen P: The runt domain
identifies a new family of heterometric transcriptional regulators.
Trends Genet. 9:338–341. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Nagata T, Gupta V, Sorce D, Kim WY, Sali
A, Chait BT, Shigesada K, Ito Y and Werner MH: Immunoglobulin motif
DNA recognition and heterodimerization of the PEBP2/CBF Runt
domain. Nat Struct Biol. 6:615–619. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Williams AF and Barclay AN: The
immunoglobulin superfamily-domains for cell surface recognition.
Annu Rev Immunol. 6:381–405. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Javed A, Guo B, Hiebert S, Choi JY, Green
J, Zhao SC, Osborne MA, Stifani S, Stein JL, Lian JB, et al:
Groucho/TLE/R-esp proteins associate with the nuclear matrix and
repress RUNX (CBF(alpha)/AML/PEBP2(alpha)) dependent activation of
tissue-specific gene transcription. J Cell Sci. 113:2221–2231.
2000.PubMed/NCBI
|
|
51
|
Imai Y, Kurokawa M, Tanaka K, Friedman AD,
Ogawa S, Mitani K, Yazaki Y and Hirai H: TLE, the human homolog of
groucho, interacts with AML1 and acts as a repressor of
AML1-induced transactivation. Biochem Biophys Res Commun.
252:582–589. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Zaidi SK, Javed A, Choi JY, van Wijnen AJ,
Stein JL, Lian JB and Stein GS: A specific targeting signal directs
Runx2/Cbfa1 to subnuclear domains and contributes to
transactivation of the osteocalcin gene. J Cell Sci. 114:3093–3102.
2001.PubMed/NCBI
|
|
53
|
Stein GS, Lian JB, Stein JL, van Wijnen
AJ, Choi JY, Pratap J and Zaidi SK: Temporal and spatial parameters
of skeletal gene expression: Targeting RUNX factors and their
coregulatory proteins to subnuclear domains. Connect Tissue Res. 44
(Suppl 1):S149–S153. 2003. View Article : Google Scholar
|
|
54
|
Harrington KS, Javed A, Drissi H, McNeil
S, Lian JB, Stein JL, Van Wijnen AJ, Wang YL and Stein GS:
Transcription factors RUNX1/AML1 and RUNX2/Cbfa1 dynamically
associate with stationary subnuclear domains. J Cell Sci.
115:4167–4176. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Kanno T, Takahashi T, Tsujisawa T,
Ariyoshi W and Nishihara T: Mechanical stress-mediated Runx2
activation is dependent on Ras/ERK1/2 MAPK signaling in
osteoblasts. J Cell Biochem. 101:1266–1277. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Terry A, Kilbey A, Vaillant F, Stewart M,
Jenkins A, Cameron E and Neil JC: Conservation and expression of an
alternative 3′exon of Runx2 encoding a novel proline-rich
C-terminal domain. Gene. 336:115–125. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Tahirov TH, Inoue-Bungo T, Morii H,
Fujikawa A, Sasaki M, Kimura K, Shiina M, Sato K, Kumasaka T,
Yamamoto M, et al: Structural analyses of DNA recognition by the
AML1/Runx-1 Runt domain and its allosteric control by CBFbeta.
Cell. 104:755–767. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Pozner A, Goldenberg D, Negreanu V, Le S,
Elroy-stein O, Levanon D and Groner Y: Transcription-coupled
translation control of AML1/RUNX1 is mediated by Cap-and internal
ribosome entry site-dependent mechanisms. Mol Cell Biol.
20:2297–2307. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Xiao ZS, Simpson LG and Quarles LD:
IRES-dependent translational control of Cbfa1/Runx2 expression. J
Cell Biochem. 88:493–505. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Tang X, Sun L, Wang G, Chen B and Luo F:
RUNX1: A regulator of NF-kB signaling in pulmonary diseases. Curr
Protein Pept Sci. 19:172–178. 2018.PubMed/NCBI
|
|
61
|
Webber BR, Iacovino M, Choi SH, Tolar J,
Kyba M and Blazar BR: DNA methylation of Runx1 regulatory regions
correlates with transition from primitive to definitive
hematopoietic potential in vitro and in vivo. Blood. 122:2978–2986.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Sood R, Kamikubo Y and Liu P: Role of
RUNX1 in hematological malignancies. Blood. 129:2070–2082. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Jonason JH, Xiao G, Zhang M, Xing L and
Chen D: Post-translational Regulation of Runx2 in Bone and
Cartilage. J Dent Res. 88:693–703. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Rojas A, Aguilar R, Henriquez B, Lian JB,
Stein JL, Stein GS, van Wijnen AJ, van Zundert B, Allende ML and
Montecino M: Epigenetic control of the bone-master Runx2 gene
during osteoblast-lineage commitment by the histone demethylase
JARID1B/KDM5B. J Biol Chem. 290:28329–28342. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
O'Riordan M and Grosschedl R:
Transcriptional regulation of early B-lymphocyte differentiation.
Immunol Rev. 175:94–103. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Leiden JM and Thompson CB: Transcriptional
regulation of T-cell genes during T-cell development. Curr Opin
Immunol. 6:231–237. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Kurklu B, Whitehead RH, Ong EK, Minamoto
T, Fox JG, Mann JR, Judd LM, Giraud AS and Menheniott TR:
Lineage-specific RUNX3 hypomethylation marks the preneoplastic
immune component of gastric cancer. Oncogene. 34:2856–2866. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Appel E, Weissmann S, Salzberg Y, Orlovsky
K, Negreanu V, Tsoory M, Raanan C, Feldmesser E, Bernstein Y,
Wolstein O, et al: An ensemble of regulatory elements controls
Runx3 spatiotemporal expression in subsets of dorsal root ganglia
proprioceptive neurons. Genes Dev. 30:2607–2622. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Levanon D, Lotem J, Negreanu V, et al:
Transcription regulation in development and disease. Life Sci Open
Day. 1–3. 2008.
|
|
70
|
Kojo S, Tanaka H, Endo TA, Muroi S, Liu Y,
Seo W, Tenno M, Kakugawa K, Naoe Y, Nair K, et al: Priming of
lineage-specifying genes by Bcl11b is required for lineage choice
in post-selection thymocytes. Nat Commun. 8:7022017. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Cho JY, Akbarali Y, Zerbini LF, Gu X,
Boltax J, Wang Y, Oettgen P, Zhang DE and Libermann TA: Isoforms of
the Ets transcription factor NERF/ELF-2 physically interact with
AML1 and mediate opposing effects on AML1-mediated transcription of
the B cell-specific blk gene. J Biol Chem. 279:19512–19522. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
72
|
D'Alonzo RC, Selvamurugan N, Karsenty G
and Partridge NC: Physical interaction of the activator protein-1
factors c-Fos and c-Jun with Cbfa1 for collagenase-3 promoter
activation. J Biol Chem. 277:816–822. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Pelletier N, Champagne N, Stifani S and
Yang XJ: MOZ and MORF histone acetyltransferases interact with the
Runt-domain transcription factor Runx2. Oncogene. 21:2729–2740.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Levanon D, Glusman G, Bangsow T, Ben-Asher
E, Male DA, Avidan N, Bangsow C, Hattori M, Taylor TD, Taudien S,
et al: Architecture and anatomy of the genomic locus encoding the
human leukemia-associated transcription factor RUNX1/AML1. Gene.
262:23–33. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Aho TL, Sandholm J, Peltola KJ, Ito Y and
Koskinen PJ: Pim-1 kinase phosphorylates RUNX family transcription
factors and enhances their activity. BMC Cell Biol. 7:212006.
View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Kim HR, Oh BC, Choi JK and Bae SC: Pim-1
kinase phosphorylates and stabilizes RUNX3 and alters its
subcellular localization. J Cell Biochem. 105:1048–1058. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Blyth K, Terry A, Mackay N, Vaillant F,
Bell M, Cameron ER, Neil JC and Stewart M: Runx2: A novel oncogenic
effector revealed by in vivo complementation and retroviral
tagging. Oncogene. 20:295–302. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Tanaka T, Kurokawa M, Ueki K, Tanaka K,
Imai Y, Mitani K, Okazaki K, Sagata N, Yazaki Y, Shibata Y, et al:
The extracellular signal-regulated kinase pathway phosphorylates
AML1, an acute myeloid leukemia gene product, and potentially
regulates its transactivation ability. Mol Cell Biol. 16:3967–3979.
1996. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Imai Y, Kurokawa M, Yamaguchi Y, Izutsu K,
Nitta E, Mitani K, Satake M, Noda T, Ito Y and Hirai H: The
corepressor mSin3A regulates phosphorylation-induced activation,
intranuclear location, and stability of AML1. Mol Cell Biol.
24:1033–1043. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Wee HJ, Voon DC, Bae SC and Ito Y:
PEBP2-beta/CBF-beta dependent phosphorylation of RUNX1 and p300 by
HIPK2: Implications for leukemogenesis. Blood. 112:3777–3787. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Aikawa Y, Nguyen LA, Isono K, Takakura N,
Tagata Y, Schmitz ML, Koseki H and Kitabayashi I: Roles of HIPK1
and HIPK2 in AML1-and p300-dependent transcription, hematopoiesis
and blood vessel formation. EMBO J. 25:3955–3965. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Seo W, Tanaka H, Miyamoto C, Levanon D,
Groner Y and Taniuchi I: Roles of VWRPY motif-mediated gene
repression by Runx proteins during T-cell development. Immunol Cell
Biol. 90:827–830. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Westendorf JJ, Zaidi SK, Cascino JE,
Kahler R, van Wijnen AJ, Lian JB, Yoshida M, Stein GS and Li X:
Runx2 (Cbfa1, AML-3) interacts with histone deacetylase 6 and
represses the p21(CIP1/WAF1) promoter. Mol Cell Biol. 22:7982–7992.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Durst KL and Hiebert SW: Role of RUNX
family members in transcriptional repression and gene silencing.
Oncogene. 23:4220–4224. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Kasahara K, Shiina M, Fukuda I, Ogata K
and Nakamura H: Molecular mechanisms of cooperative binding of
transcription factors Runx1-CBFβ-Ets1 on the TCRα gene enhancer.
PLoS One. 12:e01726542017. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Hassig CA, Fleischer TC, Billin AN,
Schreiber SL and Ayer DE: Histone deacetylase activity is required
for full transcriptional repression by mSin3A. Cell. 89:341–347.
1997. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Gelmetti V, Zhang J, Fanelli M, Minucci S,
Pelicci PG and Lazar MA: Aberrant recruitment of the nuclear
receptor corepressor-histone deacetylase complex by the acute
myeloid leukemia fusion partner ETO. Mol Cell Biol. 18:7185–7191.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Huang G, Shigesada K, Ito K, Wee HJ,
Yokomizo T and Ito Y: Dimerization with PEBP2beta protects
RUNX1/AML1 from ubiquitin-proteasome-mediated degradation. EMBO J.
20:723–733. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Zelent A, Greaves M and Enver T: Role of
the TEL-AML1 fusion gene in the molecular pathogenesis of childhood
acute lymphoblastic leukaemia. Oncogene. 23:4275–4283. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Bakshi R, Hassan MQ, Pratap J, Lian JB,
Montecino MA, van Wijnen AJ, Stein JL, Imbalzano AN and Stein GS:
The human SWI/SNF complex associates with RUNX1 to control
transcription of hematopoietic target genes. J Cell Physiol.
225:569–576. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Henriquez B, Hepp M, Merino P, Sepulveda
H, van Wijnen AJ, Lian JB, Stein GS, Stein JL and Montecino M:
C/EBPβ binds the P1 promoter of the Runx2 gene and up-regulates
Runx2 transcription in osteoblastic cells. J Cell Physiol.
226:3043–3052. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
De Bruijn M and Dzierzak E: Runx
transcription factors in the development and function of the
definitive hematopoietic system. Blood. 129:2061–2069. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Inoue K, Ozaki S, Shiga T, Ito K, Masuda
T, Okado N, Iseda T, Kawaguchi S, Ogawa M, Bae SC, et al: Runx3
controls the axonal projection of proprioceptive dorsal root
ganglion neurons. Nat Neurosci. 5:946–954. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Senzaki K, Ozaki S, Yoshikawa M, Ito Y and
Shiga T: Runx3 is required for the specification of TrkC-expressing
mechanoreceptive trigeminal ganglion neurons. Mol Cell Neurosci.
43:296–307. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Woolf E, Brenner O, Goldenberg D, Levanon
D and Groner Y: Runx3 regulates dendritic epidermal T cell
development. Dev Biol. 303:703–714. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Kamikubo Y: Genetic compensation of RUNX
family transcription factors in leukemia. Cancer Sci.
109:2358–2363. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Ross K, Sedello AK, Todd GP,
Paszkowski-Rogacz M, Bird AW, Ding L, Grinenko T, Behrens K, Hubner
N, Mann M, et al: Polycomb group ring finger 1 cooperates with
Runx1 in regulating differentiation and self-renewal of
hematopoietic cells. Blood. 119:4152–4161. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Yokomizo T, Ogawa M, Osato M, Kanno T,
Yoshida H, Fujimoto T, Fraser S, Nishikawa S, Okada H, Satake M, et
al: Requirement of Runx1/AML1/PEBP2alphaB for the generation of
haematopoietic cells from endothelial cells. Genes Cells. 6:13–23.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Tanaka K, Tanaka T, Ogawa S, Kurokawa M,
Mitani K, Yazaki Y and Hirai H: Increased expression of AML1 during
retinoic-acid-induced differentiation of U937 cells. Biochem
Biophys Res Commun. 211:1023–1030. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Pratap J, Galindo M, Zaidi SK, Vradii D,
Bhat BM, Robinson JA, Choi JY, Komori T, Stein JL, Lian JB, et al:
Cell growth regulatory role of Runx2 during proliferative expansion
of preosteoblasts. Cancer Res. 63:5357–5362. 2003.PubMed/NCBI
|
|
101
|
Galindo M, Pratap J, Young DW,
Hovhannisyan H, Im HJ, Choi JY, Lian JB, Stein JL, Stein GS and van
Wijnen AJ: The bone-specific expression of Runx2 oscillates during
the cell cycle to support a G 1-related Antiproliferative function
in osteoblasts. J Biol Chem. 280:20274–20285. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Stein GS, Lian JB, Van Wijnen AJ, Stein
JL, Montecino M, Javed A, Zaidi SK, Young DW, Choi JY and Pockwinse
SM: Runx2 control of organization, assembly and activity of the
regulatory machinery for skeletal gene expression. Oncogene.
23:4315–4329. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Krege JH, Hodgin JB, Couse JF, Enmark E,
Warner M, Mahler JF, Sar M, Korach KS, Gustafsson JA and Smithies
O: Generation and reproductive phenotypes of mice lacking estrogen
receptor beta. Proc Natl Acad Sci USA. 95:15677–15682. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Ehrhardt GR, Hijikata A, Kitamura H, Ohara
O, Wang JY and Cooper MD: Discriminating gene expression profiles
of memory B cell subpopulations. J Exp Med. 205:1807–1817. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Vaillant F, Blyth K, Andrew L, Neil JC and
Cameron ER: Enforced expression of Runx2 perturbs T cell
development at a stage coincident with beta-selection. J Immunol.
169:2866–2874. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Taniuchi I, Osato M, Egawa T, Sunshine MJ,
Bae SC, Komori T, Ito Y and Littman DR: Differential requirements
for Runx proteins in CD4 repression and epigenetic silencing during
T lymphocyte development. Cell. 111:621–633. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Bauer O, Sharir A, Kimura A, Hantisteanu
S, Takeda S and Groner Y: Loss of osteoblast Runx3 produces severe
congenital osteopenia. Mol Cell Biol. 35:1097–109. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Wotton S, Terry A, Kilbey A, Jenkins A,
Herzyk P and Neil JC: UKPMC funders group gene array analysis
reveals a common Runx transcriptional program controlling cell
adhesion and survival. Oncogene. 27:5856–5866. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Zaidi SK, Pande S, Pratap J, Gaur T,
Grigoriu S, Ali SA, Stein JL, Lian JB, van Wijnen AJ and Stein GS:
Runx2 deficiency and defective subnuclear targeting bypass
senescence to promote immortalization and tumorigenic potential.
Proc Natl Acad Sci USA. 104:19861–19866. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Ghali O, Chauveau C, Hardouin P, Broux O
and Devedjian JC: TNF-alpha's effects on proliferation and
apoptosis in human mesenchymal stem cells depend on RUNX2
expression. J Bone Miner Res. 25:1616–1626. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Scheitz CJ, Lee TS, McDermitt DJ and
Tumbar T: Defining a tissue stem cell-driven Runx1/Stat3 signalling
axis in epithelial cancer. EMBO J. 31:4124–4139. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Speck NA and Gilliland DG: Core-binding
factors in haematopoiesis and leukaemia. Nat Rev Cancer. 2:502–513.
2002. View
Article : Google Scholar : PubMed/NCBI
|
|
113
|
Look AT: Oncogenic transcription factors
in the human acute leukemias. Science. 278:1059–1064. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Regha K, Assi SA, Tsoulaki O, Gilmour J,
Lacaud G and Bonifer C: Developmental-stage-dependent
transcriptional response to leukaemic oncogene expression. Nat
Commun. 6:72032015. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Golub TR, Barker GF, Bohlander SK, Hiebert
SW, Ward DC, Bray-Ward P, Morgan E, Raimondi SC, Rowley JD and
Gilliland DG: Fusion of the TEL gene on 12p13 to the AML1 gene on
21q22 in acute lymphoblastic leukemia. Proc Natl Acad Sci USA.
92:4917–4921. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Nucifora G and Rowley JD: AMLl and the
8;21 and 3;21 translocations in acute and chronic myeloid leukemia.
Blood. 86:1–14. 1995.PubMed/NCBI
|
|
117
|
Mitani K, Ogawa S, Tanaka T, Miyoshi H,
Kurokawa M, Mano H, Yazaki Y, Ohki M and Hirai H: Generation of the
AML1-EVI-1 fusion gene in the t(3;21)(q26;q22) causes blastic
crisis in chronic myelocytic leukemia. EMBO J. 13:504–510. 1994.
View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Krygier A, Szmajda D, Żebrowska M, Jeleń A
and Balcerczak E: Expression levels of the runt-related
transcription factor 1 and 3 genes in the development of acute
myeloid leukemia. Oncol Lett. 15:6733–6738. 2018.PubMed/NCBI
|
|
119
|
Wotton S, Stewart M, Blyth K, Vaillant F,
Kilbey A, Neil JC and Cameron ER: Proviral insertion indicates a
dominant oncogenic role for Runx1/AML-1 in T-cell lymphoma. Cancer
Res. 62:7181–7185. 2002.PubMed/NCBI
|
|
120
|
Osato M, Asou N, Abdalla E, Hoshino K,
Yamasaki H, Okubo T, Suzushima H, Takatsuki K, Kanno T, Shigesada K
and Ito Y: Biallelic and heterozygous point mutations in the runt
domain of the AML1/PEBP2alphaB gene associated with myeloblastic
leukemias. Blood. 93:1817–1824. 1999.PubMed/NCBI
|
|
121
|
Song WJ, Sullivan MG, Legare RD, Hutchings
S, Tan X, Kufrin D, Ratajczak J, Resende IC, Haworth C, Hock R, et
al: Haploinsufficiency of CBFA2 causes familial thrombocytopenia
with propensity to develop acute myelogenous leukaemia. Nat Genet.
23:166–175. 1999. View
Article : Google Scholar : PubMed/NCBI
|
|
122
|
Fonatsch C: The role of chromosome 21 in
hematology and oncology. Genes Chromosomes Cancer. 49:497–508.
2010.PubMed/NCBI
|
|
123
|
Banerji S, Cibulskis K, Rangel-Escareno C,
Brown KK, Carter SL, Frederick AM, Lawrence MS, Sivachenko AY,
Sougnez C, Zou L, et al: Sequence analysis of mutations and
translocations across breast cancer subtypes. Nature. 486:405–409.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Ellis MJ, Ding L, Shen D, Luo J, Suman VJ,
Wallis JW, Van Tine BA, Hoog J, Goiffon RJ, Goldstein TC, et al:
Whole-genome analysis informs breast cancer response to aromatase
inhibition. Nature. 486:353–360. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
125
|
Morita K, Suzuki K, Maeda S, Matsuo A,
Mitsuda Y, Tokushige C, Kashiwazaki G, Taniguchi J, Maeda R, Noura
M, et al: Genetic regulation of the RUNX transcription factor
family has antitumor effects. J Clin Invest. 127:2815–2828. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
126
|
Illendula A, Gilmour J, Grembecka J,
Tirumala VSS, Boulton A, Kuntimaddi A, Schmidt C, Wang L, Pulikkan
JA, Zong H, et al: Small molecule inhibitor of CBFβ-RUNX binding
for RUNX transcription factor driven cancers. EBioMedicine.
8:117–131. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Pelassa I, Corà D, Cesano F, Monje FJ,
Montarolo PG and Fiumara F: Association of polyalanine and
polyglutamine coiled coils mediates expansion disease-related
protein aggregation and dysfunction. Hum Mol Genet. 23:3402–3420.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Stewart M, Terry A, Hu M, O'Hara M, Blyth
K, Baxter E, Cameron E, Onions DE and Neil JC: Proviral insertions
induce the expression of bone-specific isoforms of PEBP2alphaA
(CBFA1): Evidence for a new myc collaborating oncogene. Proc Natl
Acad Sci USA. 94:8646–8651. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
129
|
Martin JW, Zielenska M, Stein GS, Van
Wijnen AJ and Squire JA: The role of RUNX2 in osteosarcoma
oncogenesis. Sarcoma. 2011:2827452011. View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Li N, Luo D, Hu X, Luo W, Lei G, Wang Q,
Zhu T, Gu J, Lu Y and Zheng Q: RUNX2 and Osteosarcoma. Anticancer
Agents Med Chem. 15:881–887. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
131
|
Pratap J, Lian JB, Javed A, Barnes GL, van
Wijnen AJ, Stein JL and Stein GS: Regulatory roles of Runx2 in
metastatic tumor and cancer cell interactions with bone. Cancer
Metastasis Rev. 25:589–600. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
132
|
Rucci N and Teti A: Osteomimicry: How
tumor cells try to deceive the bone. Front Biosci (Schol Ed).
2:907–915. 2010.PubMed/NCBI
|
|
133
|
Niu DF, Kondo T, Nakazawa T, Oishi N,
Kawasaki T, Mochizuki K, Yamane T and Katoh R: Transcription factor
Runx2 is a regulator of epithelial-mesenchymal transition and
invasion in thyroid carcinomas. Lab Invest. 92:1181–1190. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
134
|
Lim M, Zhong C, Yang S, Bell AM, Cohen MB
and Roy-Burman P: Runx2 regulates survivin expression in prostate
cancer cells. Lab Invest. 90:222–233. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
135
|
Pratap J, Javed A, Languino LR, van Wijnen
AJ, Stein JL, Stein GS and Lian JB: The Runx2 osteogenic
transcription factor regulates matrix metalloproteinase 9 in bone
metastatic cancer cells and controls cell invasion. Mol Cell Biol.
25:8581–8591. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
136
|
Onodera Y, Miki Y, Suzuki T, Takagi K,
Akahira J, Sakyu T, Watanabe M, Inoue S, Ishida T, Ohuchi N and
Sasano H: Runx2 in human breast carcinoma: Its potential roles in
cancer progression. Cancer Sci. 101:2670–2675. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
137
|
Underwood KF, D'Souza DR, Mochin-Peters M,
Pierce AD, Kommineni S, Choe M, Bennett J, Gnatt A, Habtemariam B,
MacKerell AD Jr and Passaniti A: Regulation of RUNX2 transcription
factor-DNA interactions and cell proliferation by vitamin D3
(cholecalciferol) prohormone activity. J Bone Miner Res.
27:913–925. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
138
|
Kim MS, Gernapudi R, Choi EY, Lapidus RG
and Passaniti A: Characterization of CADD522, a small molecule that
inhibits RUNX2-DNA binding and exhibits antitumor activity.
Oncotarget. 8:70916–70940. 2017.PubMed/NCBI
|
|
139
|
Tandon M, Gokul K, Ali SA, Chen Z, Lian J,
Stein GS and Pratap J: Runx2 mediates epigenetic silencing of the
bone morphogenetic protein-3B (BMP-3B/GDF10) in lung cancer cells.
Mol Cancer. 11:272012. View Article : Google Scholar : PubMed/NCBI
|
|
140
|
Lotem J, Levanon D, Negreanu V, Bauer O,
Hantisteanu S, Dicken J and Groner Y: Runx3 at the interface of
immunity, inflammation and cancer. Biochim Biophys Acta.
1855:131–143. 2015.PubMed/NCBI
|
|
141
|
Ito K, Liu Q, Salto-Tellez M, Yano T, Tada
K, Ida H, Huang C, Shah N, Inoue M, Rajnakova A, et al: RUNX3, a
novel tumor suppressor, is frequently inactivated in gastric cancer
by protein mislocalization. Cancer Res. 65:7743–7750. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
142
|
Goh YM, Cinghu S, Hong ET, Lee YS, Kim JH,
Jang JW, Li YH, Chi XZ, Lee KS, Wee H, et al: Src kinase
phosphorylates RUNX3 at tyrosine residues and localizes the protein
in the cytoplasm. J Biol Chem. 285:10122–10129. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
143
|
Lau QC, Raja E, Salto-Tellez M, Liu Q, Ito
K, Inoue M, Putti TC, Loh M, Ko TK, Huang C, et al: RUNX3 is
frequently inactivated by dual mechanisms of protein
mislocalization and promoter hypermethylation in breast cancer.
Cancer Res. 66:6512–6520. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
144
|
Chen LF: Tumor suppressor function of
RUNX3 in breast cancer. J Cell Biochem. 113:1470–1477.
2012.PubMed/NCBI
|
|
145
|
Li QL, Ito K, Sakakura C, Fukamachi H,
Inoue KI, Chi XZ, Lee KY, Nomura S, Lee CW, Han SB, et al: Causal
relationship between the loss of RUNX3 expression and gastric
cancer. Cell. 109:113–124. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
146
|
Fujii S, Ito K, Ito Y and Ochiai A:
Enhancer of zeste homologue 2 (EZH2) down-regulates RUNX3 by
increasing histone H3 methylation. J Biol Chem. 283:17324–17332.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
147
|
Tsang YH Lamb A and Chen LF: New insights
into the inactivation of gastric tumor suppressor RUNX3: The role
of H. pylori infection. J Cell Biochem. 112:381–386. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
148
|
Katayama Y, Takahashi M and Kuwayama H:
Helicobacter pylori causes runx3 gene methylation and its loss of
expression in gastric epithelial cells, which is mediated by nitric
oxide produced by macrophages. Biochem Biophys Res Commun.
388:496–500. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
149
|
Yong WP; National University Hospital,
Singapore, : The effect of preoperative docetaxel, cisplatin and
capecitabine on serum RUNX3 hypermethylation status in patients
with gastric and lower oesophagus adenocarcinoma. Clin Trials.
12017.
|
|
150
|
Hor YT, Voon DC, Koo JK, Wang H, Lau WM,
Ashktorab H, Chan SL and Ito Y: A role for RUNX3 in
inflammation-induced expression of IL23A in gastric epithelial
cells. Cell Rep. 8:50–58. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
151
|
National Institutes of Health: GDC data
portal: TCGA-HNSC project. US Dep Heal Hum Serv|Natl Institutes
Heal. 12018.
|
|
152
|
Bae SC, Kolinjivadi AM and Ito Y:
Functional relationship between p53 and RUNX proteins. J Mol Cell
Biol. Dec 11–2018.(Epub ahead of print). doi: 10.1093/jmcb/mjy076.
PubMed/NCBI
|
|
153
|
Lee JW, van Wijnen A and Bae SC: RUNX3 and
p53: How two tumor suppressors cooperate against oncogenic ras? Adv
Exp Med Biol. 962:321–332. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
154
|
Saha A and Robertson ES: Epstein-barr
virus-associated B-cell lymphomas: Pathogenesis and clinical
outcomes. Clin Cancer Res. 17:3056–3063. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
155
|
Gunnell A, Webb HM, Wood CD, McClellan MJ,
Wichaidit B, Kempkes B, Jenner RG, Osborne C, Farrell PJ and West
MJ: RUNX super-enhancer control through the Notch pathway by
Epstein-Barr virus transcription factors regulates B cell growth.
Nucleic Acids Res. 44:4636–4650. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
156
|
Spender LC, Cornish GH, Sullivan A and
Farrell PJ: Expression of transcription factor AML-2 (RUNX3,
CBF(alpha)-3) Is Induced by Epstein-Barr Virus EBNA-2 and
correlates with the B-cell activation phenotype. J Virol.
76:4919–4927. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
157
|
Levanon D, Bernstein Y, Negreanu V, Bone
KR, Pozner A, Eilam R, Lotem J, Brenner O and Groner Y: Absence of
Runx3 expression in normal gastrointestinal epithelium calls into
question its tumour suppressor function. EMBO Mol Med. 3:593–604.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
158
|
Lotem J, Levanon D, Negreanu V and Groner
Y: The false paradigm of RUNX3 function as tumor suppressor in
gastric cancer. J Cancer Ther. 4:16–25. 2013. View Article : Google Scholar
|
|
159
|
Levanon D, Negreanu V, Lotem J, Bone KR,
Brenner O, Leshkowitz D and Groner Y: Transcription Factor Runx3
regulates interleukin-15-dependent natural killer cell activation.
Mol Cell Biol. 34:1158–1169. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
160
|
Whittle MC, Izeradjene K, Rani PG, Feng L,
Carlson MA, DelGiorno KE, Wood LD, Goggins M, Hruban RH, Chang AE,
et al: RUNX3 controls a metastatic switch in pancreatic ductal
adenocarcinoma. Cell. 161:1345–1360. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
161
|
Whittle MC and Hingorani SR: RUNX3 defines
disease behavior in pancreatic ductal adenocarcinoma. Mol Cell
Oncol. 3:e10765882015. View Article : Google Scholar : PubMed/NCBI
|
|
162
|
Jian Z, Cheng T, Zhang Z, Raulefs S, Shi
K, Steiger K, Maeritz N, Kleigrewe K, Hofmann T, Benitz S, et al:
Glycemic variability promotes both local invasion and metastatic
colonization by pancreatic ductal adenocarcinoma. Cell Mol
Gastroenterol Hepatol. 6:429–449. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
163
|
Lotem J, Levanon D, Negreanu V, Bauer O,
Hantisteanu S, Dicken J and Groner Y: Runx3 in immunity,
inflammation and cancer. Adv Exp Med Biol. 962:369–393. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
164
|
Berger AH, Knudson AG and Pandolfi PP: A
continuum model for tumour suppression. Nature. 476:163–169. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
165
|
Tsunematsu T, Kudo Y, Iizuka S, Ogawa I,
Fujita T, Kurihara H, Abiko Y and Takata T: RUNX3 has an oncogenic
role in head and neck cancer. PLoS One. 4:e58922009. View Article : Google Scholar : PubMed/NCBI
|
|
166
|
Manandhar S and Lee YM: Emerging role of
RUNX3 in the regulation of tumor microenvironment. BMB Rep.
51:174–181. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
167
|
Pattabiraman DR and Weinberg RA: Tackling
the cancer stem cells-what challenges do they pose? Nat Rev Drug
Discov. 13:497–512. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
168
|
Gopal S, Saraswati S, Sanyal M, Bulbule A,
Ramdasi A, Kumar D, Behera R, Ahmed M, Chakraborty G, Kumar V, et
al: Therapeutic targeting of osteopontin in breast cancer cells.
In: Breast cancer-current and alternative therapeutic modalities.
InTech. 23–36. 2011.
|
|
169
|
Osorio KM, Lilja KC and Tumbar T: Runx1
modulates adult hair follicle stem cell emergence and maintenance
from distinct embryonic skin compartments. J Cell Biol.
193:235–250. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
170
|
Fortini ME: Notch Signaling: The core
pathway and its posttranslational regulation. Dev Cell. 16:633–647.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
171
|
Yuan X, Wu H, Han N, Xu H, Chu Q, Yu S,
Chen Y and Wu K: Notch signaling and EMT in non-small cell lung
cancer: Biological significance and therapeutic application. J
Hematol Oncol. 7:872014. View Article : Google Scholar : PubMed/NCBI
|
|
172
|
Thouverey C and Caverzasio J: Focus on the
p38 MAPK signaling pathway in bone development and maintenance.
Bonekey Rep. 4:7112015. View Article : Google Scholar : PubMed/NCBI
|
|
173
|
Franceschi RT, Xiao G, Jiang D,
Gopalakrishnan R, Yang S and Reith E: Multiple signaling pathways
converge on the Cbfa1/Runx2 transcription factor to regulate
osteoblast differentiation. Connect Tissue Res. 44 (Suppl
1):S109–S116. 2003. View Article : Google Scholar
|
|
174
|
Nikitovic D, Kavasi RM, Berdiaki A,
Papachristou DJ, Tsiaoussis J, Spandidos DA, Tsatsakis AM and
Tzanakakis GN: Parathyroid hormone/parathyroid hormone-related
peptide regulate osteosarcoma cell functions: Focus on the
extracellular matrix (Review). Oncol Rep. 36:1787–1792. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
175
|
Cohen-Solal KA, Boregowda RK and Lasfar A:
RUNX2 and the PI3K/AKT axis reciprocal activation as a driving
force for tumor progression. Mol Cancer. 14:1372015. View Article : Google Scholar : PubMed/NCBI
|
|
176
|
Petcherski AG and Kimble J: Mastermind is
a putative activator for Notch. Curr Biol. 10:R471–R473. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
177
|
Nakagawa M, Ichikawa M, Kumano K, Goyama
S, Kawazu M, Asai T, Ogawa S, Kurokawa M and Chiba S: AML1/Runx1
rescues Notch1-null mutation-induced deficiency of para-aortic
splanchnopleural hematopoiesis. Blood. 108:3329–3334. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
178
|
Burns CE, Traver D, Mayhall E, Shepard JL
and Zon LI: Hematopoietic stem cell fate is established by the
Notch-Runx pathway. Genes Dev. 19:2331–2342. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
179
|
Meier-Stiegen F, Schwanbeck R, Bernoth K,
Martini S, Hieronymus T, Ruau D, Zenke M and Just U: Activated
Notch1 target genes during embryonic cell differentiation depend on
the cellular context and include lineage determinants and
inhibitors. PLoS One. 5:e114812010. View Article : Google Scholar : PubMed/NCBI
|
|
180
|
Ann EJ, Kim HY, Choi YH, Kim MY, Mo JS,
Jung J, Yoon JH, Kim SM, Moon JS, Seo MS, et al: Inhibition of
Notch1 signaling by Runx2 during osteoblast differentiation. J Bone
Miner Res. 26:317–330. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
181
|
Shen Q and Christakos S: The vitamin D
receptor, Runx2, and the notch signaling pathway cooperate in the
transcriptional regulation of osteopontin. J Biol Chem.
280:40589–40598. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
182
|
Fu YX, Chang AC, Fournier M, Chang L,
Niessen K and Karsan A: RUNX3 maintains the mesenchymal phenotype
after termination of the notch signal. J Biol Chem.
286:11803–11813. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
183
|
Cadigan KM and Peifer M: Wnt signaling
from development to disease: Insights from model systems. Cold
Spring Harb Perspect Biol. 1:a0028812009. View Article : Google Scholar : PubMed/NCBI
|
|
184
|
Chen F, Liu X, Bai J, Pei D and Zheng J:
The emerging role of RUNX3 in cancer metastasis (Review). Oncol
Rep. 35:1227–1236. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
185
|
Medina MA, Ugarte GD, Vargas MF, Avila ME,
Necuñir D, Elorza AA, Gutiérrez SE and De Ferrari GV: Alternative
RUNX1 promoter regulation by Wnt/β-catenin signaling in leukemia
cells and human hematopoietic progenitors. J Cell Physiol.
231:1460–1467. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
186
|
Moore A, Tahinci E, Lee E and Hiebert S:
RUNX1-ETO stimulates wnt signaling by inhibiting the function of
ETO family member proteins. Blood. 104:25542004.
|
|
187
|
Dong YF, Soung do Y, Schwarz EM, O'Keefe
RJ and Drissi H: Wnt induction of chondrocyte hypertrophy through
the Runx2 transcription factor. J Cell Physiol. 208:77–86. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
188
|
Gaur T, Lengner CJ, Hovhannisyan H, Bhat
RA, Bodine PV, Komm BS, Javed A, van Wijnen AJ, Stein JL, Stein GS
and Lian JB: Canonical WNT signaling promotes osteogenesis by
directly stimulating Runx2 gene expression. J Biol Chem.
280:33132–33140. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
189
|
Attisano L and Labbé E: TGFbeta and Wnt
pathway cross-talk. Cancer Metastasis Rev. 23:53–61. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
190
|
Ju X, Ishikawa TO, Naka K, Ito K, Ito Y
and Oshima M: Context-dependent activation of Wnt signaling by
tumor suppressor RUNX3 in gastric cancer cells. Cancer Sci.
105:418–424. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
191
|
Chimge NO and Frenkel B: The RUNX family
in breast cancer: Relationships with estrogen signaling. Oncogene.
32:2121–2130. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
192
|
Mitsuda Y, Morita K, Kashiwazaki G,
Taniguchi J, Bando T, Obara M, Hirata M, Kataoka TR, Muto M, Kaneda
Y, et al: RUNX1 positively regulates ErbB2/HER2 signaling pathway
through modulating the expression of SOS1 in gastric cancer cells.
Sci Rep. 8:64232018. View Article : Google Scholar : PubMed/NCBI
|
|
193
|
Khalid O, Baniwal SK, Purcell DJ, Leclerc
N, Gabet Y, Stallcup MR, Coetzee GA and Frenkel B: Modulation of
Runx2 activity by estrogen receptor-alpha: Implications for
osteoporosis and breast cancer. Endocrinology. 149:5984–5995. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
194
|
Chimge NO, Baniwal SK, Little GH, Chen YB,
Kahn M, Tripathy D, Borok Z and Frenkel B: Regulation of breast
cancer metastasis by Runx2 and estrogen signaling: The role of
SNAI2. Breast Cancer Res. 13:R1272011. View Article : Google Scholar : PubMed/NCBI
|
|
195
|
Teplyuk NM, Galindo M, Teplyuk VI, Pratap
J, Young DW, Lapointe D, Javed A, Stein JL, Lian JB, Stein GS and
van Wijnen AJ: Runx2 regulates G protein-coupled signaling pathways
to control growth of osteoblast progenitors. J Biol Chem.
283:27585–27597. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
196
|
Berry NB, Fan M and Nephew KP: Estrogen
Receptor-alpha Hinge-Region Lysines 302 and 303 regulate receptor
degradation by the proteasome. Mol Endocrinol. 22:1535–1551. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
197
|
Subramanian K, Jia D, Kapoor-Vazirani P,
Powell DR, Collins RE, Sharma D, Peng J, Cheng X and Vertino PM:
Regulation of estrogen receptor alpha by the SET7 lysine
methyltransferase. Mol Cell. 30:336–347. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
198
|
Fan M, Park A and Nephew KP: CHIP
(Carboxyl Terminus of Hsc70-Interacting Protein) promotes basal and
geldanamycin-induced degradation of estrogen receptor-alpha. Mol
Endocrinol. 19:2901–2914. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
199
|
Duong V, Boulle N, Daujat S, Chauvet J,
Bonnet S, Neel H and Cavaillès V: Differential regulation of
estrogen receptor alpha turnover and transactivation by Mdm2 and
stress-inducing agents. Cancer Res. 67:5513–5521. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
200
|
Huang B, Qu Z, Ong CW, Tsang YH, Xiao G,
Shapiro D, Salto-Tellez M, Ito K, Ito Y and Chen LF: RUNX3 acts as
a tumor suppressor in breast cancer by targeting estrogen receptor
α. Oncogene. 31:527–534. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
201
|
Yano T, Ito K, Fukamachi H, Chi XZ, Wee
HJ, Inoue K, Ida H, Bouillet P, Strasser A, Bae SC and Ito Y: The
RUNX3 tumor suppressor upregulates Bim in gastric epithelial cells
undergoing transforming growth factor beta-induced apoptosis. Mol
Cell Biol. 26:4474–4488. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
202
|
Krishnan V, Chong YL, Tan TZ, Kulkarni M,
Bin Rahmat MB, Tay LS, Sankar H, Jokhun DS, Ganesan A, Chuang LSH,
et al: TGFβ promotes genomic instability after loss of RUNX3.
Cancer Res. 78:88–102. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
203
|
Chi XZ, Yang JO, Lee KY, Ito K, Sakakura
C, Li QL, Kim HR, Cha EJ, Lee YH, Kaneda A, et al: RUNX3 suppresses
gastric epithelial cell growth by inducing p21(WAF1/Cip1)
expression in cooperation with transforming growth factor
{beta}-activated SMAD. Mol Cell Biol. 25:8097–8107. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
204
|
Ito K, Lim AC, Salto-Tellez M, Motoda L,
Osato M, Chuang LS, Lee CW, Voon DC, Koo JK, Wang H, et al: RUNX3
Attenuates beta-catenin/T cell factors in intestinal tumorigenesis.
Cancer Cell. 14:226–237. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
205
|
Tanaka S, Shiraha H, Nakanishi Y, Nishina
S, Matsubara M, Horiguchi S, Takaoka N, Iwamuro M, Kataoka J,
Kuwaki K, et al: Runt-related transcription factor 3 reverses
epithelial-mesenchymal transition in hepatocellular carcinoma. Int
J Cancer. 131:2537–2546. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
206
|
Voon DC, Wang H, Koo JK, Nguyen TA, Hor
YT, Chu YS, Ito K, Fukamachi H, Chan SL, Thiery JP and Ito Y: Runx3
protects gastric epithelial cells against epithelial-mesenchymal
transition-induced cellular plasticity and tumorigenicity. Stem
Cells. 30:2088–2099. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
207
|
Lee YS, Lee JW, Jang JW, Chi XZ, Kim JH,
Li YH, Kim MK, Kim DM, Choi BS, Kim EG, et al: Runx3 Inactivation
is a crucial early event in the development of lung adenocarcinoma.
Cancer Cell. 24:603–616. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
208
|
Kilbey A, Blyth K, Wotton S, Terry A,
Jenkins A, Bell M, Hanlon L, Cameron ER and Neil JC: Runx2
disruption promotes immortalization and confers resistance to
oncogene-induced senescence in primary murine fibroblasts. Cancer
Res. 67:11263–11271. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
209
|
Cameron ER, Blyth K, Hanlon L, Kilbey A,
Mackay N, Stewart M, Terry A, Vaillant F, Wotton S and Neil JC: The
Runx genes as dominant oncogenes. Blood Cells Mol Dis. 30:194–200.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
210
|
Aronson BD, Fisher AL, Blechman K, Caudy M
and Gergen JP: Groucho-dependent and -independent repression
activities of Runt domain proteins. Mol Cell Biol. 17:5581–5587.
1997. View Article : Google Scholar : PubMed/NCBI
|
|
211
|
Fukushima-Nakase Y, Naoe Y, Taniuchi I,
Hosoi H, Sugimoto T and Okuda T: Shared and distinct roles mediated
through C-terminal subdomains of acute myeloid
leukemia/runt-related transcription factor molecules in murine
development. Blood. 105:4298–4307. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
212
|
Goyama S, Yamaguchi Y, Imai Y, Kawazu M,
Nakagawa M, Asai T, Kumano K, Mitani K, Ogawa S, Chiba S, et al:
The transcriptionally active form of AML1 is required for
hematopoietic rescue of the AML1-deficient embryonic para-aortic
splanchnopleural (P-Sp) region. Blood. 104:3558–3564. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
213
|
Javed A, Barnes GL, Jasanya BO, Stein JL,
Gerstenfeld L, Lian JB and Stein GS: Runt homology domain
transcription factors (Runx, Cbfa, and AML) mediate repression of
the bone sialoprotein promoter: Evidence for promoter
context-dependent activity of Cbfa proteins. Mol Cell Biol.
21:2891–2905. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
214
|
Neil JC, Gilroy K, Borlan G, Hay J, Terry
A and Kilbey A: The RUNX genes as conditional oncogenes: Insights
from retroviral targeting and mouse models. Adv Exp Med Biol.
962:247–264. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
215
|
Miething C, Grundler R, Mugler C, Brero S,
Hoepfl J, Geigl J, Speicher MR, Ottmann O, Peschel C and Duyster J:
Retroviral insertional mutagenesis identifies RUNX genes involved
in chronic myeloid leukemia disease persistence under imatinib
treatment. Proc Natl Acad Sci USA. 104:4594–4599. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
216
|
Burillo-Sanz S, Vargas MT, Morales-Camacho
RM, Caballero-Velázquez T, Sánchez J, García-Lozano JR, Pérez de
Soto I, Prats-Martín C, Bernal R and Pérez-Simón JA: RUNX1
amplification in AML with myelodysplasia-related changes and ring
21 chromosomes. Hematol Oncol. 35:894–899. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
217
|
Ito Y: Oncogenic potential of the RUNX
gene family: ‘Overview.’. Oncogene. 23:4198–4208. 2004. View Article : Google Scholar : PubMed/NCBI
|