Introduction
According to the 2018 report of the World Health
Organization (WHO), lung cancer remains the most common cancer type
and is considered as a leading cause of cancer-related mortality in
patients worldwide. Non-small cell lung cancer (NSCLC) accounts for
80–85% of all human lung cancer cases (1). According to the histological
characteristics, NSCLC can be further divided into several
subtypes, including adenocarcinoma, squamous cell carcinoma and
large cell carcinoma (2). Over 65% of
NSCLC cases are diagnosed as stage III and IV cancers that
represent locally advanced malignancy and metastasis status,
respectively (3). Enhanced invasive
and migratory abilities have been reported to be associated with
the epithelial-to-mesenchymal transition (EMT) (4). In addition, the expression level of the
EMT-associated transcription factor Twist has been proposed as a
poor prognostic marker in lung and breast cancer (5–7).
Several lines of evidence indicate that remodeling
of the actin cytoskeleton can induce or regulate EMT in various
human cancer types (8–11). Recent reports have also proposed that
the expression levels of actin-binding proteins can regulate actin
cytoskeleton reorganization and dynamics for the modulation of EMT
(12,13). Cofilin-1 (~19 kDa) is an actin-binding
protein that belongs to a member of the actin depolymerizing factor
(ADF)/cofilin family. This protein has been shown to accelerate
actin dynamics required for cell chemotaxis and increase the
migration of non-muscle cells (13).
In addition, the activity of cofilin-1 is regulated by the
phosphorylation of the protein on serine-3 by the Rho/LIM kinase
enzymes (14). Although cofilin-1 is
a ubiquitously expressed biomolecule, its expression levels are
cell type-dependent. Although cofilin-1 levels are usually
increased in advanced human cancers, the etiology and mechanism of
these processes remain unclear (15).
Recently, the cofilin-1 signaling pathway has been reported to
mediate EMT by promoting actin cytoskeletal reorganization and
cell-cell adhesion in colorectal and gastric cancer (16,17). The
overexpression of cofilin-1 has been reported to induce
let-7 microRNA expression and suppress the growth of NSCLC
cells via the downregulation of TWIST1 (18). In addition, let-7 microRNA
levels were found to regulate EMT by inhibiting the expression
levels of high mobility group A2 (HMGA2) and Twist-1 proteins that
decrease the development of EMT in cancer cells (19,20). Since
overexpression of cofilin-1 can influence the expression level of
Twist-1 in cultured cells, it is of considerable interest to
further investigate the expression pattern of cofilin-1 and Twist-1
proteins in cancer tissues.
In the present study, the expression levels of
cofilin-1 and Twist-1 proteins were examined in human NSCLC tissue
arrays and in clinicopathological lung cancer tissue sections by
immunohistological staining (IHC). The data demonstrated that 67.4%
of lung cancer tissue spots expressed reciprocal levels of
cofilin-1 and Twist-1 proteins and 80% of these tissue samples
exhibited high levels of cofilin-1 and low levels of Twist-1. The
inverse expression levels of cofilin-1 and Twist-1 were also noted
in 8 of 15 NSCLC cancer cell lines. Furthermore, overexpression of
cofilin-1 directly suppressed Twist-1, whereas disruption of actin
cytoskeleton by cytochalasin B did not cause the same effect.
Therefore, the reciprocal expression levels of cofilin-1 and
Twist-1 proteins are an important characteristic noted in NSCLC
tissues, suggesting that cofilin-1 may be a novel factor that
influences the expression level of Twist-1.
Materials and methods
Cell lines
Several NSCLC cell lines used in the present study
included CL1-0, CL1-5, H661, H596, H1975, H1299, A549, H460, H1563,
H2122, H441, PC9, H1355, H23 and H157. The H157 cell line is
identical to H1264 as reported by American Type Culture Collection
(ATCC) (https://web.expasy.org/cellosaurus/CVCL_0463). These
cell lines were maintained in culture media (DMEM or RPMI, can be
provided upon request) supplemented with 10% fetal bovine serum and
2 mM L-glutamine (Sigma-Aldrich; Merck KGaA). The protein lysates
of these differernt cell lines were extracted for western blot
analysis. The addition of 0.1 mg/ml of hygromycine B (Invitrogen;
Thermo Fisher Scientific, Inc.) in DMEM medium was used to culture
H1299 cells harboring a tetracycline inducible gene expression
system for overexpression of cofilin-1 cDNA (HCOXP).
Doxycycline (1 µg/ml) was added to the cells for 24 h in order to
induce cofilin-1 expression. The cells were collected for cell
lysis and western blot analysis following an additional 4 days of
incubation. All of the cell lines were maintained in a humidified
incubator with 5% CO2 at 37°C and passaged every 48
h.
Reagents
Cytochalasin B was purchased from
Sigma-Aldrich/Merck KGaA. The reagents were dissolved in dimethyl
sulfoxide (DMSO) to obtain concentrations of 100 mM as stock
solutions. The working concentration was 10 mM in the culture
medium, and the cells were treated for 24 h prior to
extraction.
Tissue arrays, clinicopathological
tissue sections, and IHC staining
The lung cancer tissue arrays BC041115b (120
cases/120 cores including 10 normal tissue spots) and
1-OD-CT-RsLug03-002 (62 cases/62 cores including 31 cancer tissues
and 31 matched normal adjacent tissue spots) were purchased from
the US Biomax Inc.. The two proteins (cofilin-1 and Twist-1) were
examined and two pieces of each tissue array type were subjected to
IHC staining. IHC staining was further used for the examination of
the protein expression in clinicopathological tissue sections. For
clinicopathological tissue sections, 25 lung cancer sections were
collected from the Division of Pathology, Tao-Yuan General
Hospital, Ministry of Health and Welfare, Taiwan from January to
December 2016 (Table I). The present
study was approved by the Institutional Review Board of Tao-Yuan
General Hospital (TYGH104504). The paraffin-embedded tissue
sections were maintained at 60°C for 1 h, and subsequently
deparaffinization in xylene (Sigma-Aldrich; Merck KGaA) was
performed. The tissue sections were rehydrated in graded ethanol
(from 95 to 75%) and finally immersed in phosphate-buffered
solution with 0.05% Tween-20 (PBST). For antigen retrieval, the
tissue slides were heated in 10 mM citric acid buffer with 0.05%
Tween-20 (pH 6.0) for 3 min at 121°C using a pressure cooker.
Following air-cooling, the tissue sections were incubated with
peroxidase blocking reagent [RTU, EnVision™+Dual Link System-HRP
(DAB+); cat. no. K4065, Dako; Agilent Technologies, Inc.] for 5 min
and subsequently blocked with goat serum for an additional 30 min.
Thereafter, the tissue sections were incubated with anti-cofilin-1
(1:100 dilution; cat. no. GTX102156) and anti-Twist-1 antibodies
(1:250 dilution; cat. no. GTX60776; GeneTex Inc.) at 4°C overnight
followed by horseradish peroxidase (HRP)-conjugated secondary
antibodies. The sections were rinsed with PBST, developed in
3′,3′-diaminobenzidine (DAB) substrate chromogen [EnVision™+Dual
Link System-HRP (DAB+); cat. no. K4065, Dako; Agilent Technologies,
Inc.] and finally counterstained with Mayer's hematoxylin (ScyTek
Laboratories). All sections were scanned using the Aperio Digital
Pathology Slide Scanner (Leica Biosystems). Lepidic growth was
excluded in all examined cases of adenocarcinoma containing
invasive area for evaluation of the IHC score. Quantification of
IHC scores was determined by multiplying the staining intensity (on
a scale of 0–3) by the positivity of the staining factor (on a
scale of 1–4: 0–25%, 1; 26–50%, 2; 51–75%, 3; and 76–100%, 4). All
of the IHC staining tissues were examined and scored by 2 to 3
different individuals in a blinded manner.
 | Table I.Features of the clinical resected
lung cancer tissue samples examined by immunohistochemistry
(IHC). |
Table I.
Features of the clinical resected
lung cancer tissue samples examined by immunohistochemistry
(IHC).
| Case no. | Stage | Age (years) | Sex | Subtype |
|---|
| 1 | 1A | 54 | F | Adenocarcinoma |
| 2 | 2A | 74 | F | Adenocarcinoma |
| 3 | 3B | 77 | F | Adenocarcinoma |
| 4 | 3B | 50 | M | Adenocarcinoma |
| 5 | 3B | 71 | M | Metastatic squamous
cell carcinomain |
| 6 | 4 | 66 | F | Mesothelioma |
| 7 | 4 | 69 | F | Adenocarcinoma |
| 8 | 4 | 61 | M | Adenocarcinoma |
| 9 | 4 | 71 | M | Non-small cell
carcinoma favored |
| 10 | 4 | 71 | M | Small cell lung
cancer |
| 11 | 4 | 68 | F | Adenocarcinoma |
| 12 | 4 | 67 | M | SCC |
| 13 | 4 | 64 | F | SCC |
| 14 | 4 | 50 | F | Mixed
adenocarcinoma and rhabdomyosarcoma |
| 15 | 4 | 63 | M | Adenocarcinoma |
| 16 | 4 | 59 | M | Adenocarcinoma |
| 17 | 4 | 38 | M | Adenocarcinoma |
| 18 | 4 | 75 | M | Adenocarcinoma |
| 19 | 4 | 53 | M | Adenocarcinoma |
| 20 | 4 | 80 | F | SCC |
| 21 | 4 | 61 | M | SCC |
| 22 | 4 | 62 | F | Adenocarcinoma |
| 23 | 4 | 68 | M | High grade
adenosquamous |
| 24 | 4 | 53 | M | Pleomorphic
carcinoma |
| 25 | 4 | 49 | M | Adenocarcinoma |
Western blot analysis and
antibodies
The procedures of lysate extraction, protein
electrophoresis and blotting were as described in our previous
research (21). For preparation of
cell lysates, the monolayers were rinsed with phosphate-buffered
saline and subsequently scraped in lysis buffer (0.5% NP-40, 50 mM
Tris HCl, 120 mM NaCl, and 1% phenylmethylsulfonyl fluoride). The
band intensity was determined using densitometry (ImageJ Software
version 1.x; National Institutes of Health, Bethesda, MD, USA). The
primary antibodies were purchased from GeneTex Inc. and included
anti-cofilin-1 (1:1,000 dilution; cat. no. GTX102156), anti-Twist-1
(1:500 dilution; cat. no. GTX60776), anti-SNAIL (1:500 dilution;
cat. no. GTX100754), and anti-tubulin (1:1,000 dilution; cat. no.
GTX112141) antibodies. Anti-glyceraldehyde-3-phosphate
dehydrogenase (GAPDH) was purchased from Thermo Fisher Scientific
Inc. (1:4,000 dilution; cat. no. MA515738).
Statistical analysis
The t-test was used for statistical analysis between
two groups. The survival probability was determined using an
on-line Kaplan-Meier plotter (kmplot.com) tool,
which is a meta-analysis based biomarker assessment for 54,675
genes based on the Affymetrix probe set IDs (22). With regard to lung cancer, the maximum
sample size of the available patients was 1,926. These data were
used for Kaplan-Meier survival and Cox regression analyses.
Significant parameters were derived using univariate amd
multivariate analyses according to the KM plotter website (23). The gene_ID of cofilin-1 and Twist-1
proteins on the Affymetrix chip were 200021_at and 213943_at,
respectively. Cox regression was used to analyze the association
between the expression levels of cofilin-1 and Twist-1 proteins and
NSCLC according to the results of the Kaplan-Meier method. The
significance was determined by the log-rank test. The correlation
analysis and scatter diagram were drawn using the
MedCalc® software version 18.2.1 (Ostend). A P-value
less than 0.05 (P<0.05) was considered to indicate a
statistically significant difference.
Results
Immunohistological staining of
cofilin-1 and Twist-1 proteins in human NSCLC tissue arrays
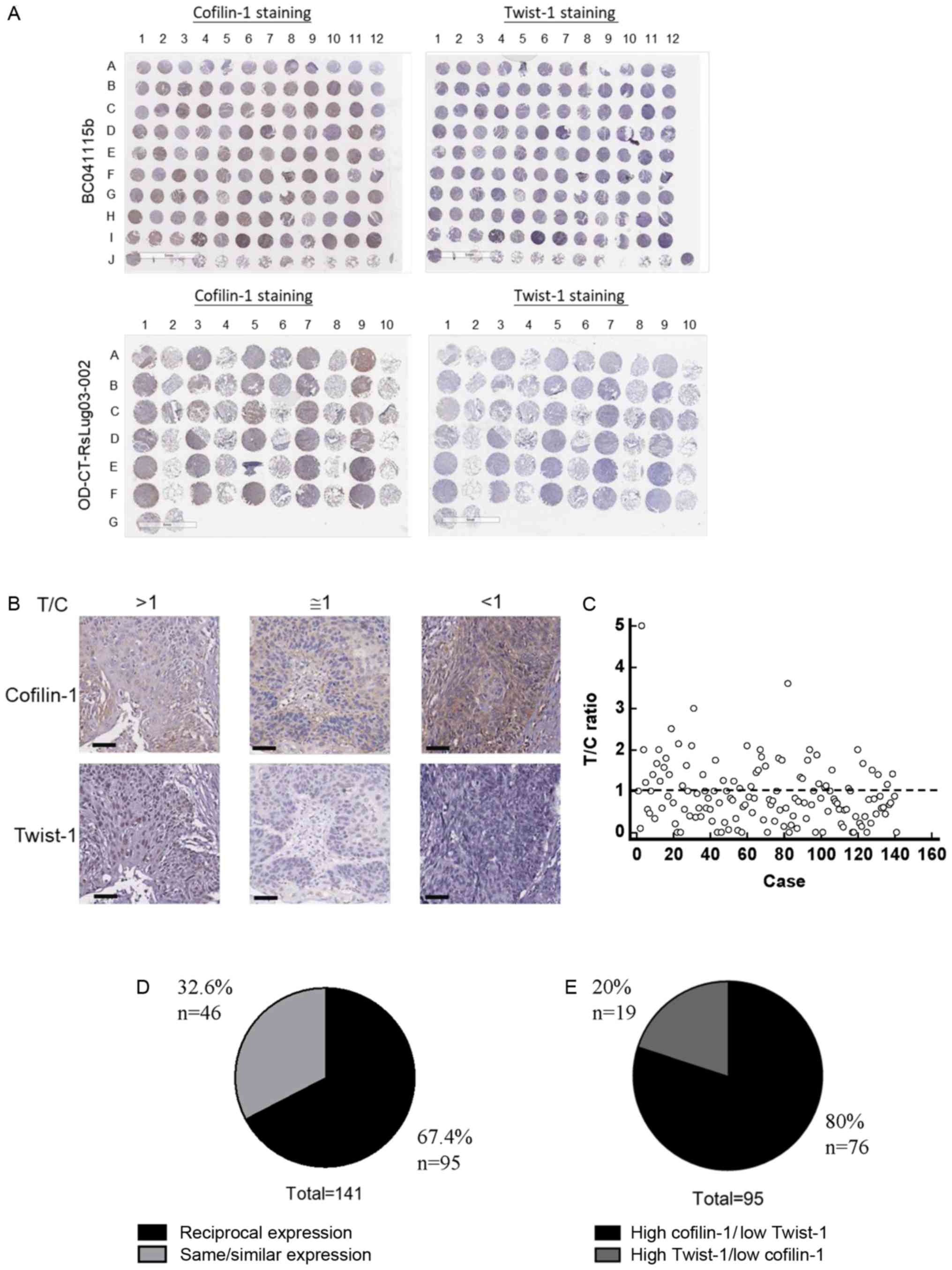
Human lung cancer tissue arrays (see Materials and
methods) were subjected to IHC staining. A total of 2 tissue arrays
with identical orders of tissue spots were analyzed using
anti-cofilin-1 or anti-Twist-1 antibodies followed by pathological
slide scanning. The complete results of IHC staining on these
tissues arrays are demonstrated in Fig.
1A. Microscopical investigation was also used to visualize the
IHC staining results of cofilin-1 and Twist-1 proteins. T/C was the
ratio of Twist-1 staining score divided by cofilin-1 staining
scores for each tissue spot (Fig.
1B). The scores of cofilin-1 were compared with those of
Twist-1. Both scores exhibited similar levels when the T/C ratio
was 1±0.5. The T/C ratios of all lung tumor tissue spots (n=141)
are presented as a scatter diagram (Fig.
1C). The results were further summarized and indicated that
67.4% of lung cancer tissue spots expressed reciprocal levels of
cofilin-1 and Twist-1 proteins (Fig.
1D). In these tissue spots, 80% of the samples expressed higher
levels of cofilin-1 than those of Twist-1 (Fig. 1E).
Comparison of cofilin-1 and Twist-1
expression levels in different stages and subtypes of lung cancer
tissues
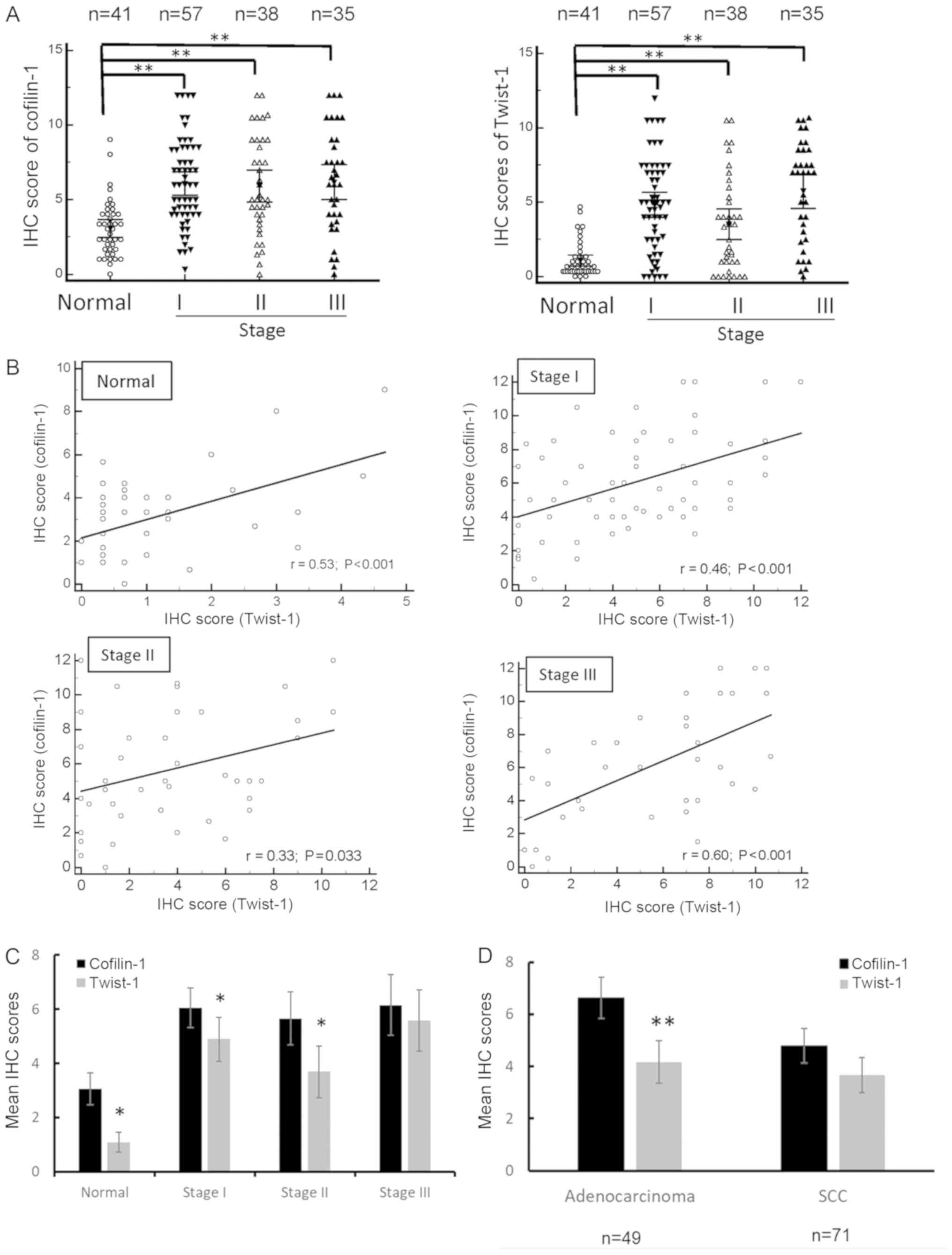
Subsequently, we compared the results of cofilin-1
and Twist-1 staining in NSCLC samples of different tumor stages and
subtypes by calculating the immunostaining scores. The expression
levels of both cofilin-1 and Twist-1 proteins were relatively high
in stage I, II and III lung cancer tissues when compared with the
normal tissues (Fig. 2A). A total of
only 130 tissue spots belonging to stage I to III were counted, as
the tumor stage of 10 tissue spots in the 1-OD-CT-RsLug03-002
tissue array was mixed or not described. A total of 1 tissue spot
was defined as stage IV, so it was not included in the statistics.
A correlation analysis was further performed with regard to the
regression of cofilin-1 and Twist-1 expression levels in normal
lung tissue spots and lung tumor tissue spots. Positive
corellations were noted in all of these tissue spots, whereas stage
III lung tumor tissues exhibited higher correlation with the
expression of the corresponding proteins (Fig. 2B). In contrast to these observations,
the mean IHC score of cofilin-1 was higher than that of Twist-1 in
normal tissues and in stage I and/or II of lung tumor tissues
(Fig. 2C). It is interesting to note
that the mean IHC scores of cofilin-1 were higher than those of
Twist-1 in the adenocarcinoma subtypes, although this finding was
not noted in the squamous cell carcinoma subtype of the lung tumor
tissues (Fig. 2D). A considerably low
Twist-1/cofilin-1 ratio was further noted in large cell cancer,
bronchioloalveolar carcinoma (or lepidic predominant
adenocarcinoma), and mucinous adenocarcinoma, although the total
sample size of these subtypes was considerably small (n=4). Taken
together, these results suggested that the expression levels of
cofilin-1 and Twist-1 proteins were correlated, while the mean
expression level of cofilin-1 were higher than those of Twist-1 in
normal tissues, low tumor stage tissues, and adenocarcinoma subtype
tissues.
Detection of cofilin-1 and Twist-1
expression in resected lung cancer tissues
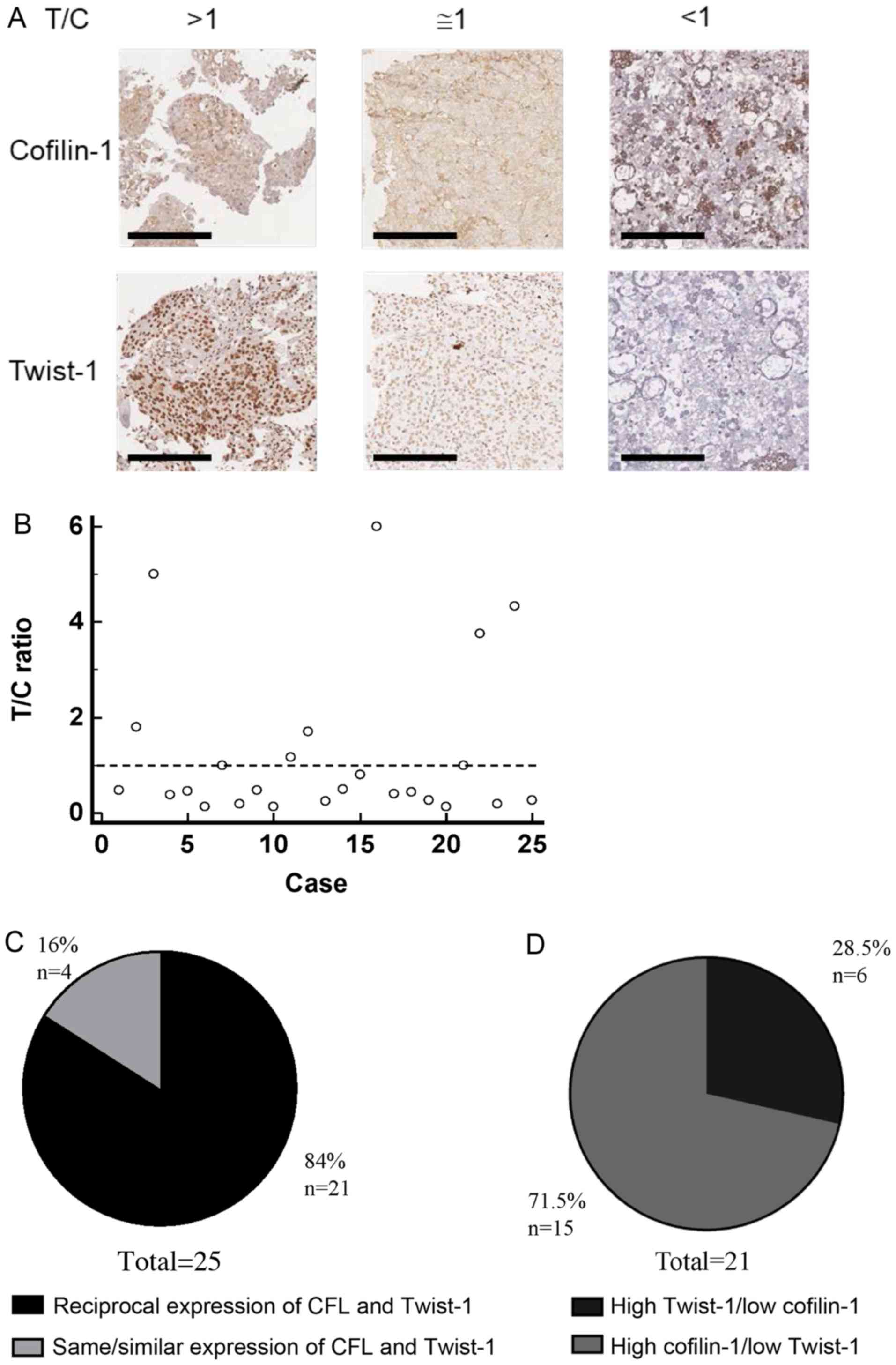
Hospital-based lung cancer tissue sections were also
examined for the expression of cofilin-1 and Twist-1 proteins. A
total of 25 lung cancer tissue samples with consecutive sections
were collected and compared for their individual cofilin-1 and
Twist-1 IHC scores in order to obtain the T/C ratio as mentioned
above (Fig. 3A). The T/C ratio of
each tumor tissue section (n=25) was depicted by a scatter diagram
(Fig. 3B). A total of 21 of these
samples (84%) exhibited reciprocal levels of cofilin-1 and Twist-1
(Fig. 3C). Furthermore, 15 out of 21
(71.5%) exhibited high expression of cofilin-1 and low expression
of Twist-1, corresponding to a low T/C ratio (Fig. 3D). In addition, 14 surgical cases were
of adenocarcinoma and 4 of squamous cell carcinoma origin,
respectively. The remaining 7 cases included various types of lung
cancer, such as mesothelioma, metastatic squamous cell carcinoma,
mixed adenocarcinoma and rhabdomyosarcoma, high grade
adenosquamous, pleomorphic carcinoma, and small cell lung cancer
(Table I). Furthermore, the mean IHC
score of cofilin-1 was significantly higher than that of Twist-1 in
adenocarcinoma, whereas this was not noted in squamous cell
carcinoma of these clinicopathological tissues (data not shown).
Although the sample size of the clinicopathological tissue sections
was small, the expression pattern of cofilin-1 and Twist-1 proteins
seemed similar with that noted in the lung cancer tissue
arrays.
Comparison of cofilin-1 and Twist-1
expression in human NSCLC cell lines
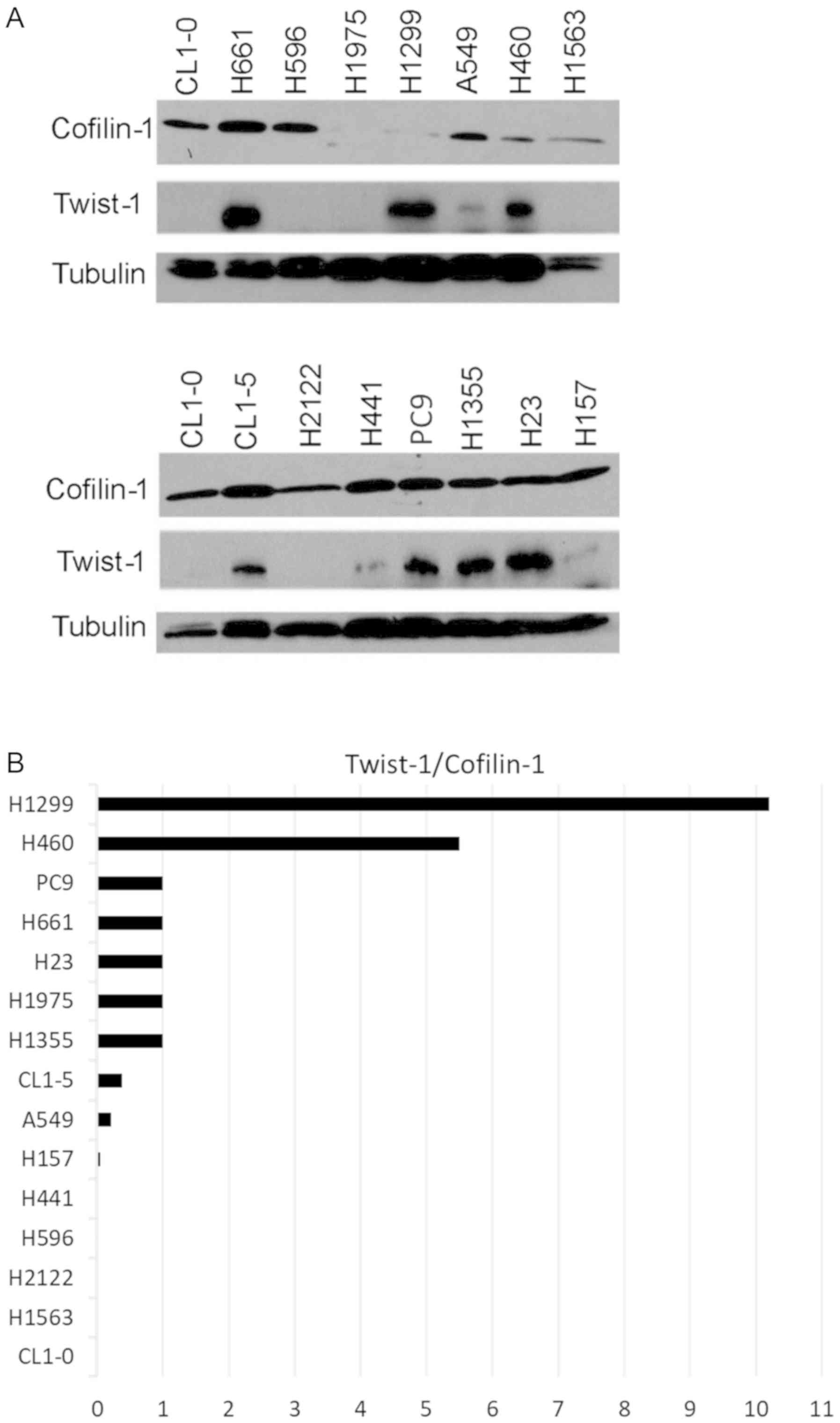
In addition to NSCLC tissues, we further
investigated whether reciprocal expression of cofilin-1 and Twist-1
could be detected in various cell lines. A total of 15 NSCLC cell
lines were collected for western blot analysis. The results
indicated that cofilin-1 and Twist-1 were differentially expressed
in these cell lines (Fig. 4A). The
expression levels of cofilin-1 in H1975 cells and H1299 cells were
significantly lower than those of the other NSCLC cell lines. In
contrast to these observations, half of these cell lines exhibited
high levels of Twist-1 protein, whereas the other half indicated
extremely low expression levels of Twist-1 (Fig. 4A). The T/C ratio was estimated in each
cell line using densitometric analysis. A total of 10 out of 15
cell lines exhibited reciprocal expression of cofilin-1 and Twist-1
proteins, whereas a low Twist-1/cofilin-1 ratio was noted in 8
NSCLC cell lines (Fig. 4B).
Therefore, the reciprocal expression of cofilin-1 and Twist-1
proteins was also detectable in cultured NSCLC cell lines.
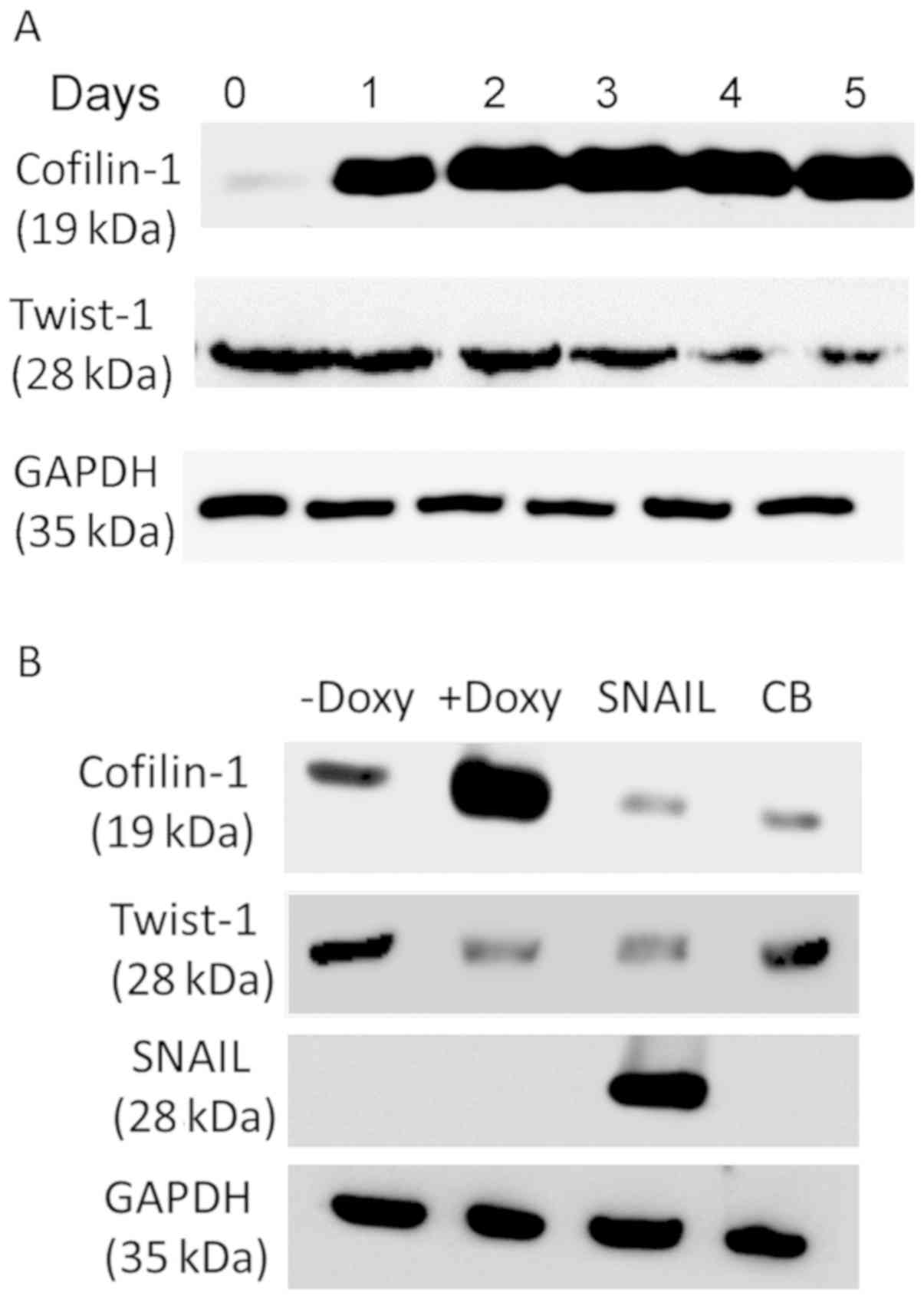
Overexpression of cofilin-1 suppresses
Twist-1 levels in NSCLC cells
Since high levels of cofilin-1 were associated with
low levels of Twist-1, the potential of cofilin-1 overexpression to
suppress the expression level of Twist-1 protein was examined in
HCOXP cells (24). The results
indicated a time-dependent suppression of Twist-1 due to the
overexpression of cofilin-1 using doxycycline induction (Fig. 5A). In contrast to cofilin-1,
disruption of the actin cytoskeleton by cytochalasin B (CB), an
actin inhibitor did not influence the expression levels of Twist-1
(Fig. 5B). Notably, overexpression of
the SNAIL transcription factor suppressed Twist-1 level, whereas
the cofilin-1 level was also decreased. These results suggested
that cofilin-1 mediates the expression level of Twist-1, although
this may not be associated with the destabilization of the actin
cytoskeleton.
Effects of cofilin-1 and Twist-1 gene
expression on the survival fraction of NSCLC patients
Although the protein expression levels of cofilin-1
and Twist-1 were compared in tissue arrays and clinicopathological
tissue sections, the survival data used to analyze the role of
these two proteins on the survival of NSCLC patients are limited.
Therefore, we adopted the public microarray database and an on-line
Kaplan-Meier plot analytical tool in order to evaluate the gene
expression levels of cofilin-1 (CFL1) and Twist-1
(TWIST1) genes on the survival of lung cancer patients
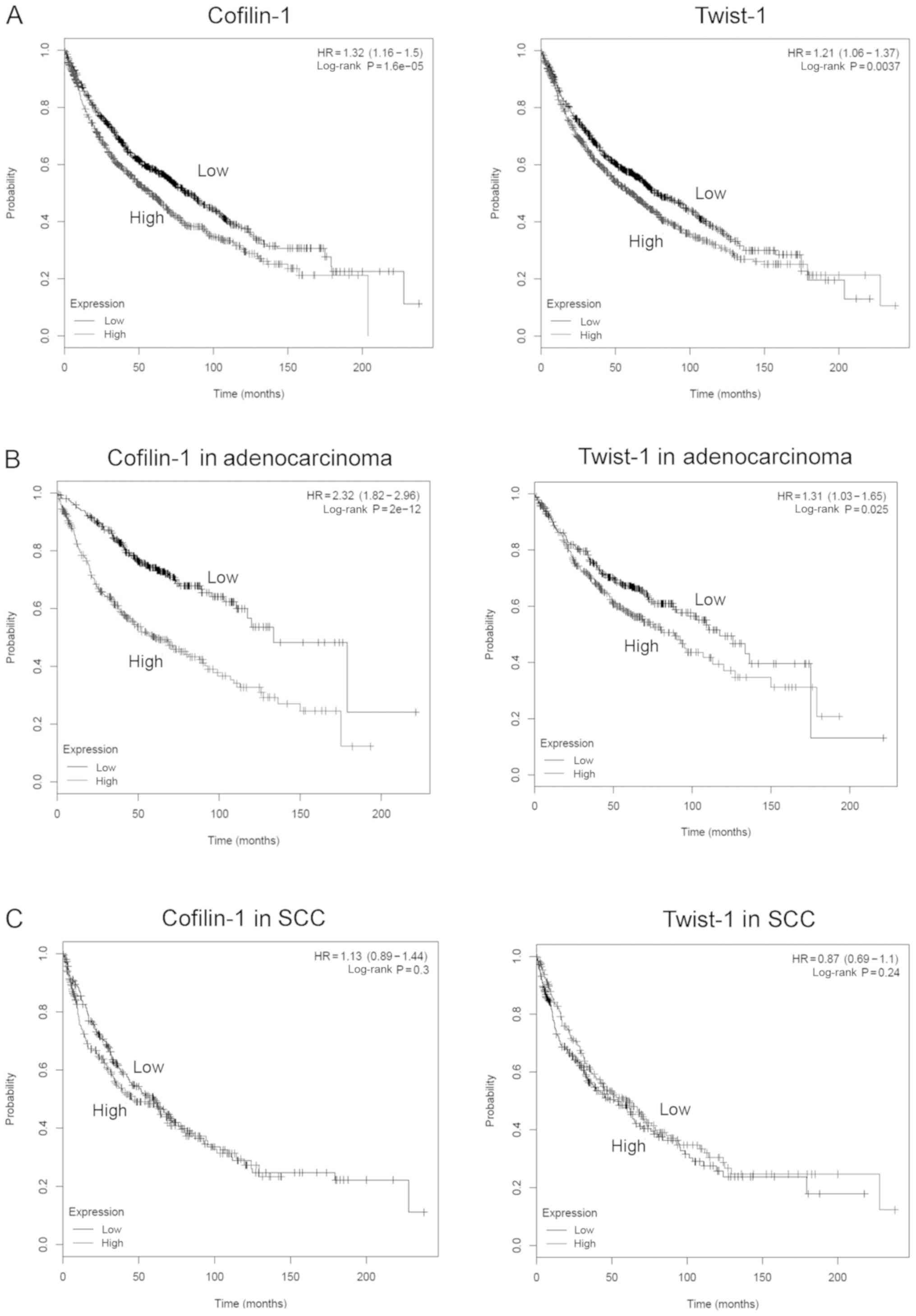
(23). High expression levels of the
CFL1 and TWIST1 genes indicated lower survival rates
in NSCLC patients with a univariate Cox regression HR of 1.32 [95%
confidence interval (CI): 1.16–1.5] and 1.21 (95% CI, 1.06–1.37),
respectively (Fig. 6A). High
cofilin-1 and Twist-1 levels were further associated with
significantly low survival in the adenocarcinoma subtype of NSCLC
patients (HR=2.32 with 95% CI, 1.82–2.96 for cofilin-1, and HR=1.31
with 95% CI, 1.03–1.65 for Twist-1). However, this was not noted in
squamous cell carcinoma (SCC) lung cancer subtypes (Fig. 6B and C). Therefore, the expression
levels of the CFL1 and TWIST1 genes may influence the
survival rate of patients with different subtypes of NSCLC.
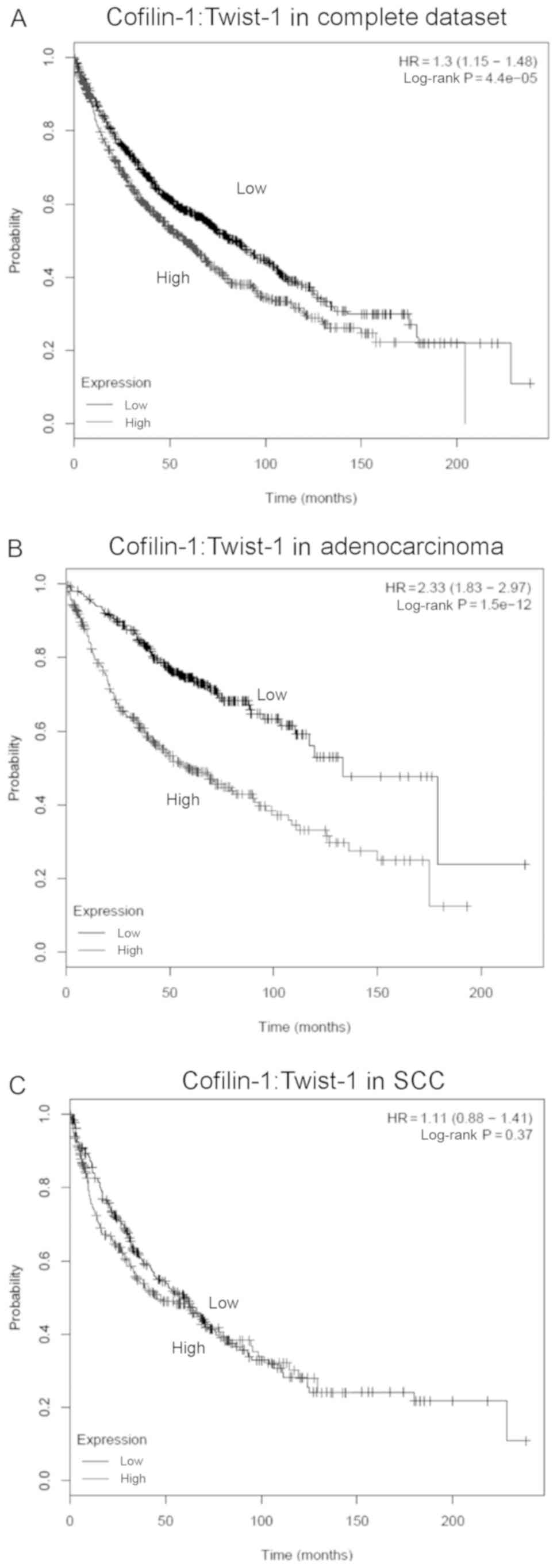
To evaluate the effects of the co-expressed
CFL1 and TWIST1 genes on the survival of NSCLC
patients, a multigene classifer with the Cox regression was applied
for further anaylsis based on the results of the Kaplan-Meier
method. Using the same public microarray database, high expression
levels of both cofilin-1 and Twist-1 were associated with reduced
survival fraction as determined by a univariate Cox regression HR
of 1.3 (95% CI, 1.15–1.48; Fig. 7A).
A similar result was also found in the adenocarcinoma subtype
samples with a multivariate Cox regression HR of 2.33 (95% CI,
1.83–2.97; Fig. 7B). However, no
significant differences were noted in the survival rate of squamous
cell carcinoma (SCC) patients with high or low expression of both
CFL1 and TWIST1 genes (Fig.
7C).
Discussion
Increased cell migration is an important feature of
cancer invasion and metastasis. Epithelial-mesenchymal transition
(EMT) is responisble for the development of cancer malignancy.
However, the mechanisms involved in the association between cancer
migration and EMT are not fully understood. We previously showed
that overexpression of cofilin-1 could suppress cell invasion of
non-small cell lung cancer (NSCLC) cells (25). Cofilin-1 was found to suppress the
Twist-1 level following its overexpression in NSCLC cells, whereas
overexpression of Twist-1 did not significantly influence the
expression level of cofilin-1 (18).
To fully understand whether the expression of cofilin-1 and Twist-1
proteins were associated with the clinicopathological
characteristics of lung cancer, their expression levels were
examined in normal and malignant lung tissues using
immunohistochemistry (IHC). It was found that in the majority of
the cases, the IHC score of cofilin-1 was higher than that of
Twist-1. However, a positive correlation of cofilin-1 and Twist-1
was also detected in these tissues, including normal lung tissues.
These findings suggested that even though the co-expression of
these two molecules was positively associated, cofilin-1 levels
remained higher than Twist-1 in the majority of the tissue samples.
This phenomenon was observed not only in lung tumor tissue samples
but also in normal lung tissues. Therefore, the expression levels
of cofilin-1 were significantly higher than those of Twist-1 in
normal lung tissues, and stage I and II lung cancer tissues.
However, these findings were not noted in stage III cancer samples
(Fig. 2C). The results of the tissue
array experiments suggested that reciprocal expression of cofilin-1
and Twist-1 could be detected with regard to different
co-expression levels of these proteins, notably in normal and early
stages of lung tumor tissues.
The differences noted with regard to the high levels
of cofilin-1 expression and the low levels of Twist-1 (low T/C
ratio) were observed in the majority of the tissue spots (80%) with
regard to the NSCLC adenocarcinoma subtype (Fig. 2B). It has been reported that squamous
cell carcinoma expresses higher levels of Twist-1 than those noted
in adenocarcinoma (5,26). Nevertheless, high gene expression
levels of both CFL1 and TWIST1 were found to only
account for the low survival rate of patients with adenocarcinoma
and not of patients with squamous cell carcinoma. Since reciprocal
expression levels of cofilin-1 and Twist-1 have been mainly
detected in adenocarcinoma but not in squamous cell carcinoma
tissues, the survival rate of adenocarcinoma may be altered if both
genes are expressed inversely. Moreover, the levels of cofilin-1
detected at different tumor stages (I to III) were similar, and
they were all higher than those noted in normal lung tissues. Since
only one tissue spot was represented as stage IV in this commercial
tissue array, this was not included for statistical analysis. In
the present tissue spot, the stage IV samples expressed low
cofilin-1 and high Twist-1 levels (data not shown). In contrast to
these findings, a recent report indicated that sputum cofilin-1
levels were higher in T4 and N stage of lung cancer patients
(27). Serum cofilin-1 levels were
further reported to be increased in the advanced stage of patients
with lung cancer (28). A total of 16
out of 20 stage IV clinicopathological sections exhibited
reciprocal expression levels of cofilin-1 and Twist-1, whereas in
10 out of 16 sections the pattern of high cofilin-1/low Twist-1
ratio was noted (Table I). Although
the sample size used in the present study was small, the samples
were derived from different sources and it appeared that the
inverse expression of cofilin-1 and Twist-1 could occur in
different cancer stages. The investigation of additional stage IV
tumor samples is important to confirm the primary expression
pattern of cofilin-1 and Twist-1 proteins in NSCLC tissues.
Since high cofilin-1/low Twist-1 ratio is a
predominant phenomenon in the reciprocal expression of cofilin-1
and Twist-1, we further demonstrated that overexpression of
cofilin-1 suppressed Twist-1 levels in H1299 cells. Overexpression
of cofilin-1 and reduced Twist-1 levels have been reported to
induce let-7 and inhibit tumor growth, invasion and motility
in vivo and in vitro (18). This is in part consistent with a
previous report suggesting that Twist-1 could interact with the
BMI1 oncogene to suppress let-7i. expression. This
interaction was associated with increased tumor invasiveness and
poor survival outcome in cancer patients (29). Although overexpression of cofilin-1
can destabilize the actin cytoskeleton (30), cytochalasin B-mediated disruption of
the actin cytoskeleton did not reduce the expression levels of
Twist-1. Therefore, overexpression of cofilin-1 caused reduction in
Twist-1 levels and this effects was not directly associated with
actin cytoskeletal destabilization. Moreover, it was previously
demonstrated that ectopic expression of Twist-1 did not influence
the expression of cofilin-1 (18). In
the present study, we further examined whether overexpression of
SNAIL could affect the expression levels of cofilin-1. The data
indicated that ectopic expression of SNAIL suppressed the
expression levels of cofilin-1 and Twist-1 proteins. These
differences may be attributed to the different signaling pathways
of Twist-1 and SNAIL.
To the best of our knowledge, little is known with
regard to the interaction of cofilin-1 and expression of Twist-1.
The expression level of Twist-1 can be modulated by a series of
upstream regulators, such as the tumor necrosis factor (TNF)-α, the
WNT, the receptor tyrosine kinase, transforming growth factor
(TGF)-β, the Notch and the hypoxia pathways (31). These pathways are directly or
indirectly involved in the remodeling of the actin cytoskeleton
(32–37). Since the upregulation of cofilin-1 is
essential for the reorganization of actin cytoskeleton, the
expression level of Twist-1 may be modulated partially by the
activation of these pathways. In contrast to the expression level
of the total form of the protein, cofilin-1 phosphorylation is
known to be controlled by the Rho small GTPase signaling pathway
(38). The inhibitors of Rho kinases
have been reported to suppress the nuclear accumulation of Twist-1
(39). Therefore, the Rho signaling
pathway may be also associated with the interaction between
cofilin-1 and Twist-1. However, the detailed mechanisms of these
processes remain to be studied.
The association of CFL1 and TWIST1
gene expression with the survival rate of lung cancer patients was
analyzed using a public microarray database, since data on the
survival information of tissue arrays were insufficient. The
expression levels of both genes accounted for the poor prognosis of
lung cancer patients, which was consistent with previous studies
(40,41). However, in a previous study, the
overexpression of cofilin-1 led to the suppression of NSCLC cell
grwoth, which was contradictory to our findings (25). Yap et al demonstrated that the
overexpression of cofilin could either promote or suppress the
motility of U373MG glioblastoma tumor cells in a
concentration-dependent manner (42).
The optimal amount of cofilin-1 overexpression required to promote
cell motility was 4.5 times higher than that noted in the control
cells. Overexpression of cofilin-1 suppressed the invasion of NSCLC
cells at 4.5-fold compared with that of the control cells (25). Using tissue arrays, the mean cofilin-1
IHC score of the NSCLC stage I–IV samples was only 2 times higher
than that of normal lung tissues (Fig. 2A
and C). Therefore, this discrepancy may be associated with the
levels of enforced cofilin-1 overexpression in cell lines and the
endogenous levels of cofilin-1 in cancer tissues. However, further
investigation is essential in order to interpret these
findings.
In summary, the expression levels of cofilin-1 and
Twist-1 were investigated using paired lung cancer tissue arrays
with consecutive tissue spots. The data demonstrated that 66.6% of
the tissue spots exhibited reciprocal expression levels of
cofilin-1 and Twist-1. Although high cofilin-1 and low Twist-1
levels were the major characteristics of the pattern of reciprocal
expression in normal lung and lung tumor tissues, the results
indicated that this pattern may be more useful for the detection of
early stage lung adenocarcinoma. Overexpression of cofilin-1 was
able to regulate the expression level of Twist-1, whereas this
effect was not associated with destabilization of the actin
cytoskeleton. Accordiing to the survival analysis of the public
microarray dataset, high expression levels of both CFL1 and
TWIST1 genes were associated with reduced survival of the
NSCLC patients. Whether reciprocal expression of cofilin-1 and
Twist-1 is able to alter the survival period of NSCLC patients is
yet to be discovered.
Acknowledgements
We thank the support from the Cancer Progression
Research Center, National Yang-Ming University, from the Featured
Areas Research Center Program within the framework of the Higher
Education Sprout Project by the Ministry of Education (MOE) in
Taiwan. Additionally, we are thankful for the technical support of
Ms. Man-Chun Liao.
Funding
The present study was supported by a united grant of
National Yang-Ming University and Tao-Yuan General Hospital,
Ministry of Health and Welfare (PTH10509), a grant from the
Ministry of Science and Technology of Taiwan
(105-2628-B-010-013-MY3), and the Department of Health, Taipei City
Government (10501-62-014).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
CYC conducted the experiments and analyzed the data.
SLC provided the clinicopathological tissue sections and organized
the demographic data. JDL reviewed the manuscript and provided
consultant on lung cancer. YCC and MH conceived and performed the
western blot analysis. LTL provided background information of lung
cancer for manuscript writing and editing. HNL and YJL conceived,
designed the study, and wrote the manuscript. All authors read and
approved the manuscript and agree to be accountable for all aspects
of the research in ensuring that the accuracy or integrity of any
part of the work are appropriately investigated and resolved.
Ethics approval and consent to
participate
The present study was approved by the Institutional
Review Board of Tao-Yuan General Hospital (TYGH104504). All of the
informed consents had been signed by the tissue donors and stored
in Tao-Yuan General Hospital.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Devesa SS, Bray F, Vizcaino AP and Parkin
DM: International lung cancer trends by histologic type:
Male:Female differences diminishing and adenocarcinoma rates
rising. Int J Cancer. 117:294–299. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Thomas A, Liu SV, Subramaniam DS and
Giaccone G: Refining the treatment of NSCLC according to
histological and molecular subtypes. Nat Rev Clin Oncol.
12:511–526. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Morgensztern D, Ng SH, Gao F and Govindan
R: Trends in stage distribution for patients with non-small cell
lung cancer: A National Cancer Database survey. J Thorac Oncol.
5:29–33. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Mahmood MQ, Ward C, Muller HK, Sohal SS
and Walters EH: Epithelial mesenchymal transition (EMT) and
non-small cell lung cancer (NSCLC): A mutual association with
airway disease. Med Oncol. 34:452017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zeng J, Zhan P, Wu G, Yang W, Liang W, Lv
T and Song Y: Prognostic value of Twist in lung cancer: Systematic
review and meta-analysis. Transl Lung Cancer Res. 4:236–241.
2015.PubMed/NCBI
|
|
6
|
Zhang YQ, Wei XL, Liang YK, Chen WL, Zhang
F, Bai JW, Qiu SQ, Du CW, Huang WH and Zhang GJ: Over-expressed
twist associates with markers of epithelial mesenchymal transition
and predicts poor prognosis in breast cancers via ERK and Akt
activation. PLoS One. 10:e01358512015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Škovierová H, Okajčeková T, Strnádel J,
Vidomanová E and Halašová E: Molecular regulation of
epithelial-to-mesenchymal transition in tumorigenesis (Review). Int
J Mol Med. 41:1187–1200. 2018.PubMed/NCBI
|
|
8
|
Morris HT and Machesky LM: Actin
cytoskeletal control during epithelial to mesenchymal transition:
Focus on the pancreas and intestinal tract. Br J Cancer.
112:613–620. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Shankar J and Nabi IR: Correction: Actin
cytoskeleton regulation of epithelial mesenchymal transition in
metastatic cancer cells. PLoS One. 10:e01327592015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Peng JM, Bera R, Chiou CY, Yu MC, Chen TC,
Chen CW, Wang TR, Chiang WL, Chai SP, Wei Y, et al: Actin
cytoskeleton remodeling drives epithelial-mesenchymal transition
for hepatoma invasion and metastasis in mice. Hepatology.
67:2226–2243. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Haynes J, Srivastava J, Madson N, Wittmann
T and Barber DL: Dynamic actin remodeling during
epithelial-mesenchymal transition depends on increased moesin
expression. Mol Biol Cell. 22:4750–4764. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Huang D, Cao L and Zheng S: CAPZA1
modulates EMT by regulating actin cytoskeleton remodelling in
hepatocellular carcinoma. J Exp Clin Cancer Res. 36:132017.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Izdebska M, Zielińska W, Grzanka D and
Gagat M: The role of actin dynamics and actin-binding proteins
expression in epithelial-to-mesenchymal transition and its
association with cancer progression and evaluation of possible
therapeutic targets. BioMed Res Int. 2018:45783732018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Prunier C, Prudent R, Kapur R, Sadoul K
and Lafanechère L: LIM kinases: Cofilin and beyond. Oncotarget.
8:41749–41763. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wang W, Eddy R and Condeelis J: The
cofilin pathway in breast cancer invasion and metastasis. Nat Rev
Cancer. 7:429–440. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang H, Tao L, Jin F, Gu H, Dai X, Ni T,
Feng J, Ding Y, Xiao W, Qian Y and Liu Y: Cofilin 1 induces the
epithelial-mesenchymal transition of gastric cancer cells by
promoting cytoskeletal rearrangement. Oncotarget. 8:39131–39142.
2017.PubMed/NCBI
|
|
17
|
Sousa-Squiavinato ACM, Rocha MR,
Barcellos-de-Souza P, de Souza WF and Morgado-Diaz JA: Cofilin-1
signaling mediates epithelial-mesenchymal transition by promoting
actin cytoskeleton reorganization and cell-cell adhesion regulation
in colorectal cancer cells. Biochim Biophys Acta Mol Cell Res.
1866:418–429. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Tsai CH, Lin LT, Wang CY, Chiu YW, Chou
YT, Chiu SJ, Wang HE, Liu RS, Wu CY, Chan PC, et al:
Over-expression of cofilin-1 suppressed growth and invasion of
cancer cells is associated with up-regulation of let-7 microRNA.
Biochim Biophys Acta. 1852:851–861. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Tan EJ, Thuault S, Caja L, Carletti T,
Heldin CH and Moustakas A: Regulation of transcription factor Twist
expression by the DNA architectural protein high mobility group A2
during epithelial-to-mesenchymal transition. J Biol Chem.
287:7134–7145. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Morishita A, Zaidi MR, Mitoro A,
Sankarasharma D, Szabolcs M, Okada Y, D'Armiento J and Chada K:
HMGA2 is a driver of tumor metastasis. Cancer Res. 73:4289–4299.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wang CY, Tsai CH, Chang CY, Liao MJ, Liu
RS and Lee YJ: Over-expression of cofilin-1 suppressed mobility of
lung cancer cells is associated with down-regulation of SNAIL-1 and
induction of Let-7. Clin Oncol. 1:10152016.
|
|
22
|
Györffy B, Lanczky A, Eklund AC, Denkert
C, Budczies J, Li Q and Szallasi Z: An online survival analysis
tool to rapidly assess the effect of 22,277 genes on breast cancer
prognosis using microarray data of 1,809 patients. Breast Cancer
Res Treat. 123:725–731. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Győrffy B, Surowiak P, Budczies J and
Lánczky A: Online survival analysis software to assess the
prognostic value of biomarkers using transcriptomic data in
non-small-cell lung cancer. PLoS One. 8:e822412013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Tsai CH, Chiu SJ, Liu CC, Sheu TJ, Hsieh
CH, Keng PC and Lee YJ: Regulated expression of cofilin and the
consequent regulation of p27(kip1) are essential for G(1) phase
progression. Cell Cycle. 8:2365–2374. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lee YJ, Mazzatti DJ, Yun Z and Keng PC:
Inhibition of invasiveness of human lung cancer cell line H1299 by
over-expression of cofilin. Cell Biol Int. 29:877–883. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Tran PT, Shroff EH, Burns TF, Thiyagarajan
S, Das ST, Zabuawala T, Chen J, Cho YJ, Luong R, Tamayo P, et al:
Twist1 suppresses senescence programs and thereby accelerates and
maintains mutant Kras-induced lung tumorigenesis. PLoS Genet.
8:e10026502012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Rangel MP, Antonangelo L, Acencio MMP,
Faria CS, de Sá VK, Leão PS, Farhat C, Fabro AT, Longatto Filho A,
Reis RM, et al: Detection of sputum cofilin-1 as indicator of
malignancy. Braz J Med Biol Res. 51:e71382018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zheng Y, Fang Y, Li S and Zheng B:
Detection of plasma cofilin protein for diagnosis of lung cancer.
Nan Fang Yi Ke Da Xue Xue Bao. 33:1551–1553. 2013.(In Chinese).
PubMed/NCBI
|
|
29
|
Yang WH, Lan HY, Huang CH, Tai SK, Tzeng
CH, Kao SY, Wu KJ, Hung MC and Yang MH: RAC1 activation mediates
Twist1-induced cancer cell migration. Nat Cell Biol. 14:366–374.
2012. View
Article : Google Scholar : PubMed/NCBI
|
|
30
|
Lee YJ and Keng PC: Studying the effects
of actin cytoskeletal destabilization on cell cycle by cofilin
overexpression. Mol Biotechnol. 31:1–10. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhao Z, Rahman MA, Chen ZG and Shin DM:
Multiple biological functions of Twist1 in various cancers.
Oncotarget. 8:20380–20393. 2017.PubMed/NCBI
|
|
32
|
Boland S, Boisvieux-Ulrich E, Houcine O,
Baeza-Squiban A, Pouchelet M, Schoëvaërt D and Marano F: TGF beta 1
promotes actin cytoskeleton reorganization and migratory phenotype
in epithelial tracheal cells in primary culture. J Cell Sci.
109:2207–2219. 1996.PubMed/NCBI
|
|
33
|
Wójciak-Stothard B, Entwistle A, Garg R
and Ridley AJ: Regulation of TNF-alpha-induced reorganization of
the actin cytoskeleton and cell-cell junctions by Rho, Rac, and
Cdc42 in human endothelial cells. J Cell Physiol. 176:150–165.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Akiyama T and Kawasaki Y: Wnt signalling
and the actin cytoskeleton. Oncogene. 25:7538–7544. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ménard L, Parker PJ and Kermorgant S:
Receptor tyrosine kinase c-Met controls the cytoskeleton from
different endosomes via different pathways. Nat Commun. 5:39072014.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Britton GJ, Ambler R, Clark DJ, Hill EV,
Tunbridge HM, McNally KE, Burton BR, Butterweck P, Sabatos-Peyton
C, Hampton-O'Neil LA, et al: PKCθ links proximal T cell and Notch
signaling through localized regulation of the actin cytoskeleton.
Elife. 6:e200032017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zieseniss A: Hypoxia and the modulation of
the actin cytoskeleton-emerging interrelations. Hypoxia (Auckl).
2:11–21. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Duan X, Liu J, Dai XX, Liu HL, Cui XS, Kim
NH, Wang ZB, Wang Q and Sun SC: Rho-GTPase effector ROCK
phosphorylates cofilin in actin-meditated cytokinesis during mouse
oocyte meiosis. Biol Reprod. 90:372014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Alexander NR, Tran NL, Rekapally H,
Summers CE, Glackin C and Heimark RL: N-cadherin gene expression in
prostate carcinoma is modulated by integrin-dependent nuclear
translocation of Twist1. Cancer Res. 66:3365–3369. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Müller CB, de Barros RL, Castro MA, Lopes
FM, Meurer RT, Roehe A, Mazzini G, Ulbrich-Kulczynski JM,
Dal-Pizzol F, Fernandes MC, et al: Validation of cofilin-1 as a
biomarker in non-small cell lung cancer: Application of
quantitative method in a retrospective cohort. J Cancer Res Clin
Oncol. 137:1309–1316. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Li M, Zhang X, Xu X, Wu J, Hu K, Guo X and
Zhang P: Clinicopathological and prognostic significance of Twist
overexpression in NSCLC. Oncotarget. 9:14642–14651. 2018.PubMed/NCBI
|
|
42
|
Yap CT, Simpson TI, Pratt T, Price DJ and
Maciver SK: The motility of glioblastoma tumour cells is modulated
by intracellular cofilin expression in a concentration-dependent
manner. Cell Motil Cytoskeleton. 60:153–165. 2005. View Article : Google Scholar : PubMed/NCBI
|