Introduction
Ovarian cancer is the leading cause of death among
all gynecologic malignancies and platinum compounds are the
standard first-line agents for the treatment of ovarian cancer
(1). However, nearly 75% of patients
who are highly responsive to cisplatin treatment experience
recurrence within 2 years and fail to respond to available
treatments due to acquired resistance (2,3).
Therefore, uncovering the mechanisms of cisplatin resistance and
seeking new therapeutic targets are critical for prolonging the
survival of patients.
Gap junctions (GJs) are composed of two
hemichannels, each of which consists of six connexin (Cx) monomers,
enabling the electrical coupling and sharing of ions and signaling
molecules (4). Almost all tissues and
cells are affected by this communication junction. Defective gap
junction intercellular communication (GJIC) has been observed in
carcinogenic progression. A lager number of studies have proved
that GJs and Cx are potential targets for tumor therapy (5,6).
However, emerging evidence has been found in recent
years showing that the increased expression of Cx may lead to
tumors with more aggressive phenotypes (7,8). The
cytoplasmic distribution of Cx also exerts an advantageous effect
on tumor progression. Kawasaki et al and Li et al
reported that cytoplasmic accumulation of Cx32 expanded the cancer
stem cell population and enhanced the motility and metastatic
ability of human hepatoma cells (9,10). Studies
performed by our team demonstrated that upregulated Cx32 in
cervical ovarian and liver cancer cells was mainly localized in the
cytoplasm and produced anti-apoptotic and pro-tumor effects in a
GJ-independent manner (11–14). Nevertheless, the mechanism of the
upregulation and internalization of Cx32 remains unknown.
The ubiquitin-proteasome system (UPS) is an
important pathway for the degradation of unnecessary proteins and
maintenance of protein homeostasis. This system comprises the 26S
proteasome and a sequence of enzymes (E1, E2 and E3) capable of
activating ubiquitin residues and adding them to the target protein
(15,16). In contrast, deubiquitinases (DUBs)
remove ubiquitin (Ub) chains from the target protein prior to
degradation. It has been reported that ubiquitination is involved
in the life cycle and localization of Cx (17,18). E3
ubiquitin ligase neural precursor cell-expressed developmentally
downregulated gene 4 (NEDD4) catalyzed the ubiquitination and
endocytosis of Cx43, which decreased GJIC (19,20). Sun
et al showed that ubiquitin-specific protease 8 (USP8)
reduced both multiple monoubiquitination and polyubiquitination of
Cx43 to prevent autophagy-mediated degradation (21). However, these studies mostly focused
on Cx43, and little research has been performed in regards to
Cx32.
Ubiquitin-specific protease 14 (USP14) is one of the
three DUBs associated with the 19S regulatory particle in mammalian
cells. USP14 disassembles the ubiquitin chain from its
substrate-distal tip and serves as a quality control component to
rescue proteins from degradation (22). Upregulation of USP14 is involved in
the progression of non-small cell lung cancer (23), breast cancer (24), ovarian cancer (25) and gastric cancer (26). USP14 knockdown was found to suppress
the proliferation and induced apoptosis of cancer cells. Therefore,
USP14 is proposed to be a target for cancer therapy. Furthermore,
many studies have shown that ubiquitination is closely related to
chemotherapy resistance (27–29). However, the role of USP14 in acquired
cisplatin resistance of ovarian cancer remains unrevealed.
Kaplan-Meier analysis demonstrated that the low expression of USP14
in ovarian cancer patients predicts poorer progression-free
survival (PFS) (Fig. 1A). Whether
USP14 is related with acquired cisplatin resistance and Cx32
internalization in ovarian cancer is unclear.
In the present study, lower expression and activity
of USP14 was found in ovarian cancer A2780-CDDP
(cisplatin-resistant) cells when compared with these parameters in
A2780 (cisplatin-sensitive) cells. USP14 inhibition induced
cisplatin resistance in A2780 cells. In addition, USP14 inhibition
upregulated Cx32 expression without changing the levels of Cx32
mRNA and ubiquitination. Cisplatin insensitivity by USP14
inhibition was abrogated when Cx32 expression was knocked down in
A2780 cells. Furthermore, upregulated Cx32 protein was
internalized, and GJIC was decreased. In conclusion, the present
study revealed that USP14 inhibition modulated Cx32 localization
and decreased cisplatin sensitivity in ovarian cancer cells,
indicating that Cx32 internalization regulated by USP14 may serve
as a marker of chemosensitivity.
Materials and methods
Analysis of Kaplan-Meier survival
tool
The Kaplan-Meier Plotter (http://kmplot.com/analysis/) was used to analyze the
association between USP14 expression and progression-free survival
(PFS) of patients with ovarian cancer. The following datasets were
used for the analysis: GSE14764 (30), GSE15622 (31), GSE18520 (32), GSE19829 (33), GSE23554 (34), GSE26193 (35,36),
GSE26712 (37,38), GSE27651 (39), GSE30161 (40), GSE3149 (41), GSE51373 (42), GSE63885 (43), GSE65986 (44), GSE9891 (45) and TCGA. ‘USP14’ was input in Affy
id/Gene symbol and chose ‘Affy ID: 201671_x_at’. The median
expression level of USP14 mRNA was regarded as the dividing line
and 1435 patients were divided into low expression group and high
expression group. Other parameters remain unchanged by default and
draw Kaplan-Meier plot. P-values <0.05 were considered to
indicate a statistically significant result.
Reagents
Cisplatin (cat. no. P4394) was obtained from
Sigma-Aldrich; Merck KGaA. IU1
[1-[1-(4-fluorophenyl)-2,5-dimethylpyrrol-3-yl]-2-pyrrolidin-1-ylethanone]
(cat. no. S7134) (a specific small-molecule inhibitor of USP14) was
purchased from Selleck Chemicals (Houston, TX, USA).
Antibodies
The mouse monoclonal anti-Cx32 (cat. no. 59948)
antibody was purchased from Santa Cruz Biotechnology, Inc. (Dallas,
Texas, USA). The mouse monoclonal anti-β-tubulin (cat. no. 86298),
mouse monoclonal anti-β-actin (cat. no. 3700), rabbit monoclonal
anti-USP14 (cat. no. 11931), rabbit monoclonal anti-caspase-3 (cat.
no. 9662), rabbit monoclonal anti-cleaved caspase-3 (cat. no.
9664), rabbit anti-caspase-9 (cat. no. 9502), rabbit
anti-Na,K-ATPase (cat. no. 3010) and mouse monoclonal IgG1 (cat.
no. 5415S) antibodies were purchased from Cell Signaling
Technology, Inc. (Danvers, MA, USA). Peroxidase-AffiniPure goat
anti-mouse IgG (H+L) (cat. no. 115-035-003) and
Peroxidase-AffiniPure goat anti-rabbit IgG (H+L) (cat. no.
111-035-003) were obtained from Jackson (West Grove, PA, USA). The
rabbit monoclonal anti-ubiquitin (cat. no. EPR8830) was purchased
from Abcam (Cambridge, UK). The secondary antibody IPKine HRP,
mouse anti-rabbit IgG LCS (cat. no. A25002) was purchased from
Abbkine Scientific Co., Ltd. (Wuhan, China).
Cell lines and cell culture
A2780 cisplatin-sensitive cells (A2780) were
obtained from the American Type Culture Collection (ATCC; Manassas,
VA, USA). The A2780 cisplatin-resistant cells (A2780-CDDP) were
established by a previously described method (11). Both A2780 and A2780-CDDP cell lines
were cultured in Dulbecco's modified Eagle's medium (DMEM) (cat.
no. 12800017; Gibco; Thermo Fisher Scientific, Inc.) containing 10%
fetal bovine serum (FBS) (Gibco; Thermo Fisher Scientific, Inc.)
and 1% penicillin/streptomycin at 37°C in an atmosphere containing
5% CO2.
siRNA and shRNA transfection
experiments
After growing A2780 cells to 30–50% confluence, 50
nM of non-specific siRNA (#siN0000001-1-5) and siRNAs targeting the
human USP14 gene were transfected into cells with Lipofectamine
3000 transfection reagent (cat. no. L3000015; Invitrogen; Thermo
Fisher Scientific, Inc.) according to the manufacturer's protocol.
The sequences of the synthesized siRNAs targeting USP14 (Guangzhou
RiboBio, Co., Ltd., Guangzhou, China) were as follows: siUSP14-1,
5′-CTGGCATATCGCTTACGTT-3′; siUSP14-2, 5′-TCCAGTATTCCACCTATTA-3′;
and siUSP14-3: 5′-TTGCCGAGAAAGGTGAACA-3′.
To generate stably transfected cells, A2780 cells
were transfected with lentiviral plasmids containing the shCx32
sequence or with the negative control vector
(pLVX-shRNA-tdTomato-Puro), which were constructed by Landbiology,
Co., Ltd. (Guangzhou, Guangdong, China). Lentiviral particles were
prepared by transfecting 293T cells with the lentiviral plasmids,
PAX2 and pMD2.G at a defined ratio. A2780 cells were incubated with
medium containing the virus and Polybrene for 48 h. After
infection, cells were selected by culturing with puromycin (2
µg/ml) for 2 weeks. The sequences of the short hairpin RNA
targeting Cx32 (shCx32) were as follows:
5′-GCTGCAACAGCGTTTGCTACTCGAGTAGCAAACGCTGTTGCAGCTTTTTTT-3′. shRNA
with non-specific sequences
(5′-TTCTCCGAACGTGTCACGTTTCTCGAGAAACGTGACACGTTCGGAGAA-3′) was used
as the control scrambled RNA.
Cell viability assay
Cell Counting Kit-8 (CCK-8) (Dojindo Molecular
Technologies Inc., Kumamoto, Japan) was used to examine cell
viability according to the manufacturer's instructions. Cells were
seeded in 96-well plates at a suitable density of 5,000 cells/well.
Then, siUSP14-1 was added for 24 h and the cells were treated with
cisplatin for 48 h or the cells were cotreated with IU1 and
cisplatin for 48 h. CCK-8 solution diluted with FBS-free DMEM at a
ratio of 1:9 was added in a total volume of 100 µl. The 96-well
plates were incubated at 37°C for 1–2 h. The optical density (OD)
was determined at 450 nm using an Epoch microplate
spectrophotometer (BioTek; Winooski, VT, USA). The concentration of
cisplatin resulting in 50% growth inhibition (IC50) was
calculated using GraphPad Prism 6.0 software (GraphPad Software,
Inc.).
Western blot analysis
Cells were lysed at 4°C in buffer [1 mM
β-glycerophosphate, 2.5 mM sodium pyrophosphate, 20 mM Tris-HCl (pH
7.4), 1% Triton X-100, 150 mM NaCl, 1 mM EGTA, 1 mM EDTA and 1 mM
Na3VO4] supplemented with protease inhibitor
cocktail (cat. no. P8340; Merck KGaA) (1:1,000). After
ultrasonication, cell lysates were cleared at 12,000 × g for 30 min
at 4°C. The protein concentration was determined with a BCA protein
assay kit (Thermo Fisher Scientific, Inc.). A total of 20 µg of
protein from each sample was separated using SDS-PAGE and
transferred to PVDF membranes. Membranes were blocked with 5% (w/v)
skim milk in wash buffer TSBT (TBS and 0.05% Tween-20) for 1 h and
were then incubated with specific antibodies against Cx32 (dilution
1:2,000), USP14 (dilution 1:1,000), cleaved caspase-3 (dilution
1:1,000), caspase-3 (dilution 1:1,000), caspase-9 (dilution
1:1,000), Na,K-ATPase (dilution 1:1,000), β-actin (dilution
1:10,000) and β-tubulin (dilution 1:10,000) overnight at 4°C. After
being washed for three times with TBST, membranes were incubated
with HRP-conjugated secondary antibodies (dilution 1:10,000) for 2
h at room temperature. The membranes were then washed with TBST
before being visualized using a Chemiluminescent HRP Substrate Kit
(Millipore; Billerica, MA, USA) and scanned using an ImageQuant LAS
4000™ (GE Healthcare). β-tubulin and β-actin were used as loading
control, and band densities were quantified using ImageJ software
(NIH, National Institutes of Health, Bethesda, MD, USA).
DUB trap assay
DUB activity can be inhibited irreversibly using
ubiquitin-vinylsulfone (Ub-VS), which forms an adduct with the
active site cysteine in DUB of the thiol protease class (22). Cells were lysed via mild sonication in
ice-cold buffer containing 50 mM Tris (pH 7.4), 5 mM
MgCl2, 250 mM sucrose, 1 mM DTT, 2 mM ATP and 1 mM PMSF.
Lysates were cleared by centrifugation at 12,000 × g for 30 min at
4°C, and 30 µg of protein extract was incubated for 15 min at 37°C
with 1 µM HA-Ub-VS (cat. no. U-212; Boston Biochem, Cambridge, MA,
USA). After boiling in loading buffer, labeled cell lysates were
subjected to immunoblot analysis. After transferring to PVDF
membranes, HA-Ub-VS labeled USP14 was immunodetected using the
USP14 antibody. The probe group was a blank control and β-actin was
used as a loading control. The band densities of USP14-Ub-VS were
quantification for the activity of USP14 (46).
Flow cytometry apoptosis detection
assay
A2780 cells were seeded in 6-well plates and
incubated with IU1 (50 µM), cisplatin (8 µg/ml) or cotreated with
IU1 (50 µM) and cisplatin (8 µg/ml) for 48 h. Afterward, the cells
were trypsinized and washed twice with cold PBS. After the cells
were re-suspended in binding buffer, Annexin V-FITC and propidium
iodide (PI) (cat. no. KGA105-KGA108; Keygen; Nanjing, Jiangsu,
China) were used to stain cells for 15 min away from light at room
temperature. Subsequently, the cells were immediately analyzed with
FACScan (Beckman Instruments, Fullerton, CA, USA) and cell
apoptosis was analyzed using FlowJo 7.6 software (FlowJo LLC,
Ashland, OR, USA).
Real-time qPCR assay
Approximately 1.5×105 A2780 cells were
seeded into 6-well plates. After adherence, the cells were
incubated with IU1 or siUSP14-1 for 48 h. Total RNA was extracted
using TRIzol (cat. no. A33252; Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's instructions. The
concentration of RNA was measured by NanoDrop 2000
Spectrophotometer (Thermo Fisher Scientific, Inc.). Then 1 µg total
RNA was reverse transcribed using a Transcriptor cDNA Synthesis kit
(cat. no. 4896866001; Roche; Basel, Switzerland) by C1000 Thermal
Cycler (Bio-Rad Laboratories, Inc.; Hercules, CA, USA). The
resulting cDNA was subjected to real-time qPCR in a final reaction
volume of 20 µl using FastStart Universal SYBR Green Master (Rox)
(cat. no. 04913914001; Roche) by the ABI Applied Biosystems StepOne
Quantitative Real-Time PCR System (Thermo Fisher Scientific, Inc.).
The thermocycling conditions were as follows: 10 min at 95°C,
followed by denaturation at 95°C for 15 sec; annealing and
extension at 60°C for 60 sec. Primers were as follows: USP14
(forward, 5′-GGCTTCAGCGCAGTATATTA-3′ and reverse,
5′-CAGATGAGGAGTCTGTCTCT-3′); Cx32 (forward,
5′-ACACCTTGCTCAGTGGCGTGA-3′ and reverse,
5′-GGGACCACAGCCGCACATGG-3′); and GAPDH (forward,
5′-GGAGCGAGATCCCTCCAAAAT-3′ and reverse,
5′-GGCTGTTGTCATACTTCTCATGG-3′). The analysis for USP14 and
normalization control gene GAPDH was performed according to the
2−ΔΔq method (47).
Immunoprecipitation
A2780 cells were treated with IU1 (50 µM for 48 h)
and then incubated with 10 µM MG132 (cat. no. S2619; Selleck
Chemicals) for 4 h prior to harvesting in buffer [1 mM
β-glycerophosphate, 2.5 mM sodium pyrophosphate, 20 mM Tris-HCl (pH
7.4), 1% Triton X-100, 150 mM NaCl, 1 mM EGTA, 1 mM EDTA and 1 mM
Na3VO4] supplemented with protease inhibitor
cocktail (1:1,000). The samples were then centrifuged at 12,000 × g
for 30 min, and the supernatants were used for immunoprecipitation.
Briefly, 1–2 µl of mouse monoclonal anti-Cx32 was added to
500–1,000 µg of protein and incubated for 3 h at 4°C. Non-specific
antibodies were used as controls. Then, 20 µl of Protein G
plus/Protein A agarose suspension (cat. no. IP05; Merck KGaA) was
added to the mixture. Following incubation for more than 12 h at
4°C, the samples were centrifuged at 12,000 × g for 3 min, and the
protein G-Sepharose precipitate was washed 5 times in an
appropriate wash buffer (500 mM NaCl, 50 mM Tris-HCl, 6 mM EDTA and
1% Triton X-100, pH 8.3), resuspended in 4X loading buffer, and
denatured at 100°C for 5 min. The input represented 10% of the
total amount of protein in the lysates before immunoprecipitation.
All samples were subjected to immunoblot analysis. After separation
by SDS-PAGE, the proteins samples were transferred to PVDF
membrane. Membranes were blocked with 5% (w/v) skim milk in wash
buffer (TBS and 0.05% Tween-20) for 1 h and were then incubated
with specific antibodies against ubiquitin (dilution 1:1,000). The
secondary antibody IPKine HRP, mouse anti-rabbit IgG LCS (dilution
1:5,000) was used to incubated the membrane for eliminating heavy
chain interference.
Parachute dye-coupling assay
GJIC was evaluated by a ‘parachute’ dye coupling
assay, as described by Goldberg et al (48) and Koreen et al (49). Calcein-AM (cat. no. C3100MP) and
CM-DiI (cat. no. C3100MP) were purchased from Invitrogen (Thermo
Fisher Scientific, Inc.). Cells were seeded in 12-well plates at an
appropriate density of 50,000 cells/well. After confluence, the
donor cells were labeled with both Calcein-AM (green fluorescence,
GJ permeable) and CM-DiI (red fluorescence, non-permeable) at 37°C
for 30 min. After being rinsed and trypsinized, 500 donor cells
were seeded onto the receiver cells per well and incubated at 37°C
for 4–6 h. Signal intensity was observed using a fluorescence
microscope (Olympus IX71; Tokyo, Japan). The average number of
receiver cells (green fluorescence) around every donor cell (both
green and red fluorescence) was recorded as an index of GJIC.
Membrane protein extraction
Approximately 2–3×106 A2780 cells were
seeded in a 10-cm culture dish. IU1 or siUSP14-1 was administered
for 48 h when the confluence was 30–40%. Membrane-bound and
cytoplasmic proteins were extracted by using a ProteoExtract
Transmembrane Protein Extraction Kit (cat. no. 71772; Merck KGaA)
according to the manufacturer's instructions. Cells were collected
in PBS and centrifuged at 1,000 × g for 5 min at 4°C. Cells were
resuspended in Extraction Buffer 1 containing protease inhibitor
cocktail and incubated for 10 min at 4°C. The cytosolic (soluble)
protein fraction was harvested after centrifugation at 1,000 × g
for 5 min at 4°C. The pellet was resuspended in Extraction Buffer 2
containing TM-PEK reagent B and protease inhibitor cocktail. The
mixture was incubated for 45 min at 4°C with gentle agitation, and
membrane proteins were obtained in the supernatant.
Statistical analysis
Every in vitro experiment was performed with
a minimum of three independent cell cultures. The data were
statistically analyzed by Student's t-test (2 groups), one-way
ANOVA (>2 groups), followed by Tukey's multiple comparison test
or two-way ANOVA (>2 groups), followed by Bonferroni post test
with GraphPad Prism 6.0 software. Histograms or scatter diagrams
were constructed with the GraphPad Prism 6.0 software (GraphPad
Software, Inc.). The results are expressed as the mean ± SE, and
P<0.05 was considered indicative of statistical
significance.
Results
The decreased USP14 expression
predicts poor prognosis in ovarian cancer
A cohort of 1435 cases of ovarian cancer of
following datasets were used for Kaplan-Meier analysis: GSE14764
(30), GSE15622 (31), GSE18520 (32), GSE19829 (33), GSE23554 (34), GSE26193 (35,36),
GSE26712 (37,38), GSE27651 (39), GSE30161 (40), GSE3149 (41), GSE51373 (42), GSE63885 (43), GSE65986 (44), GSE9891 (45) and TCGA. The results showed that
decreased USP14 expression is associated with poorer
progression-free survival (PFS) of patients with ovarian cancer
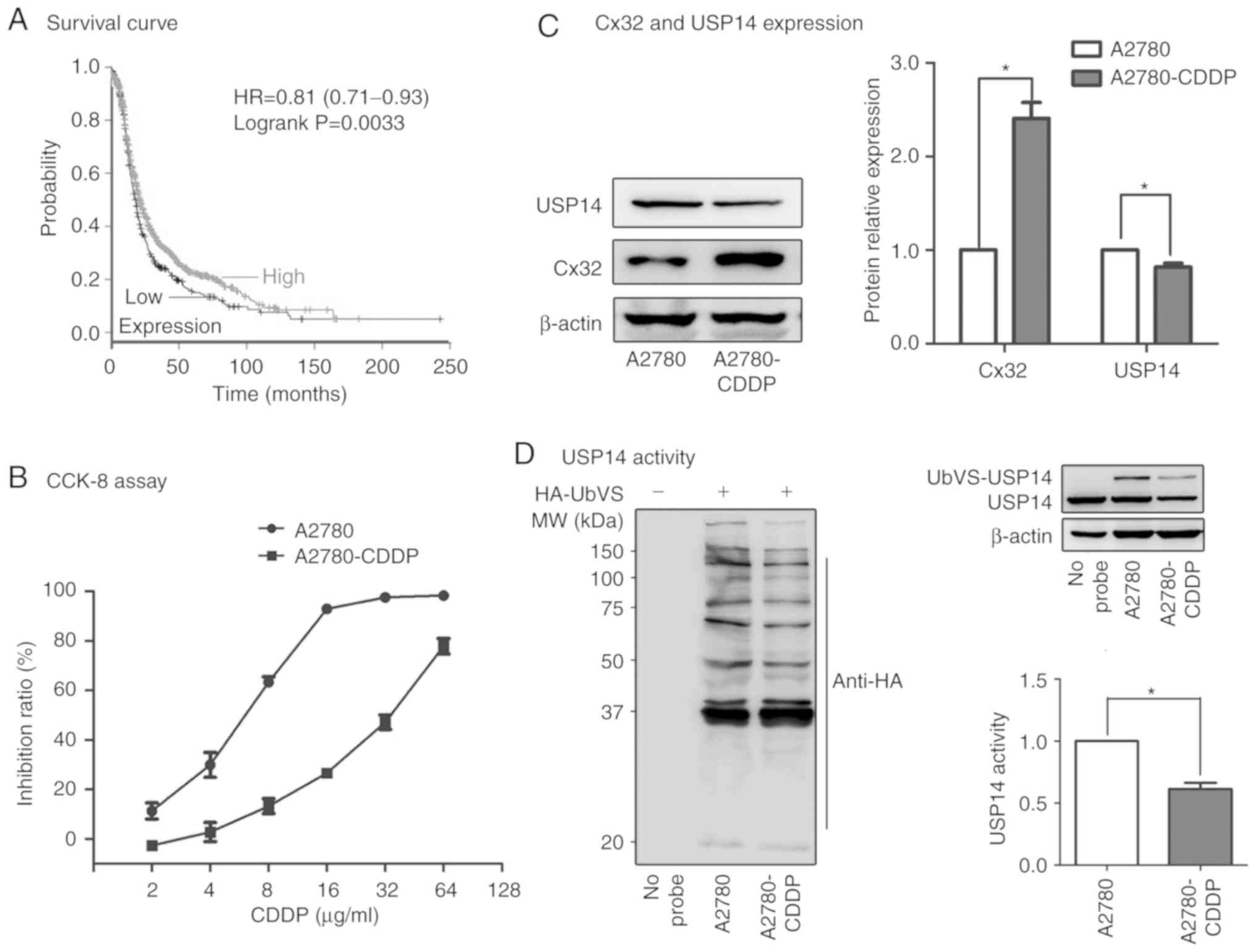
(P<0.001; Fig. 1A). Then we
performed experiments to explore the role of USP14 in cisplatin
resistance of ovarian cancer.
Activity and expression of USP14 were decreased in
A2780-CDDP cells. The A2780-CDDP cells were established by a
previously described method (11).
The IC50 of the A2780 cells used in present study was
5.946±0.61 µg/ml, and the IC50 of the A2780-CDDP cells
was 32.45±2.52 µg/ml. The resistance index (RI) was 5.46.
A2780-CDDP cells were moderately resistant to cisplatin and
qualified for subsequent experiments (Fig. 1B). As showed in Fig. 1C, the expression of Cx32 was
significantly higher while the expression of USP14 was
significantly lower in the A2780-CDDP cells when compared to the
expression levels in A2780 cells. Furthermore, the results of DUB
trap assay showed that USP14 activity was significantly decreased
in the A2780-CDDP cells (Fig.
1D).
Inhibition of USP14 in A2780 cells
decreases cisplatin cytotoxicity
Next we sought to determine whether USP14 is
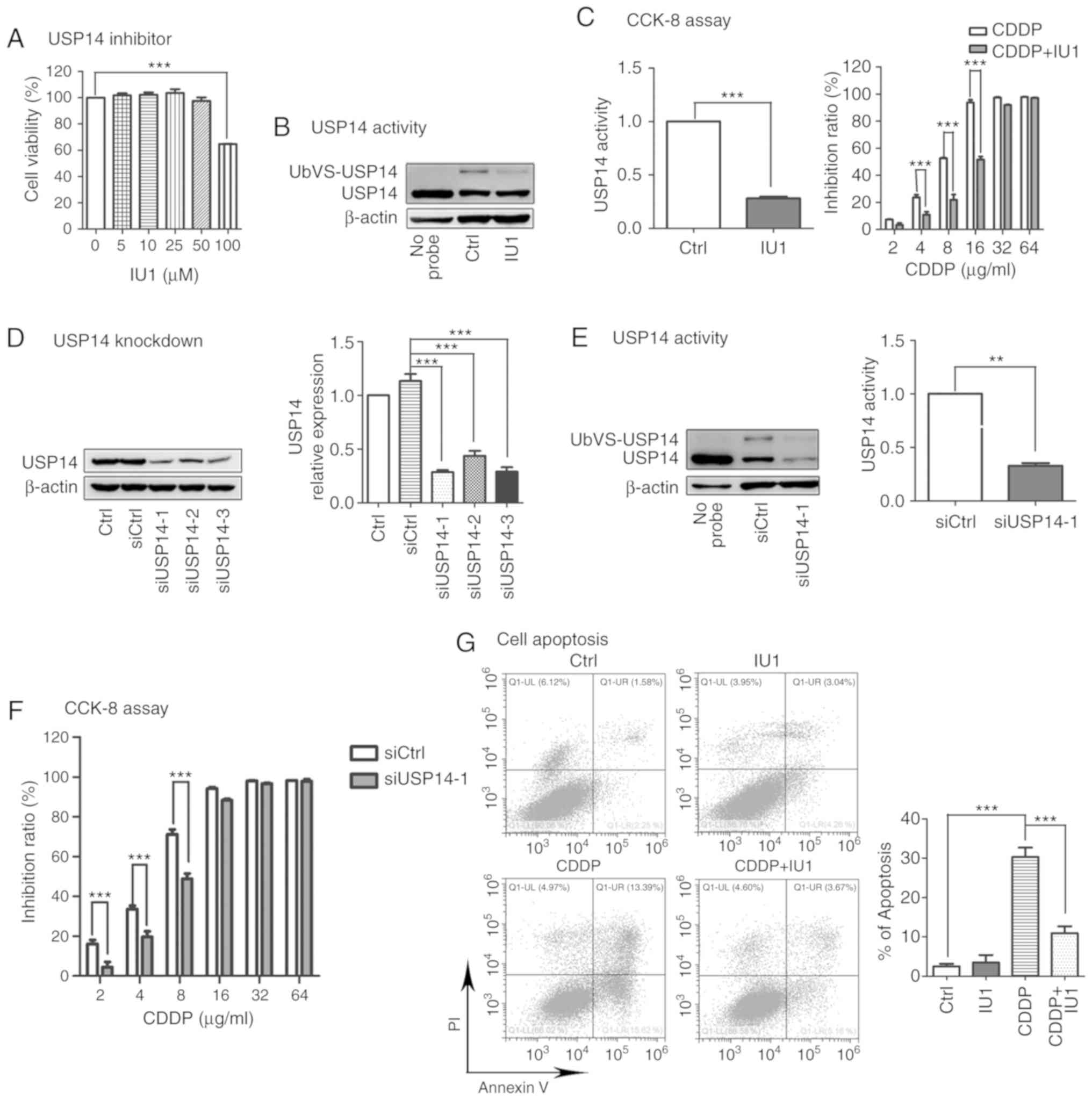
involved in cisplatin resistance. It has been reported that IU1
specifically inhibits USP14 by preventing its docking on the
proteasome (22). Therefore IU1 was
used as a USP14 inhibitor. A2780 cells were incubated with gradient
concentrations (5, 10, 25, 50 and 100 µM) of IU1 for 48 h. The
results of CCK-8 assay indicated that 100 µM IU1 significantly
inhibited A2780 cell proliferation while 0–50 µM IU1 had no effect
(Fig. 2A). Hence, we chose 50 µM IU1
for subsequent studies. As expected, 50 µM IU1 clearly inhibited
the activity of USP14 in A2780 cells without changing its protein
level (Fig. 2B). The results of CCK-8
assay revealed that IU1 markedly counteracted cisplatin
cytotoxicity (Fig. 2C). To make the
experiments more convincing, siRNAs were used to inhibit USP14
expression. siUSP14-1 showed the greatest efficiency in silencing
USP14 expression (Fig. 2D). Moreover,
siUSP14-1 decreased both the protein level and activity of USP14
(Fig. 2E). Then A2780 cells were
transfected with siUSP14-1 for 24 h, followed by cisplatin
administration for 48 h. Similarly, USP14 knockdown decreased
cisplatin cytotoxicity in A2780 cells (Fig. 2F).
Additionally, the apoptosis rate of the cells was
examined by flow cytometry, and the levels of cleaved caspase-9 and
cleaved caspase-3, two well-known protein markers of apoptosis,
were assessed by western blotting. As showed in Fig. 2G-I, significant apoptosis of A2780
cells was induced by cisplatin (8 µg/ml, 48 h) but was
significantly suppressed by cotreatment with the USP14 inhibitor
IU1 (50 µM, 48 h; Fig. 2G and H) or
siUSP14-1 (Fig. 2I). Taken together,
these results indicated that USP14 inhibition decreased
cisplatin-induced apoptosis.
Inhibition of USP14 increases
expression of Cx32 without altering its mRNA and ubiquitination
levels
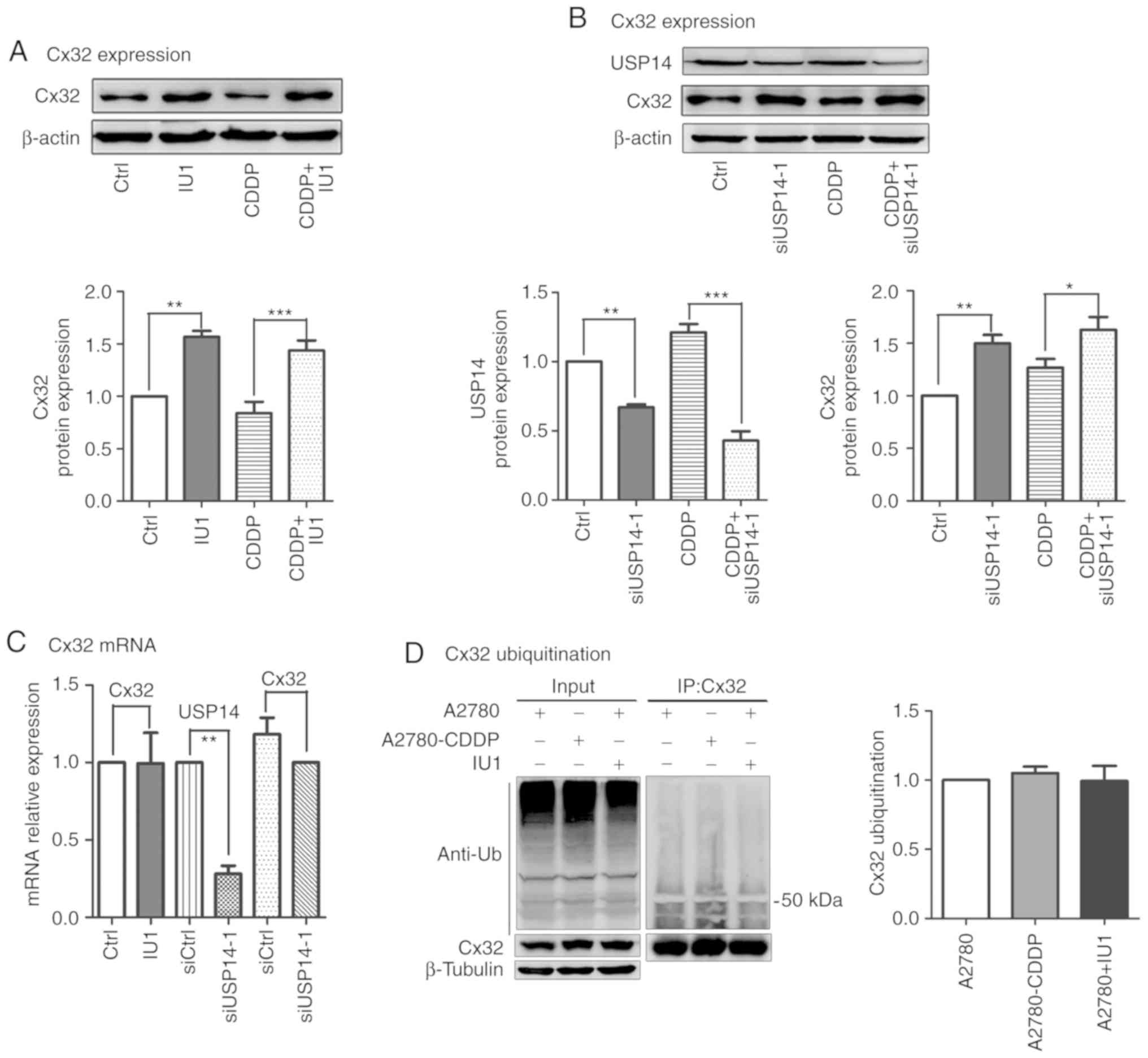
Our previous studies reported that the
anti-apoptotic effect of USP14 inhibition was closely related to
the upregulation of Cx32 expression in cervical and ovarian cancer
(11,13). The present results showed that
cisplatin cytotoxicity was also reduced by USP14 inhibition in
A2780 cells. Then, we aimed to ascertain whether USP14 inhibition
could affect the expression of Cx32. As showed in Fig. 3A and B, IU1 (50 µM, 48 h) and
siUSP14-1 alone or in combination with cisplatin upregulated Cx32
expression. To further explore this mechanism, qPCR and
immunoprecipitation were performed. The results demonstrated that
the mRNA and ubiquitination level of Cx32 remained unchanged.
Moreover, the ubiquitination level of Cx32 in A2780 cells was
consistent with that in A2780-CDDP cells (Fig. 3C and D). Taken together, USP14
inhibition increased the expression of Cx32 without changing its
mRNA and ubiquitination levels.
Cx32 knockdown counteracts USP14
downregulation-induced cisplatin resistance in A2780 cells
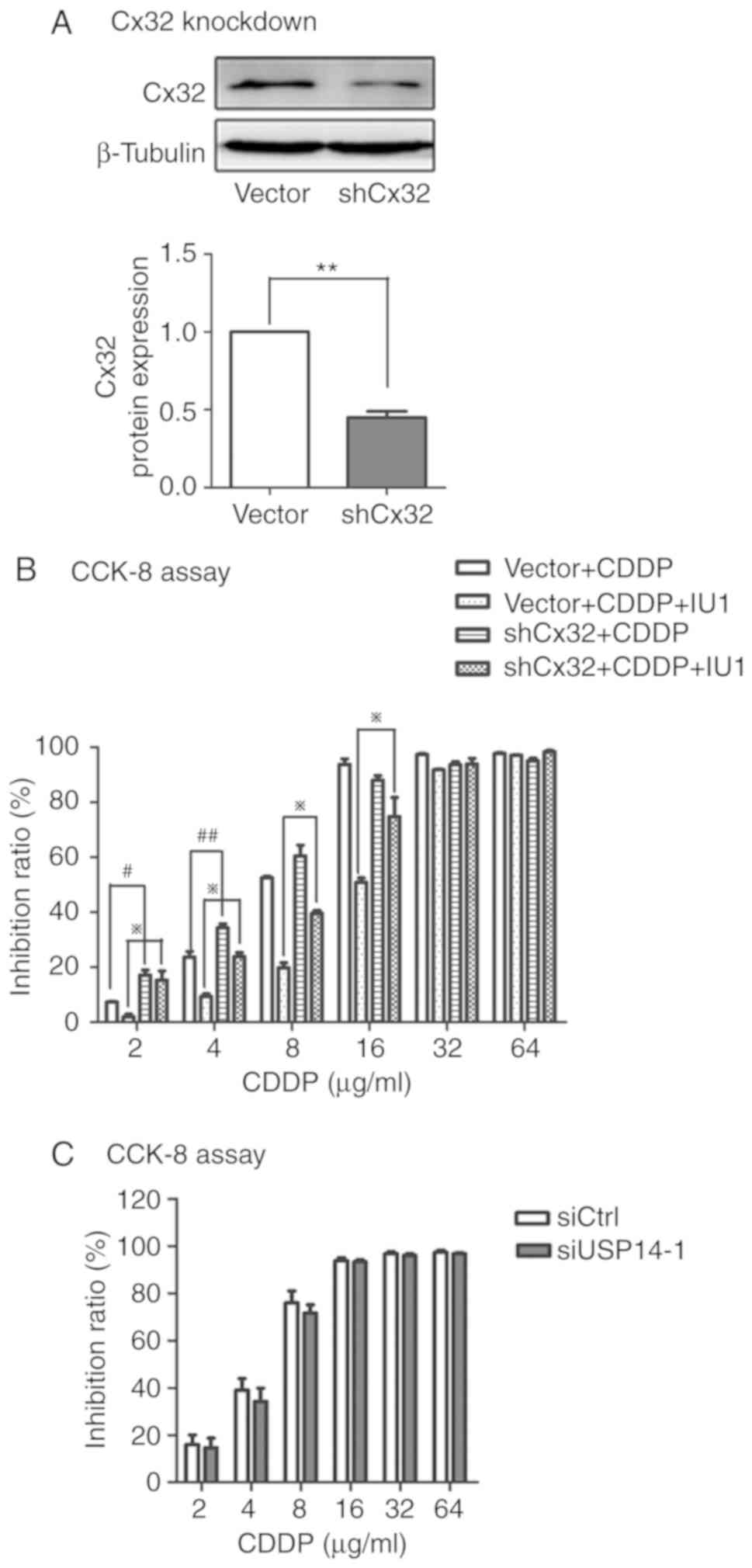
The above results showed that IU1 and siUSP14-1
alone or in combination with cisplatin upregulated Cx32 expression.
To determine the role of Cx32 in the anti-apoptotic effect of USP14
inhibition, Cx32 expression was knocked down with shRNA in A2780
cells (Fig. 4A). As shown in Fig. 4B, cytotoxicity was increased in the
Cx32-knockdown cells compared with that in A2780 cells after
administration of 2 or 4 µg/ml cisplatin. Similarly, cytotoxicity
was increased in the Cx32-knockdown cells compared with that in
A2780 cells after co-administration. Although the anti-apoptotic
effect of cotreatment remained after Cx32 knockdown, the extent was
indeed lessened compared to that in A2780 cells when cisplatin was
administered at 2, 4, 8 or 16 µg/ml. Consistent with the above
results, Cx32 knockdown in A2780 cells partially abrogated the
cisplatin resistance induced by USP14 silencing (Fig. 4C). These findings suggest that Cx32
plays an important role in USP14 inhibition-induced cisplatin
resistance.
Inhibition of USP14 induces Cx32
internalization and impairs GJIC
The upregulation of Cx32 induced by USP14 inhibition
was found to contribute to cisplatin insensitivity. We aimed to
ascertain whether GJIC and Cx32 localization are affected by USP14
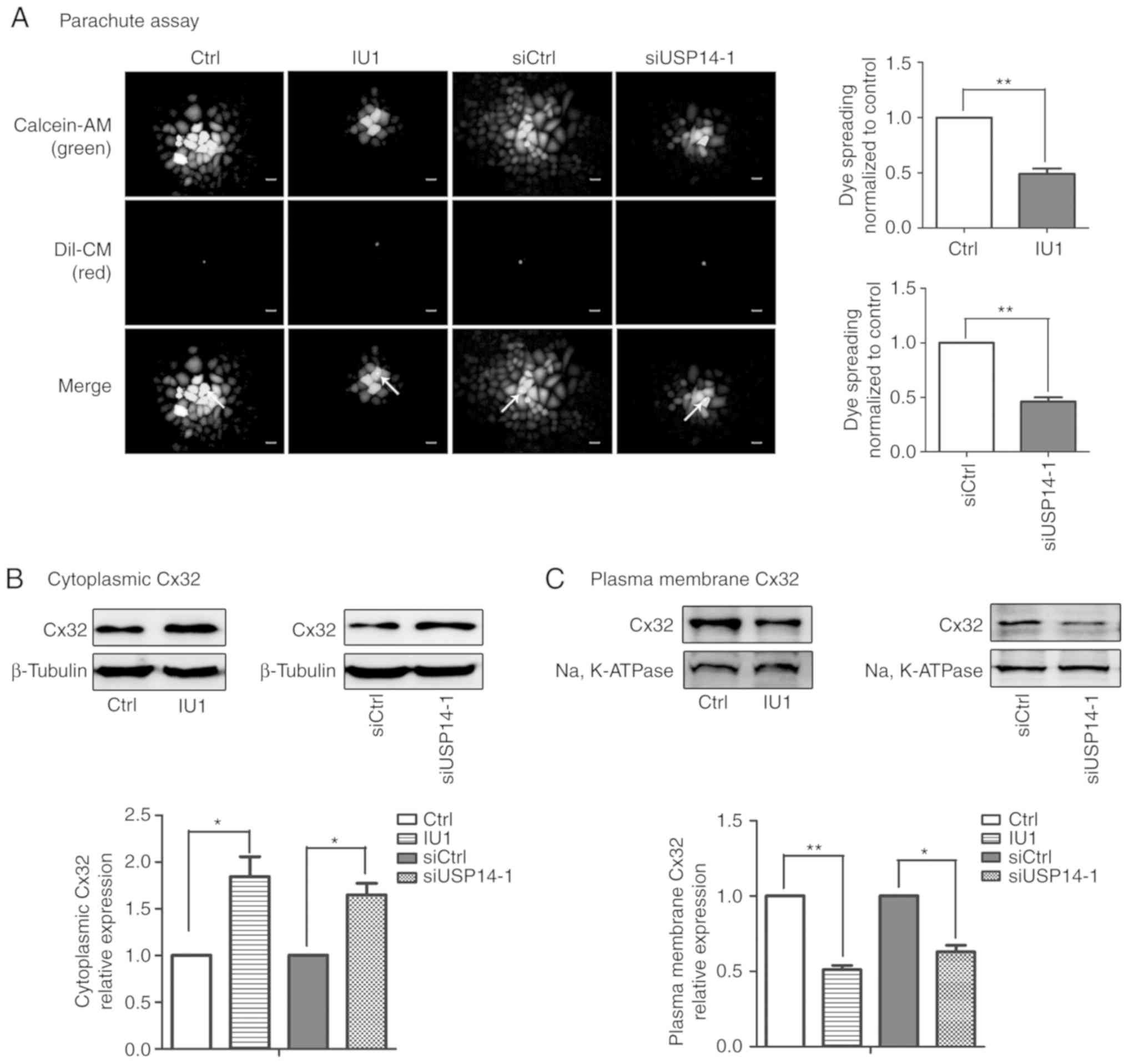
inhibition. As depicted in Fig. 1A,
USP14 suppression caused a marked reduction in GJIC (Fig. 5A). In addition, membrane and
cytoplasmic proteins were extracted after administration of IU1 or
siUSP14-1. As expected, the results demonstrated that Cx32 was
predominantly distributed in the cytoplasm and the Cx32 that
localized on the membrane was decreased (Fig. 5B and C). In summary, Cx32
internalization caused by USP14 inhibition caused the cisplatin
insensitivity in a non-gap junction-mediated manner.
Discussion
The results of the present study indicated that
decreased expression and activity of USP14 in A2780-CDDP cells
resulted in upregulation and internalization of Cx32, which
contributed to cisplatin resistance.
Previous studies have reported that expression and
activity of USP14 are elevated in gastric carcinoma, ovarian cancer
and melanoma. Knockdown of this 19S proteasome-associated DUB
sensitizes cancer to chemotherapeutics and induces apoptosis
(25,26,50). To
the best of ou knowledge, this evidence is the first to show that
the expression and activity of USP14 are downregulated in
A2780-CDDP cells and that the reduction in the expression and
activity of USP14 is associated with cisplatin insensitivity. These
findings suggest that USP14 is not only a target of oncotherapy but
also a marker of chemoresistance.
Studies indicate that the cytoplasmic localization
of Cx32 serves as a pro-tumor factor in a number of cancer types
(11,14). Similarly, in the present study, it was
shown that the internalization of Cx32 and reduction in GJIC
induced by USP14 inhibition contributed to cisplatin resistance.
This effect is not limited to Cx32. Cx26 was found to be
cytoplasmic in human invasive carcinomas of the breast (51). Intracellular accumulation of Cx43 and
Cx32 was observed in human prostate cancer cells and facilitated
their malignant phenotype (52).
Furthermore, it is important to note that even nuclear localization
of Cx32 has been reported and suppresses
streptonigrin/cisplatin-induced apoptosis (13). Taken together, these findings indicate
that the upregulation and mis-localization of Cx play a pro-tumor
role in a non-junctional manner.
Regarding the reasons for the augmentation of Cx32
protein expression by USP14 inhibition, the qPCR results
demonstrated that the Cx32 mRNA level remained unchanged,
indicating that the increase in Cx32 protein expression was not due
to an upregulated transcription level. Toler et al noted
that mRNA of Cx was detected without corresponding levels in Cx
protein expression (53). In this
study, posttranslational modifications may be responsible for the
increased expression and cytoplasmic localization of Cx32 based on
the fact that USP14 is a DUB modulating ubiquitination and
degradation. The E3 ubiquitin ligase, AMSH (associated molecule
with the SH3 domain of STAM) and a number of ubiquitin-binding
proteins such as EGF receptor pathway substrate 15 (Eps15),
hepatocyte growth factor-regulated tyrosine kinase substrate (Hrs)
and tumor susceptibility gene 101 (TSG101) were implicated in
modulating Cx43 localization and GJIC (20,54–56). Cx26
and Cx32 have also been shown to undergo ubiquitination (57,58).
However, in this study, the ubiquitination level of Cx32 remained
unchanged when USP14 was inhibited, suggesting that Cx32 was not
directly affected by USP14. It has been reported that Wnt signaling
(59), NF-κB signaling (60) and the estrogen receptor (ER) signaling
pathway (24) are mediated by USP14.
Furthermore, Xu et al reported that USP14 regulates
autophagy by suppressing K63 ubiquitination of Beclin 1 (61). Cx32 may be regulated by USP14
inhibition via the signaling pathways mentioned above.
As for the mechanism of cisplatin resistance by
USP14 inhibition, we demonstrated that Cx32 knockdown partially
decreased the cisplatin resistance induced by USP14 inhibition.
Although Cx32 upregulation by USP14 inhibition was found to be an
important factor to counteract cisplatin cytotoxicity, there still
may exist other mechanisms. Lee et al showed that inhibition
of USP14 by IU1 enhanced the proteasome activity and accelerated
the degradation of oxidized proteins (22). Sharma et al reported that USP14
inhibition stabilized RNF168 and restored downstream DNA damage
response (DDR) signaling, thus protecting prostate cancer cells
from radiotherapy (62). Furthermore,
USP14 was found to modulate hippocampal short-term plasticity and
long-term memory formation independent of its deubiquitinating
activity (63,64). In the present study, eliminating toxic
proteins, facilitating DNA repair and other signaling pathways
regulated by USP14 may also have accounted for the observed
cisplatin insensitivity. Although many studies have demonstrated
the importance of USP14 in cell physiology and diseases, the global
substrates of USP14 are still to be elucidated, representing a
major ‘bottleneck’ to uncover the functional characterization of
USP14 and understand the complexity of proteasome-associated
deubiquitination events (65,66).
The tumor-promoting effect of USP14 inhibition is
strongly supported by in vitro data, but only one cell line
was used in this research, thus, this may be a limitation of this
study and further investigation using more cell lines to determine
the underlying mechanism of USP14 is warranted in the future.
Moreover, clinical pathology and in vivo experiments are
needed for definite confirmation. Our experimental results support
the hypothesis that USP14 inhibition results in the abnormal
distribution of Cx32 and the dysfunction of GJIC, while Cx32 is not
a direct target of USP14, and the mechanism underlying Cx32
internalization requires further study.
In conclusion, the expression and activity of USP14
were markedly decreased, accompanied by cisplatin resistance, in
A2780-CDDP cells. USP14 inhibition led to the upregulation and
internalization of Cx32 and the impairment of GJIC, which mediates
cisplatin resistance in A2780 cells. Recovering the mechanism of
Cx32 internalization and trafficking may be a potential therapy
against tumors and chemoresistance.
Acknowledgements
Not applicable.
Funding
This study was funded by the National Natural
Science Foundation of China (grant no. 81473234), the Joint Fund of
the National Natural Science Foundation of China (grant no.
U1303221), the Fundamental Research Funds for the Central
Universities (grant no. 16ykjc01) and a grant from the Department
of Science and Technology of Guangdong Province (grant no.
20160908).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
LT, QW and GH conceived and designed the study. HL,
XW performed the siRNA and shRNA transfection experiments, CCK-8
assay and western blot. NZ performed the flow cytometry
experiments, collected and analyse the data. YF was involved in
cell culture and parachute dye-coupling assay. FP made substantial
contributions in the acquisition and interpretation of the data. HL
and HG wrote the manuscript. LT and XW revised and amended the
draft. QW contributed to final approval of the version to be
published. All authors read and approved the final manuscript and
agree to be accountable for all aspects of the research in ensuring
that the accuracy or integrity of any part of the work are
appropriately investigated and resolved.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
USP14
|
ubiquitin-specific protease 14
|
|
Cx
|
connexin
|
|
Cx32
|
connexin 32
|
|
GJIC
|
gap junction intercellular
communication
|
|
UPS
|
ubiquitin-proteasome system
|
|
NEDD4
|
neural precursor cell-expressed
developmentally downregulated gene 4
|
|
USP8
|
ubiquitin-specific protease 8
|
|
Cx43
|
connexin 43
|
|
siRNA
|
small interfering RNA
|
|
Eps15
|
EGF receptor pathway substrate 15
|
|
AMSH
|
associated molecule with the SH3
domain of STAM
|
|
Hrs
|
hepatocyte growth factor-regulated
tyrosine kinase substrate
|
|
TSG101
|
tumor susceptibility gene 101
|
|
TP53BP1
|
tumor protein p53 binding protein
1
|
|
RNF168
|
ring finger protein 168
|
|
DDR
|
DNA damage response
|
References
|
1
|
Mikula-Pietrasik J, Witucka A, Pakula M,
Uruski P, Begier-Krasińska B, Niklas A, Tykarski A and Książek K:
Comprehensive review on how platinum- and taxane-based chemotherapy
of ovarian cancer affects biology of normal cells. Cell Mol Life
Sci. 76:681–697. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Norouzi-Barough L, Sarookhani MR, Sharifi
M, Moghbelinejad S, Jangjoo S and Salehi R: Molecular mechanisms of
drug resistance in ovarian cancer. J Cell Physiol. 233:4546–4562.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kujawa KA and Lisowska KM: Ovarian
cancer-from biology to clinic. Postepy Hig Med Dosw (Online).
2:1275–1290. 2015.(In Polish). View Article : Google Scholar
|
|
4
|
Aasen T, Mesnil M, Naus CC, Lampe PD and
Laird DW: Gap junctions and cancer: Communicating for 50 years. Nat
Rev Cancer. 16:775–788. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ruch RJ: Connexin43 suppresses lung cancer
stem cells. Cancers (Basel). 11(pii): E1752019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Suzhi Z, Liang T, Yuexia P, Lucy L,
Xiaoting H, Yuan Z and Qin W: Gap junctions enhance the
antiproliferative effect of MicroRNA-124-3p in glioblastoma cells.
J Cell Physiol. 230:2476–2488. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Polusani SR, Kalmykov EA, Chandrasekhar A,
Zucker SN and Nicholson BJ: Cell coupling mediated by connexin 26
selectively contributes to reduced adhesivity and increased
migration. J Cell Sci. 129:4399–4410. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chen Q, Boire A, Jin X, Valiente M, Er EE,
Lopez-Soto A, Jacob L, Patwa R, Shah H, Xu K, et al:
Carcinoma-astrocyte gap junctions promote brain metastasis by cGAMP
transfer. Nature. 533:493–498. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kawasaki Y, Omori Y, Li Q, Nishikawa Y,
Yoshioka T, Yoshida M, Ishikawa K and Enomoto K: Cytoplasmic
accumulation of connexin32 expands cancer stem cell population in
human HuH7 hepatoma cells by enhancing its self-renewal. Int J
Cancer. 128:51–62. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Li Q, Omori Y, Nishikawa Y, Yoshioka T,
Yamamoto Y and Enomoto K: Cytoplasmic accumulation of connexin32
protein enhances motility and metastatic ability of human hepatoma
cells in vitro and in vivo. Int J Cancer. 121:536–546. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wu W, Fan L, Bao Z, Zhang Y, Peng Y, Shao
M, Xiang Y, Zhang X, Wang Q and Tao L: The cytoplasmic
translocation of Cx32 mediates cisplatin resistance in ovarian
cancer cells. Biochem Biophys Res Commun. 487:292–299. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhang Y, Tao L, Fan LX, Huang K, Luo HM,
Ge H, Wang X and Wang Q: Cx32 mediates cisplatin resistance in
human ovarian cancer cells by affecting drug efflux transporter
expression and activating the EGFR-Akt pathway. Mol Med Rep.
19:2287–2296. 2019.PubMed/NCBI
|
|
13
|
Zhao Y, Lai Y, Ge H, Guo Y, Feng X, Song
J, Wang Q, Fan L, Peng Y, Cao M, et al: Non-junctional Cx32
mediates anti-apoptotic and pro-tumor effects via epidermal growth
factor receptor in human cervical cancer cells. Cell Death Dis.
8:e27732017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Xiang Y, Wang Q, Guo Y, Ge H, Fu Y, Wang X
and Tao L: Cx32 exerts anti-apoptotic and pro-tumor effects via the
epidermal growth factor receptor pathway in hepatocellular
carcinoma. J Exp Clin Cancer Res. 38:1452019. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Saeki Y: Ubiquitin recognition by the
proteasome. J Biochem. 161:113–124. 2017.PubMed/NCBI
|
|
16
|
Kwon YT and Ciechanover A: The ubiquitin
code in the ubiquitin-proteasome system and autophagy. Trends
Biochem Sci. 42:873–886. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Salat-Canela C, Munoz MJ, Sese M, Ramon y
Cajal S and Aasen T: Post-transcriptional regulation of connexins.
Biochem Soc Trans. 43:465–470. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Aasen T, Johnstone S, Vidal-Brime L, Lynn
KS and Koval M: Connexins: Synthesis, Post-translational
modifications, and trafficking in health and disease. Int J Mol
Sci. 19(pii): E12962018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Spagnol G, Kieken F, Kopanic JL, Li H,
Zach S, Stauch KL, Grosely R and Sorgen PL: Structural studies of
the Nedd4 WW domains and their selectivity for the Connexin43
(Cx43) Carboxyl terminus. J Biol Chem. 291:7637–7650. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Totland MZ, Bergsland CH, Fykerud TA,
Knudsen LM, Rasmussen NL, Eide PW, Yohannes Z, Sørensen V, Brech A,
Lothe RA and Leithe E: The E3 ubiquitin ligase NEDD4 induces
endocytosis and lysosomal sorting of connexin 43 to promote loss of
gap junctions. J Cell Sci. 130:2867–2882. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Sun J, Hu Q, Peng H, Peng C, Zhou L, Lu J
and Huang C: The ubiquitin-specific protease USP8 deubiquitinates
and stabilizes Cx43. J Biol Chem. 293:8275–8284. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lee BH, Lee MJ, Park S, Oh DC, Elsasser S,
Chen PC, Gartner C, Dimova N, Hanna J, Gygi SP, et al: Enhancement
of proteasome activity by a small-molecule inhibitor of USP14.
Nature. 467:179–184. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chen X, Yang Q, Chen J, Zhang P, Huang Q,
Zhang X, Yang L, Xu D, Zhao C, Wang X and Liu J: Inhibition of
proteasomal deubiquitinase by silver complex induces apoptosis in
non-small cell lung cancer cells. Cell Physiol Biochem. 49:780–797.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Xia X, Liao Y, Guo Z, Li Y, Jiang L, Zhang
F, Huang C, Liu Y, Wang X, Liu N, et al: Targeting
proteasome-associated deubiquitinases as a novel strategy for the
treatment of estrogen receptor-positive breast cancer. Oncogenesis.
7:752018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang Y, Wang J, Zhong J, Deng Y, Xi Q, He
S, Yang S, Jiang L, Huang M, Tang C and Liu R: Ubiquitin-specific
protease 14 (USP14) regulates cellular proliferation and apoptosis
in epithelial ovarian cancer. Med Oncol. 32:3792015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Fu Y, Ma G, Liu G, Li B, Li H, Hao X and
Liu L: USP14 as a novel prognostic marker promotes cisplatin
resistance via Akt/ERK signaling pathways in gastric cancer. Cancer
Med. 7:5577–5588. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Hu X, Meng Y, Xu L, Qiu L, Wei M, Su D, Qi
X, Wang Z, Yang S, Liu C and Han J: Cul4 E3 ubiquitin ligase
regulates ovarian cancer drug resistance by targeting the
antiapoptotic protein BIRC3. Cell Death Dis. 10:1042019. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Lu Y, Han D, Liu W, Huang R, Ou J, Chen X,
Zhang X, Wang X, Li S, Wang L, et al: RNF138 confers cisplatin
resistance in gastric cancer cells via activating Chk1 signaling
pathway. Cancer Biol Ther. 19:1128–1138. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Li L, Liu T, Li Y, Wu C, Luo K, Yin Y,
Chen Y, Nowsheen S, Wu J, Lou Z and Yuan J: The deubiquitinase
USP9X promotes tumor cell survival and confers chemoresistance
through YAP1 stabilization. Oncogene. 37:2422–2431. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Denkert C, Budczies J, Darb-Esfahani S,
Györffy B, Sehouli J, Könsgen D, Zeillinger R, Weichert W, Noske A,
Buckendahl AC, et al: A prognostic gene expression index in ovarian
cancer-validation across different independent data sets. J Pathol.
218:273–280. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ahmed AA, Mills AD, Ibrahim AE, Temple J,
Blenkiron C, Vias M, Massie CE, Iyer NG, McGeoch A, Crawford R, et
al: The extracellular matrix protein TGFBI induces microtubule
stabilization and sensitizes ovarian cancers to paclitaxel. Cancer
Cell. 12:514–527. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Mok SC, Bonome T, Vathipadiekal V, Bell A,
Johnson ME, Wong KK, Park DC, Hao K, Yip DK, Donninger H, et al: A
gene signature predictive for outcome in advanced ovarian cancer
identifies a survival factor: Microfibril-associated glycoprotein
2. Cancer Cell. 16:521–532. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Konstantinopoulos PA, Spentzos D, Karlan
BY, Taniguchi T, Fountzilas E, Francoeur N, Levine DA and Cannistra
SA: Gene expression profile of BRCAness that correlates with
responsiveness to chemotherapy and with outcome in patients with
epithelial ovarian cancer. J Clin Oncol. 28:3555–3561. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Marchion DC, Cottrill HM, Xiong Y, Chen N,
Bicaku E, Fulp WJ, Bansal N, Chon HS, Stickles XB, Kamath SG, et
al: BAD phosphorylation determines ovarian cancer chemosensitivity
and patient survival. Clin Cancer Res. 17:6356–6366. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Mateescu B, Batista L, Cardon M, Gruosso
T, de Feraudy Y, Mariani O, Nicolas A, Meyniel JP, Cottu P,
Sastre-Garau X and Mechta-Grigoriou F: miR-141 and miR-200a act on
ovarian tumorigenesis by controlling oxidative stress response. Nat
Med. 17:1627–1635. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Gentric G, Kieffer Y, Mieulet V, Goundiam
O, Bonneau C, Nemati F, Hurbain I, Raposo G, Popova T, Stern MH, et
al: PML-regulated mitochondrial metabolism enhances
chemosensitivity in human ovarian cancers. Cell Metab.
29:156–173.e110. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Bonome T, Levine DA, Shih J, Randonovich
M, Pise-Masison CA, Bogomolniy F, Ozbun L, Brady J, Barrett JC,
Boyd J and Birrer MJ: A gene signature predicting for survival in
suboptimally debulked patients with ovarian cancer. Cancer Res.
68:5478–5486. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Vathipadiekal V, Wang V, Wei W, Waldron L,
Drapkin R, Gillette M, Skates S and Birrer M: Creation of a human
secretome: A novel composite library of human secreted proteins:
Validation using ovarian cancer gene expression data and a virtual
secretome array. Clin Cancer Res. 21:4960–4969. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
King ER, Tung CS, Tsang YT, Zu Z, Lok GT,
Deavers MT, Malpica A, Wolf JK, Lu KH, Birrer MJ, et al: The
anterior gradient homolog 3 (AGR3) gene is associated with
differentiation and survival in ovarian cancer. Am J Surg Pathol.
35:904–912. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Ferriss JS, Kim Y, Duska L, Birrer M,
Levine DA, Moskaluk C, Theodorescu D and Lee JK: Multi-gene
expression predictors of single drug responses to adjuvant
chemotherapy in ovarian carcinoma: Predicting platinum resistance.
PLoS One. 7:e305502012. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Bild AH, Yao G, Chang JT, Wang Q, Potti A,
Chasse D, Joshi MB, Harpole D, Lancaster JM, Berchuck A, et al:
Oncogenic pathway signatures in human cancers as a guide to
targeted therapies. Nature. 439:353–357. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Koti M, Gooding RJ, Nuin P, Haslehurst A,
Crane C, Weberpals J, Childs T, Bryson P, Dharsee M, Evans K, et
al: Identification of the IGF1/PI3K/NF κB/ERK gene signalling
networks associated with chemotherapy resistance and treatment
response in high-grade serous epithelial ovarian cancer. BMC
Cancer. 13:5492013. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Lisowska KM, Olbryt M, Dudaladava V,
Pamuła-Piłat J, Kujawa K, Grzybowska E, Jarząb M, Student S,
Rzepecka IK, Jarząb B and Kupryjańczyk J: Gene expression analysis
in ovarian cancer-faults and hints from DNA microarray study. Front
Oncol. 4:62014. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Uehara Y, Oda K, Ikeda Y, Koso T, Tsuji S,
Yamamoto S, Asada K, Sone K, Kurikawa R, Makii C, et al: Integrated
copy number and expression analysis identifies profiles of
whole-arm chromosomal alterations and subgroups with favorable
outcome in ovarian clear cell carcinomas. PLoS One.
10:e01280662015. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Tothill RW, Tinker AV, George J, Brown R,
Fox SB, Lade S, Johnson DS, Trivett MK, Etemadmoghadam D, Locandro
B, et al: Novel molecular subtypes of serous and endometrioid
ovarian cancer linked to clinical outcome. Clin Cancer Res.
14:5198–5208. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Zhao C, Chen X, Zang D, Lan X, Liao S,
Yang C, Zhang P, Wu J, Li X, Liu N, et al: Platinum-containing
compound platinum pyrithione is stronger and safer than cisplatin
in cancer therapy. Biochem Pharmacol. 116:22–38. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Goldberg GS, Bechberger JF and Naus CC: A
pre-loading method of evaluating gap junctional communication by
fluorescent dye transfer. Biotechniques. 3:490–497. 1995.
|
|
49
|
Koreen IV, Elsayed WA, Liu YJ and Harris
AL: Tetracycline-regulated expression enables purification and
functional analysis of recombinant connexin channels from mammalian
cells. Biochem J. 383:111–119. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Didier R, Mallavialle A, Ben Jouira R,
Domdom MA, Tichet M, Auberger P, Luciano F, Ohanna M,
Tartare-Deckert S and Deckert M: Targeting the
proteasome-associated deubiquitinating enzyme USP14 impairs
melanoma cell survival and overcomes resistance to MAPK-targeting
therapies. Mol Cancer Ther. 17:1416–1429. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Jamieson S, Going JJ, D'Arcy R and George
WD: Expression of gap junction proteins connexin 26 and connexin 43
in normal human breast and in breast tumours. J Pathol. 184:37–43.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Govindarajan R, Zhao S, Song XH, Guo RJ,
Wheelock M, Johnson KR and Mehta PP: Impaired Trafficking of
connexins in androgen-independent human prostate cancer cell lines
and its mitigation by alpha-catenin. J Biol Chem. 277:50087–50097.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Toler CR, Taylor DD and Gercel-Taylor C:
Loss of communication in ovarian cancer. Am J Obstet Gynecol.
194:e27–e31. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Ribeiro-Rodrigues TM, Catarino S, Marques
C, Ferreira JV, Martins-Marques T, Pereira P and Girão H:
AMSH-mediated deubiquitination of Cx43 regulates internalization
and degradation of gap junctions. FASEB J. 28:4629–4641. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Girao H, Catarino S and Pereira P: Eps15
interacts with ubiquitinated Cx43 and mediates its internalization.
Exp Cell Res. 315:3587–3597. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Auth T, Schluter S, Urschel S, Kussmann P,
Sonntag S, Höher T, Kreuzberg MM, Dobrowolski R and Willecke K: The
TSG101 protein binds to connexins and is involved in connexin
degradation. Exp Cell Res. 315:1053–1062. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Alaei SR, Abrams CK, Bulinski JC,
Hertzberg EL and Freidin MM: Acetylation of C-terminal lysines
modulates protein turnover and stability of Connexin-32. BMC Cell
Biol. 19:222018. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Xiao D, Chen S, Shao Q, Chen J, Bijian K,
Laird DW and Alaoui-Jamali MA: Dynamin 2 interacts with connexin 26
to regulate its degradation and function in gap junction formation.
Int J Biochem Cell Biol. 55:288–297. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Jung H, Kim BG, Han WH, Lee JH, Cho JY,
Park WS, Maurice MM, Han JK, Lee MJ, Finley D and Jho EH:
Deubiquitination of Dishevelled by Usp14 is required for Wnt
signaling. Oncogenesis. 2:e642013. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Meng Q, Cai C, Sun T, Wang Q, Xie W, Wang
R and Cui J: Reversible ubiquitination shapes NLRC5 function and
modulates NF-κB activation switch. J Cell Biol. 211:1025–1040.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Xu D, Shan B, Sun H, Xiao J, Zhu K, Xie X,
Li X, Liang W, Lu X, Qian L and Yuan J: USP14 regulates autophagy
by suppressing K63 ubiquitination of Beclin 1. Genes Dev.
30:1718–1730. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Sharma A, Alswillah T, Singh K, Chatterjee
P, Willard B, Venere M, Summers MK and Almasan A: USP14 regulates
DNA damage repair by targeting RNF168-dependent ubiquitination.
Autophagy. 14:1976–1990. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Walters BJ, Hallengren JJ, Theile CS,
Ploegh HL, Wilson SM and Dobrunz LE: A catalytic independent
function of the deubiquitinating enzyme USP14 regulates hippocampal
synaptic short-term plasticity and vesicle number. J Physiol.
592:571–586. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Jarome TJ, Kwapis JL, Hallengren JJ,
Wilson SM and Helmstetter FJ: The ubiquitin-specific protease 14
(USP14) is a critical regulator of long-term memory formation.
Learn Mem. 21:9–13. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Lee BH, Lu Y, Prado MA, Shi Y, Tian G, Sun
S, Elsasser S, Gygi SP, King RW and Finley D: USP14 deubiquitinates
proteasome-bound substrates that are ubiquitinated at multiple
sites. Nature. 532:398–401. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Liu B, Jiang S, Li M, Xiong X, Zhu M, Li
D, Zhao L, Qian L, Zhai L, Li J, et al: Proteome-wide analysis of
USP14 substrates revealed its role in hepatosteatosis via
stabilization of FASN. Nat Commun. 9:47702018. View Article : Google Scholar : PubMed/NCBI
|