Introduction
Renal cell carcinoma (RCC) is a common cancer that
accounts for 2–3% of all cancers worldwide, and clear cell renal
cell carcinoma (ccRCC) is the most common histological subtype.
Notably, ccRCC accounts for ~75% of all RCCs and is characterized
by a loss of chromosome 3p in over 90% of all cases (1). After surgical resection, local
recurrence or metastases occurs in ~20–50% of patients diagnosed
with localized RCC tumors, and the 5-year survival rate is <10%
(2). Currently, the molecular
mechanisms underlying the progression of RCC are largely unknown.
Thus, studies designed to elucidate the mechanisms of RCC and
identify novel biomarkers are important to improve the clinical
outcomes of RCC.
MicroRNAs (miRNAs) are a class of endogenous small
non-coding RNAs that post-transcriptionally repress the expression
of target genes by inhibiting the translation or promoting the
degradation of mRNAs (3).
Dysregulation of miRNAs has been shown to play vital roles in the
tumorigenesis and development of various types of cancer, including
ccRCC (4). Screening of the miRNA
expression profile is one of the advanced strategies used to study
tumor-associated molecules. Notably, miRNAs are highly conserved
among different species and regulate various biological functions
in an epigenetic manner (5).
Importantly, miRNAs function as oncogenes or tumor-suppressor genes
in various types of cancer and play significant roles in tumor
development, suppression, metastasis and sensitivity or resistance
to chemotherapy (6). Therefore,
investigation of the biological functions of dysregulated miRNAs in
RCC may contribute to the identification of novel biomarkers and
therapeutic strategies for patients with RCC.
Recently, miR-205-5p has been reported to be
involved in the tumorigenesis of various types of cancer, such as
colon, prostate and hepatocellular carcinomas (7–10). For
example, miR-205-5p has been reported to inhibit zinc finger E-box
binding homeobox 1 (ZEB1), a transcription factor, in prostate
cancer (10). Additionally,
miR-205-5p was found to downregulate PTEN expression and thereby
contribute to cisplatin resistance in ovarian cancer cells
(11). However, the expression and
function of miR-205-5p in RCC remain elusive. In the present study,
miR-205-5p was found to be downregulated in RCC tissues and cell
lines compared to corresponding normal tissues and cells. According
to the results of the Kaplan-Meier analysis, low levels of
miR-205-5p predicted a poor prognosis for patients with RCC.
Overexpression of miR-205-5p inhibited the proliferation,
migration, invasion and EMT of RCC cells. Bioinformatic analyses
revealed that vascular endothelial growth factor A (VEGFA) is a
direct downstream target of miR-205-5p. The expression of VEGFA in
RCC tissues was negatively correlated with the miR-205-5p level.
Furthermore, the PI3K/Akt/mTOR signaling pathway was inhibited in
cells overexpressing miR-205-5p. In vivo experiments also
confirmed that miR-205-5p inhibited the growth of xenograft tumors
in mice. Based on our findings, miR-205-5p suppresses the
tumorigenicity of RCC cells by targeting VEGFA and suppressing the
PI3K/Akt/mTOR signaling pathway.
Materials and methods
Tissue collection
Twenty-five pairs of human RCC and adjacent normal
tissues were surgically collected from patients at Ningbo Urology
and Nephrology Hospital from March, 2015 to December 2017. Among
the 25 enrolled patients, 12 were male, 13 were female and the mean
age was 62.4±5.5 years. Before surgery, none of the patients
received any chemotherapy or radiotherapy. The clinicopathological
features were recorded based on the American Joint Committee on
Cancer (AJCC) standards (12). All
patients provided written informed consent and the study was
approved by the Ethics Committee of Ningbo Urology and Nephrology
Hospital (Ningbo, China).
Cell culture and cell viability
assay
Cells (293) were purchased from the Shanghai
Institute for Biological Sciences (Shanghai, China). Human RCC
cells (786-O, ACHN and Caki-1) were obtained from the American Type
Culture Collection (ATCC; Manassas, VA, USA). 786-O and Caki-1
cells were maintained in RPMI-1640 medium (Gibco; Thermo Fisher
Scientific, Inc., Waltham, MA, USA). ACHN and 293 cells were
maintained in Dulbecco's modified Eagle's medium (DMEM; Gibco;
Thermo Fisher Scientific, Inc.). Media were supplemented with 10%
fetal bovine serum (FBS; Gibco Thermo Fisher Scientific, Inc.), 1%
antibiotics (100 µl/ml penicillin and 100 mg/ml streptomycin
sulfate; Gibco; Thermo Fisher Scientific, Inc.) and 1% glutamine
(Gibco; Thermo Fisher Scientific, Inc.). Cells were cultured in a
humidified incubator containing 5% CO2 at a temperature
of 37°C. The Cell Counting Kit-8 (CCK-8) assay was performed to
assess cell viability. Briefly, cells (5,000 cells/well) were
seeded in a 96-well plate. Twenty-four hours after transfection, 10
µl of CCK-8 solution (Beyotime Institute of Biotechnology,
Shanghai, China) was added, and 1 h later, the optical density (OD)
value of each well was measured with an ELISA microplate reader
(Bio-Rad Laboratories Inc., Hercules, CA, USA) at a wavelength of
595 nm. Wells without cells were used as blanks. The experiments
were performed in triplicate and repeated at least three times.
RNA purification and RT-PCR
Total RNA was extracted from the tissue samples and
cells using TRIzol (Invitrogen; Thermo Fisher Scientific, Inc.),
and purified with the RNeasy Maxi kit (Qiagen, Inc., Santa Clarita,
CA, USA) according to the manufacturer's guidelines. RNA
concentrations were measured using a NanoDrop 2000/2000c
spectrophotometer (Thermo Fisher Scientific, Inc.). Reverse
transcription to prepare cDNA templates was performed using a
TaqMan MicroRNA Reverse Transcription kit (Applied Biosystems;
Thermo Fisher Scientific, Inc.). Then, qPCR was performed with a
miScript SYBR-Green PCR kit (Qiagen) and the LightCycler 480
Real-Time PCR system (Roche Diagnostics, Basel, Switzerland). GAPDH
and U6 were used as internal controls for VEGFA and miR-205-5p,
respectively. The following thermocycling conditions were used:
95°C for 1 min, then 40 cycles of 95°C for 15 sec, 55°C for 30 sec
and 70°C for 30 sec. The expression levels in tissues and cells
were calculated using the 2−ΔΔCq method (13).
Cell transfection
Synthesized miR-205-5p mimics
(5′-UCCUUCAUUCCACCGGAGUCUG-3′) or the negative control (miR-NC)
(5′-UCACAACCUCCUAGAAAGAGUAGA-3′) were purchased from Suzhou
GenePharma Co., Ltd. (Suzhou, China). The myr-Akt vector and empty
vector were generous gifts from Dr Rui Yu (Ningbo University,
Ningbo, China). Cells (2×105) were transfected with 20
µM miRNA mimics or 2 µg vector plasmid. Twenty-four hours after
transfection, the cells were collected and assayed. Transfection
was performed using Lipofectamine 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.), according to the manufacturer's
instructions.
Wound healing and cell invasion
assays
Cells were seeded in a 6-well plate and allowed to
grow to 90% confluence to assess migration in vitro.
Twenty-four hours after transfection, an artificial wound was
created with a 200-µl pipette tip in the center of the confluent
cell monolayer. Then, the cells were cultured for another 24 h and
the closure of the wound in each group was evaluated at the
magnification of ×2,000 under an inverted microscope (Olympus
Corp., Tokyo, Japan). For the invasion assay, 1×105
cells were suspended in 200 µl of serum-free medium and plated in
upper Transwell chambers (Costar; Corning Inc., Corning, NY, USA)
coated with Matrigel (BD Biosciences, Franklin Lakes, NJ, USA). Six
hundred microliters of serum-containing medium were added to the
bottom well. After culture in a humidified atmosphere containing 5%
CO2 at 37°C for 24 h, the cells that had migrated and
adhered to the lower surface of the membrane were fixed with 70%
methanol and stained with 1% crystal violet for 15 min. Finally,
the number of cells that migrated across the membrane were counted.
Migration and invasion of cells was evaluated with an inverted
Olympus phase-contrast microscope (×200 magnification) (Olympus
Corp.). Each experiment was performed in triplicate.
Apoptosis assay
The percentage of apoptotic cells was measured using
Nucleosome ELISA kit (cat. no. 11544675001; Roche Diagnostics),
according to the manufacturer's instructions. Briefly, cells were
seeded into 24-well plates and transfected. Twenty-four hours
later, the cells were collected and lysed. A biotin-labeled mouse
antibody against histone 3 that specifically binds to the histone
component of captured nucleosomes derived from apoptotic cells was
used in this assay. The bound antibody was detected following an
incubation with horseradish peroxidase (HRP)-conjugated
streptavidin at room temperature for 1 h. HRP catalyzes the
conversion of the colorless tetramethylbenzidine (TMB) to produce a
blue color. The addition of stop solution turns the solution
yellow, and the intensity was proportional to the number of
nucleosomes in the sample. Each experiment was performed in
triplicate.
Western blot assay
Cells were lysed in RIPA buffer (Beyotime Institute
of Biotechnology). The protein concentration was determined by the
Bradford assay kit according to the manufacturer's guidelines
(Beyotime Institute of Biotechnology). Lysates were centrifuged at
12,000 × g 5 min at 4°C and stored at −70°C. Equal amounts (20 µg)
of protein lysates were separated on 10% SDS-PAGE gels and
electrophoretically transferred onto polyvinylidene difluoride
(PVDF) membranes (Millipore, Minneapolis, MN, USA). PVDF membranes
were blocked with 10% skim milk powder dissolved in TBST buffer at
room temperature for 1 h. Then, PVDF membranes were stripped and
washed with TBST, and then incubated with a primary antibody
(dilution of 1:1,000) in TBST containing 5% bovine serum albumin
(BSA) overnight at 4°C. Primary antibodies against the following
proteins were used: MMP-7 (cat. no. ab207299), MMP-9 (cat. no.
ab73734), Snail (cat. no. ab53519) (all from Abcam, Cambridge, MA,
USA), N-cadherin (cat. no. 4061), caspase-3 (cat. no. 9662), Bcl-2
(cat. no. 3498), Bcl-xL (cat. no. 2762), p-PI3K (cat. no. 4228),
t-PI3K (cat. no. 4225), p-Akt (cat. no. 4058), t-Akt (cat. no.
4691), p-mTOR (cat. no. 5536), t-mTOR (cat. no. 2983) (Cell
Signaling Technology, Danvers, USA) and GAPDH (Sigma-Aldrich; Merck
KGaA, Darmstadt, Germany). Next, membranes were washed with TBST
three times and incubated with an HRP-conjugated anti-rabbit (cat.
no. RABHRP1) or anti-mouse secondary antibody (cat. no. RABHRP2)
(Sigma-Aldrich; Merck KGaA) for 1 h at room temperature. Signals
were visualized using an ECL reagent (Pierce; Thermo Fisher
Scientific, Inc.). All western blots were replicated in three times
and the intensity of the western blot signals was analyzed using
ImageJ software version 1.48 (NIH; National Institutes of Health,
Bethesda, MD, USA).
Luciferase reporter assay
The potential miR-205-5p binding sites in the VEGFA
3′-UTR were predicted using TargetScan 7.1 software (www.targetscan.org/). Sequences containing the
wild-type (VEGFA-wt) or mutant (VEGFA-mut) seed region of VEGFA
were synthesized and cloned into a luciferase reporter plasmid
(pMIR-REPORT) (Promega Corporation, Madison, WI, USA). The VEGFA-wt
or VEGFA-mut plasmids and miR-205-5p or miR-NC were co-transfected
using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific,
Inc.), according to the manufacturer's instructions. Twenty-four
hours after transfection, firefly and Renilla luciferase
activities were measured using the Dual-Luciferase assay (Promega
Corporation) according to the manufacturer's instructions. All
luciferase assays were performed in triplicate.
In vivo experiments
The animal study was approved by the Ethics
Committee on Animal Research of Ningbo Urology and Nephrology
Hospital (Ningbo, China). Male nude mice (BALB/c, 4–5 weeks old)
were purchased from the Shanghai Laboratory Animal Center, Shanghai
Institute for Life Sciences, Chinese Academy of Sciences (Shanghai,
China). The total number of mice were 50 and the weight of the mice
was 20±5 g. The mice were housed in a facility at 23–24°C, and the
light-dark cycle was set at 12-h intervals. The
miR-205-5p-overexpressing 786-O cells or miR-NC-overexpressing
786-O cells were prepared by transfecting the cells with a
recombinant lentivirus carrying the miR-205-5p precursor sequence
or a scrambled control, respectively (Shanghai GenePharma Co.,
Ltd., Shanghai, China). Cells were suspended in 100 µl of
phosphate-buffered saline (PBS) and subcutaneously injected into
the flank of a nude mouse. The width and length of the tumor
nodules were measured every three days. The volume of subcutaneous
tumors was calculated as tumor volume = length width × width/2. At
31 days post-injection, the mice were removed from their cages and
gently restrained while resting on the benchtop. Cervical
dislocation was performed manually and resulted in euthanasia
within ~10 sec and the tumors were excised for use in subsequent
experiments.
Statistical analysis
All statistical analyses were conducted using the
SPSS 20.0 software (IBM Corp., Armonk, NY, USA). All measurement
data are presented as means ± standard deviations (SDs). Univariate
Cox proportional hazards regression analyses were performed to
identify any significant variables predicting survival status.
Multivariate Cox proportional hazard analyses were performed to
assess the independent predictors of survival. One-way analysis of
variance (ANOVA) with post hoc Tukey's test was chosen to compare
data between multiple groups and paired Student's t-test was used
to compare data between two groups. P<0.05 was considered
indicative of a significant difference.
Results
Expression of miR-205-5p is
downregulated in RCC and correlates with the outcomes of patients
with RCC
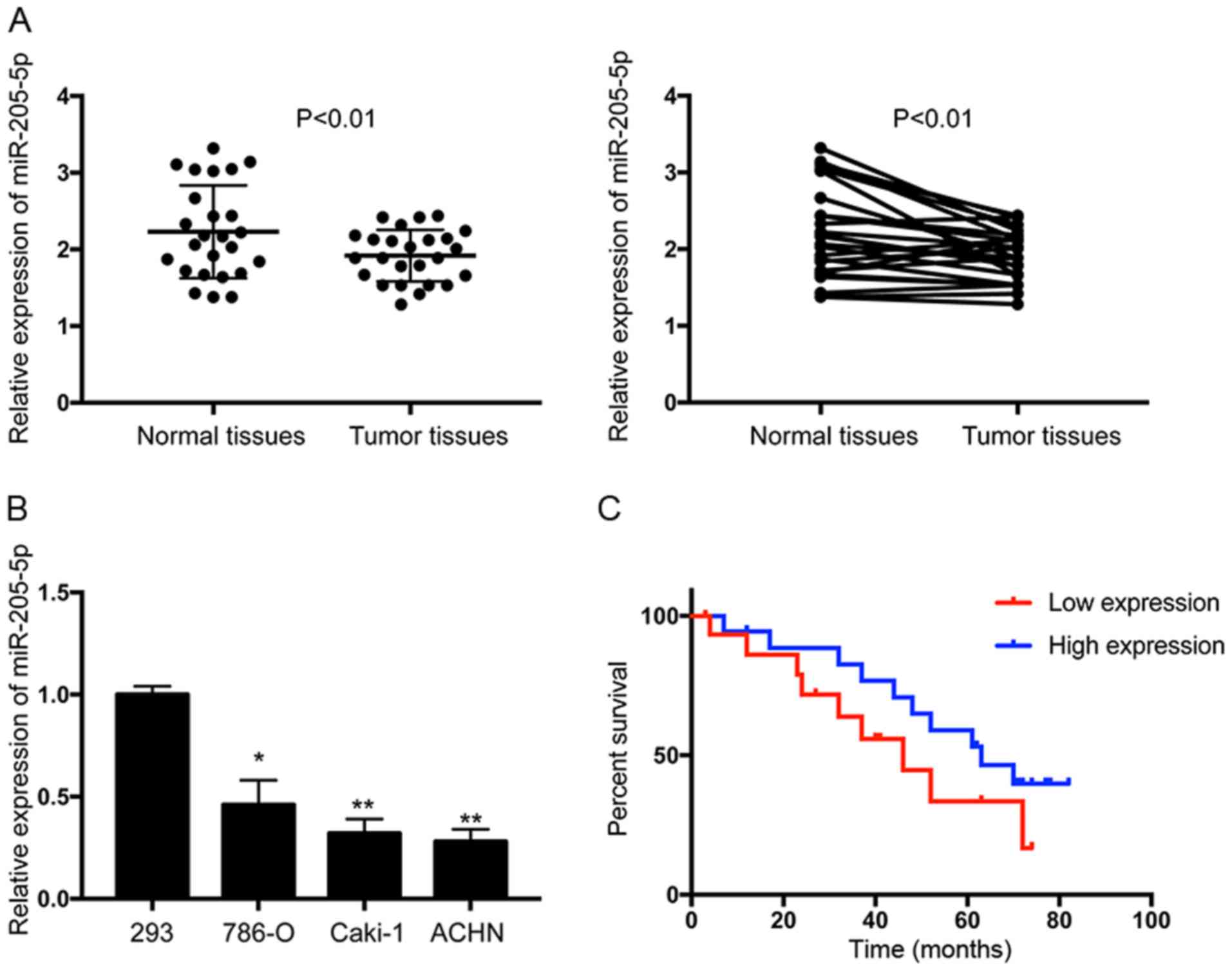
We examined the endogenous expression of miR-205-5p
in RCC tissues (n=25) and adjacent normal kidney tissues (n=25) to
determine whether miR-205-5p is dysregulated in RCC. Significantly
lower expression of miR-205-5p was detected in RCC tumor tissues
than that observed in the normal tissues (P<0.01; Fig. 1A). In addition, we measured the
expression of miR-205-5p in the cell line 293 and RCC cell lines
786-O, Caki-1 and ACHN. Significantly lower expression of
miR-205-5p was observed in RCC cell lines than in the 293 cells
(Fig. 1B). Then, we divided the 25
samples into a high miR-205-5p expression group (n=13) and low
miR-205-5p expression group (n=12) according the median expression
to further evaluate the relationship between miR-205-5p expression
and the clinicopathological features of RCC. As shown in Table I, a lower miR-205-5p level was
associated with an advanced tumor stage, Fuhrman stage and more
lymph node metastasis, but was not associated with age, sex or
tumor size. In addition, the Kaplan-Meier analysis revealed that
downregulation of miR-205-5p was associated with a poor prognosis,
indicating that miR-205-5p may serve as a promising candidate
prognostic biomarker for patients with RCC (mean follow-up time of
42 months) (Fig. 1C). A univariate
analysis using the Cox proportional hazard regression model
revealed statistically significant correlations between the overall
survival of patients with miR-205-5p expression (P=0.017) and tumor
stage (P=0.023) (Table II). A
multivariate analysis including miR-205-5p expression and the tumor
stage showed that miR-205-5p (P=0.024) and tumor stage (P=0.034)
were independent prognostic factors for the overall survival of
patients with RCC (Table II).
 | Table I.Correlation between miR-205-5p
expression and the clinicopathological features of the RCC
patients. |
Table I.
Correlation between miR-205-5p
expression and the clinicopathological features of the RCC
patients.
|
|
| miR-205-5p
levels |
|
|---|
|
|
|
|
|
|---|
| Parameters | Cases | Low | High | P-value |
|---|
| Sex |
|
|
| 0.320 |
|
Male | 13 | 8 | 5 |
|
|
Female | 12 | 5 | 7 |
|
| Age (years) |
|
|
| 0.513 |
|
<60 | 10 | 6 | 4 |
|
|
≥60 | 15 | 7 | 8 |
|
| Tumor size
(cm) |
|
|
| 0.561 |
|
<4 | 11 | 5 | 6 |
|
| ≥4 | 14 | 8 | 6 |
|
| Tumor stage |
|
|
| 0.009 |
|
T1-T2 | 12 | 3 | 9 |
|
|
T3-T4 | 13 | 10 | 3 |
|
| Fuhrman grade |
|
|
| 0.025 |
|
I–II | 9 | 2 | 7 |
|
|
III–IV | 16 | 11 | 5 |
|
| Lymph node
metastasis |
|
|
| 0.008 |
|
Yes | 11 | 9 | 2 |
|
| No | 14 | 4 | 10 |
|
 | Table II.Univariate and multivariate analyses
of the clinicopathological factors for overall survival in the RCC
cases. |
Table II.
Univariate and multivariate analyses
of the clinicopathological factors for overall survival in the RCC
cases.
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|---|
| Variables | HR | 95% CI | P-value | HR | 95% CI | P-value |
|---|
| miR-205-5p
expression (high vs. low) | 0.382 | 0.147–0.683 | 0.017 | 0.398 | 0.215–0.842 | 0.024 |
| Sex (male vs.
female) | 1.732 | 0.422–2.531 | 0.142 |
|
|
|
| Age (<60 vs. ≥60
years) | 0.973 | 0.684–2.023 | 0.783 |
|
|
|
| Tumor size (<4
vs. ≥4 cm) | 2.054 | 0.563–3.127 | 0.426 |
|
|
|
| Tumor stage (T1-T2
vs. T3-T4) | 2.128 | 1.378–5.241 | 0.023 | 2.351 | 1.942–4.336 | 0.034 |
| Fuhrman grade (I–II
vs. III–IV) | 2.52 | 0.768–3.082 | 0.613 |
|
|
|
| Lymph node
metastasis (yes vs. no) | 3.83 | 0.542–4.117 | 0.532 |
|
|
|
miR-205-5p inhibits the proliferation
and migration of RCC cells
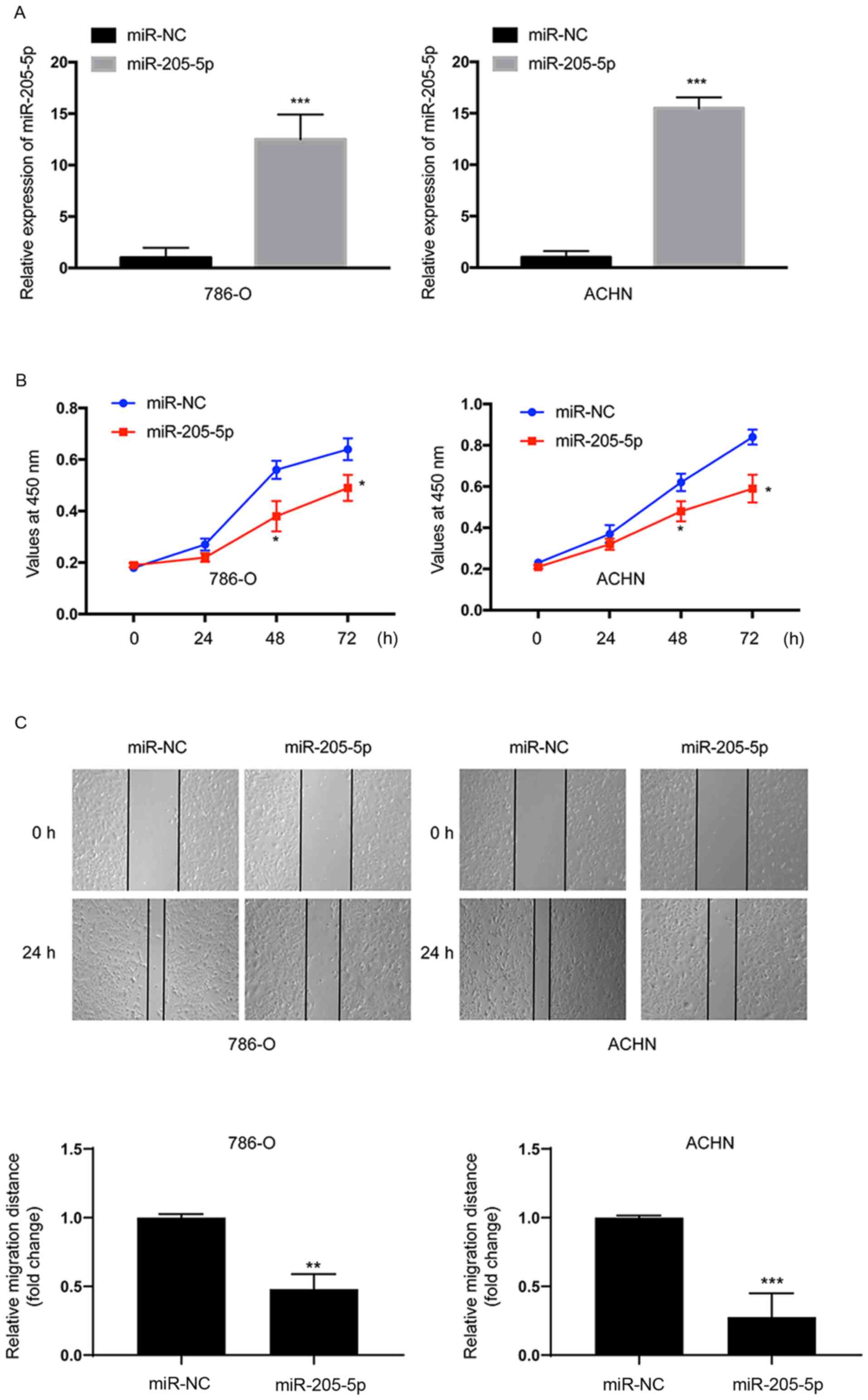
The 786-O and ACHN cells were transfected with the
miR-205-5p mimic or miR-NC to investigate the function of
miR-205-5p in RCC. We performed RT-qPCR to analyze the miR-205-5p
expression in the transfected cells. The expression of miR-205-5p
was significantly upregulated in both 786-O (P<0.001) and ACHN
(P<0.001) cells at 24 h after transfection (Fig. 2A). The CCK-8 assay was employed to
evaluate cell proliferation. Overexpression of miR-205-5p
significantly suppressed the proliferation of both 786-O and ACHN
cells compared with cells transfected with miR-NC (Fig. 2B). Thus, miR-205-5p suppressed the
proliferation of RCC cells. Then, a scratch assay was performed to
assess cell migration. Based on the results of the scratch assay,
the migratory distance was markedly reduced in both cell lines
transfected with the miR-205-5p mimic (Fig. 2C). Thus, miR-205-5p suppresses the
migration of RCC cells.
Overexpression of miR-205-5p inhibits
invasion and epithelial-mesenchymal transition (EMT) and induces
apoptosis in RCC cells
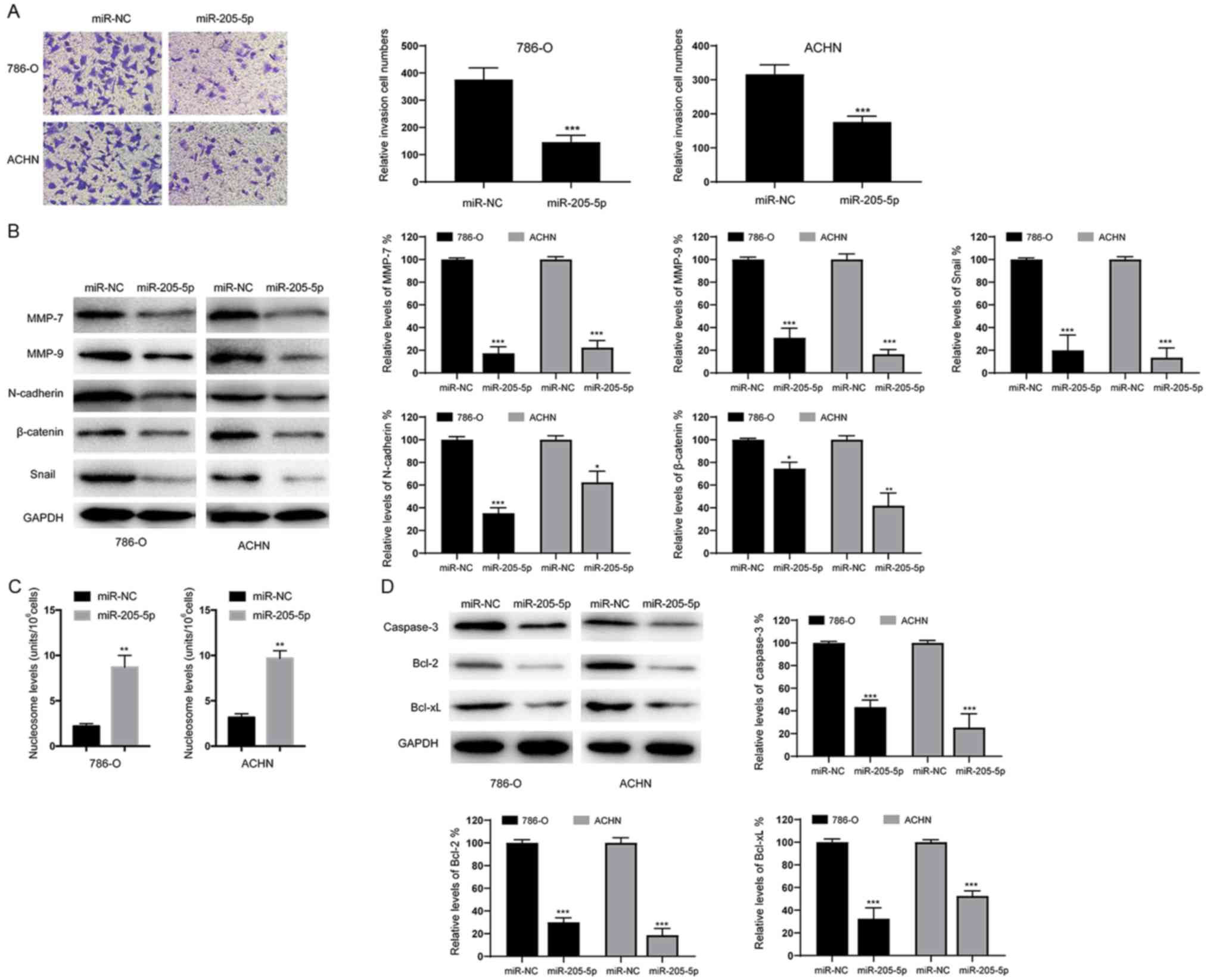
Next, we assessed the effects of miR-205-5p on the
invasion of RCC cells using Transwell assays. As shown in Fig. 3A, overexpression of miR-205-5p
decreased the invasive ability of both 786-O and ACHN cell lines.
Meanwhile, the levels of invasion and migration marker proteins
(MMP-7 and MMP-9) were also obviously decreased in cells
overexpressing miR-205-5p (Fig. 3B).
Since epithelial-mesenchymal transition (EMT) is an indispensable
process for tumor cell invasion and migration (14), we examined whether miR-205-5p affects
EMT in RCC cells. As shown in Fig.
3B, overexpression of miR-205-5p significantly decreased the
protein levels of N-cadherin, β-catenin and Snail. Based on these
data, miR-205-5p may inhibit the migration of RCC cells by
suppressing the EMT. We also analyzed the numbers of apoptotic
cells and found that overexpression of miR-205-5p significantly
increased the percentage of apoptotic 786-O and ACHN cells
(Fig. 3C). Furthermore, the levels of
apoptosis-related proteins were examined using western blotting. As
indicated in Fig. 3D, caspase-3,
Bcl-2 and Bcl-xL levels were reduced in cells overexpressing
miR-205-5p. Therefore, miR-205-5p likely suppresses the
proliferation of RCC cells by inducing apoptosis.
miR-205-5p increases the
chemosensitivity of RCC cells
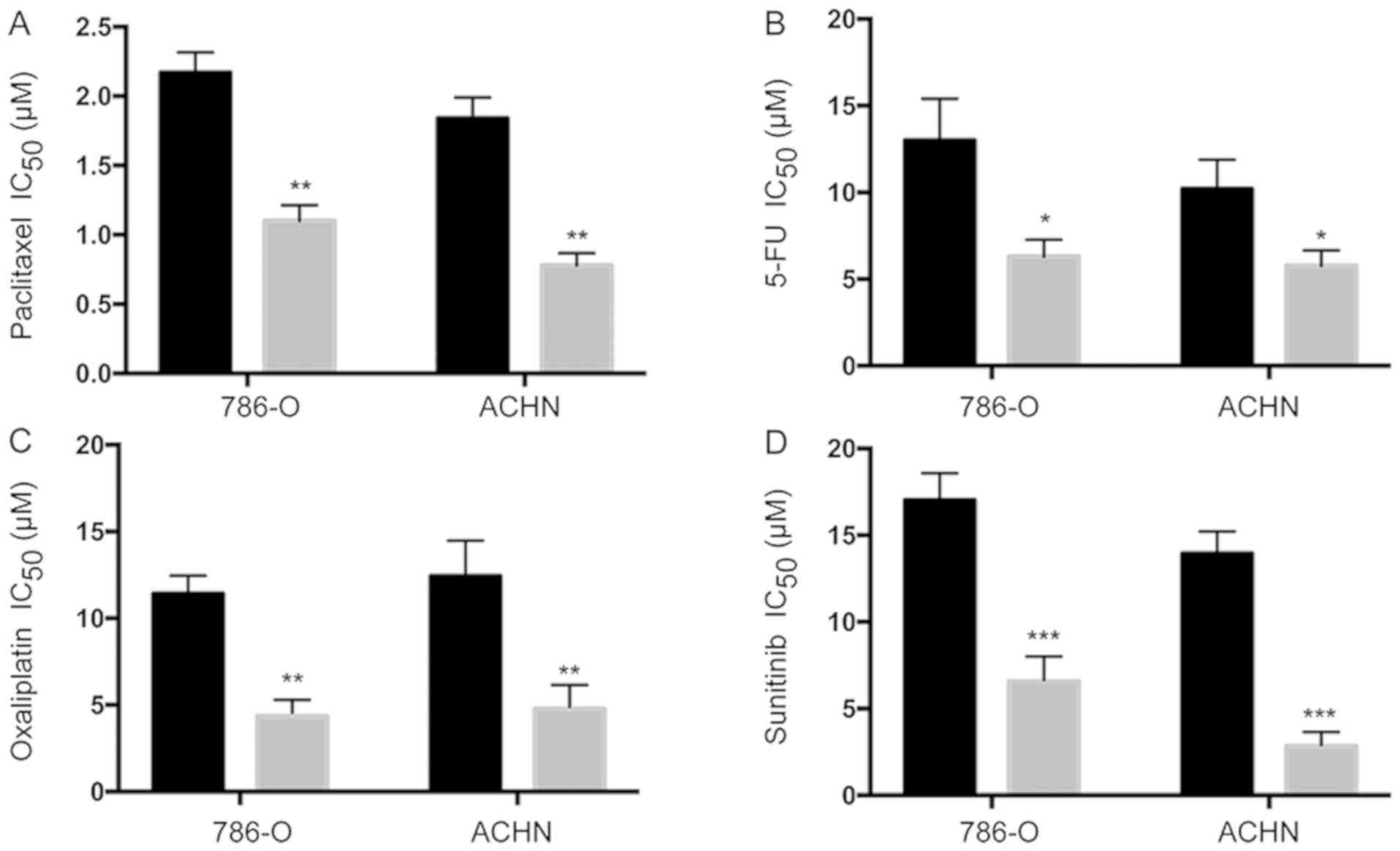
RCC is normally resistant to conventional cytotoxic
chemotherapies. Therefore, we investigated the effect of miR-205-5p
on the responses of RCC cells to various chemotherapeutic agents.
First, we tested paclitaxel, an agent known to inhibit cell
division and thereby induce cell death. Measurements of the half
maximal inhibitory concentration (IC50) showed an
increased sensitivity of 786-O and ACHN cells to paclitaxel
(Fig. 4A). Similar results were also
obtained with 5-FU and oxaliplatin, agents that block DNA
replication (Fig. 4B and C). We also
tested sunitinib, a receptor tyrosine kinase (RTK) inhibitor used
to treat patients with advanced ccRCC (15). Notably, overexpression of miR-205-5p
also increased the sensitivity of 786-O and ACHN to sunitinib
(Fig. 4D). Therefore, miR-205-5p is
also involved in the response of RCC cells to chemotherapy.
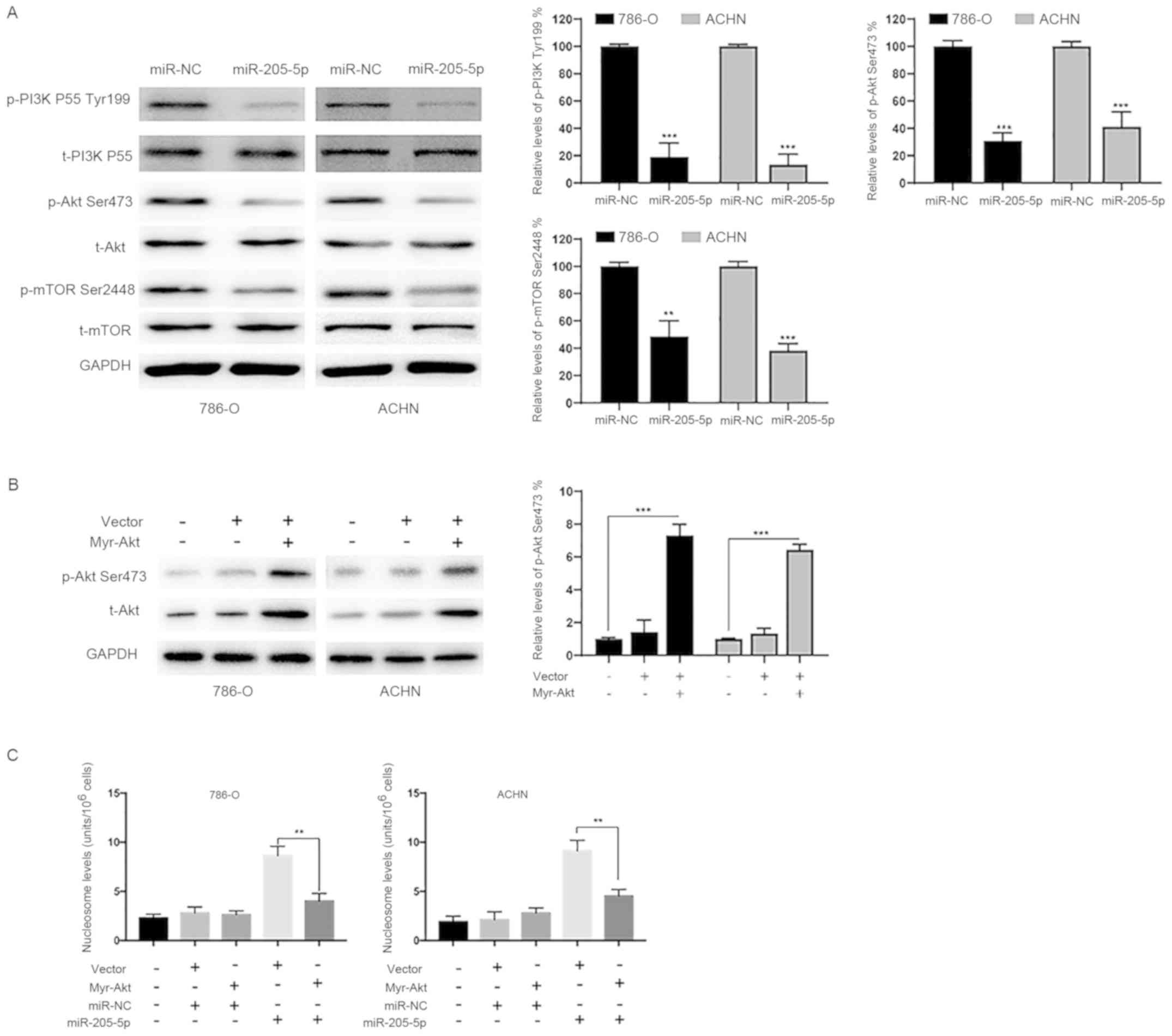
miR-205-5p inactivates the
PI3K/Akt/mTOR signaling pathway in RCC cells
The PI3K/Akt/mTOR signaling pathway exerts
significant effects on the proliferation, invasion and apoptosis of
RCC cells (16). We, therefore,
investigated the effects of miR-205-5p on the PI3K/Akt signaling
pathway. As indicated in Fig. 5A,
overexpression of miR-205-5p markedly decreased p-PI3K, p-Akt and
p-mTOR levels. Constitutively active Akt (myr-Akt) was
overexpressed as described in a previous study to further elucidate
the role of the PI3K/Akt pathway in the antitumor effects of
miR-205-5p (17). Overexpression of
myr-Akt successfully mimicked the activation of Akt (Fig. 5B). Myr-Akt also significantly
suppressed miR-205-5p-induced apoptosis (Fig. 5C). Based on these data, the tumor
suppressor function of miR-205-5p in RCC is likely and at least
partially mediated by its repression of the PI3K/Akt/mTOR
pathway.
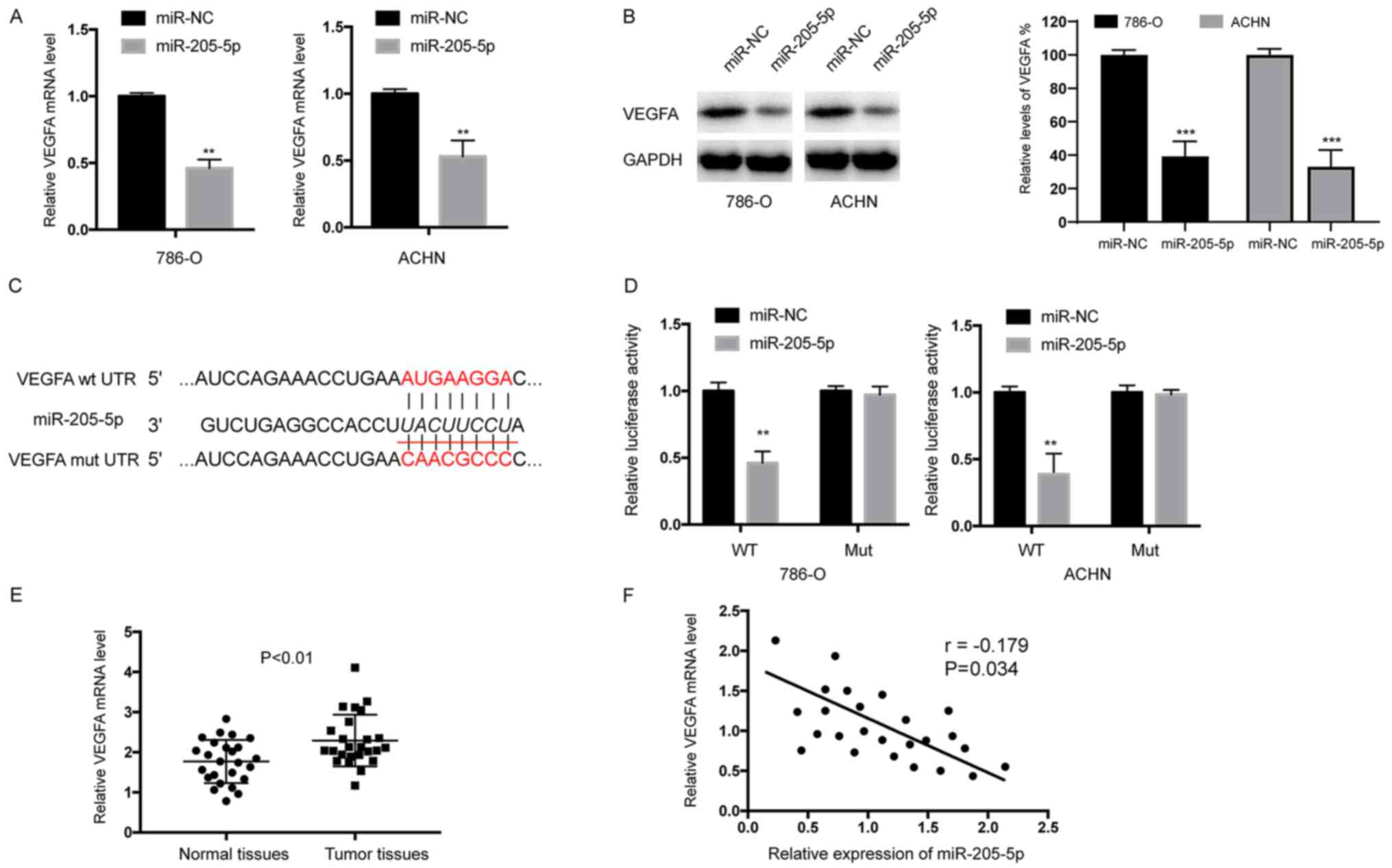
VEGFA is a direct target of
miR-205-5p
According to bioinformatic analysis tools
(TargetScan; www.targetscan.org and miRanda; www.microrna.org), we speculated that VEGFA is a
candidate target of miR-205-5p. As shown in Fig. 6A, the RT-qPCR analysis revealed
significantly decreased levels of the VEGFA mRNA in 786-O and ACHN
cells transfected with miR-205-5p. Western blot analyses also
showed decreased levels of the VEGFA protein in 786-O and ACHN
cells transfected with the miR-205-5p mimic (Fig. 6B). We performed a Dual-Luciferase
assay to further confirm that VEGFA is a target of miR-205-5p. The
wild-type (WT) and mutant (Mut) VEGFA 3-untranslated region
(3′-UTR) constructs were subcloned into the pMIR reporter plasmid.
Overexpression of miR-205-5p decreased luciferase activity in both
786-O and ACHN cells transfected with the wild-type (WT) 3′-UTR of
VEGFA, but not the mutant (Mut) 3′-UTR (Fig. 6D). Levels of the VEGFA mRNA in RCC and
normal adjacent tissues were examined using qRT-PCR to elucidate
the clinical relevance of miR-205-5p-mediated targeting of VEGFA in
RCC. As shown in Fig. 6E, the VEGF
mRNA was expressed at much higher levels in RCC tumor tissues than
that noted in normal tissues. Furthermore, an inverse correlation
was observed between the expression of miR-205-5p and VEGFA
expression in RCC tissues (r=−0.179) (Fig. 6F). These results further confirmed
that VEGFA expression is negatively regulated by miR-205-5p in RCC.
In summary, VEGFA is a target of miR-205-5p in RCC.
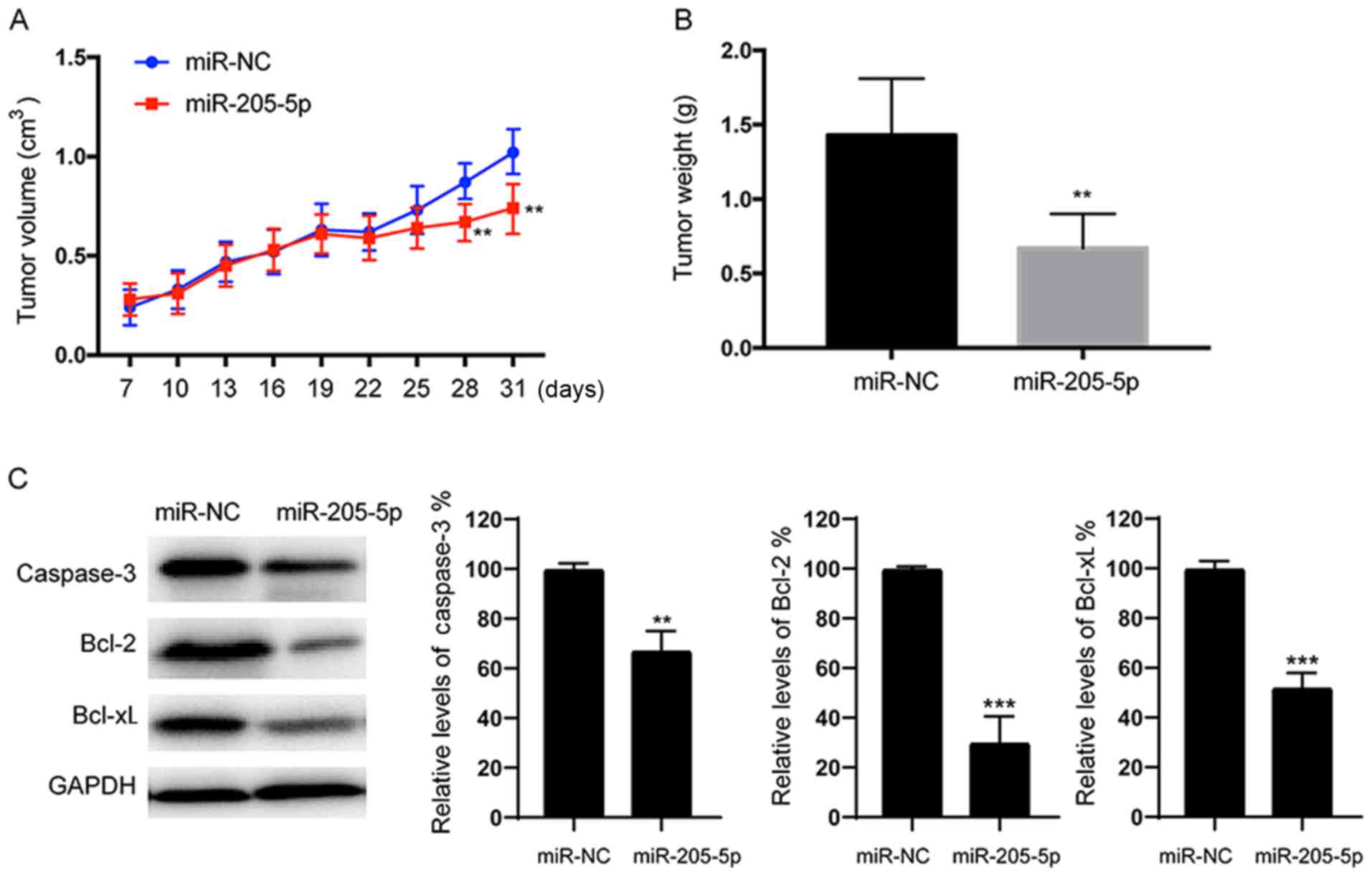
miR-205-5p suppresses RCC cell
tumorigenesis in vivo
We stably expressed miR-NC or miR-205-5p in 786-O
cells and then subsequently implanted these cells into both
posterior flanks of nude mice to investigate whether overexpression
of miR-205-5p attenuates the progression of RCC in vivo.
Tumor sizes were measured after 7 days. From the 22nd to the 31st
day, the miR-NC group developed significantly larger tumors than
the miR-205-5p group (Fig. 7A).
Meanwhile, the final tumor weight of the miR-NC group was much
greater than the miR-205-5p group (Fig.
7B). Consistent with the results from the in vitro
studies, lower levels of the caspase-3, Bcl-2 and Bcl-xL proteins
were detected in the tumor tissues from the miR-205-5p-expressing
group than in the miR-NC-expressing group (Fig. 7C). Based on these data, miR-205-5p
also inhibits tumor growth in vivo.
Discussion
In the present study, we identified a novel
molecular mechanism by which miR-205-5p modulates renal cell
carcinoma (RCC) through the suppression of VEGFA expression and the
PI3K/Akt/mTOR signaling pathway. Based on accumulating evidence,
microRNAs (miRNAs) play vital roles in tumorigenesis, and the
expression of various miRNAs correlates with clinical
characteristics and outcomes. Numerous studies have investigated
the biological roles of miRNAs in RCC. For example, miR-30a-5p was
found to suppress the proliferation and promote the apoptosis of
RCC cells by targeting GRP78 (18).
Moreover, miR-720 targets the E-cadherin-α/E-catenin complex and
plays a clinically significant role in RCC (19). In addition, miR-30b-5p inhibits the
proliferation, metastasis and epithelial to mesenchymal transition
(EMT) of RCC cells by repressing G-protein subunit α-13 (20). Thus, miRNAs play important roles in
RCC progression by regulating various biological activities of
cells.
In the present study, we confirmed significantly
lower miR-205-5p expression in RCC tissues and cell lines than
these levels in their normal counterparts. Notably, miR-205-5p has
been shown to be downregulated and function as a tumor suppressor
in various types of cancer. For example, miR-205-5p expression was
found to be significantly decreased in prostate cancer tissues and
to inhibit cancer cell aggressiveness by targeting HMGB3 (9). In a recent study, miR-205-5p
significantly suppressed the migration and invasion of oral
squamous carcinoma cells by inhibiting TIMP-2 expression (21). However, miR-205-5p expression is
significantly increased in non-small cell lung cancer tissues, and
it functions as an oncogene by downregulating erbB3 expression
(22). Notably, overexpression of
miR-205-5p increased the chemosensitivity of RCC cells to various
agents, such as paclitaxel, 5-FU, oxaliplatin and sunitinib, in the
present study. Meanwhile, overexpression of miR-205-5p was found to
mediate the resistance of hepatocellular carcinoma cells to 5-FU
(8). These discrepancies may be due
to the differences in cancer types. Since miRNAs regulate the
expression of different target genes depending on specific cellular
and disease context (3),
investigation of the functions of miR-205-5p in additional cancer
types is warranted.
Overexpression of miR-205-5p inhibited the
proliferation, migration and invasion of RCC cells. Moreover,
overexpression of miR-205-5p induced apoptosis, accompanied by the
downregulation of caspase-3, Bcl-2 and Bcl-xL. Two different
pathways lead to the apoptosis, namely, the extrinsic and intrinsic
pathways (23). The intrinsic pathway
is strictly regulated by Bcl-2 family members (24). Our data are consistent with a previous
study showing that miR-205-5p modulated the levels of Bcl-2
proteins in melanoma (25). Thus,
miR-205-5p may induce tumor cell apoptosis via the intrinsic
pathway.
Multiple signaling pathways participate in
modulating tumor progression, including the EMT signaling pathway
(26). EMT plays a vital role in the
metastasis of tumor cells. During the EMT process, the expression
of E-cadherin, tight junction proteins and other epithelial markers
are decreased in tumor cells, causing them to lose their epithelial
characteristics (27). Meanwhile, the
expression of mesenchymal markers, such as vimentin and N-cadherin,
are upregulated during EMT, leading to an increase in migratory and
invasive behaviors (28). Based on
accumulating evidence, miRNAs are key modulators of EMT in many
types of cancer. Recently, overexpression of miR-205-5p was shown
to inhibit EMT in colon cancer (7).
According to Elgamal et al, overexpression of miR-205
decreases the protein levels of the mesenchymal markers N-cadherin,
vimentin and ZEB1 in breast cancer cells (29). These findings are consistent with our
findings. In the present study, miR-205-5p decreased the levels of
mesenchymal marker proteins, such as N-cadherin, Snail and
β-catenin. Therefore, miR-205-5p may suppress the invasion and
migration of RCC cells by inhibiting EMT.
The PI3K/Akt/mTOR signaling pathway plays important
roles in various significant biological processes, such as
proliferation, development and apoptosis (30). Activity of the PI3K/Akt signaling
pathway is relatively high in ccRCC among all cancer types, as
indicated by increased levels of phosphorylated Akt and Akt
substrates (31). Therefore,
treatments targeting the PI3K/Akt/mTOR pathway represent a
promising strategy to inhibit ccRCC (32). Recently, miRNAs have emerged as a new
class of important regulators of the PI3K/Akt pathway. For example,
miR-182-5p has been identified as a negative regulator of AKT, and
downregulation of miR-182-5p results in AKT activation and
subsequent RCC proliferation (33).
In contrast, miR-122 is a positive regulator of the PI3K/Akt
signaling pathway, promoting the proliferation, invasion and
migration of RCC cells (34). In the
present study, we investigated the effects of miR-205-5p on the
PI3K/Akt/mTOR pathway and observed decreased phosphorylation of
proteins involved in the PI3K/Akt/mTOR pathway. Notably, forced
expression of constitutively activated Akt decreased
miR-205-5p-induced apoptosis, suggesting that miR-205-5p at least
partially exerts its antitumor effects by inhibiting the
PI3K/Akt/mTOR signaling pathway. The inhibition of the
PI3K/Akt/mTOR pathway was likely mediated by several possible
mechanisms. For example, phosphatase and tensin homologue (PTEN)
dephosphorylates phosphatidylinositol (3,4,5)-triphosphate (PIP3) and therefore inhibits
the activation of the PI3K/Akt/mTOR signaling pathway. In the
present study, VEGFA expression was downregulated by miR-205-5p.
VEGFA activates the PI3K/Akt pathway (35,36). Thus,
we hypothesized that one mechanism by which miR-205-5p inactivated
the PI3K/Akt pathway was through inhibition of VEGFA
expression.
We applied bioinformatics tools to identify the
potential target genes of miR-205-5p and clarify the mechanisms by
which miR-205-5p inhibited tumor growth. Among the putative
targets, we focused on VEGFA, since accumulating evidence indicates
that VEGFA is associated with the proliferation and metastasis of
RCC (37). VEGFA plays an important
role in the response to angiogenesis during tumorigenesis (38). Consistent with a previous study, the
study also validated VEGFA as a target of miR-205-5p using a
luciferase reporter assay (39).
Overexpression of miR-205-5p inhibited the expression of both the
VEGFA mRNA and protein in 786-O and ACHN cells. Moreover, VEGFA
expression was found to be inversely correlated with miR-205-5p
expression in RCC tissues. Based on these data, VEGFA is a direct
target gene of miR-205-5p in RCC. To note, we did not investigate
the effects of VEGFA on RCC cells in our study, as previous studies
have already investigated the functions of VEGFA which could exert
antitumor effects against the RCC (40,41).
According to previous studies, VEGFA has been described as an
important determinant of the increase in the tumorigenicity of
cells that undergo EMT in solid cancers (42,43).
Moreover, VEGFA has been demonstrated to increase the
tumor-initiating stem cell population, to induced EMT (44). Furthermore, inhibition of VEGFA could
lead to a decrease in EMT markers in cancer stem cells (45). Based on these findings, we
hypothesized that VEGFA plays an essential role in the EMT process.
Thus, the change in expression of EMT-related proteins in this
study may be due to the inhibition of VEGFA by miR-205a and we will
test this in future research.
In summary, miR-205-5p expression was downregulated
in RCC tissues and cells. Ectopic expression of miR-205-5p
inhibited the proliferation, migration and invasion of RCC cells
and promoted apoptosis. Overexpression of miR-205-5p repressed the
PI3K/Akt/mTOR pathway. Moreover, miR-205-5p inhibited RCC growth
in vivo in a mouse xenograft model. In addition, we
identified VEGFA as a target of miR-205-5p in RCC. To note, other
possible targets of miR-205-5p may exist, and VEGFA could also be
subject to the regulation of other miRNAs, since we found that
overexpression of miR-205-5p could only partially inhibit the
expression of VEGFA. To the best of our knowledge, this study is
the first to explore the role of miR-205-5p in RCC. However, the
regulatory mechanism of miR-205-5p remains elusive. Further studies
of miR-205-5p in RCC are required to elucidate the complicated
mechanisms of tumorigenesis.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used in the present study are available
from the corresponding author upon reasonable request.
Authors' contributions
JH performed most of the experiments and drafted the
manuscript; XW performed some of the experiments; GW analyzed the
data; YR designed the study and reviewed the manuscript. All
authors read and approved the manuscript and agree to be
accountable for all aspects of the research in ensuring that
accuracy or integrity of any parts of the work are appropriately
investigated and resolved.
Ethics approval and consent to
participate
The clinicopathological features were recorded based
on the American Joint Committee on Cancer (AJCC) standards. All
patients provided written informed consent and the study was
approved by the Ethics Committee of Ningbo Urology and Nephrology
Hospital (Ningbo, China). The animal study was approved by the
Ethics Committee on Animal Research of Ningbo Urology and
Nephrology Hospital (Ningbo, China).
Patient consent for publication
Not applicable.
Competing interests
The authors state that they have no competing
interests.
References
|
1
|
van den Berg E and Dijkhuizen T:
Classification of renal cell cancer based on (cyto)genetic
analysis. Contrib Nephrol. 128:51–61. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Capitanio U and Montorsi F: Renal cancer.
Lancet. 387:894–906. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zimmerman AL and Wu S: MicroRNAs, cancer
and cancer stem cells. Cancer Lett. 300:10–19. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Tutar Y: miRNA and cancer; Computational
and experimental approaches. Curr Pharm Biotechnol. 15:4292014.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Gulei D, Magdo L, Jurj A, Raduly L,
Cojocneanu-Petric R, Moldovan A, Moldovan C, Florea A, Pasca S, Pop
LA, et al: The silent healer: miR-205-5p up-regulation inhibits
epithelial to mesenchymal transition in colon cancer cells by
indirectly up-regulating E-cadherin expression. Cell Death Dis.
9:662018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Shao P, Qu WK, Wang CY, Tian Y, Ye ML, Sun
DG, Sui JD, Wang LM, Fan R and Gao ZM: MicroRNA-205-5p regulates
the chemotherapeutic resistance of hepatocellular carcinoma cells
by targeting PTEN/JNK/ANXA3 pathway. Am J Transl Res. 9:4300–4307.
2017.PubMed/NCBI
|
|
9
|
Yamada Y, Nishikawa R, Kato M, Okato A,
Arai T, Kojima S, Yamazaki K, Naya Y, Ichikawa T and Seki N:
Regulation of HMGB3 by antitumor miR-205-5p inhibits
cancer cell aggressiveness and is involved in prostate cancer
pathogenesis. J Hum Genet. 63:195–205. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Li L and Li S: miR-205-5p inhibits cell
migration and invasion in prostatic carcinoma by targeting ZEB1.
Oncol Lett. 16:1715–1721. 2018.PubMed/NCBI
|
|
11
|
Shi X, Xiao L, Mao X, He J, Ding Y, Huang
J, Peng C and Xu Z: miR-205-5p mediated downregulation of PTEN
contributes to cisplatin resistance in C13K human ovarian cancer
cells. Front Genet. 9:5552018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Edge SB and Compton CC: The American joint
committee on cancer: The 7th edition of the AJCC cancer staging
manual and the future of TNM. Ann Surg Oncol. 17:1471–1474. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2−ΔΔCT method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Gonzalez DM and Medici D: Signaling
mechanisms of the epithelial-mesenchymal transition. Sci Signal.
7:re82014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Rodriguez-Vida A, Hutson TE, Bellmunt J
and Strijbos MH: New treatment options for metastatic renal cell
carcinoma. ESMO Open. 2:e0001852017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Duran I, Lambea J, Maroto P,
González-Larriba JL, Flores L, Granados-Principal S, Graupera M,
Sáez B, Vivancos A and Casanovas O: Resistance to targeted
therapies in renal cancer: The importance of changing the mechanism
of action. Target Oncol. 12:19–35. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yu R, Yu BX, Chen JF, Lv XY, Yan ZJ, Cheng
Y and Ma Q: Anti-tumor effects of Atractylenolide I on bladder
cancer cells. J Exp Clin Cancer Res. 35:402016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wang C, Cai L, Liu J, Wang G, Li H, Wang
X, Xu W, Ren M, Feng L, Liu P, et al: MicroRNA-30a-5p inhibits the
growth of renal cell carcinoma by modulating GRP78 expression. Cell
Physiol Biochem. 43:2405–2419. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Bhat NS, Colden M, Dar AA, Saini S, Arora
P, Shahryari V, Yamamura S, Tanaka Y, Kato T, Majid S, et al:
MicroRNA-720 regulates E-cadherin-αE-catenin complex and
promotes renal cell carcinoma. Mol Cancer Ther. 16:2840–2848. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Liu W, Li H, Wang Y, Zhao X, Guo Y, Jin J
and Chi R: MiR-30b-5p functions as a tumor suppressor in cell
proliferation, metastasis and epithelial-to-mesenchymal transition
by targeting G-protein subunit α−13 in renal cell
carcinoma. Gene. 626:275–281. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Nagai H, Hasegawa S, Uchida F, Terabe T,
Ishibashi Kanno N, Kato K, Yamagata K, Sakai S, Kawashiri S, et al:
MicroRNA-205-5p suppresses the invasiveness of oral squamous cell
carcinoma by inhibiting TIMP2 expression. Int J Oncol. 52:841–850.
2018.PubMed/NCBI
|
|
22
|
Jiang M, Zhong T, Zhang W, Xiao Z, Hu G,
Zhou H and Kuang H: Reduced expression of miR-205-5p promotes
apoptosis and inhibits proliferation and invasion in lung cancer
A549 cells by upregulation of ZEB2 and downregulation of erbB3. Mol
Med Rep. 15:3231–3238. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Fulda S: Therapeutic opportunities based
on caspase modulation. Semin Cell Dev Biol. 82:150–157. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kale J, Osterlund EJ and Andrews DW: BCL-2
family proteins: Changing partners in the dance towards death. Cell
Death Differ. 25:65–80. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Dahmke IN, Backes C, Rudzitis-Auth J,
Laschke MW, Leidinger P, Menger MD, Meese E and Mahlknecht U:
Curcumin intake affects miRNA signature in murine melanoma with
mmu-miR-205-5p most significantly altered. PLoS One. 8:e811222013.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Brabletz T: EMT and MET in metastasis:
Where are the cancer stem cells? Cancer Cell. 22:699–701. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Brabletz T, Kalluri R, Nieto MA and
Weinberg RA: EMT in cancer. Nat Rev Cancer. 18:128–134. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kraljevic Pavelic S, Sedic M, Bosnjak H,
Spaventi S and Pavelic K: Metastasis: New perspectives on an old
problem. Mol Cancer. 10:222011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Elgamal OA, Park JK, Gusev Y,
Azevedo-Pouly AC, Jiang J, Roopra A and Schmittgen TD: Tumor
suppressive function of mir-205 in breast cancer is linked to HMGB3
regulation. PLoS One. 8:e764022013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
LoRusso PM: Inhibition of the
PI3K/AKT/mTOR pathway in solid tumors. J Clin Oncol. 34:3803–3815.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Akbani R, Ng PK, Werner HM, Shahmoradgoli
M, Zhang F, Ju Z, Liu W, Yang JY, Yoshihara K, Li J, et al: A
pan-cancer proteomic perspective on The Cancer Genome Atlas. Nat
Commun. 5:38872014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Porta C, Paglino C and Mosca A: Targeting
PI3K/Akt/mTOR signaling in cancer. Front Oncol. 4:642014.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Xu X, Wu J, Li S, Hu Z, Xu X, Zhu Y, Liang
Z, Wang X, Lin Y, Mao Y, et al: Downregulation of microRNA-182-5p
contributes to renal cell carcinoma proliferation via activating
the AKT/FOXO3a signaling pathway. Mol Cancer. 13:1092014.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Lian JH, Wang WH, Wang JQ, Zhang YH and Li
Y: MicroRNA-122 promotes proliferation, invasion and migration of
renal cell carcinoma cells through the PI3K/Akt signaling pathway.
Asian Pac J Cancer Prev. 14:5017–5021. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ruan GX and Kazlauskas A: Axl is essential
for VEGF-A-dependent activation of PI3K/Akt. EMBO J. 31:1692–1703.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Chen CH, Lai JM, Chou TY, Chen CY, Su LJ,
Lee YC, Cheng TS, Hong YR, Chou CK, Whang-Peng J, et al: VEGFA
upregulates FLJ10540 and modulates migration and invasion of lung
cancer via PI3K/AKT pathway. PLoS One. 4:e50522009. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Vachhani P and George S: VEGF inhibitors
in renal cell carcinoma. Clin Adv Hematol Oncol. 14:1016–1028.
2016.PubMed/NCBI
|
|
38
|
Comunanza V and Bussolino F: Therapy for
cancer: Strategy of combining anti-angiogenic and target therapies.
Front Cell Dev Biol. 5:1012017. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
An G, Liang S, Sheng C, Liu Y and Yao W:
Upregulation of microRNA-205 suppresses vascular endothelial growth
factor expression-mediated PI3K/Akt signaling transduction in human
keloid fibroblasts. Exp Biol Med. 242:275–285. 2017. View Article : Google Scholar
|
|
40
|
Zeng FC, Zeng MQ, Huang L, Li YL, Gao BM,
Chen JJ, Xue RZ and Tang ZY: Downregulation of VEGFA inhibits
proliferation, promotes apoptosis, and suppresses migration and
invasion of renal clear cell carcinoma. Onco Targets Ther.
9:2131–2141. 2016.PubMed/NCBI
|
|
41
|
Chen YS, Meng F, Li HL, Liu QH, Hou PF,
Bai J and Zheng JN: Dicer suppresses MMP-2-mediated invasion and
VEGFA-induced angiogenesis and serves as a promising prognostic
biomarker in human clear cell renal cell carcinoma. Oncotarget.
7:84299–84313. 2016.PubMed/NCBI
|
|
42
|
Fantozzi A, Gruber DC, Pisarsky L, Heck C,
Kunita A, Yilmaz M, Meyer-Schaller N, Cornille K, Hopfer U,
Bentires-Alj M and Christofori G: VEGF-mediated angiogenesis links
EMT-induced cancer stemness to tumor initiation. Cancer Res.
74:1566–1575. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Gonzalez-Moreno O, Lecanda J, Green JE,
Segura V, Catena R, Serrano D and Calvo A: VEGF elicits
epithelial-mesenchymal transition (EMT) in prostate intraepithelial
neoplasia (PIN)-like cells via an autocrine loop. Exp Cell Res.
316:554–567. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Kim LC, Cook RS and Chen J: mTORC1 and
mTORC2 in cancer and the tumor microenvironment. Oncogene.
36:2191–2201. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Besharat ZM, Sabato C, Po A, Gianno F,
Abballe L, Napolitano M, Miele E, Giangaspero F, Vacca A, Catanzaro
G, et al: Low expression of miR-466f-3p sustains epithelial to
mesenchymal transition in Sonic hedgehog medulloblastoma stem cells
through Vegfa-Nrp2 signaling pathway. Front Pharmacol. 9:12812018.
View Article : Google Scholar : PubMed/NCBI
|