Introduction
Breast cancer (BC) is the most common cancer in
women, and was the second leading cause of cancer-related mortality
worldwide in 2015, second only to lung cancer (1,2). China has
many patients with BC, and a large number of these patients suffer
from personal and family problems caused by the disease (3). BC patients often have metastasis and
recurrence despite the surgical removal of primary tumors (4). Previous studies have found that specific
molecular targeted therapy can improve the quality of life of
patients and bring new hope (5,6). Although
many reliable molecular targets for the treatment of BC have been
discovered, current molecular targeted therapies for BC are
expensive, are not effective for all patients and most patients
develop resistance (7).
microRNAs (miRNAs/miRs) are a class of
single-stranded non-coding small RNAs 21–23 nt in length (8). Mature miRNAs are integrated into the
RNA-induced silencing complex and bind to specific sites in the
non-coding 3′untranslated region (3′UTR) of target mRNA in an
incomplete complementary manner, to mediate the degradation of RNA
or inhibit the translation of proteins, thus playing a key role in
the silencing of RNA and the regulation of post-transcriptional
expression (9,10). A large number of studies have found
that miRNAs are abnormally expressed in a variety of malignant
tumors and are associated with malignant tumor growth (11–14).
Expression of miR-425-5p is upregulated in renal cell carcinoma and
promotes cell viability, invasion and migration (15). In prostate cancer, high miR-425-5p
expression can significantly promote the proliferation, migration
and invasion of tumor cells (16).
However, the role of miR-425-5p in BC remains unknown, and the
underlying functional mechanism of the effects of miR-425-5p in BC
progression remains to be further studied.
In the present study, the expression of miR-425-5p
in BC was determined by RT-qPCR, and the association of miR-425-5p
expression with the clinicopathological parameters and prognosis of
patients with BC was analyzed. A downstream target of miR-425-5p
was also investigated. The current study may provide a scientific
basis for exploring the role of miR-425-5p in BC, thus providing
new molecular targets for BC treatment.
Materials and methods
Clinical specimens
A total of 77 samples of BC tumor tissue and paired
adjacent tissue were collected from patients with BC at the First
People's Hospital of Yibin (Yibin, China). Tissue specimens were
collected from patients (age range, 30–82 years; all patients were
female) with primary BC who underwent surgical resection between
September 2010 and December 2012. All patients were histologically
diagnosed with invasive ductal breast cancer after surgery. Patient
information on age, tumor size, clinical stage, lymph node
metastasis, distant metastasis, and information on the expression
of estrogen receptor, progesterone receptor and human epidermal
growth factor receptor 2 was collected (17,18). This
study was conducted in accordance with the Helsinki Declaration and
was approved by the Ethics Review Committee of the Yibin First
People's Hospital and the Ethics Review Committee of Southwest
Medical University (Luzhou, China). Written informed consent was
obtained from all patients for the use of their tissue in this
study.
Cell culture and transfection
Five BC cell lines (MCF-7, SKBR-3, MDA-MB-231,
MDA-MB-468 and BT-20) and a normal human mammary epithelial cell
line (MCF-10A) were obtained from the Institute of Biochemistry and
Cell Biology (Chinese Academy of Sciences). All cell lines were
cultured in DMEM containing 10% fetal bovine serum (FBS) and
incubated at 37°C in 5% CO2.
MCF-7 and MDA-MB-231 cells (3×105) were
transfected with pre-miR-425-5p (miR-425-5p), mimic negative
control (NC), miR-425-5p inhibitor, inhibitor NC or pcDNA-PTEN
(Shanghai GenePharma Co., Ltd.). pcDNA plasmid without PTEN was
used as a negative control. The sequences of the miRNA mimic,
inhibitor and controls are presented in Table SI. The concentrations used for mimic,
inhibitor and pcDNA were 50, 100 nM and 1000 ng/ul, respectively)
Cells were transfected using Lipofectamine 3000 reagent
(Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's instructions. Subsequent experiments were performed
24–72 h after transfection.
Reverse transcription quantitative PCR
(RT-qPCR)
Total RNA was extracted from BC tissues, paired
adjacent tissue and cell lines using TRIzol reagent (Takara Bio,
Inc.). PrimeScript RT Reagent Kit (Takara Bio, Inc.) was used for
the reverse transcription of cDNA. The temperature protocol for RT
was as follows: 35°C for 5 min, followed by 42°C for 40 min and
75°C for 5 min. Next, qPCR was performed with SYBR Premix Ex Taq II
(Takara Bio, Inc.) and a LightCycler system (Roche Diagnostics
GmbH). The results were analyzed using the 2−ΔΔCq method
(19). U6 was used as an internal
control for microRNA, and GAPDH was used for mRNA. The sequences of
the primers used for each gene are presented in Table SII. The thermocycling conditions for
qPCR were as follows: Initial activation step at 95°C for 15 min
followed by 40 cycles of denaturation at 94°C for 15 sec, annealing
at 55°C for 30 sec and extension at 72°C for 30 sec.
Western blot analysis
Total protein from BC cell lines, a normal human
mammary epithelial cell line (1×106) and tissues was
extracted using RIPA lysis buffer (Beyotime Institute of
Biotechnology Co., Ltd.). Protein was quantified using the Bradford
protein assay (Bio-Rad Laboratories, Inc.) with a Nanodrop
spectrophotometer, and an equal amount (30 µg) was added to each
well of 12% gels and resolved by SDS-PAGE. Next, proteins were
transferred to PVDF membranes and were blocked by incubation for 1
h at room temperature with 5% non-fat powdered milk. The membranes
were probed at 4°C overnight with antibodies against PTEN (1:5,000;
ab32199; Abcam) and anti-GAPDH (1:5,000; ab181602; Abcam).
Subsequently, the PVDF membranes were incubated with horseradish
peroxidase-conjugated secondary antibody (1:5,000; ab6721; Abcam)
at room temperature for 1 h. Protein detection was performed using
incubation with an enhanced chemiluminescence solution (EMD
Millipore), and imaged using a ChemiDoc Imaging System (Bio-Rad
Laboratories, Inc.). GAPDH was used as the internal loading control
in the western blot. The protein bands were quantified using
QuantityOne software v4.6.7 (Bio-Rad Laboratories, Inc.), and the
values are expressed relative to GAPDH. Each experiment was
repeated three times.
Cell-Counting Kit-8 assay (CCK-8)
Following transfection, cells were seeded onto
96-well plates and incubated at 37°C for 24, 48 or 72 h before the
addition of CCK-8 reagent. Next, 10 µl CCK-8 reagent was added to
each well, and after 2 h of incubation at 37°C, absorption was
determined at 450 nm was determined by microplate
spectrophotometer.
Colony-forming assay
Transfected cells were seeded into 6-well plates and
cultured for 14 days. At that time, the colonies that had formed
were fixed with 4% paraformaldehyde for 30 min at room temperature
and stained with 0.1% crystal violet for 15 min at room
temperature. Finally, the cell colonies were counted with a light
microscope (Olympus Corporation; magnification, ×40).
Transwell migration and invasion
assays
For the invasion assay, Tranwell inserts were
precoated with Matrigel (1 mg/ml) at 37°C for 30 min. Transwells
were uncoated for the migration assay. Transfected cells were
seeded into the upper chamber with serum-free media. In the bottom
chamber, 500 µl DMEM containing 10% FBS functioning as a
chemoattractant was added. After 48 h of incubation, the migrated
or invaded cells were fixed with 4% paraformaldehyde for 30 min at
room temperature and stained with 0.1% crystal violet for 15 min at
room temperature. Finally, the migrated or invaded cells were
counted with a Leica DM IL LED inverted light microscope (Leica
Microsystems, Inc.; magnification, ×100). Five microscopic fields
of each well were randomly selected and the number of cells in
microscopic fields was counted.
5-Ethynyl-2′-deoxyuridine (EdU)
assay
Following transfection, cells were seeded into
96-well plates and incubated at 37°C until the cells reached 30%
confluence. An EdU assay was performed using a Cell-Light EdU
Apollo 567 in vitro kit (cat. no. 100T; Ruibo Biotechnology
Co., Ltd.). According to manufacturer's instructions, the cells
were incubated with EdU (50 µM) for 120 min, 0.5% Triton X and
ApolloR reaction cocktail (100 µl) for 30 min, and Hoechst 33342
(100 µl) for 30 min sequentially. Cell proliferation was analyzed
using the mean number of the cells in three fields for each sample
using a fluorescence microscope (Lionheart; BioTek Instruments,
Inc.; magnification, ×100).
Luciferase reporter assay
The online miRNA databases Oncomir (http://www.oncomir.org/), MiRanda (http://www.microrna.org/microrna/home.do), miRWalk
(http://mirwalk.umm.uni-heidelberg.de/) and TargetScan
(http://www.targetscan.org) were used to
identify downstream target genes of miR-425-5p. The wild-type (WT)
PTEN 3′ 3′UTR and mutant (MUT) PTEN 3′UTR oligonucleotides
containing the putative binding site of miR-425-5p were cloned into
the firefly luciferase-expressing pMIR-REPORT vector (Obio
Technology Corp., Ltd.). These constructs were co-transfected with
inhibitor NC or miR-425-5p inhibitor into MCF-7 and MDA-MB-231
cells. After 48 h of transfection, luciferase activity was
determined using the Dual-Luciferase Reporter Assay kit (Promega
Corporation) according to the manufacturer's protocol. The ratio of
Renilla luciferase activity to firefly luciferase activity was
calculated.
Statistical analysis
The statistical data were analyzed using SPSS
version 22.0 software (IBM Corp.) and GraphPad Prism version 6.0
software (GraphPad Software Inc.). The differences between groups
were analyzed using paired or unpaired t test and one-way analysis
of variance, followed by the Newman-Keuls test. Kaplan-Meier and
log-rank tests were used to assess recurrence-free survival (RFS)
and disease-specific survival (DSS) times. For Kaplan-Meier curves,
patients were divided into high and low expression groups using the
mean expression (0.2) as the cut-off value. Correlation analysis
was performed using Spearman's rank correlation test. The
χ2 test was used to analyze the association of
miR-425-5p expression with the clinicopathological characteristics
of BC. Univariate and multivariate Cox regression analyses were
performed to analysis the prognostic significance of miR-425-5p.
P<0.05 were considered to indicate a statistically significant
difference.
Results
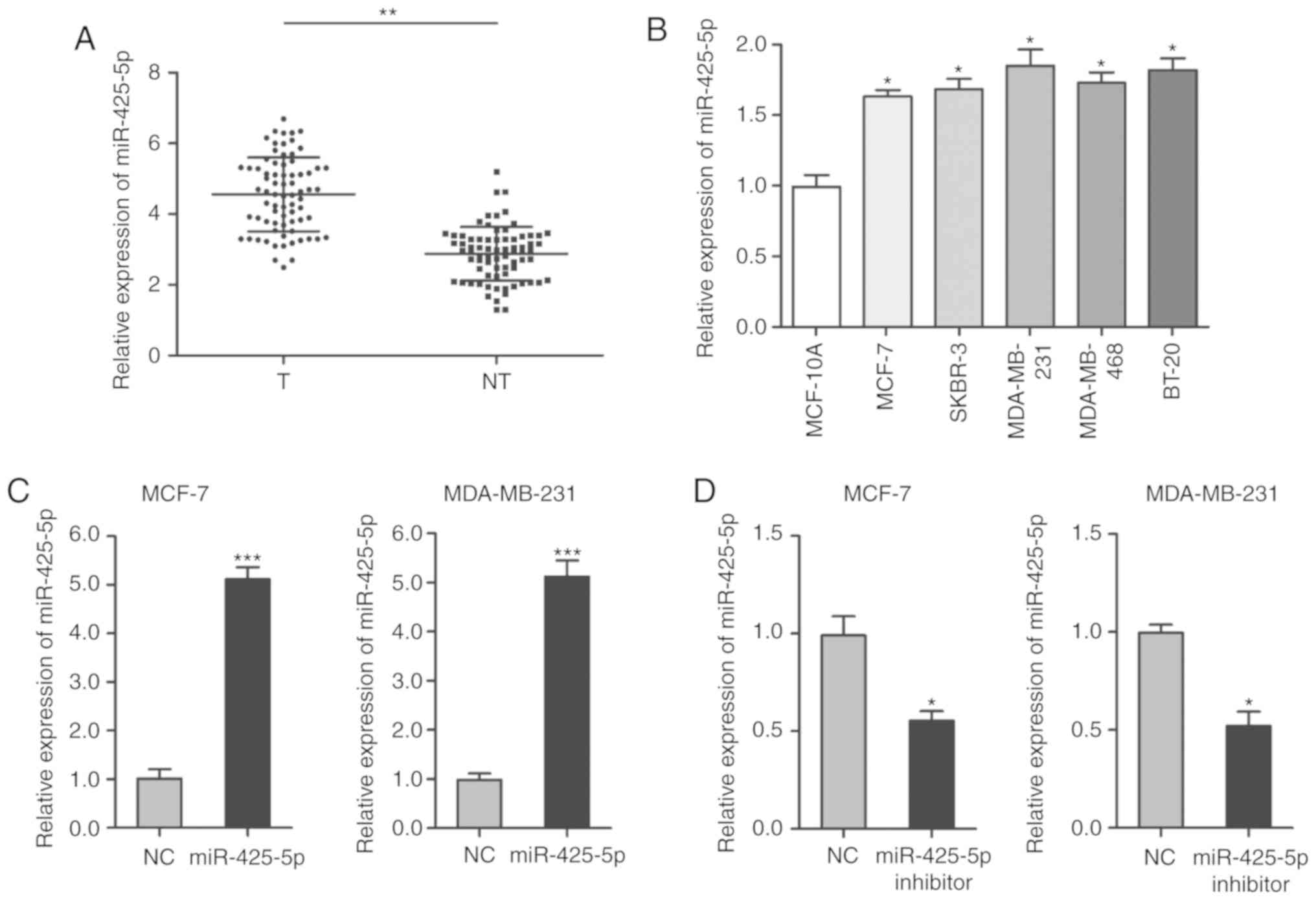
miR-425-5p is upregulated in BC
tissues and cell lines
RT-qPCR was performed to detect the expression of
miR-425-5p in BC tumor tissue, paired adjacent tissue and cell
lines. The results showed that miR-425-5p was higher in BC tissues
compared with in paired adjacent tissue (Fig. 1A). The expression of miR-425-5p was
also higher in BC cell lines compared with human mammary epithelial
cells (Fig. 1B). To further explore
the role of miR-425-5p in BC, MCF-7 and MDA-MB-231 cells were
transfected with pre-miR-425-5p to increase the expression of
miR-425-5p (Fig. 1C), and miR-425-5p
inhibitor to knockdown the expression of miR-425-5p (Fig. 1D).
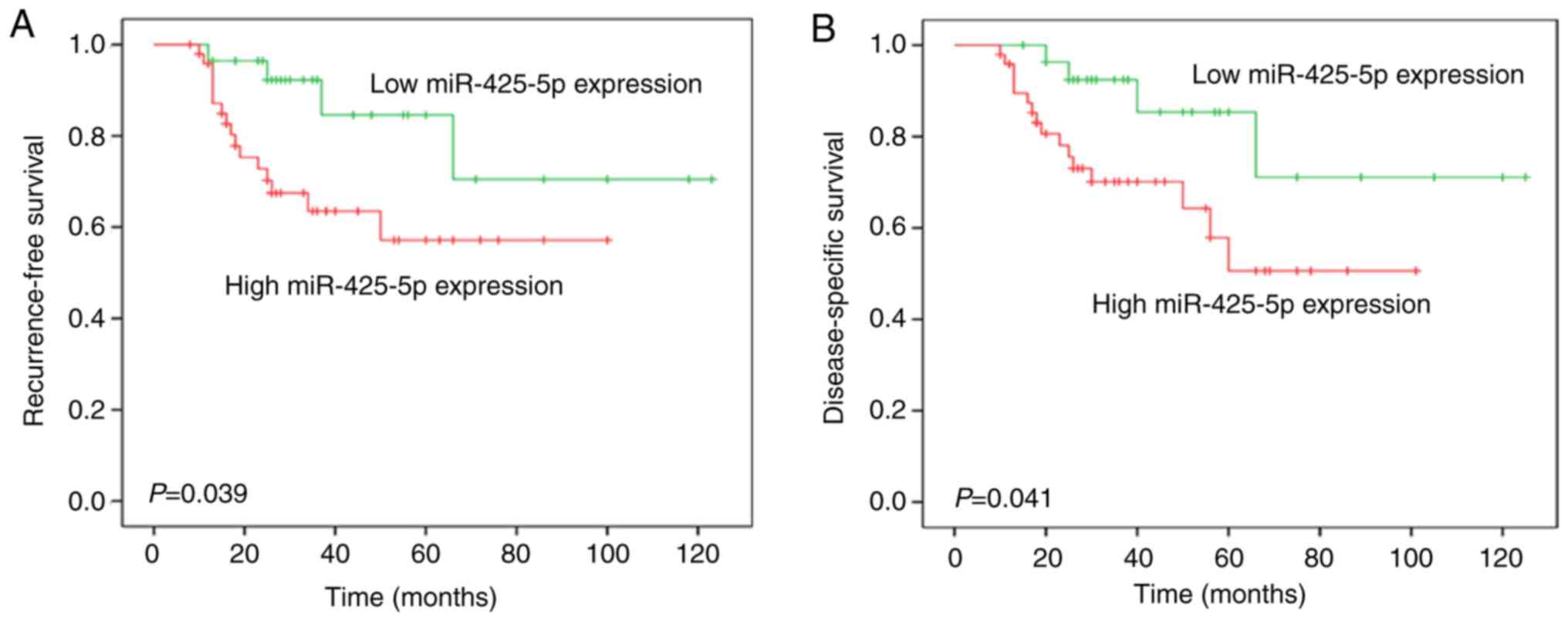
High expression of miR-425-5p is
associated with aggressive clinicopathological features and poor
prognosis in patients with BC
The association between miR-425-5p expression and
the clinicopathological characteristics of patients with BC was
analyzed. High miR-425-5p expression was significantly associated
with tumor size, clinical stage, lymph node metastasis and distant
metastasis (P<0.05; Table I).
Kaplan-Meier analysis revealed that patients with high miR-425-5p
expression had significantly shorter RFS and DSS times compared
with those with low miR-425-5p expression (Fig. 2). Univariate analysis for RFS in
patients with BC is presented in Table
II. Subsequent multivariate analysis demonstrated that tumor
size (HR, 1.184; P=0.017), clinical stage (HR, 1.418; P=0.033),
lymph node metastasis (HR, 1.524; P=0.020), distant metastasis (HR,
0.887; P=0.009) and miR-425-5p level (HR, 1.771; P=0.013) were
independent prognostic factors for RFS in patients with BC
(Table II).
 | Table I.Association between miR-425-5p and
clinicopathological features of patients with breast cancer. |
Table I.
Association between miR-425-5p and
clinicopathological features of patients with breast cancer.
|
|
| miR-425-5p
expression, n |
|
|---|
|
|
|
|
|
|---|
| Variables | Cases, n | Low (n=28) | High (n=49) | P-value |
|---|
| Age, years |
|
|
| 0.659 |
|
<45 | 30 | 10 | 20 |
|
|
≥45 | 47 | 18 | 29 |
|
| Tumor size, cm |
|
|
| 0.003 |
| ≤2 | 35 | 19 | 16 |
|
|
>2 | 42 | 9 | 33 |
|
| Clinical stage |
|
|
| <0.001 |
|
I/II | 39 | 22 | 17 |
|
|
III/IV | 38 | 6 | 32 |
|
| Lymph node
metastasis |
|
|
| 0.006 |
| ≤3 | 39 | 20 | 19 |
|
|
>3 | 38 | 8 | 30 |
|
| Distant
metastasis |
|
|
| 0.010 |
|
Presence | 45 | 11 | 34 |
|
|
Absence | 32 | 17 | 15 |
|
| Estrogen receptor
status |
|
|
| 0.575 |
|
Positive | 38 | 15 | 23 |
|
|
Absence | 39 | 13 | 26 |
|
| Progesterone
receptor status |
|
|
| 0.659 |
|
Presence | 47 | 18 | 29 |
|
|
Absence | 30 | 10 | 20 |
|
| Epidermal growth
factor receptor 2 status |
|
|
| 0.862 |
|
Presence | 43 | 16 | 27 |
|
|
Absence | 34 | 12 | 22 |
|
 | Table II.Univariate and multivariate analysis
of the prognostic variables influencing recurrence-free survival in
patients with breast cancer. |
Table II.
Univariate and multivariate analysis
of the prognostic variables influencing recurrence-free survival in
patients with breast cancer.
|
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|
|---|
| Variables | Cases, n | HR (95% CI) | P-value | HR (95% CI) | P-value |
|---|
| Age, years |
| 0.348
(0.210–1.364) | 0.470 |
|
|
|
<45 | 30 |
|
|
|
|
|
≥45 | 47 |
|
|
|
|
| Tumor size, cm |
| 1.044
(0.531–2.017) | 0.014 | 1.184
(0.821–2.647) | 0.017 |
| ≤2 | 35 |
|
|
|
|
|
>2 | 42 |
|
|
|
|
| Clinical stage |
| 1.504
(1.047–3.647) | 0.029 | 1.418
(0.889–2.691) | 0.033 |
|
I/II | 39 |
|
|
|
|
|
III/IV | 38 |
|
|
|
|
| Lymph node
metastasis |
| 1.773
(0.941–2.097) | 0.026 | 1.524
(0.862–3.394) | 0.020 |
| ≤3 | 39 |
|
|
|
|
|
>3 | 38 |
|
|
|
|
| Distant
metastasis |
| 0.943
(0.430–1.813) | 0.004 | 0.887
(0.397–1.530) | 0.009 |
|
Presence | 45 |
|
|
|
|
|
Absence | 32 |
|
|
|
|
| Estrogen receptor
status |
| 1.009
(0.473–1.820) | 0.679 |
|
|
|
Presence | 38 |
|
|
|
|
|
Absence | 39 |
|
|
|
|
| Progesterone
receptor status |
| 0.873
(1.047–3.974) | 0.407 |
|
|
|
Presence | 47 |
|
|
|
|
|
Absence | 30 |
|
|
|
|
| Epidermal growth
factor receptor 2 status |
| 1.340
(1.873–3.911) | 0.637 |
|
|
|
Presence | 43 |
|
|
|
|
|
Absence | 34 |
|
|
|
|
| microRNA-425-5p
expression |
| 1.833
(0.947–3.075) | 0.007 | 1.771
(0.994–2.647) | 0.013 |
|
High | 49 |
|
|
|
|
|
Low | 28 |
|
|
|
|
In addition, univariate analysis was performed for
DSS in patients with BC, and results are presented in Table III. Subsequent multivariate analysis
showed that tumor size (HR, 1.190; P=0.037), clinical stage (HR,
1.938; P=0.030), lymph node metastasis (HR, 1.619; P=0.019),
distant metastasis (HR, 1.473; P=0.019) and miR-425-5p level (HR,
1.074; P=0.015) were independent prognostic factors for DSS in
patients with BC (Table III).
 | Table III.Univariate and multivariate analysis
of the prognostic variables influencing disease-specific survival
in patients with breast cancer. |
Table III.
Univariate and multivariate analysis
of the prognostic variables influencing disease-specific survival
in patients with breast cancer.
|
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|
|---|
| Variables | Cases, n | HR (95% CI) | P-value | HR (95% CI) | P-value |
|---|
| Age, years |
| 0.631
(0.510–1.881) | 0.607 |
|
|
|
<45 | 30 |
|
|
|
|
|
≥45 | 47 |
|
|
|
|
| Tumor size, cm |
| 1.204
(0.765–1.947) | 0.030 | 1.190
(0.843–2.108) | 0.037 |
| ≤2 | 35 |
|
|
|
|
|
>2 | 42 |
|
|
|
|
| Clinical stage |
| 1.881
(1.238–3.705) | 0.027 | 1.938
(1.047–3.447) | 0.030 |
|
I/II | 39 |
|
|
|
|
|
III/IV | 38 |
|
|
|
|
| Lymph node
metastasis |
| 1.077
(0.647–1.849) | 0.013 | 1.619
(0.731–1.667) | 0.019 |
| ≤3 | 39 |
|
|
|
|
|
>3 | 38 |
|
|
|
|
| Distant
metastasis |
| 1.647
(0.840–2.661) | 0.020 | 1.473
(0.845–2.304) | 0.019 |
|
Presence | 45 |
|
|
|
|
|
Absence | 32 |
|
|
|
|
| Estrogen receptor
status |
| 0.660
(0.843–2.614) | 0.431 |
|
|
|
Presence | 38 |
|
|
|
|
|
Absence | 39 |
|
|
|
|
| Progesterone
receptor status |
| 1.243
(0.842–1.994) | 0.377 |
|
|
|
Presence | 47 |
|
|
|
|
|
Absence | 30 |
|
|
|
|
| Epidermal growth
factor receptor 2 status |
| 1.084
(1.234–2.941) | 0.740 |
|
|
|
Presence | 43 |
|
|
|
|
|
Absence | 34 |
|
|
|
|
| microRNA-425-5p
expression |
| 1.453
(0.534–1.882) | 0.019 | 1.074
(0.947–2.334) | 0.015 |
|
High | 49 |
|
|
|
|
|
Low | 28 |
|
|
|
|
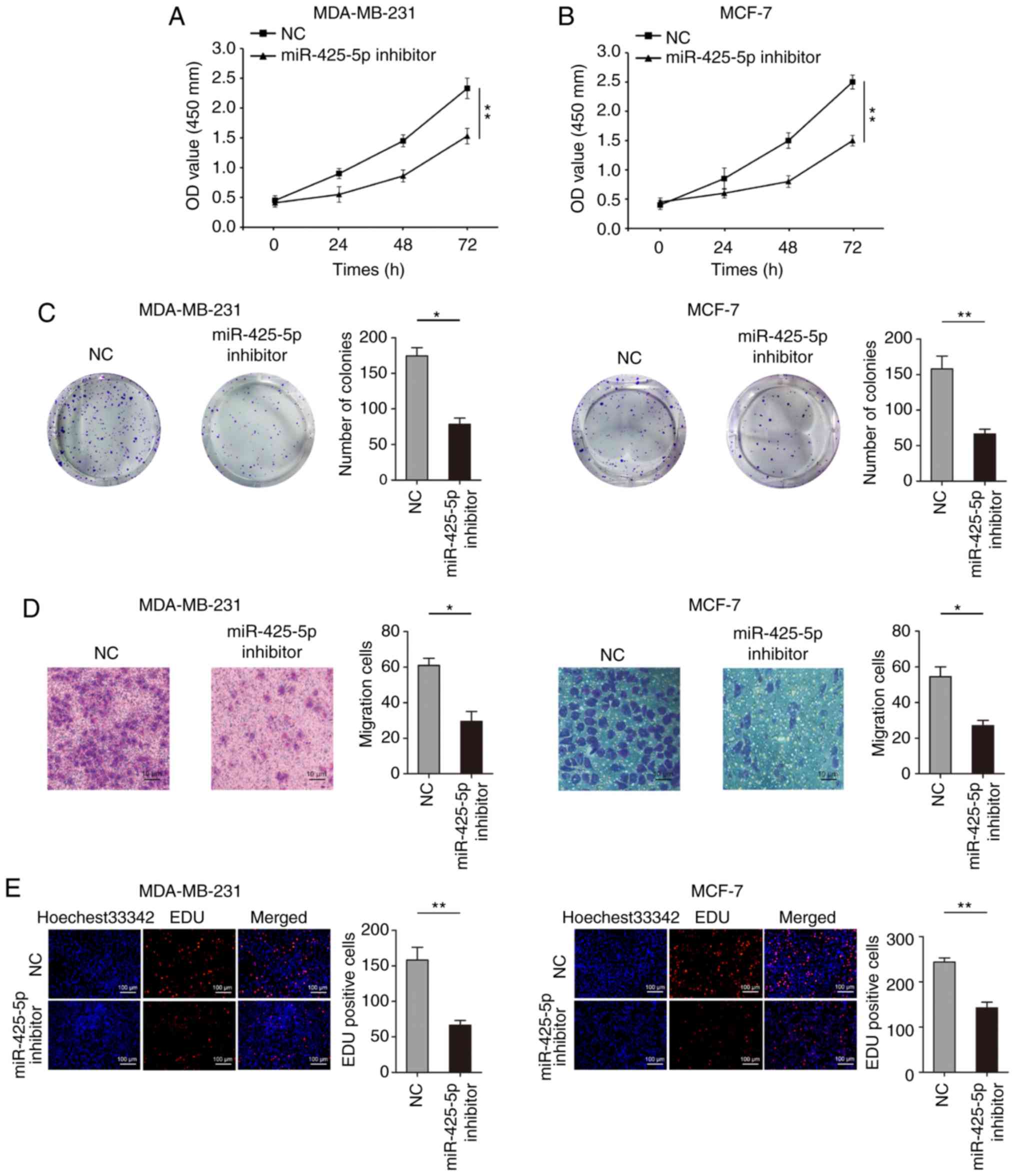
miR-425-5p promotes cell proliferation
and migration in BC
Among the BC cell lines, the expression of
miR-425-5p was lowest in MCF-7 cells and highest in MDA-MB-231
cells (Fig. 1B). Therefore, these two
cell lines were selected for subsequent experiments. To investigate
the biological role of miR-425-5p on proliferation of BC cells,
CCK-8 assays showed that knockdown of miR-425-5p expression
significantly inhibited MDA-MB-231 and MCF-7 cell proliferation
(Fig. 3A and B). Colony formation
assays also revealed that reducing the expression of miR-425-5p
resulted in reduced colony number in MDA-MB-231 and MCF-7 cells
(Fig. 3C). Transwell assays indicated
that knockdown of miR-425-5p inhibited migration in MDA-MB-231 and
MCF-7 cells (Fig. 3D). However,
knockdown of miR-425-5p had no significant effect on the invasive
ability of MDA-MB-231 and MCF-7 cells (Fig. S1). Moreover, decreased miR-425-5p
expression significantly inhibited EdU uptake, representing the
proliferation of cells, in MDA-MB-231 and MCF-7 cells (Fig. 3E).
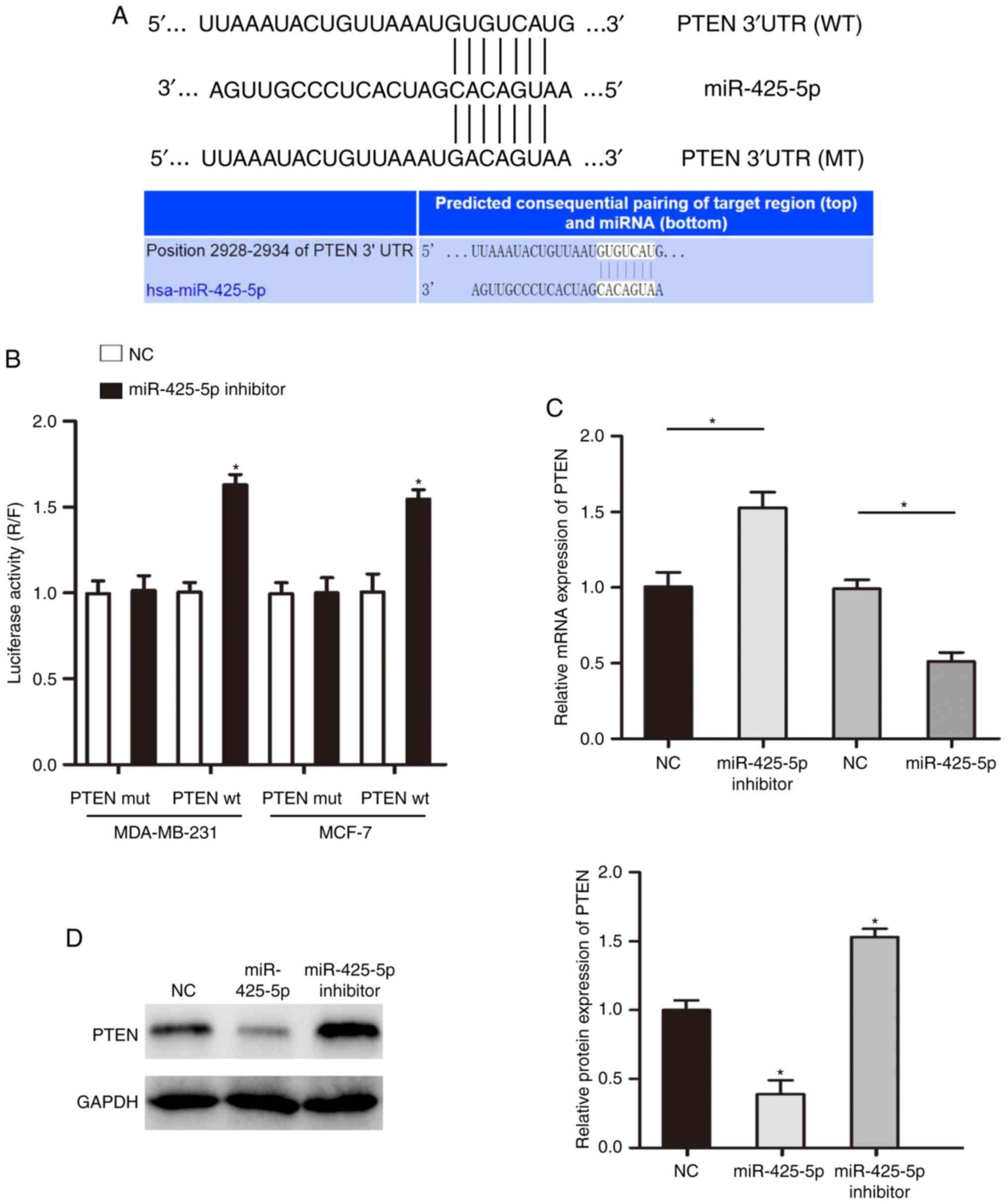
PTEN is a direct target of
miR-425-5p
Using an online target prediction tool, it was found
that PTEN may be a direct target of miR-425-5p (Figs. 4A and S2). To further verify that miR-425-5p
biologically targets PTEN, a luciferase reporter assay was
performed. The results indicated that co-transfection with a
miR-425-5p inhibitor significantly promoted luciferase activity in
cells transfected with WT PTEN 3′UTR. However, increase no
luciferase activity was observed in cells co-transfected with MUT
PTEN 3′UTR (Fig. 4B). Additionally,
in cells with decreased expression of miR-425-5p, the expression of
PTEN was significantly increased (Fig. 4C
and D).
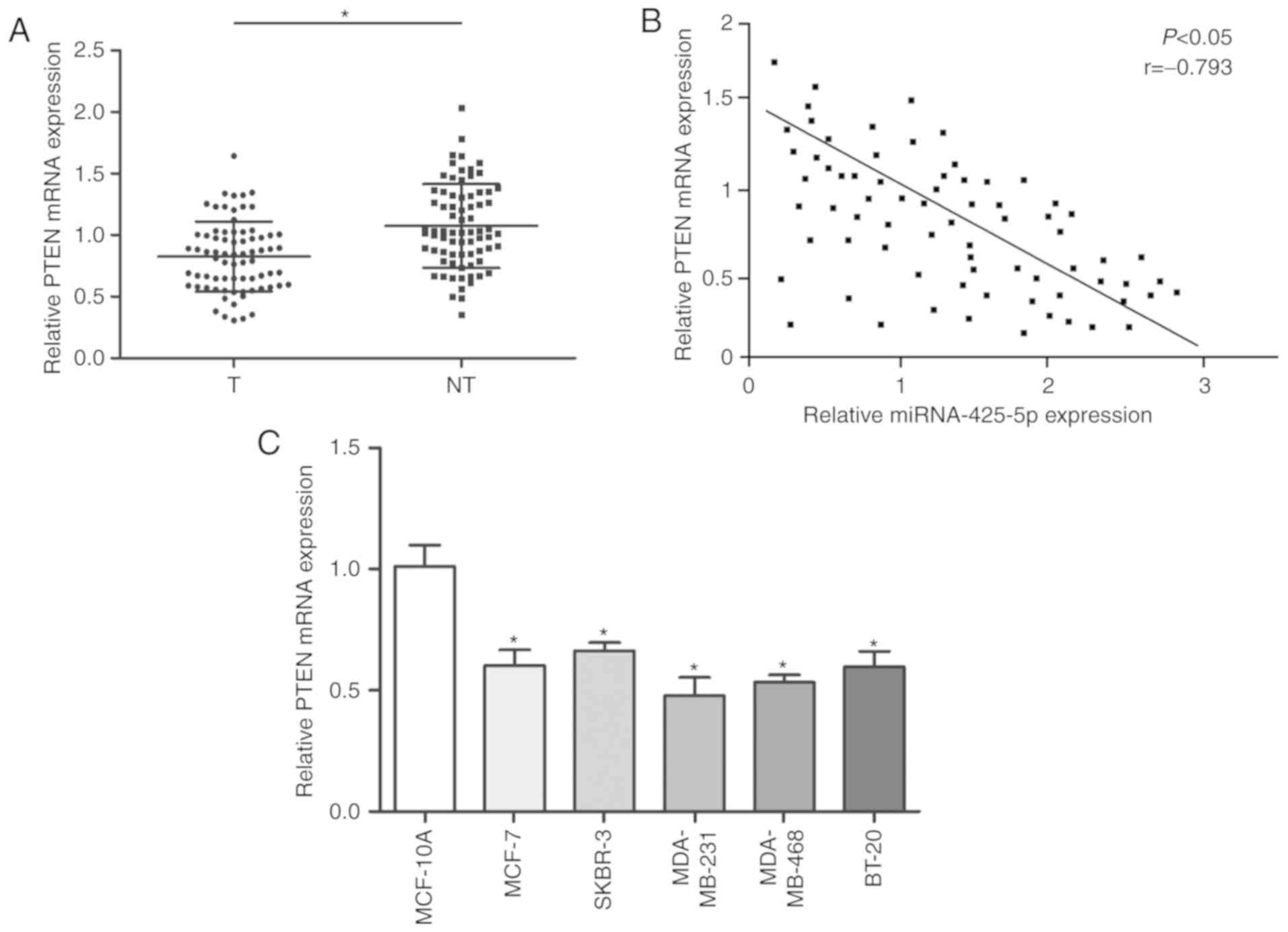
Furthermore, PTEN mRNA was lower in BC tissues
compared with that in paired adjacent tissue (Fig. 5A). PTEN mRNA expression levels
inversely correlated with expression of miR-425-5p (Fig. 5B). Similarly, the expression of PTEN
was found to be lower in BC cells compared with normal mammary
epithelial cells (Fig. 5C).
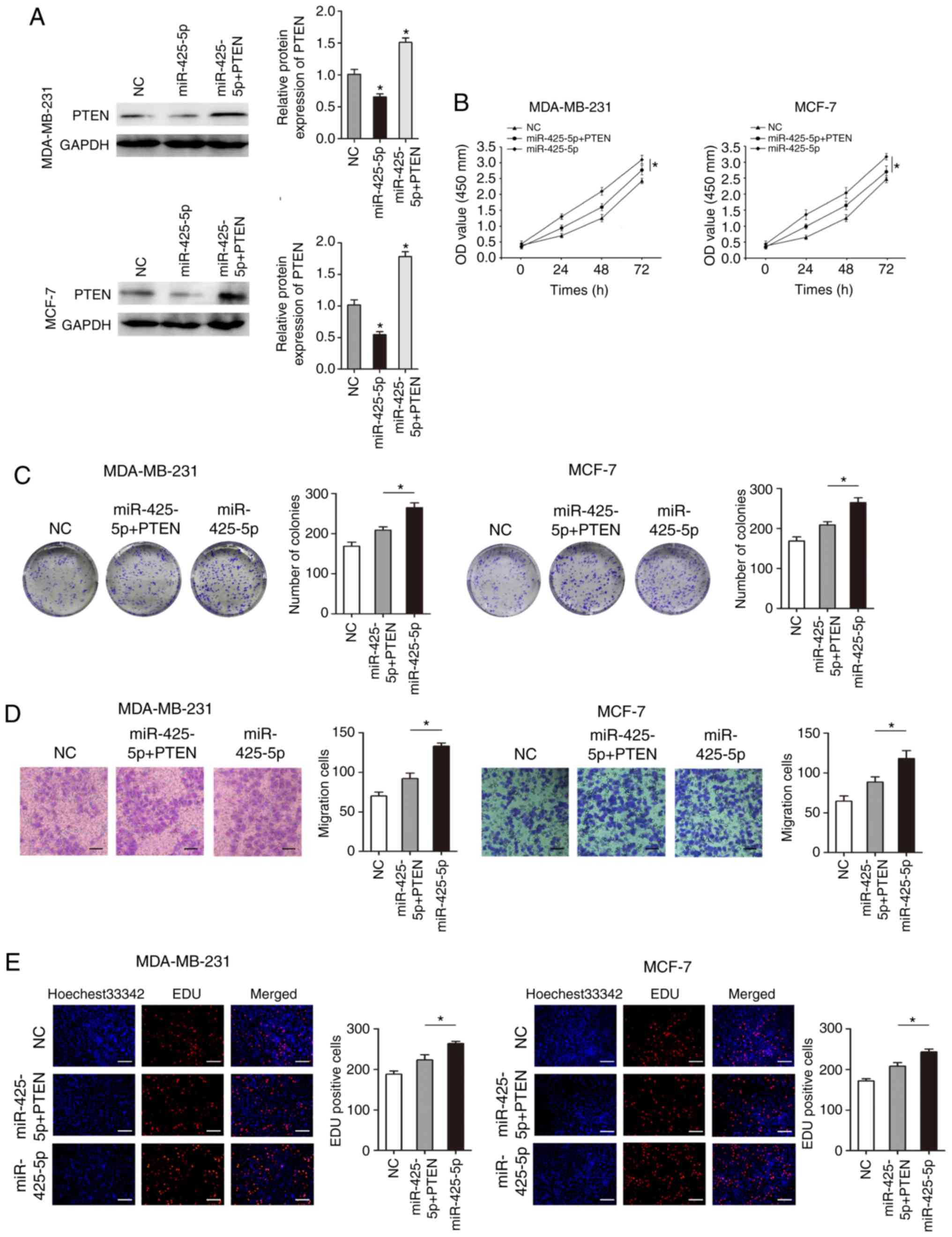
Overexpression of PTEN partially
rescues miR-425-5p-mediated effects on BC cells
To further confirm that PTEN is a functional target
gene of miR-425-5p, the expression of PTEN was restored by
transfecting MDA-MB-231 and MCF-7 cells with a PTEN-containing
plasmid (pcDNA-PTEN). Overexpression of PTEN compared to controls
(untransfected cells and empty vector) was confirm by
RT-qPCR(Fig. S3). Co-transfection
with pre-miR-425-5p and pcDNA-PTEN resulted in increased PTEN
protein levels (Fig. 6A).
Overexpression of miR-425-5p significantly promoted cell
proliferation and migration in MDA-MB-231 and MCF-7 (Fig. 6B-E). Overexpression of PTEN
significantly inhibited the promotional effects of miR-425-5p on
proliferation and migration in MDA-MB-231 and MCF-7 cells (Fig. 6B-E). These results suggest that
miR-425-5p plays a biological role through PTEN.
Discussion
BC is the most common type of tumor in women
worldwide (1). A previous study found
that there are ~300,000 new patients with BC in China each year
(3). The incidence of BC in China is
lower than that in western countries (3); however, due to the large population of
China, the number and growth rate of the disease are among the
highest in the world, and there are many young patients (20,21).
Therefore, effective molecular targets for the diagnosis and
treatment of BC are urgently required. The results of the current
study indicate that miR-425-5p is upregulated in BC, and high
expression of miR-425-5p is associated with poor prognosis of
patients with BC. Therefore, miR-425-5p may be a potential
therapeutic target for BC.
As the roles of miRNAs have been investigated more
thoroughly, miR-425-5p has been shown to play an important role in
the progression of various types of cancer. FMR1 autosomal homolog
1 regulates the expression of miR-425-5p and MIR17HG, thereby
promoting the malignant development of glioma cells (22). miR-425-5p has also been found to
increase the resistance of colorectal cancer to chemotherapeutic
drugs by regulating the expression of PDCD10, suggesting that
miR-425-5p may have potential as a new target to enhance the
sensitivity of colorectal cancer to chemotherapeutic drugs
(23). In osteosarcoma cells,
increased expression of miR-425-5p can significantly increase the
expression of MALAT1 and TUG1 and activate the Wnt/β-catenin
signaling pathway and promote the development of the malignant
biological processes of the osteosarcoma process (24). The findings of the current study are
consistent with these previous studies. Overexpression of
miR-425-5p was found to promote proliferation and migration of BC
cells, and decreased expression of miR-425-5p inhibited the
proliferation and migration of BC cells. PTEN was identified as a
target of miR-425-5p, and overexpression of PTEN partially
inhibited the promotional effect of miR-425-5p in BC cells.
Previous studies suggest that four of the five
miR-425-5p targets identified in the current study are not closely
associated with biological functions of cancer cells or their
specific functions are unknown (25,26).
However, PTEN is a protein phosphatase that may be involved in
cellular regulation through dephosphorylation (27,28).
Phosphorylation and dephosphorylation are important ways to
regulate cellular activity, and many oncogene products stimulate
cell growth through phosphorylation (29–31).
Through miRNA database prediction and experimental verification,
PTEN predicted found to be a target of miR-425-5piIn the current
study. In gastric cancer, PDZ domain-containing 1 inhibits the
phosphorylation of PTEN at the S380/T382/T383 cluster by direct
interaction with PTEN, further enhancing the ability of PTEN to
inhibit the activation of PI3K/AKT (32). A previous study found that zinc finger
HIT-type containing 1 affected the malignant development of BC by
upregulating PTEN and inactivating the PI3K/AKT/mTOR pathway
(33). In the present study, PTEN
expression was revealed to be reduced in BC tissues, and miR-425-5p
was shown to negative regulate PTEN expression. Additionally,
restoration of PTEN expression in BC cells with miR-425-5p
overexpression partially reversed the effects of the miRNA on
proliferation and migration.
In conclusion, the findings of the present study
demonstrate that miR-425-5p regulates BC cell proliferation by
influencing PTEN expression. This provides new information
regarding the occurrence and development of BC, and provides a
potential novel target for molecular therapy for patients with
BC.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
This study was supported by the Key Research
Projects on Application Foundation of Sichuan Science and
Technology Department (grant no. 2017JY0030).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Author's contributions
SX and MX designed and conducted the experiments and
wrote the manuscript. HZ, JL and ZW provided the research materials
and analyzed the data. All authors read and approved the manuscript
and agree to be accountable for all aspects of the research.
Ethics approval and consent to
participate
All protocols involving the use of humans were
approved by the Ethics Review Committee of the Yibin First People's
Hospital (Yibin, China) and the Ethics Review Committee of
Southwest Medical University (Luzhou, China). Written informed
consent was obtained from all patients for the use of their tissue
in this study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Davidson NE, Armstrong SA, Coussens LM,
Cruz-Correa MR, DeBerardinis RJ, Doroshow JH, Foti M, Hwu P,
Kensler TW, Morrow M, et al: AACR cancer progress report 2016. Clin
Cancer Res. 22 (Suppl 19):S1–S137. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
McGuire A, Brown JA, Malone C, McLaughlin
R and Kerin MJ: Effects of age on the detection and management of
breast cancer. Cancers (Basel). 7:908–929. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Christenson JL, Butterfield KT, Spoelstra
NS, Norris JD, Josan JS, Pollock JA, McDonnell DP, Katzenellenbogen
BS, Katzenellenbogen JA and Richer JK: MMTV-PyMT and derived Met-1
mouse mammary tumor cells as models for studying the role of the
androgen receptor in triple-negative breast cancer progression.
Horm Cancer. 8:69–77. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Gu G, Dustin D and Fuqua SA: Targeted
therapy for breast cancer and molecular mechanisms of resistance to
treatment. Curr Opin Pharmacol. 31:97–103. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Riobo-Del Galdo NA, Lara Montero Á and
Wertheimer EV: Role of hedgehog signaling in breast cancer:
Pathogenesis and therapeutic. Cells. 8:E3752019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Harbeck N and Gnant M: Breast cancer.
Lancet. 389:1134–1150. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wu H, Zhang W, Wu Z, Liu Y, Shi Y, Gong J,
Shen W and Liu C: miR-29c-3p regulates DNMT3B and LATS1 methylation
to inhibit tumor progression in hepatocellular carcinoma. Cell
Death Dis. 10:482019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Du H, Xu Q, Xiao S, Wu Z, Gong J, Liu C,
Ren G and Wu H: MicroRNA-424-5p acts as a potential biomarker and
inhibits proliferation and invasion in hepatocellular carcinoma by
targeting TRIM29. Life Sci. 224:1–11. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hausser J and Zavolan M: Identification
and consequences of miRNA-target interactions-beyond repression of
gene expression. Nat Rev Genet. 15:599–612. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Cohen A, Burgos-Aceves MA and Smith Y:
Estrogen repression of microRNA as a potential cause of cancer.
Biomed Pharmacother. 78:234–238. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Bartel DP: Metazoan microRNAs. Cell.
173:20–51. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hobert O: Gene regulation by transcription
factors and microRNAs. Science. 319:1785–1786. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Quan J, Li Y, Pan X, Lai Y, He T, Lin C,
Zhou L, Zhao L, Sun S, Ding Y, et al: Oncogenic miR-425-5p is
associated with cellular migration, proliferation and apoptosis in
renal cell carcinoma. Oncol Lett. 16:2175–2184. 2018.PubMed/NCBI
|
|
16
|
Zhang JY, Su XP, Li YN and Guo YH:
MicroRNA-425-5p promotes the development of prostate cancer via
targeting forkhead box J3. Eur Rev Med Pharmacol Sci. 23:547–554.
2019.PubMed/NCBI
|
|
17
|
Jiang C, Cao S, Li N, Jiang L and Sun T:
PD-1 and PD-L1 correlated gene expression profiles and their
association with clinical outcomes of breast cancer. Cancer Cell
Int. 19:2332019. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Jiang Y, Liu Y, Tan X, Yu S and Luo J:
TPX2 as a novel prognostic indicator and promising therapeutic
target in triple-negative breast cancer. Clin Breast Cancer.
S1526–S8209. Jun 13–2019.(Epub ahead of print).
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Turashvili G and Brogi E: Tumor
heterogeneity in breast cancer. Front Med. 4:2272017. View Article : Google Scholar
|
|
21
|
Hu L, Gao Y, Cao Y, Zhang Y, Xu M, Wang Y,
Jing Y, Guo S, Jing F, Hu X and Zhu Z: Identification of arginine
and its ‘downstream’ molecules as potential markers of breast
cancer. IUBMB Life. 68:817–822. 2016. View
Article : Google Scholar : PubMed/NCBI
|
|
22
|
Cao S, Zheng J, Liu X, Liu Y, Ruan X, Ma
J, Liu L, Wang D, Yang C, Cai H, et al: FXR1 promotes the malignant
biological behavior of glioma cells via stabilizing MIR17HG. J Exp
Clin Cancer Res. 38:372019. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhang Y, Hu X, Miao X, Zhu K, Cui S, Meng
Q, Sun J and Wang T: MicroRNA-425-5p regulates chemoresistance in
colorectal cancer cells via regulation of programmed cell death 10.
J Cell Mol Med. 20:360–369. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yang G, Zhang C, Wang N and Chen J:
miR-425-5p decreases LncRNA MALAT1 and TUG1 expressions and
suppresses tumorigenesis in osteosarcoma via Wnt/β-catenin
signaling pathway. Int J Biochem Cell Biol. 111:42–51. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Charzewska A, Rzońca S, Janeczko M, Nawara
M, Smyk M, Bal J and Hoffman-Zacharska D: A duplication of the
whole KIAA2022 gene validates the gene role in the pathogenesis of
intellectual disability and autism. Clin Genet. 88:297–299. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Melchior B, Mittapalli GK, Lai C,
Duong-Polk K, Stewart J, Güner B, Hofilena B, Tjitro A, Anderson
SD, Herman DS, et al: Tau pathology reduction with SM07883, a
novel, potent, and selective oral DYRK1A inhibitor: A potential
therapeutic for Alzheimer's disease. Aging Cell. 18:e130002019.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wu J and Chen H, Ye M, Wang B, Zhang Y,
Sheng J, Meng T and Chen H: Downregulation of long noncoding RNA
HCP5 contributes to cisplatin resistance in human triple-negative
breast cancer via regulation of PTEN expression. Biomed
Pharmacother. 115:1088692019. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhu L, Wang X, Wang T, Zhu W and Zhou X:
miR-494-3p promotes the progression of endometrial cancer by
regulating the PTEN/PI3K/AKT pathway. Mol Med Rep. 19:581–588.
2019.PubMed/NCBI
|
|
29
|
Shen F, Zheng H, Zhou L, Li W, Liu J and
Xu X: Identification of CD28 and PTEN as novel prognostic markers
for cervical cancer. J Cell Physiol. 234:7004–7011. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Liang L, Williams MD and Bell D:
Expression of PTEN, androgen receptor, HER2/neu, cytokeratin 5/6,
estrogen receptor-beta, HMGA2, and PLAG1 in salivary duct
carcinoma. Head Neck Pathol. Nov 2–2018.(Epub ahead of print).
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhu L, Zhang C and Liu Q: PTEN
S-nitrosylation by NOS1 inhibits autophagy in NPC cells. Cell Death
Dis. 10:3062019. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhao C, Tao T, Yang L, Qin Q, Wang Y, Liu
H, Song R, Yang X, Wang Q, Gu S, et al: Loss of PDZK1 expression
activates PI3K/AKT signaling via PTEN phosphorylation in gastric
cancer. Cancer Lett. 453:107–121. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Cui C, Li S and Wu D: Znhit1 inhibits
breast cancer by up-regulating PTEN to deactivate the PI3K/Akt/mTOR
pathway. Life Sci. 224:204–211. 2019. View Article : Google Scholar : PubMed/NCBI
|