Introduction
Colon cancer is one of the most common
gastrointestinal malignancies (1).
Despite marked developments in the diagnosis and treatment of colon
cancer in the past few decades, its prognosis remains poor. To
date, the treatment of colon cancer is faced with major challenges,
including the serious side effects caused by chemotherapy agents,
drug resistance and metastasis. Thus, there is still a clinical
need for the development of new treatment regimens for colon
cancer.
Although chemotherapy drugs possess serious adverse
effects, chemotherapy remains one of the major treatments for colon
cancer (2). Natural products and
their active derivates, including semi-synthetic and synthetic
analogs, comprise one of the most important sources of chemotherapy
drugs. Several plant-derived compounds, such as paclitaxel,
vincristine, camptothecin and etoposide, have already been used
against cancer for several decades (3–6).
Evodiamine (Evo), a quinolone alkaloid extracted from traditional
herbal medicine Evodia rutaecarpa (7), has multiple pharmacological actions
and could be used for obesity, inflammation, infectious and
cardiovascular diseases (8). A
growing amount of evidence has indicated that Evo exhibits
anticancer activity against various types of cancer, such as
tongue, colon and breast cancer (9–12).
According to studies, this activity of Evo may be mediated by NF-κB
(9), transforming growth factor β1
(TGF-β1) (11) and/or the
p53/p21/Rb pathway (12). However,
the explicit mechanism underlying this activity requires further
exploration.
Although the global pathogenesis of colon cancer
remains unclear, the aberrant function of several important signals
and genes, such as p53, APC, PIK3CA and Smad4 mutations, has been
identified (13–16). TGF-β is a cytokine that plays an
important role in deciding the fate of cells by regulating
proliferation and differentiation. Bone morphogenetic proteins
(BMPs) are another sub-group of TGF-β superfamily, which were
reported by Urist in 1965 as osteogenic factor (17). In addition to the development of
skeletal system, BMPs also play an important role in the
development of gastrointestinal track by regulating the stromal
microenvironment, protecting from polyposis initiation of the
colonic mesenchyme and terminal differentiation of intestinal
secretory progenitor cells (18,19).
Thus, the aberrant signal transduction of BMP may be another major
cause of colon cancer (20). Our
previous study demonstrated that BMP9 partly mediated the
anticancer activity of several natural products, such as
resveratrol (21). Although Evo
exhibited effective anticancer activity in colon cancer, it remains
unknown whether BMP9 is involved in this process.
In the present study, it was determined whether BMP9
could mediate the anticancer activity of Evo in colon cancer, and
the possible mechanism underlying this biological process was
revealed.
Materials and methods
Chemicals and cell culture
Evo was purchased from Xi'an Hao-Xuan Bio-tech Co.,
Ltd. and dissolved with dimethyl sulfoxide (DMSO) for in
vitro testing, or suspended with 0.4% carboxymethylcellulose
sodium for in vivo testing. Human colon epithelial cell line
(FHC) and human colon cancer cell lines (including HCT116, LoVo,
SW620 and SW480) were obtained from the American Type Culture
Collection (ATCC). Primary antibodies for PCNA (cat. no. sc-56;
mouse, monoclonal; 1:1,000), GAPDH (cat. no. sc-32233; mouse,
monoclonal; 1:1,000), Bad (cat. no. sc-8044; mouse, monoclonal;
1:1,000), Bcl-2 (cat. no. sc-7382; mouse, monoclonal; 1:1,000),
BMP9 (cat. no. sc-514211; rabbit, polyclonal; 1:1,000), HIF-1α
(cat. no. sc-10790; rabbit, polyclonal; 1:1,000), Smad1/5/8 (cat.
no. sc-6031-R; rabbit, polyclonal; 1:1,000), p53 (cat. no.
sc-55476; mouse, monoclonal; 1:1,000) and p-p53 (cat. no. sc-13580;
mouse, monoclonal; 1:1,000) were purchased from Santa Cruz
Biotechnology Inc. Phosphorylated (p)-Smad1/5/9 (cat. no. 13820S;
goat, monoclonal; 1:1,000) was ordered from Cell signaling
Technology. Cells were maintained in Dulbecco's modified Eagle's
medium with 10% fetal bovine serum, penicillin (100 U/ml) and
streptomycin (100 µg/ml) at 37°C in 5% CO2.
Cell viability assay
Cell viability was measured using CCK-8 kits (cat.
no. C008-2; Seven Sea Biotechnology, Shanghai China). Briefly,
subconfluent cells were placed in 96-well plates with 200 µl medium
(2,000 cells/well), and treated with different concentrations of
Evo (0.5, 1, 1.5, 2 and 2.5 µM) for 24, 48 and 72 h, according to
the experimental design. CCK-8 (10 µl/well) was added and then the
cells were incubated for another 2 h at 37°C. The optical density
of each well was measured at 450 nm using a microplate reader
(ELx800; BioTek Instruments, Inc.). Each assay was repeated at
least three times.
Crystal violet staining and colony
formation assay
Crystal violet staining was performed as previously
reported (22). In brief,
subconfluent HCT116 cells were treated with Evo (0.5, 1 or 2 µM)
for 24 h. Cells were then re-plated in 12-well cell culture plates
without Evo at 100 or 200 cells/well. Colonies were subjected to
crystal violet staining after treatment for 14 days. They were then
carefully washed with cold (4°C) phosphate-buffered saline (PBS)
and stained with PBS-buffered 0.5% crystal violet formalin solution
at room temperature for 20 min. Next, the plates were washed with
tap water and air-dried for imaging under inverted microscope
(magnification, ×40, Ti Nikon; Nikon) or scanning. There are over
450 cell colonies in each well at least. Each assay was repeated at
least three times.
Construction of recombinant
adenovirus
Recombinant adenoviruses for the present study were
constructed using the AdEasy system (23,24).
In brief, the coding sequence of human BMP9 and HIF-1α was
amplified and sub-cloned into a shuttle vector (pAdTrace-TO4);
Oligo cassettes for BMP9 or HIF-1α silencing were cloned into a
pSES1 shuttle vector. The shuttle vectors were then recombined with
pAdEasy1 in BJ5183 cells. Finally, the correct recombinant vectors
were transfected into 293 cells (obtained from ATCC) for packaging
adenoviruses, which were designated as AdBMP9, AdHIF-1α, AdsiBMP9
and AdsiHIF-1α. All recombinant adenoviruses were tagged with green
fluorescent protein (GFP) and AdGFP was used as the vector
control.
Flow cytometry for cell cycle and
apoptosis
Subconfluent cells were placed into 6-well culture
plates and treated according to the experimental design for 48 h.
For cell cycle analysis, cells were harvested and washed carefully
with PBS (4°C), fixed with cold (4°C) 70% ethanol, washed with 50%
ethanol, 30% ethanol, and PBS. Finally, the cells were stained with
propidium iodide (PI) PBS solution (20 mg/ml, containing RNase 1
mg/ml) for 30 min, followed by flow cytometric analysis (BD
FACSVantage SE; Kaluza Analysis ver 2.0). For apoptosis analysis,
cells were collected and washed with PBS (4°C), followed by
incubation with Annexin V-EGFP and PI, following the manufacturer's
instructions (cat. no. KGA104; Nanjing KeyGen Biotech Co., Ltd.).
Finally, the cells were subjected to fluorescence-activated cell
sorting. Each assay was repeated at least three times.
Protein harvest and western blot
analysis
Subconfluent HCT116 cells were seeded in 6-well
culture plates and treated with different concentrations of Evo
(0.5, 1 or 2 µM) or DMSO. At each scheduled time-point, the cells
were washed with PBS (4°C) and lysed with 300 µl lysis buffer (cat.
no. R0020; Solarbio Science and Technology Co., Ltd.). The protein
level was assessed with BCA, and lysates were boiled for 10 min.
The protein mass for each loading was 45 µg per lane, and proteins
were subjected to 10% SDS-PAGE gel separation. Then, the proteins
were transferred onto polyvinylidene fluoride membranes and blocked
with 5% bovine serum albumin (BSA) (cat. no. SW3015; Solarbio
Science and Technology Co., Ltd.) for 1 h. Finally, the membranes
were incubated with corresponding primary antibodies (GAPDH, PCNA,
Bad, Bcl-2, BMP9, HIF-1α, Smad1/5/8, p-Smad1/5/8, p53 and p-p53)
for 2 h and horseradish peroxidase-conjugated secondary antibodies
for anti-rabbit, anti-mouse or anti-goat (cat. nos. A0208, A0216
and A0181; Beyotime Institute of Biotechnology) for 1 h sequently
at room temperature. Target proteins were visualized using
SuperSignal West Pico Substrate (cat. no. 34096; Thermo Fisher
Scientific, Inc.), images were captured with chemiluminescence
imager (ChemiDoc XRS; Bio-Rad Laboratories Co., Ltd.) and
quantified using Image Lab Software (version 4.1; Bio-Rad
Laboratories Co., Ltd.). Each assay was repeated at least three
times.
Reverse transcription quantitative
polymerase chain reaction (RT-qPCR)
Subconfluent HCT116 cells were seeded in a T25
culture flask and treated according to the experimental design. At
24 and/or 48 h after treatment, total RNA was extracted using
TRIzol reagent (cat. no. 15596-026; Thermo Fisher Scientific,
Inc.). The RNA was used to generate cDNA with RT kit (cat. no.
R037A; Takara Biotechnology). Next, the cDNA products were used as
templates for the following PCR assay. SYBR-Green kit (cat. no.
B21202) was purchased from Bimake (Shanghai, China). The qPCR
assays were performed with CFX Connect Real-Time PCR detection
system (Bio-Rad Laboratories, Inc.). The thermocycling conditions
consisted of an initial denaturation, followed by 40 cycles at 95°C
for 5 sec and 60°C for 30 sec. Analysis was conducted with CFX
Connect system's software (Bio-Rad Laboratories, Inc.). GAPDH was
used as internal control for mRNA expression levels and the
relative mRNA levels were calculated with the 2−ΔΔCq
method (25). The primers used for
this study are as follows: Bad forward, 5′-CGGAGGATGAGTGACGAGTT-3′
and reverse, 5′-CGGAGGATGAGTGACGAGTT-3′; Bcl-2 forward,
5′-GGATGCCTTTGTGGAACTGT-3′ and reverse, 5′-AGCCTGCAGCTTTGTTTCAT-3′;
GAPDH forward, 5′-CAACGAATTTGGCTACAGCA-3′ and reverse,
5′-AGGGGAGATTCAGTGTGGTG-3′. Each assay was repeated at least three
times.
Xenograft tumor model of human colon
cancer
The animal experiment was approved by the
Institutional Animal Care and Use Committee of Chongqing Medical
University (Chongqing, China). Twenty athymic nude mice (female,
4–6 weeks old, 18–22 g, 5/group) were purchased from the Animal
Center of Chongqing Medical University (Chongqing, China). HCT116
cells were pretreated with AdGFP, AdBMP9, or AdBMP9 plus
AdsiHIF-1α, and then collected and resuspended in PBS (4°C). The
final density was 2×107 cells/ml. Cells in 50 µl PBS
(4°C) were injected into the flanks of athymic mice. At 3 days
following injection, animals were treated with Evo (10 mg/kg) or
the same volume of solvent intragastrically once a day. Four weeks
after injection, mice were euthanized by intraperitoneal injection
of pentobarbital sodium (180 mg/kg body weight). Animals were
sacrificed and the tumor masses were retrieved for histological
evaluation when no autonomous breathing was produced for 2–3 min
and no blink reflexes appeared.
Histological evaluation
Retrieved tumor masses were fixed in 10% formalin
and embedded with paraffin. Sections were stained with hematoxylin
and eosin (H&E) after being deparaffinized and rehydrated.
Statistical analysis
All quantitative experiments were performed in
triplicate. Data are expressed as the mean ± SD. Statistical
analysis was performed with GraphPad Prism 6 (GraphPad Software,
Inc.). A two-tailed t-test was used to compare differences between
two groups, and one-way analysis of variance with Tukey's post hoc
test was used to compare differences among multiple groups.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Effect of Evo on HCT116 cell
proliferation
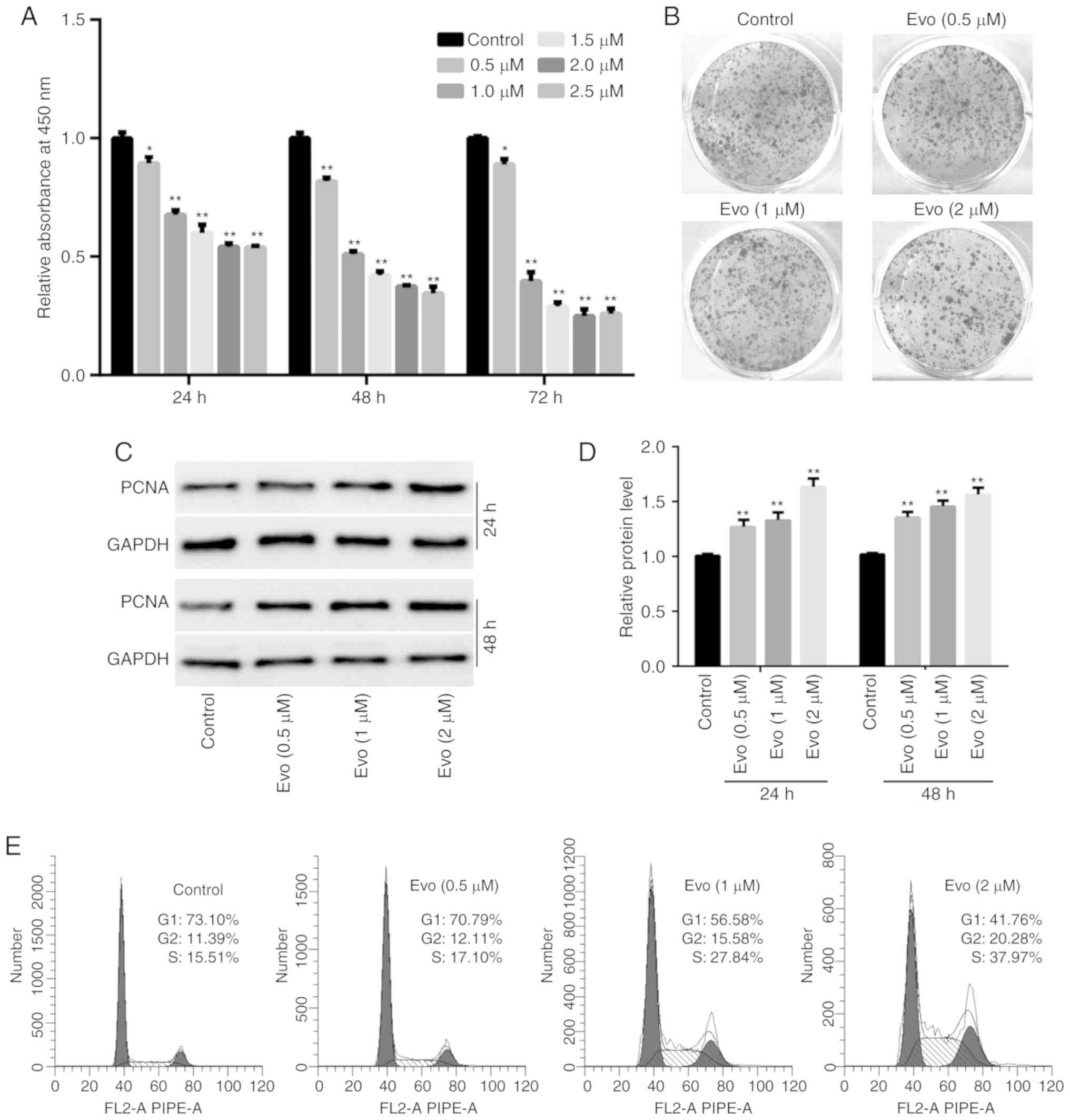
CCK-8 assay results revealed that Evo decreased the
viability of HCT116 cells in a concentration- and time-dependent
manner (Fig. 1A). Crystal violet
staining results revealed that Evo decreased the HCT116 cell colony
formation ability when the concentration of cells was >1 µM
(Fig. 1B). Western blot analysis
results revealed that Evo significantly increased the protein
expression of PCNA in HCT116 cells compared to the control
(Fig. 1C and D). Flow cytometric
results revealed that Evo markedly arrested the cell-cycle at the S
phase in HCT116 cells compared to the control (Fig. 1E). These results indicated that Evo
inhibited the proliferation of human colon cancer cells.
Effects of Evo on HCT116 cell
apoptosis
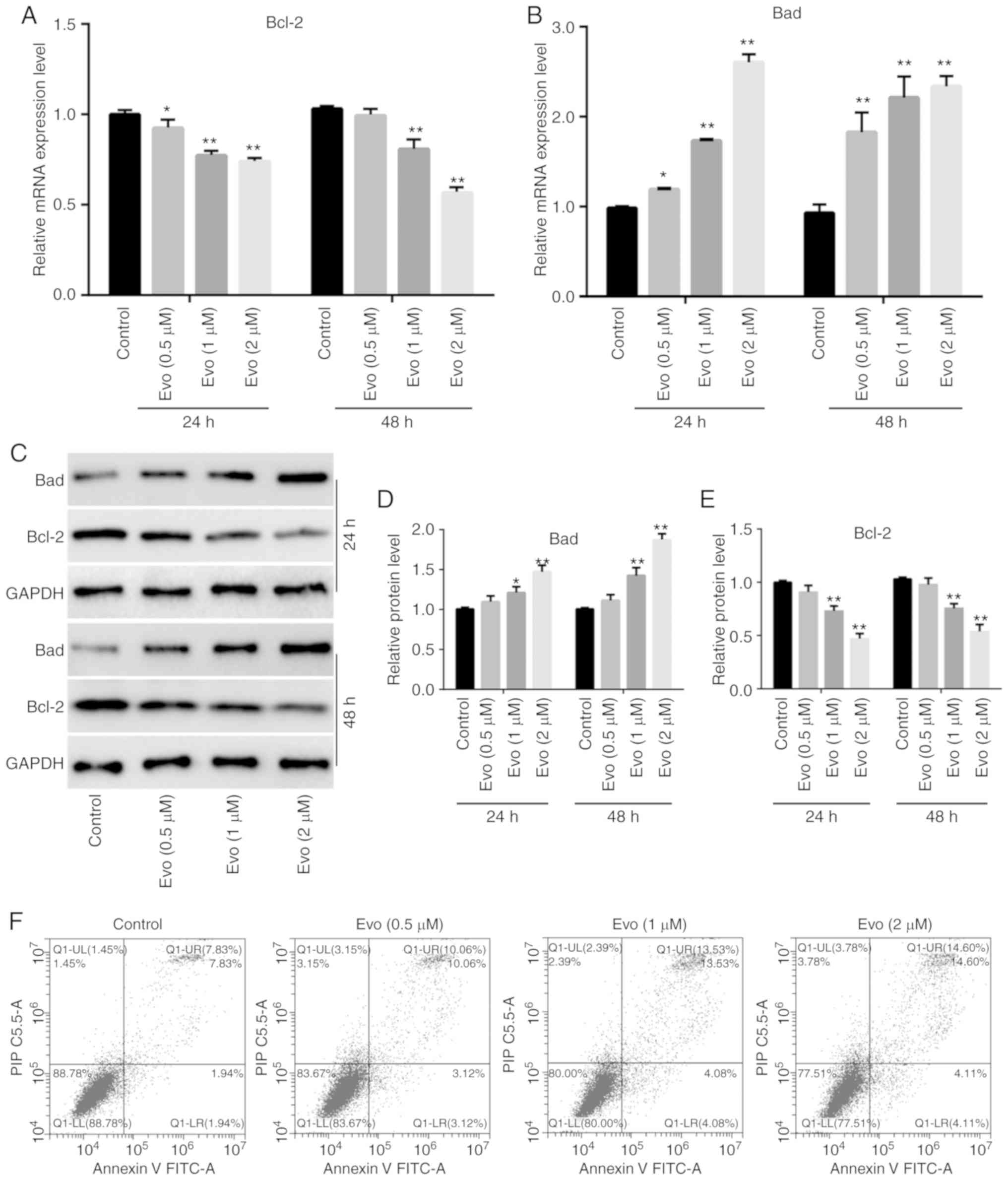
RT-qPCR assay results revealed that Evo increased
the mRNA expression of Bad, but significantly decreased that of
Bcl-2 (Fig. 2A and B). In addition,
western blot analysis results revealed that Evo (at 1 and 2 µM)
increased the expression of Bad, and significantly decreased that
of Bcl-2, which was more pronounced at 48 h compared to the control
(Fig. 2C-E). Flow cytometric
results revealed that Evo also slightly increased the percentage of
apoptotic HCT116 cells, even at a concentration of 0.5 µM (Fig. 2F). These findings indicated that Evo
induced apoptosis in HCT116 cells.
Effects of BMP9 on the anticancer
activity of Evo in HCT116 cells
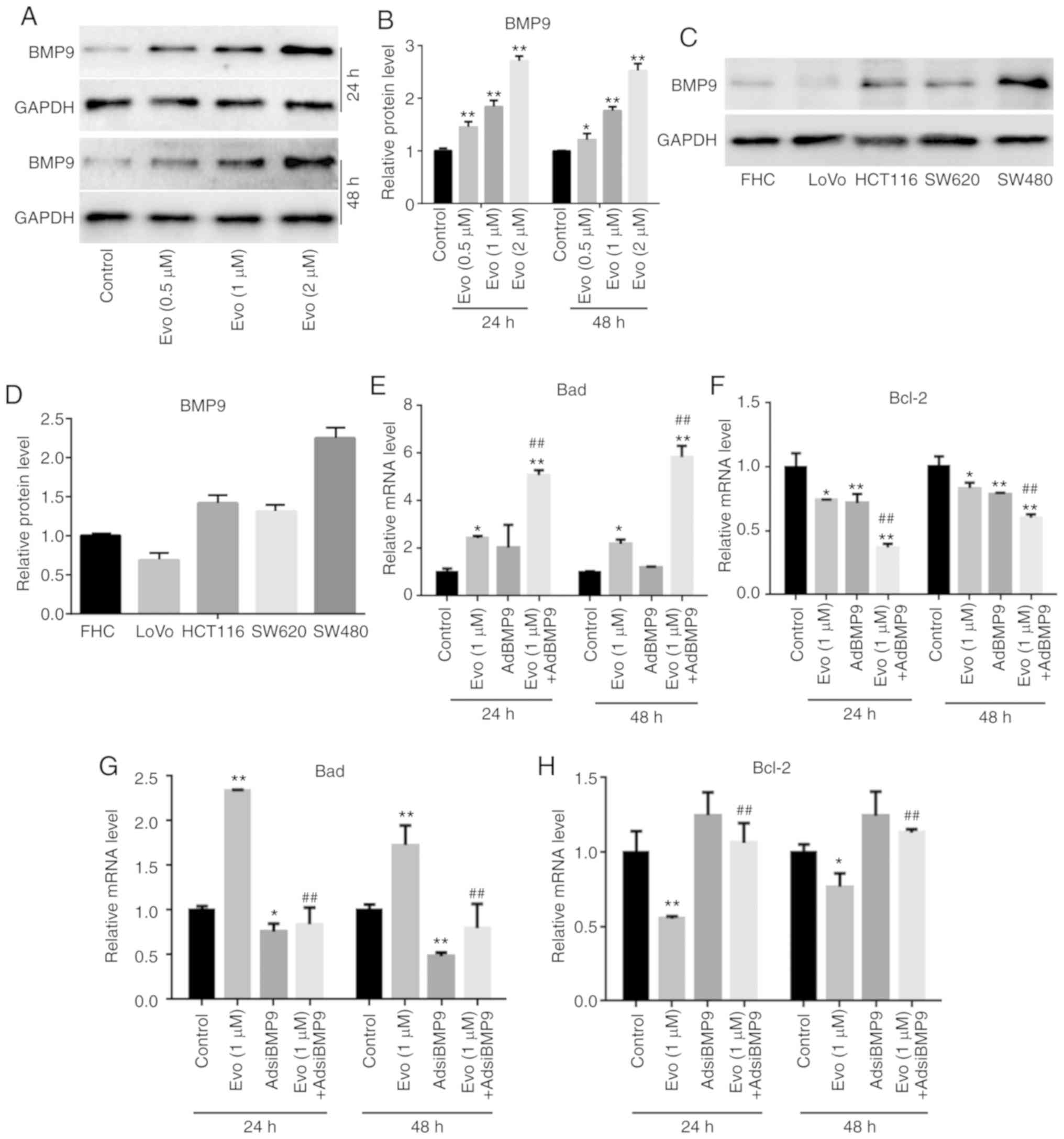
Western blot analysis results revealed that Evo
significantly increased the level of BMP9 in HCT116 cells compared
to the control (Fig. 3A and B), and
that endogenous BMP9 could be detected in the FCH cells and several
other colon cancer cell lines (Fig. 3C
and D). RT-qPCR results revealed that BMP9 could slightly
increase the mRNA expression of Bad and significantly decrease that
of Bcl-2 compared to the control, and BMP9 promoted the increasing
effect of Evo on the mRNA expression of Bad, and the reducing
effect of Evo on Bcl-2 in HCT116 cells compared to the group
treated with Evo only (Fig. 3E and
F). Conversely, BMP9 silencing slightly decreased the mRNA
expression of Bad and increased that of Bcl-2 compared to the
control. It was also revealed that BMP9 silencing decreased the
increasing effect of Evo on the mRNA expression of Bad, and
reversed the decreasing effect of Evo on the mRNA expression of
Bcl-2 in HCT116 cells compare to the group treated with Evo only
(Fig. 3G and H). These results
indicated that BMP9 may mediate the anticancer effect of Evo in
colon cancer cells.
Effects of HIF-1α on the anticancer
activity of Evo in HCT116 cells
HIF-1α has been revealed to be upregulated by BMP9
in progenitor cells. HIF-1α has also been revealed to be involved
in tumorigenesis. Therefore, it was next determined whether HIF-1α
could mediate the effect of BMP9 on the anticancer activity of Evo
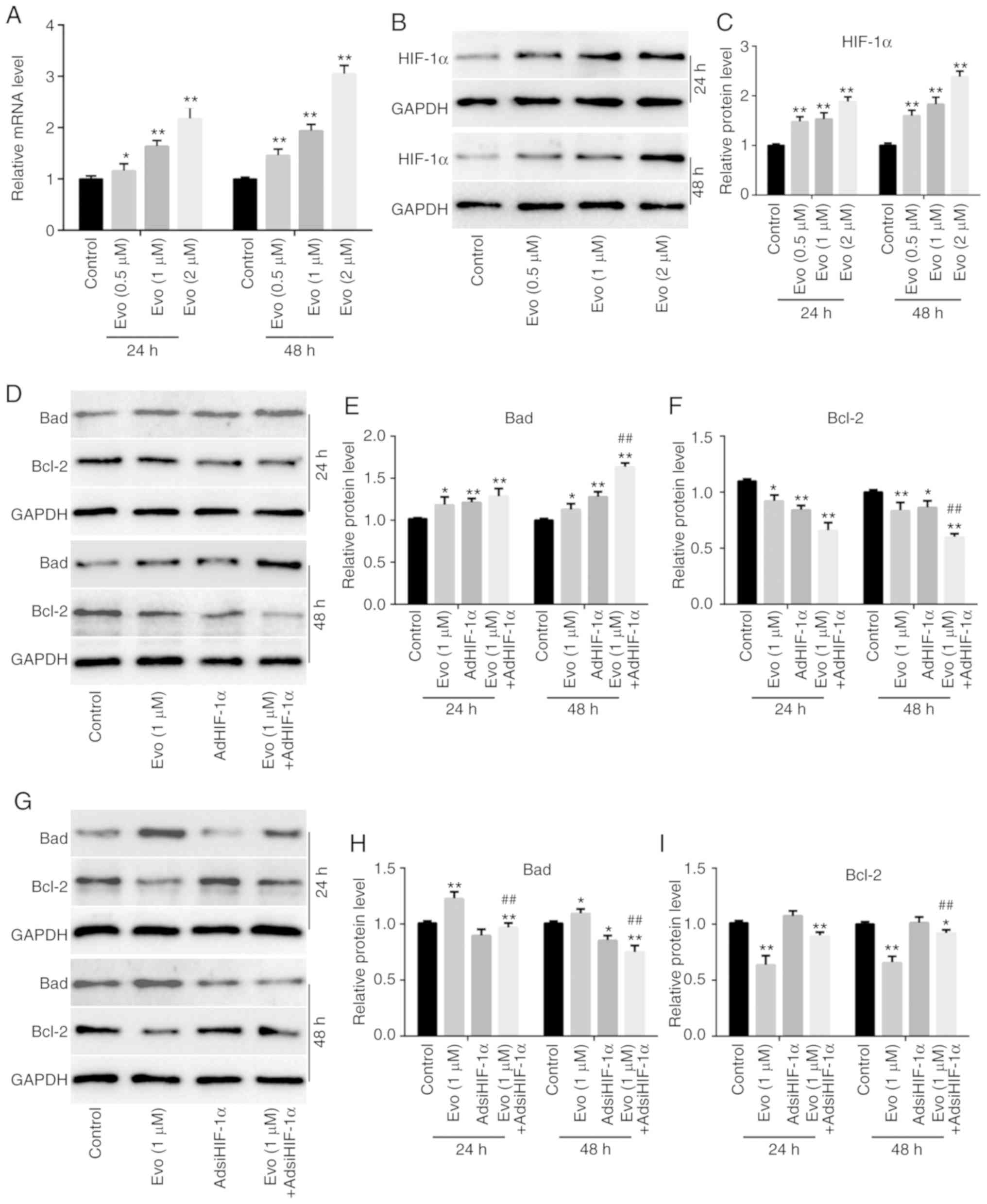
in HCT116 cells. RT-qPCR results revealed that Evo significantly
increased the mRNA expression of HIF-1α in HCT116 cells compared to
the control (Fig. 4A). Western blot
analysis results also revealed that Evo increased HIF-1α in HCT116
cells compared to the control (Fig. 4B
and C). Furthermore, AdHIF-1α-mediated exogenous HIF-1α
increased the protein level of Bad and reduced the level of Bcl-2
compared to the control, and promoted the increasing effect of Evo
on Bad and the decreasing effect of Evo on Bcl-2 compared to the
group treated with Evo only (Fig.
4D-F). Conversely, HIF-1α silencing slightly reduced the
protein level of Bad compared to the control, but attenuated the
increasing effect of Evo on Bad and the decreasing effect of Evo on
Bcl-2 in HCT116 cells compared to the group treated with Evo only
(Fig. 4G-I). In addition, our
previous study also indicated that the anticancer activity of Evo
was associated with the downregulation of HIF-1α in human colon
cancer cells (LoVo) (10). These
results indicated that HIF-1α may also participate in mediating the
anticancer activity of Evo in HCT116 cells.
Effects of BMP9 and/or HIF-1α on the
anticancer activity of Evo in human colon cancer
BMP9 and HIF-1α both affected the anticancer
activity of Evo. Thus, whether HIF-1α could mediate the effect of
BMP9 on Evo in colon cancer cells was next determined. Western blot
analysis results revealed that Evo exhibited no substantial effect
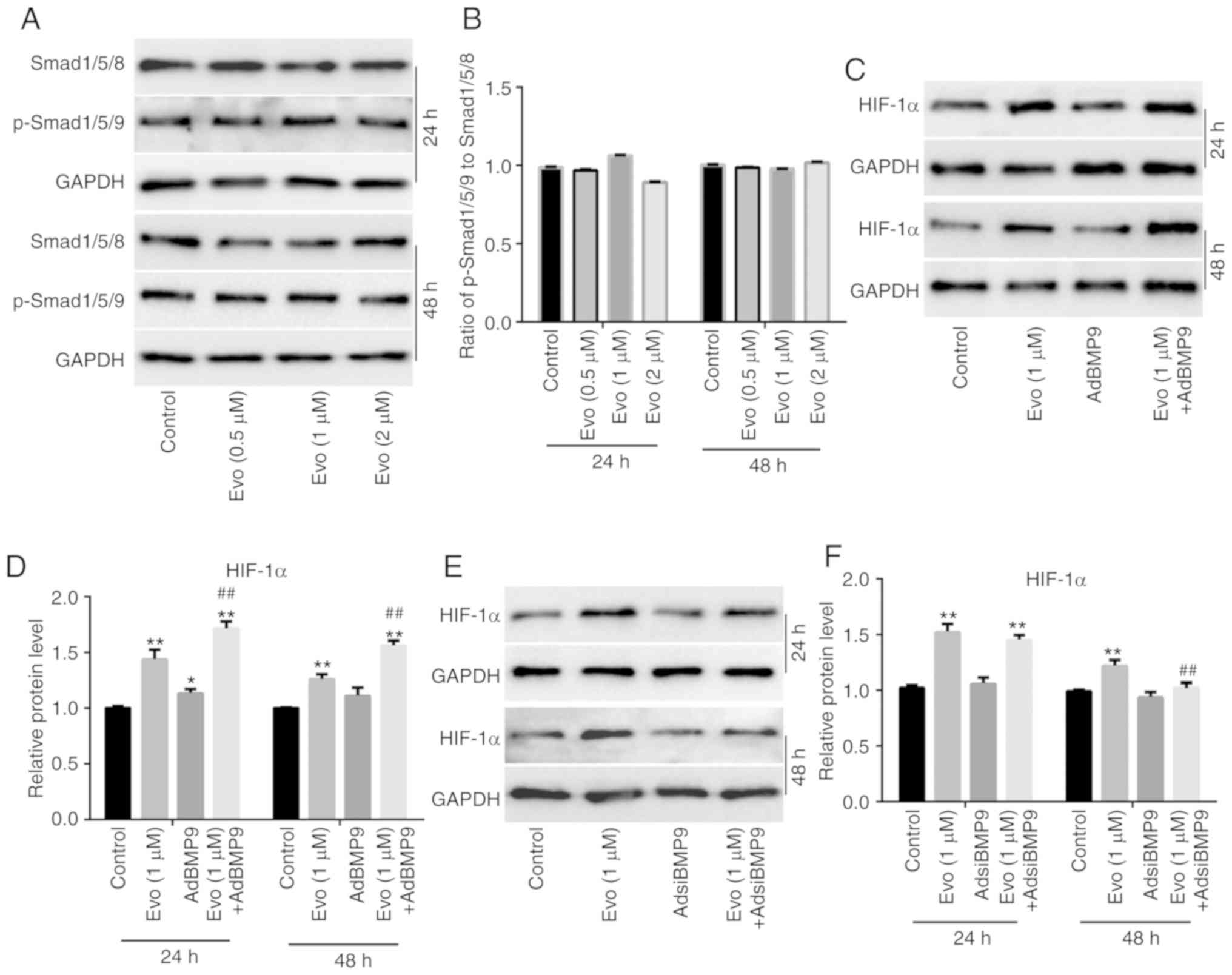
on the protein expression of Smad1/5/8 and p-Smad1/5/8 in HCT116
cells (Fig. 5A). The quantification
of western blot analysis revealed that Evo exerted no obvious
effect on the ratio of p-Smad1/5/8 to Smad1/5/8 in HCT116 cells
(Fig. 5B). Further western blot
analysis results revealed that BMP9 slightly increased HIF-1α
compared to the control, but significantly promoted the increasing
effect of Evo on HIF-1α in HCT116 cells compared to the group
treated with Evo only (Fig. 5C and
D). BMP9 silencing also exerted no substantial effect on HIF-1α
compared to the control, but significantly reduced the increasing
effect of Evo on HIF-1α in HCT116 cells compared to the group
treated with Evo only (Fig. 5E and
F). Xenograft tumor assay results revealed that Evo markedly
inhibited tumor growth compared to the control, and BMP9 enhanced
the antitumor growth effect of Evo (the largest tumor mass appeared
in the control group, and the maximum diameter of the tumor masses
was 1.1 cm), which could almost be reversed by HIF-1α silencing
(Fig. 5G). Histochemical staining
(H&E) results revealed that Evo markedly increased
karyopyknosis compared to the control, and BMP9 potentiated that
effect, which was clearly attenuated by HIF-1α silencing (Fig. 5H). These results indicated that
HIF-1α may mediate the effect of BMP9 on the anticancer activity of
Evo in colon cancer.
Effects of Evo, BMP9 and/or HIF-1α on
the activity of p53 in HCT116 cells
Finally, it was investigated how HIF-1α mediates the
effect of BMP9 on the anticancer activity of Evo in HCT116 cells.
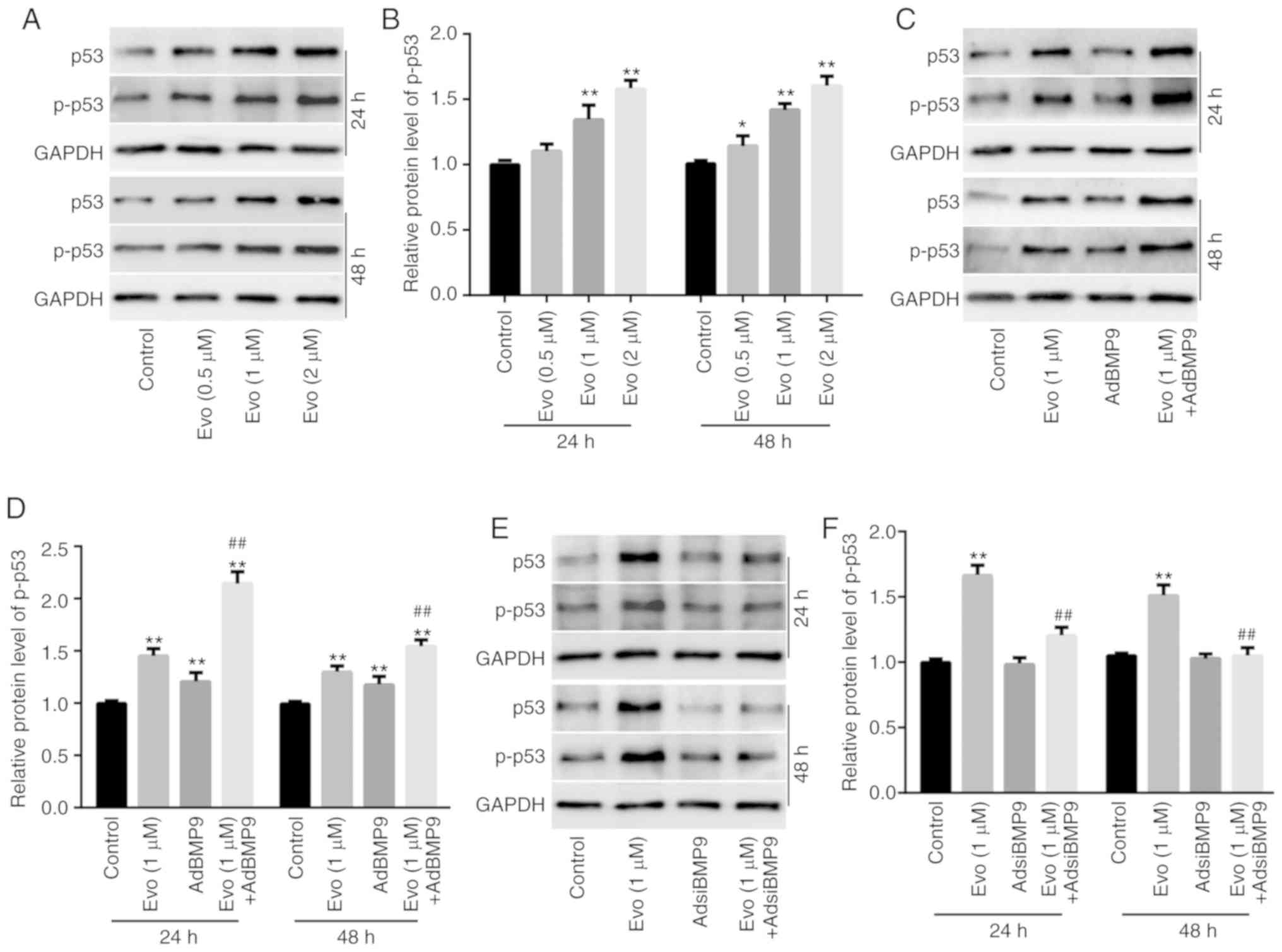
Western blot analysis results revealed that Evo (1 and 2 µM)
significantly increased the protein level of p53 and p-p53 in
HCT116 cells compared to the control (Fig. 6A and B). Evo increased the level of
p53 and p-p53 in HCT116 cells compared to the control, and BMP9
potentiated the increasing effect of Evo on the level of p53 and
p-p53 compared to the group treated with Evo only (Fig. 6C and D). Conversely, BMP9 silencing
slightly decreased the level of p53 and p-p53 compared to the
control, and significantly reduced the increasing effect of Evo on
p53 and p-p53 in HCT116 cells compared to the group treated with
Evo only (Fig. 6E and F).
Furthermore, western blot analysis results revealed that HIF-1α
slightly increased the level of p53 and p-p53 in HCT116 cells
compared to the control, but significantly promoted the increasing
effect of Evo on p53 and p-p53 in HCT116 cells compared to the
group treated with Evo only (Fig. 6G
and H). HIF-1α silencing also exerted no obvious effect on p53
and p-p53 compared to the control, but significantly decreased the
increasing effect of Evo on p53 and p-p53 in HCT116 cells compared
to the group treated with Evo only (Fig. 6I and J). These data indicated that
HIF-1α may mediate the effect of BMP9 on the anticancer activity of
Evo by partly enhancing the activity of p53 in colon cancer
cells.
Discussion
Colon cancer remains one of the most common
malignancies. In the past decades, there have been substantial
developments in diagnostic and clinical treatment regimens for
colon cancer. However, its prognosis remains poorer than expected.
Therefore, there is still a need for the development of new and
effective drugs or strategies for colon cancer treatment. In the
present study, the effective anticancer activity of Evo, which may
be mediated by BMP9 via the upregulation of HIF-1α to partly
increase the activity of p53, was demonstrated in human colon
cancer.
It has been reported that Evo exhibits great
anti-cancer activities in many types of cancer, such as tongue,
breast, colon, prostate and lung cancer (9–12,26,27).
As far as colon cancer is concerned, the anticancer activity of Evo
may be mediated by inactivating the NF-κB (9) or TGF-β1 (11), or activating the p53/p21/Rb pathway
(12). Our previous study indicated
that the anti-cancer activity of Evo was associated with the
downregulation of HIF-1α, at least in colon cancer (10). However, the explicit molecular
mechanism underlying this activity of Evo needs to be further
clarified.
TGF-β is a super-family that includes various
cytokines, which have been revealed to regulate several essential
cell physiological processes, such as cell proliferation and
differentiation (28). BMPs belong
to the TGF-β super-family, and it was reported that normal BMP
signaling is necessary for the development of colon
microenvironment and terminal differentiation of intestinal
progenitor cells (18,19). The loss or inactivation of BMP
signaling may lead to the development of gastric neoplasm,
colorectal epithelial overgrowth and polyp formation (29–31).
Therefore, the aberrant BMP/Smad signaling may be one of major
causes of colon cancer, and a potential target for its treatment.
Voorneveld et al reported that the administration of statins
following diagnosis can significantly reduce the risk of colon
cancer-associated mortality, if BMP signaling remains intact
(32). Our previous studies have
indicated that oridion and honokiol both exhibited effective
anticancer activity in human colon cancer by upregulating BMP7 to
increase p53 activity (33,34). BMP9 is another member of the BMP
family, which has been less studied than any of the other BMPs. It
was reported that BMP9 exhibited a potential to commit mesenchymal
stem cell to osteoblastic lineage, which is much stronger than that
of BMP2 or BMP7 (35). However, our
previous results demonstrated that BMP9 may also mediate the
anticancer effect of resveratrol in colon cancer in a p38
MAPK-dependent manner (21). We
therefore speculated that BMP9 may also be associated with the
anticancer activity of Evo in colon cancer. In the present study,
it was demonstrated that Evo significantly increased the expression
of BMP9. However, the endogenous BMP9 level in HCT116 cells
(Fig. 3C) was slightly higher than
that of the control group (Fig.
3A). This difference may due to the different treatments,
namely cells were treated with the same volume of DMSO as the
Evo-treated groups for the data in Fig.
3A, but no treatment was introduced for the data in Fig. 3C. Exogenous BMP9 enhanced the
anti-cancer activity of Evo in HCT116 cells, and BMP9 silencing
showed the reverse effect. Therefore, BMP9 may also contribute to
the anticancer activity of Evo in colon cancer cells.
BMP9, also named growth and differentiation factor
2, is capable of regulating cell proliferation and differentiation.
In general, BMP9 exerts its physiological function through the
BMP/Smad pathway. However, it can also realize its function through
the non-canonical BMP/Smad pathway, such as PI3K/Akt or MAPKs
(36,37). A previous study revealed that the
aberrant BMP/Smad signaling transduction is one of the pathogenic
causes of colon cancer, along with Smad4 mutation or function loss
(38). In the present study, it was
revealed that Evo exhibited no substantial effect on increasing the
activity of BMP/Smad signaling. The effect of BMP9 on the
anticancer activity of Evo may therefore not be mediated through
the BMP/Smad signaling pathway, at least in HCT116 cells. Our
previous study demonstrated that Evo could downregulate
hypoxia-inducible factor 1α (HIF-1α) in a human colon cancer cell
line (LoVo), and HIF-1α silencing potentiated the anticancer effect
of Evo (10). François et al
reported that BMP2 can induce HIF-1α in chronic wounds (39). We therefore speculated that the
effect of BMP9 on the anticancer activity of Evo may also be
associated with HIF-1α. HIFs, including HIF-1, HIF-2 and HIF-3, are
special transcriptional factors that respond to the decreased
oxygen level in cellular context. HIF-1 consists of HIF-1α and
HIF-1β subunits, which form heterodimers to regulate downstream
targets. Although HIF-1α and HIF-1β are both constitutively
expressed, the half-life of HIF-1α is so short, that the
transcriptional activity of HIF-1 is mostly determined by HIF-1α
(40). HIF-1α is critical for
angiogenesis and new vascular formation, which is very important
for tumor growth, metastasis and relapse (41). Therefore, HIF-1α may promote tumor
progression and has been reported as a promising anticancer
therapeutic target (42). Our
previous studies also indicated that the downregulation of HIF-1α
may mediate the anticancer activity of Evo in LoVo colon cancer
cells (10). However, in the
present study, it was revealed that Evo upregulated HIF-1α in
HCT116 cells, BMP9 potentiated the upregulating effect of Evo on
HIF-1α, and BMP9 silencing attenuated the upregulating effect of
Evo on HIF-1α. Furthermore, BMP9 enhanced the antitumor growth
effect of Evo in colon cancer, which could be partly reduced by
silencing of HIF-1α. Therefore, the present data indicated that
BMP9 may mediate the anticancer activity of Evo by upregulating
HIF-1α in colon cancer cells. These controversial effects of Evo
and/or HIF-1α in colon cancer cells may be dependent on the cell
type or cellular context, and mechanism that needs to be thoroughly
elucidated. In addition, the way in which HIF-1α mediated the
anticancer activity of Evo and/or BMP9 in HCT116 cells remains
unclear.
Usually, cancer cells are characteristic of
mutations or the inactivation of tumor suppressors, such as p53 and
PTEN (43,44). As one most famous tumor suppressor,
p53 is considered as a potential target for several drugs. Further
analysis indicated that Evo increased the activity of p53, which
was potentiated by BMP9 and reduced by BMP9 silencing. Furthermore,
HIF-1α potentiated the increasing effect of Evo on the activity of
p53, and HIF-1α silencing had the reverse effect in HCT116 cells.
These findings indicated that p53 may also be activated by BMP9 in
cancer cells, which may be the result of the upregulation of HIF-1α
by BMP9 in HCT116 cells. MDM2 is a specific p53 ubiquitin ligase
and transcriptional inhibitor, which negatively regulates p53
activity (45). Therefore, the
effect of BMP9 on p53 may be mediated by HIF-1α through regulation
of the expression of MDM2. The results from further analysis also
revealed that Evo decreased the expression of MDM2, which could be
enhanced by HIF-1α and reduced by HIF-1α silencing in HCT116 cells
(data not shown).
Collectively, the present study indicated that Evo
exhibits effective anticancer activity in human colon cancer, and
BMP9 may partly mediate this activity by upregulating HIF-1α to
enhance the activity of p53, at least in HCT116 cells. BMP9 was
revealed to suppress cancer cell proliferation in a p53-dependent
manner in colon cancer. However, the exact mechanism underlying
this process needs to be further studied.
Acknowledgements
The authors would like to thank Dr. T.C.-He
(University of Chicago Medical Center, Chicago, USA) for providing
all recombinant adenoviruses.
Funding
The present study was supported in part by research
grants from the Chongqing Health and Family Planning Commission
(grant no. ZY201702104 awarded to DZS) and the National Natural
Science Foundation of China (grant no. NSFC 81572226 awarded to
BCH).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author upon reasonable
request.
Authors' contributions
BCH and DZS designed the study. FSL, JH, MZC and JRZ
conducted the experiments. PPL, LL, YD and YH analyzed the data.
FSL and BCH wrote the manuscript. All authors read and approved the
manuscript and agree to be accountable for all aspects of the work
in ensuring that questions related to the accuracy or integrity of
any part of the work are appropriately investigated and
resolved.
Ethics approval and consent to
participate
All animal experiments were carried out in
accordance with the NIH Guide for the Care and Use of Laboratory
Animals and were approved by the Institutional Animal Care and Use
Committee of Chongqing Medical University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Lee JJ and Chu E: The adjuvant treatment
of stage III colon cancer: might less be more? Oncology (Williston
Park). 32:437–442, 444. 2018.PubMed/NCBI
|
|
2
|
Kang J, Chong SW, Park EJ, Baik SH and Lee
KY: Safety and feasibility of in-hospital early chemotherapy
initiation after surgery in patients with stage II–IV colon cancer.
Medicine (Baltimore). 98:e153712019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zhao J, Xue Y, Pan Y, Yao A, Wang G, Li D,
Wang T, Zhao S and Hou Y: Toll-like receptor 3 agonist poly I:C
reinforces the potency of cytotoxic chemotherapy via the
TLR3-UNC93B1-IFN-β signaling axis in paclitaxel-resistant colon
cancer. J Cell Physiol. 234:7051–7061. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Sun QL, Zhao CP, Wang TY, Hao XB, Wang XY,
Zhang X and Li YC: Expression profile analysis of long non-coding
RNA associated with vincristine resistance in colon cancer cells by
next-generation sequencing. Gene. 572:79–86. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Gokduman K: Strategies targeting DNA
topoisomerase I in cancer chemotherapy: Camptothecins, nanocarriers
for camptothecins, organic non-camptothecin compounds and metal
complexes. Curr Drug Targets. 17:1928–1939. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Yoon SB and Park HR: Arctigenin inhibits
etoposide resistance in HT-29 colon cancer cells during
microenvironmental stress. J Microbiol Biotechnol. 29:571–576.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Jiang J and Hu C: Evodiamine: A novel
anti-cancer alkaloid from Evodia rutaecarpa. Molecules.
14:1852–1859. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yu H, Jin H, Gong W, Wang Z and Liang H:
Pharmacological actions of multi-target-directed evodiamine.
Molecules. 18:1826–1843. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Guo Q, Liu Y, Zhao J and Wang J, Li Y,
Pang Y, Chen J and Wang J: Evodiamine inactivates NF-κB and
potentiates the antitumor effects of gemcitabine on tongue cancer
both in vitro and in vivo. Onco Targets Ther. 12:257–267. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Huang J, Chen ZH, Ren CM, Wang DX, Yuan
SX, Wu QX, Chen QZ, Zeng YH, Shao Y, Li Y, et al: Antiproliferation
effect of evodiamine in human colon cancer cells is associated with
IGF-1/HIF-1α downregulation. Oncol Rep. 34:3203–3211. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Huang C, Liu H, Gong XL, Wu LY and Wen B:
Effect of evodiamine and berberine on the interaction between DNMTs
and target microRNAs during malignant transformation of the colon
by TGF-β1. Oncol Rep. 37:1637–1645. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Han S, Woo JK, Jung Y, Jeong D, Kang M,
Yoo YJ, Lee H, Oh SH, Ryu JH and Kim WY: Evodiamine selectively
targets cancer stem-like cells through the p53-p21-Rb pathway.
Biochem Biophys Res Commun. 469:1153–1158. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Russo A, Bazan V, Iacopetta B, Kerr D,
Soussi T and Gebbia N; TP53-CRC Collaborative Study Group, : The
TP53 colorectal cancer international collaborative study on the
prognostic and predictive significance of p53 mutation: Influence
of tumor site, type of mutation, and adjuvant treatment. J Clin
Oncol. 23:7518–7528. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Jass JR: Pathogenesis of colorectal
cancer. Surg Clin North Am. 82:891–904. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ogino S, Lochhead P, Giovannucci E,
Meyerhardt JA, Fuchs CS and Chan AT: Discovery of colorectal cancer
PIK3CA mutation as potential predictive biomarker: Power and
promise of molecular pathological epidemiology. Oncogene.
33:2949–2955. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Huang D, Sun W, Zhou Y, Li P, Chen F, Chen
H, Xia D, Xu E, Lai M, Wu Y and Zhang H: Mutations of key driver
genes in colorectal cancer progression and metastasis. Cancer
Metastasis Rev. 37:173–187. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Urist MR: Bone: Formation by
autoinduction. Science. 150:893–899. 1965. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Allaire JM, Roy SA, Ouellet C, Lemieux É,
Jones C, Paquet M, Boudreau F and Perreault N: Bmp signaling in
colonic mesenchyme regulates stromal microenvironment and protects
from polyposis initiation. Int J Cancer. 138:2700–2712. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Auclair BA, Benoit YD, Rivard N, Mishina Y
and Perreault N: Bone morphogenetic protein signaling is essential
for terminal differentiation of the intestinal secretory cell
lineage. Gastroenterology. 133:887–896. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Bertrand FE, Angus CW, Partis WJ and
Sigounas G: Developmental pathways in colon cancer: Crosstalk
between WNT, BMP, Hedgehog and Notch. Cell cycle. 11:4344–4351.
2012. View
Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yuan SX, Wang DX, Wu QX, Ren CM, Li Y,
Chen QZ, Zeng YH, Shao Y, Yang JQ, Bai Y, et al: BMP9/p38 MAPK is
essential for the antiproliferative effect of resveratrol on human
colon cancer. Oncol Rep. 35:939–947. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
He BC, Gao JL, Zhang BQ, Luo Q, Shi Q, Kim
SH, Huang E, Gao Y, Yang K, Wagner ER, et al: Tetrandrine inhibits
Wnt/β-catenin signaling and suppresses tumor growth of human
colorectal cancer. Mol Pharmacol. 79:211–219. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
He TC, Zhou S, da Costa LT, Yu J, Kinzler
KW and Vogelstein B: A simplified system for generating recombinant
adenoviruses. Proc Natl Acad Sci USA. 95:2509–2514. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Luo J, Deng ZL, Luo X, Tang N, Song WX,
Chen J, Sharff KA, Luu HH, Haydon RC, Kinzler KW, et al: A protocol
for rapid generation of recombinant adenoviruses using the AdEasy
system. Nat Protoc. 2:1236–1247. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kan SF, Yu CH, Pu HF, Hsu JM, Chen MJ and
Wang PS: Anti-proliferative effects of evodiamine on human prostate
cancer cell lines DU145 and PC3. J Cell Biochem. 101:44–56. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Su T, Yang X, Deng JH, Huang QJ, Huang SC,
Zhang YM, Zheng HM, Wang Y, Lu LL and Liu ZQ: Evodiamine, a novel
NOTCH3 methylation stimulator, significantly suppresses lung
carcinogenesis in vitro and in vivo. Front Pharmacol. 9:4342018.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
de Araújo Farias V, Carrillo-Gálvez AB,
Martin F and Anderson P: TGF-β and mesenchymal stromal cells in
regenerative medicine, autoimmunity and cancer. Cytokine Growth
Factor Rev. 43:25–37. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Roy SAB, Allaire JM, Ouellet C,
Maloum-Rami F, Pomerleau V, Lemieux É, Babeu JP, Rousseau J, Paquet
M, Garde-Granger P, et al: Loss of mesenchymal bone morphogenetic
protein signaling leads to development of reactive stroma and
initiation of the gastric neoplastic cascade. Sci Rep. 6:327592016.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Beppu H, Mwizerwa ON, Beppu Y, Dattwyler
MP, Lauwers GY, Bloch KD and Goldstein AM: Stromal inactivation of
BMPRII leads to colorectal epithelial overgrowth and polyp
formation. Oncogene. 27:1063–1070. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Means AL, Freeman TJ, Zhu J, Woodbury LG,
Marincola-Smith P, Wu C, Meyer AR, Weaver CJ, Padmanabhan C, An H,
et al: Epithelial Smad4 deletion Up-regulates inflammation and
promotes inflammation-associated cancer. Cell Mol Gastroenterol
Hepatol. 6:257–276. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Voorneveld PW, Reimers MS, Bastiaannet E,
Jacobs RJ, van Eijk R, Zanders MMJ, Herings RMC, van Herk-Sukel
MPP, Kodach LL, van Wezel T, et al: Statin use after diagnosis of
colon cancer and patient survival. Gastroenterology. 153:470–479
e4. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Liu RX, Ma Y, Hu XL, Ren WY, Liao YP, Wang
H, Zhu JH, Wu K, He BC and Sun WJ: Anticancer effects of oridonin
on colon cancer are mediated via BMP7/p38 MAPK/p53 signaling. Int J
Oncol. 53:2091–2101. 2018.PubMed/NCBI
|
|
34
|
Liu RX, Ren WY, Ma Y, Liao YP, Wang H, Zhu
JH, Jiang HT, Wu K, He BC and Sun WJ: BMP7 mediates the anticancer
effect of honokiol by upregulating p53 in HCT116 cells. Int J
Oncol. 51:907–917. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Luu HH, Song WX, Luo X, Manning D, Luo J,
Deng ZL, Sharff KA, Montag AG, Haydon RC and He TC: Distinct roles
of bone morphogenetic proteins in osteogenic differentiation of
mesenchymal stem cells. J Orthop Res. 25:665–677. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Jiang HT, Ran CC, Liao YP, Zhu JH, Wang H,
Deng R, Nie M, He BC and Deng ZL: IGF-1 reverses the osteogenic
inhibitory effect of dexamethasone on BMP9-induced osteogenic
differentiation in mouse embryonic fibroblasts via PI3K/AKT/COX-2
pathway. J Steroid Biochem Mol Biol. 191:1053632019. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zhu JH, Liao YP, Li FS, Hu Y, Li Q, Ma Y,
Wang H, Zhou Y, He BC and Su YX: Wnt11 promotes BMP9-induced
osteogenic differentiation through BMPs/Smads and p38 MAPK in
mesenchymal stem cells. J Cell Biochem. 119:9462–9473. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Mehrvarz Sarshekeh A, Advani S, Overman
MJ, Manyam G, Kee BK, Fogelman DR, Dasari A, Raghav K, Vilar E,
Manuel S, et al: Association of SMAD4 mutation with patient
demographics, tumor characteristics, and clinical outcomes in
colorectal cancer. PLoS One. 12:e01733452017. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
François S, Eder V, Belmokhtar K, Machet
MC, Douay L, Gorin NC, Benderitter M and Chapel A: Synergistic
effect of human bone morphogenic protein-2 and mesenchymal stromal
cells on chronic wounds through hypoxia-inducible factor-1 α
induction. Sci Rep. 7:42722017. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Pugh CW, O'Rourke JF, Nagao M, Gleadle JM
and Ratcliffe PJ: Activation of hypoxia-inducible factor-1;
definition of regulatory domains within the alpha subunit. J Biol
Chem. 272:11205–11214. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Balamurugan K: HIF-1 at the crossroads of
hypoxia, inflammation, and cancer. Int J Cancer. 138:1058–1066.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Lin MC, Lin JJ, Hsu CL, Juan HF, Lou PJ
and Huang MC: GATA3 interacts with and stabilizes HIF-1α to enhance
cancer cell invasiveness. Oncogene. 36:4243–4252. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Basu S and Murphy ME: Genetic modifiers of
the p53 pathway. Cold Spring Harb Perspect Med. 6:a0263022016.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Molinari F and Frattini M: Functions and
regulation of the PTEN gene in colorectal cancer. Front Oncol.
3:3262014. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Xia M, Knezevic D, Tovar C, Huang B,
Heimbrook DC and Vassilev LT: Elevated MDM2 boosts the apoptotic
activity of p53-MDM2 binding inhibitors by facilitating MDMX
degradation. Cell Cycle. 7:1604–1612. 2008. View Article : Google Scholar : PubMed/NCBI
|