Introduction
Cervical cancer is the fourth leading cause of
tumor-related mortality and is the most common gynecological tumor
worldwide (1). Persistent infection
with a subset of human papillomaviruses (HPVs), denoted as
‘high-risk’ HPVs, plays a critical role in the initiation and
development of cervical cancer. A cascade of abnormal events occurs
during cervical carcinogenesis, including the induction of genomic
instability, dysregulation of cell proliferation, disruption of
cell cycle control mechanisms and aberrant expression of certain
oncogenes and tumor-suppressor genes (2). Upregulated phosphoinositide 3 kinase
(PI3K), AKT and the mammalian target of rapamycin (mTOR) via the
mTOR pathway have been reported in metastatic and recurrent
cervical cancer patients. Accordingly, a clinical benefit of
therapy targeted to these cell signaling pathways has been reported
in phase I and II clinical trials (3–5),
consistent with the finding that AKT inhibitors promote cell death
in cervical cancer (6).
MicroRNAs (miRNAs) are endogenous, 19-25mer, small
non-coding RNAs that regulate gene expression by base-pairing to
the 3′-untranslated region (3′-UTR) of target messenger RNAs
(mRNAs) (7). miRNAs regulate gene
expression at the post-transcriptional level, subsequently
triggering translational repression or mRNA destabilization.
MicroRNA-126-3p (miR-126-3p), which has been identified in many
types of cancer including lung, breast, gastric, colorectal,
glioblastoma, bladder and prostate, may act either as a tumor
suppressor by inhibiting oncogenes (8–17) or
as a tumor promoter by inhibiting tumor suppressors (18,19).
In cervical cancer, disparate data on the levels of miR-126-3p have
been reported, either downregulation in some studies (12,14,20,21)
while upregulation in others (15,22),
albeit under different experimental conditions. Previous studies
have shown that miR-126-3p can target a site in the PI3K subunit
p85β (PIK3R2) 3′-UTR, not only in different types of cancer
(23–26) but also in benign diseases (27,28).
p85β also acts as an important upstream regulatory factor in the
AKT-dependent signaling pathway. However, the involvement of
miR-126-3p in cervical cancer and the molecular mechanisms of its
action associated with the PI3K/PDK1/AKT pathway have not yet been
fully established. Using an in vitro model, we examined the
molecular mechanisms potentially responsible for the
tumor-suppressor activity of miR-126-3p in cervical cancer,
focusing on its action via the PI3K/PDK1/AKT signaling pathway.
Materials and methods
Cell culture
The human cervical cancer cell line HeLa was
obtained from Keio University in Japan, and cultured in Ham's F12
medium (Wako Pure Chemical, Osaka, Japan) supplemented with 10%
fetal bovine serum (FBS; Gibco Life Technologies; Thermo Fisher
Scientific, Inc.) and 1% penicillin/streptomycin (Wako Pure
Chemical). Cells were incubated at 37°C in 5% CO2. To
confirm the identity of the analyzed cell line, we performed short
tandem repeat (STR) genotyping, which revealed correspondence with
more than 80% of the markers tested. We found no mycoplasma in
cells tested with MycoAlert mycoplasma detection kit (Lonza).
Plasmid construction and mimic
miRNA
A dual luciferase reporter system, the psiCheck2
vector system (Promega) with two luciferase enzymes was employed
here (one from Renilla containing the experimental sequence
and another from firefly containing the internal control). The
psiCHECK2-miR126-3p (Assay ID: MIC0000445) vector contains the
complementary sequence of hsa-miR-126-3p (MiRBase ID MIMAT0000445)
inserted into the psiCHECK-2 vector at the 3′end of the coding
sequence of the Renilla reniformis luciferase gene in
Fig. S1 (Promega). Mature miRNA
molecules (mirVana miRNA hsa-miR-126-3p mimic; assay ID:
MC12841) and Negative Control mimic (mirVana miRNA Mimic
Negative Control #1) were purchased from Thermo Fisher Scientific,
Inc. The target sequence and its vector construct are depicted in
Fig. S1.
Transfection
HeLa cells were trypsinized, diluted with medium
without antibiotics, and seeded into 96-well culture plates
(2×104 cells/well) or 6-well culture plates
(5×105 cells/well). HeLa cells were transfected with
Lipofectamine 2000 transfection reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) at a 2:1 Lipofectamine/DNA ratio in Opti-Minimal
Essential Serum-Free medium (Opti-MEM medium; Gibco Life
Technologies; Thermo Fisher Scientific, Inc.). We then transfected
the cells with a mixture of psiCHECK2-miR126-3p and 100 nM
hsa-miR-126-3p mimic or 100 nM negative control (NC) mimic. Cells
were incubated with transfection reagent/DNA/mimic for 4 h followed
by replacement with regular culture medium. Transfection with the
psiCHECK2-miR126-3p vector was performed in every experiment in
order to assure the same conditions for transfection and to monitor
transfection efficiencies in all experiments.
Dual-luciferase reporter assay
At 24 h post-transfection, Renilla and
firefly luciferase activities were measured by luminometer in the
same plate using the Dual-Glo Luciferase Assay System kit
(Promega); 2030 ARVO X5 (Perkin Elmer). Renilla luciferase
activity values were normalized for transfection efficiency using
the corresponding firefly luciferase activity as an internal
control. Three independent transfection experiments were performed,
each in triplicate.
CCK-8 assay
Cell proliferation was measured with the Cell
Counting Kit-8 (CCK-8, Dojindo Molecular Technologies, Inc.,
Japan). After transfection, 10 µl of CCK-8 solution [WST-8:
2-(2-methoxy-4-nitrophenyl)-3-(4-nitrophenyl)-5-(2,4-disulfophenyl)-2H-tetrazolium,
mono-sodium salt], was added to each well at 24, 48 and 72 h. The
plates were then incubated for 2 h at 37°C in 5% CO2.
Absorbance was measured using a Benchmark microplate reader
(Bio-Rad Laboratories) at 450 nm. Each assay was independently
performed 3 times in triplicate. Cell proliferation was calculated
by subtracting the absorbance values of the media alone (background
level) from the samples.
Wound healing assay
HeLa cells were seeded at a concentration of
5×105 cells in 2 ml of Ham's F12 medium supplemented
with 5% FBS in 6-well plates. The following day, the plasmid vector
(4 µg) was transfected with the hsa-miR-126-3p mimic (100 nM) or
the negative control mimic (100 nM). Scratch wounds were made 24 h
after transfection on a confluent monolayer of cultured cells with
a sterile 200-µl-pipette tip and photographed at 0, 18, 24 and 48 h
using a digital camera system coupled to an Axio-Observer
microscope (Carl Zeiss Microscopy). Wound closure was measured by
ImageJ 1.60 software (NIH, National Institutes of Health) and
expressed as Wound closure rate (%)=migrated cell surface
area/total surface area ×100. At least 5 points in each of 3 random
fields per well were examined and three independent experiments
were performed. All results were recorded as the means of
triplicate assays with standard deviation.
Cell migration and invasion
assays
Cells were seeded in the upper chamber of a
Transwell vessel with a coated Matrigel membrane (to assess
invasion; Corning Costar Corp.) or without a coating (to assess
migration). At 24 h post-transfection, in both assays,
2×105 cells in 200 µl serum-free medium were reseeded
into the upper chamber, and 750 µl of Dulbecco's modified Eagle's
medium (DMEM; Wako Pure Chemical) supplemented with 20% FBS was
added to the lower chamber as a chemoattractant. After 48 h,
non-migrating and non-invading cells on the upper surface of the
membrane were mechanically removed by wiping with a moistened
cotton swab. Cells that migrated and invaded into the lower surface
of the membrane were stained with Hemacolor Stain Set (Merck,
Darmstadt, Germany) according to the manufacturer's instructions.
The membranes were then plated on glass slides with the cells on
the top. The slides were photographed at a magnification of ×100,
using a light microscope and a digital color camera (Olympus BX43
with DP70; Olympus Optical Co.). The migrating and invading cells
were counted in five randomly selected visual fields and analyzed
using NIH Image J 1.60 software. Data are expressed as the
percentage of invasion through the Matrigel matrix and membrane
relative to migration through the control membrane. Invasion
percentage was calculated as [(the mean number of cells that
invaded the Matrigel insert)/(the mean number of cells that
migrated through the non-coated insert membrane)] ×100.
Apoptosis assay
At 24 h post-transfection, HeLa cells were seeded in
triplicate at 5×103 cells/well in a 96-well plate. After
24 h, caspase 3/7 activity was measured in the same plate using a
Caspase-Glo 3/7 Assay kit (Promega) and a luminometer (Perkin
Elmer) according to the manufacturer's instructions. Luminometer
readings were taken 1 h after adding the caspase 3/7 reagent.
Background readings were determined from wells containing culture
medium without cells. The data were normalized to the controls.
Three independent experiments were performed, each in
triplicate.
Antibodies
Antibodies for immunoblotting were obtained from
Cell Signaling Technology and Santa Cruz Biotechnology. The
following primary antibodies were used in this study (all rabbit):
Anti-PI3 kinase p85 (dilution 1:500; cat. no. 4257), anti-p110α
(dilution 1:1,000; cat. no. 4249), anti-AKT (dilution 1:1,000; cat.
no. 4691), anti-p-Akt (Ser473) (dilution 1:1,000; cat. no. 4060),
anti-PDK1 (dilution 1:1,000; cat. no. 5662), anti-p-PDK1 (dilution
1:1,000; cat. no. 3438), anti-p70S6 kinase (dilution 1:1,000; cat.
no. 2708), anti-p-p70S6 kinase (dilution 1:1,000; cat. no. 9234),
anti-S6 ribosomal protein (dilution 1:1,000; cat. no. 14467),
anti-p-S6 ribosomal protein (dilution 1:1,000; cat. no. 4858),
anti-GSK3β (dilution 1:1,000; cat. no. 12456), anti-p-GSK-3β
(dilution 1:1,000; cat. no. 5558), anti-cyclin D1 (dilution 1:500;
cat. no. 2978), anti-ROCK1 (dilution 1:1,000; cat. no. 4035),
anti-PAK1 (dilution 1:1,000; cat. no. 2602), anti-p-PAK1 (dilution
1:500; cat. no. 2601), anti-PLCγ (dilution 1:1,000; no. 5690),
anti-p-PLCγ (dilution 1:500; cat. no. 14008), anti-Bad (dilution
1:1,000; cat. no. 5023), anti-Bax (dilution 1:1,000; cat. no.
9239), anti-Bcl-xL (dilution 1:1,000; cat. no. 2764), anti-β-actin
(dilution 1:2,000; cat. no. 4970), and anti-MRCKα (dilution 1:100;
cat. no. sc-374568). The secondary antibody was horseradish
peroxidase (HRP)-conjugated anti-rabbit IgG antibody (cat. no.
7074).
Western blotting
HeLa cells were transfected and harvested after 48
or 72 h in a 6-well plate at 5×105 cells/well. Cells
were washed twice with ice-cold phosphate-buffered saline (PBS) and
lysed using RIPA lysis buffer (Santa Cruz Biotechnology) containing
1 mM PMSF, 1 mM Na-orthovanadate and 1X complete protease inhibitor
cocktail (Roche Diagnostics). After a 30-min incubation on ice,
cell lysates were triturated and centrifuged at 20,630 × g for 15
min at 4°C. The clear supernatants were saved as whole-cell lysate.
Protein concentration was determined using Pierce BCA Protein Assay
Kits (Thermo Fisher Scientific, Inc.). Total cell lysates (20 µg)
in each lane were mixed with 6X SDS sample buffer containing
dithiothreitol (DTT). Samples were incubated at 96°C for 5 min
before being loaded onto 10% SDS-polyacrylamide gels (ATTO Corp.)
and electrophoresed for 90 min at 18 mA. Gels were transferred to
polyvinylidene difluoride membranes (PVDF; ATTO Corp.) at 70 V for
1 h and then blocked with blocking buffer (#AE-1476 EzBlock BSA;
ATTO Corp.) for 1 h at room temperature. All antibodies were
diluted in Tris-buffered saline containing 0.1% Tween-20 (TBS-T).
Membranes were incubated overnight at 4°C with primary antibodies
and washed with TBS-T before incubation with horseradish peroxidase
(HRP)-conjugated secondary antibody at room temperature for 1 h.
The blots were examined using an ECL Prime Western Blotting
Detection System (GE Healthcare Life Science) according to the
manufacturer's instructions, and visualized using the Image Quant
LAS 4000 Mini image analysis system (GE Healthcare Life Science).
Band quantification was performed using NIH ImageJ 1.60 software
and protein levels were normalized to β-actin levels.
Statistical analysis
Statistical analysis was performed using SPSS
22.0.0.0 software (IBM Corp.). Data were recorded as mean ± SD. The
independent samples Student t-test was used for comparison. A
P-value of ≤0.05 or less (P<0.001; P<0.01) was considered
significant.
Results
Use of the reporter plasmid to study
the function of miR-126-3p
The relative transcriptional activity (Rlu
activity/Flu activity) was 0.063±0.014 in the HeLa cells
transfected with the miR-126-3p mimic compared to 0.65±0.13 in the
NC mimic group (Fig. S2).
Renilla reporter activity was significantly reduced by the
miR-126-3p mimic compared to the NC mimic; P<0.001). This
finding documents that the miR-126-3p mimic binds to the
3′-untranslated region (3′-UTR) of the Renilla luciferase
reporter gene and thus contributes to its suppression. We then
adopted this experimental system to investigate potential
biological effects exerted by miR126-3p in HeLa cells that may be
relevant to cervical carcinogenesis.
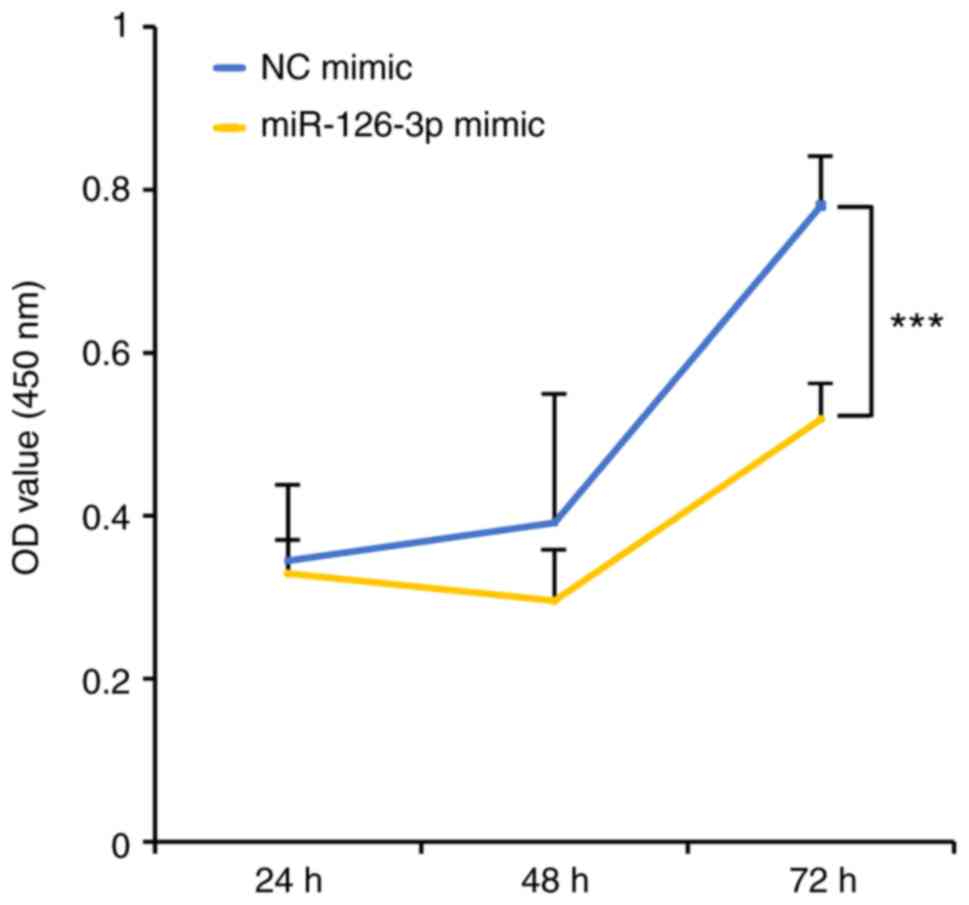
Suppression of proliferation by
transfection with miR-126-3p
To investigate the functional role of miR-126-3p in
the proliferation of HeLa cells, we used the CCK-8 assay after
transfection with the miR-126-3p mimic or the negative control (NC)
mimic. Compared with the NC, the cells transfected with the 126-3p
mimic showed lower cell growth, indicating suppression of
proliferation (Fig. 1). The degree
of suppression by the miR-126-3p mimic reached statistical
significance at 72 h (P<0.001).
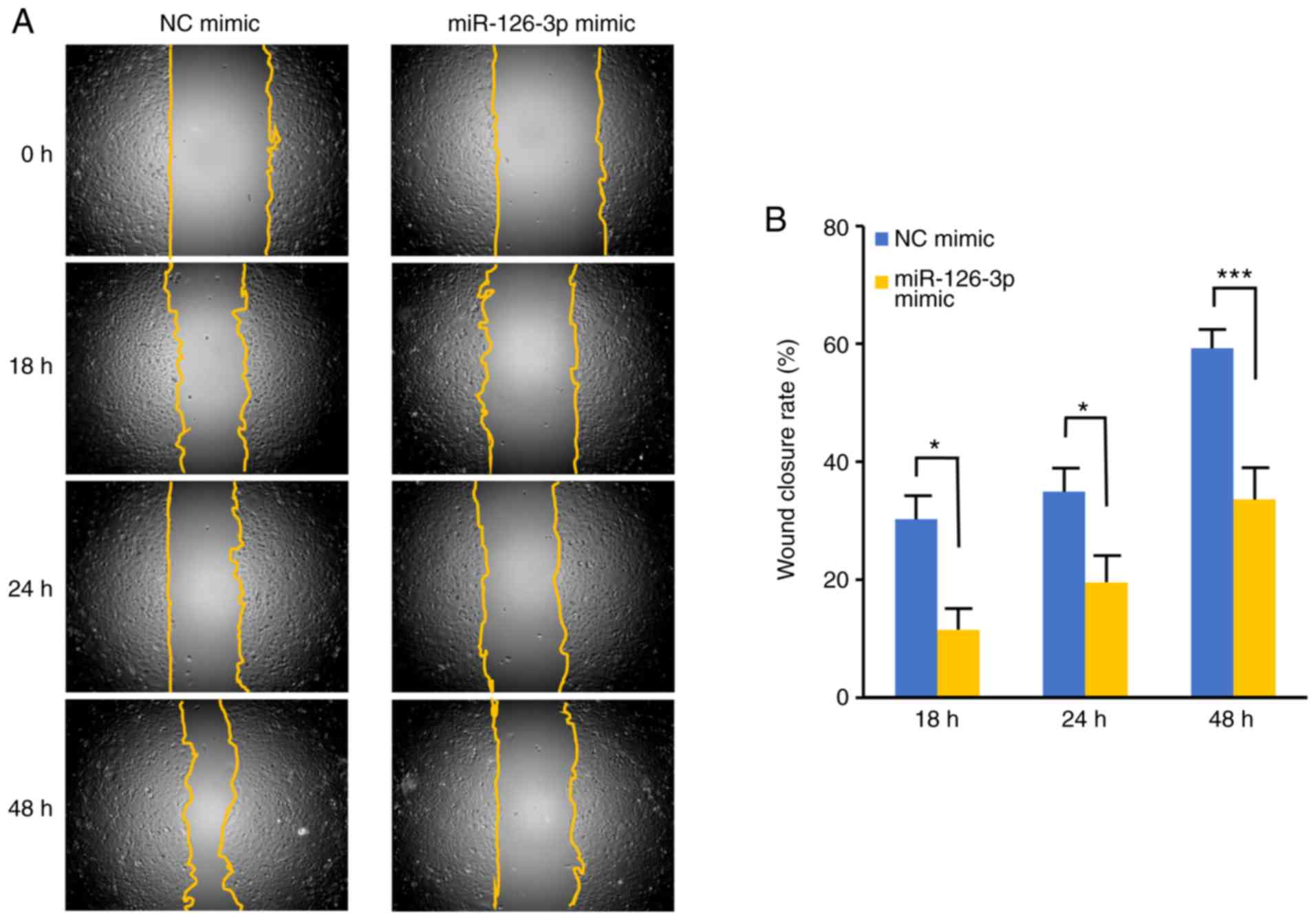
miR-126-3p suppresses migration and
invasion
To determine whether miR-126-3p also affects
migration, we used a wound healing assay to examine the effects of
miR-126-3p transfection (Fig. 2A and
B). The wound scratches healed significantly more slowly when
the HeLa cells were transfected with the miR-126-3p mimic than with
the NC mimic at 18, 24 (P<0.05) and 48 h (P<0.001).
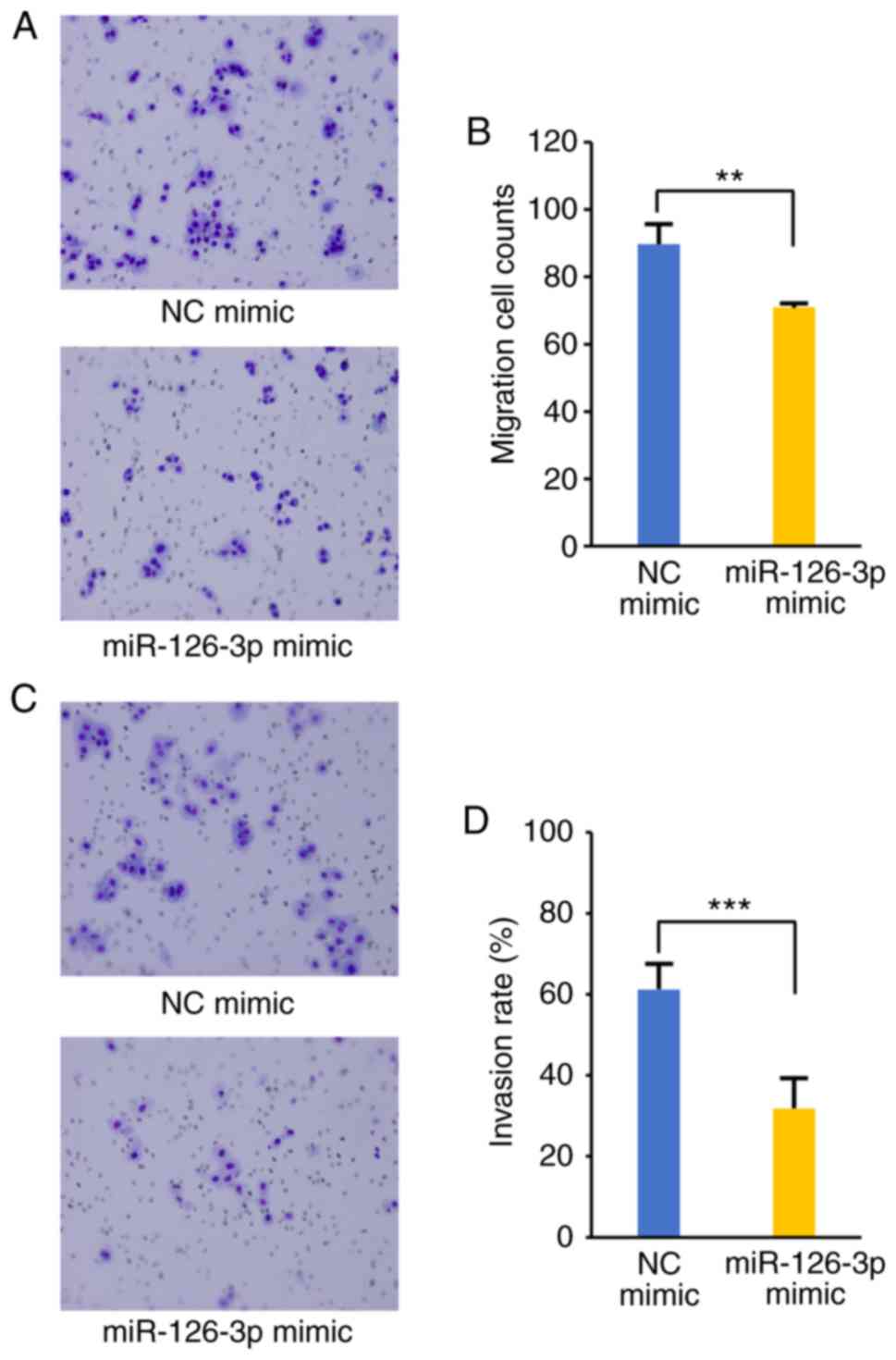
Inhibition of migration and invasion was confirmed by Transwell
assays (Fig. 3A-D). As shown in
Fig. 3B, transfection with the
miR-126-3p mimic led to significantly reduced migration capacity
than that in the NC (70.9±0.41 vs. 89.7±5.5 cells per field,
respectively; P<0.01). Significant differences in invasion were
also observed between the miR-126-3p mimic-transfected cells and
NC, as shown in Fig. 3D
(P<0.001). The invasion rate recorded was 31.8% (range
21.8–40.4%) and 61.3% (range 52.9–70.4%) at 72 h in the miR-126-3p
mimic and NC mimic transfectants, respectively.
miR-126-3p suppresses the
PI3K/PDK1/AKT pathway
We showed above that the role of miR-126-3p is as a
tumor suppressor that regulates proliferation, migration, and
invasion in HeLa cells. To elucidate the underlying molecular
mechanisms responsible for these effects of miR-126-3p, we analyzed
the expression of components of the PI3K/PDK1/AKT signaling
pathway. Western blotting revealed that transfection of the
miR-126-3p mimic in the HeLa cells decreased the expression of
multiple PI3K/PDK1/AKT pathway proteins, including p85β, p110α,
phosphorylated (p)-3-phosphoinositide-dependent protein kinase-1
(PDK1) and phosphorylated (p)-AKT, at 48 and 72 h after
transfection (Fig. 4A and B).
Expression levels of phosphorylated glycogen synthase kinase 3β
(p-GSK3β), cyclin D1, phosphorylated p70S6K (p-p70S6K) and
phosphorylated S6K (p-S6K), which are all located downstream of the
AKT pathway, were also significantly reduced relative to the NC
mimic transfected cells. We also focused on PDK1 and its downstream
pathway (Fig. 5A and B). To further
determine the contribution of PI3K/PDK1 inactivation to downstream
signaling, we examined the PDK1-related protein, myotonic
dystrophy-related CDC42-binding kinase α (MRCKα), Rho-associated
coiled-coil-containing protein kinase 1 (ROCK1), phospholipase C γ1
(PLCγ1) and p21-activated kinase 1 (PAK1). We found lower levels of
all these proteins in cells transfected with the miR-126-3p mimic
than with the NC mimic. The kinetics of the altered expression of
these proteins between 48 and 72 h is important; proteins located
upstream of the pathway, such as p85β, p110α, p-PDK1, p-AKT,
p-GSK3β, cyclin D1, MRCKα, ROCK1 and p-PLCγ1 were decreased 48 h
after transfection and gradually recovered at 72 h. In contrast,
proteins located downstream of this pathway, such as p-p70S6K, p-S6
and p-PAK1, were reduced 24 later, at 72 h post-transfection.
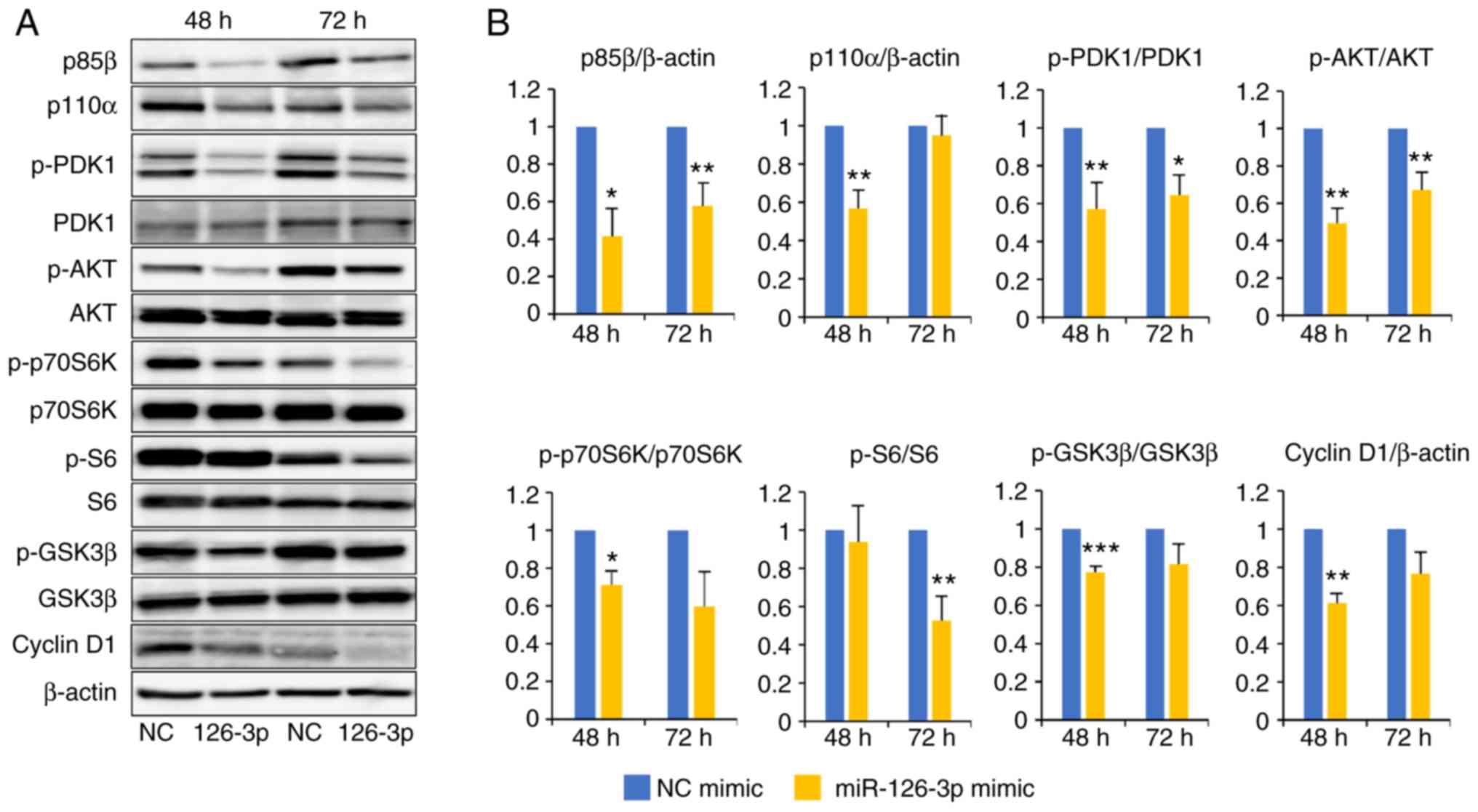 | Figure 4.Amounts of proteins in the PI3K/AKT
pathway including p85β, p110α, p-PDK1, p-AKT, p-p70S6K, p-S6,
p-GSK3β and cyclin D1 are decreased in HeLa cells following
transfection with the miR-126-3p mimic. (A) Cells were harvested 48
or 72 h after transfection with miR-126-3p (126-3p) or negative
control (NC) mimic concomitant with the reporter plasmid
psiCHECK2-miR126-3p. Representative western blots of the protein
levels of the AKT/PI3K pathway are shown. (B) Quantification of the
western blots by densitometry. Quantification of signals is
presented as ‘fold-change’ relative to levels of β-actin or
phosphorylated (p-) protein vs. total protein. All experiments were
performed in triplicate. *P<0.05, **P<0.01 and ***P<0.001,
respectively. No asterisk indicates a non-significant difference.
PI3K, phosphoinositide 3 kinase; AKT, protein kinase B; PDK1,
3-phosphoinositide-dependent protein kinase-1; GSK3β, glycogen
synthase kinase 3β. |
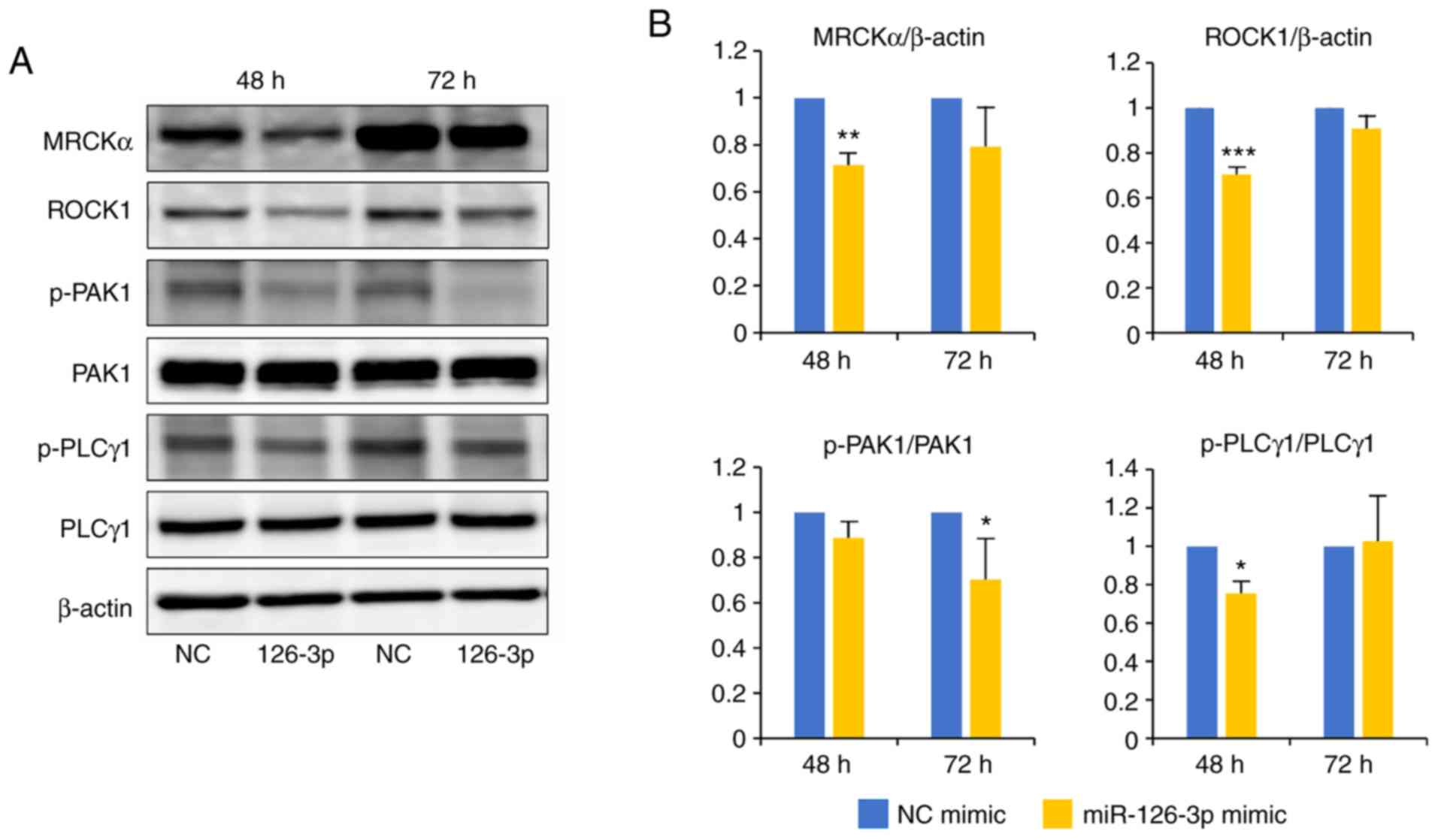 | Figure 5.Amounts of proteins in the
PI3K/PDK1/AKT pathway including MRCKα, ROCK1, PAK1 and PLCγ1 are
decreased in HeLa cells following transfection with the miR-126-3p
mimic. (A) Cells were harvested 48 or 72 h after transfection with
miR-126-3p or the negative control (NC) mimic concomitant with the
reporter plasmid psiCHECK2-miR126-3p. Representative western blot
showing levels of the proteins in the AKT/PDK1/PI3K pathway are
shown. (B) Quantification of the western blot by densitometry.
Quantification of signals presented as fold-change relative to
levels of β-actin or phosphorylated (p-) protein vs. total protein.
All experiments were performed in triplicate. *P<0.05,
**P<0.01 and ***P<0.001, respectively. No asterisk indicates
a non-significant difference. PI3K, phosphoinositide 3 kinase; AKT,
protein kinase B; PDK1, 3-phosphoinositide-dependent protein
kinase-1; MRCKα, myotonic dystrophy-related CDC42-binding kinases
α; ROCK1, Rho associated coiled-coil containing protein kinase 1;
PAK1, p21-activated kinase 1; PLCγ1, phospholipase C γ1. |
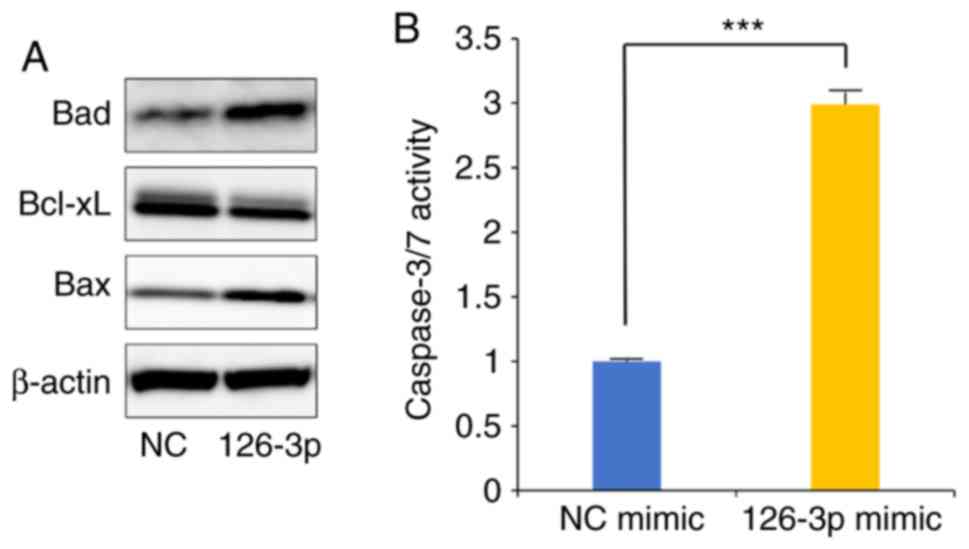
miR-126-3p-induced apoptosis
Finally, we investigated the apoptosis in HeLa cells
transfected with the miR-126-3p mimic. Western blotting
demonstrated that the BCL-2-associated agonist of cell death (Bad)
and BCL-2-associated X (Bax) proteins were upregulated, whereas
B-cell lymphoma-extra large (Bcl-xL) protein expression was
downregulated. The miR-126-3p mimic consequently increased
caspase-3/7 activity in the HeLa cells (Fig. 6A and B).
Discussion
We previously found that miR-126-3p is highly
expressed in patients with overt cervical cancer or precursor
lesions compared with normal cells (22). Accordingly, we hypothesized that
miR-126-3p promotes carcinogenesis. However, we observed that
levels of miR-126-3p in patients with cervical cancer fluctuate
(22). We also found variability in
the amounts of miR-126-3p in cervical cancer cells themselves
(Fig. S3). Hence, as expression of
miR-126-3p is not ubiquitous in vivo or in vitro in
cervical cancer, we hypothesized that endogenous miR-126-3p does
not necessarily play a critical role in carcinogenesis. Indeed, in
the present study, in contrast, we found that the results of
enforced expression of miR-126-3p reflected characteristics of
anticancer activity in vitro. We thus aimed to ascertain how
miR-126-3p is involved in cervical carcinogenesis. Initially we
used the DIANA-miRPath V3.0: miRNA pathway analysis program via a
web-server (29), which indicated
that miR-126-3p is associated with tPIK3R2 and AKT1 in the mTOR
signaling pathway, a candidate pathway for targeted therapy in
cervical cancer (30–32). Therefore, we investigated the
function of miR-126-3p in relation to the PI3K/PDK1/AKT signaling
pathway in the cervical cancer cell line HeLa. The highest
frequency of mutation of the PI3K gene in cervical cancer is
20–30%, and its mutation is a late event during cervical
carcinogenesis (33,34). In the present study, we used the
cervical cancer cell line HeLa since these cells are suitable for
transfection experiments due to their ability to highly express the
luciferase gene with a reporter plasmid; 10.3-fold increase
(Fig. S2). This also indicated
that enforced miR-126-3p was biologically active in the cells,
despite that direct overexpression of miR-126-3p by RT-PCR was not
shown. There is no mutation in, nor amplification of, the
PI3K gene in HeLa cells (6,35,36).
Here, we report that enforced expression of miR-126-3p inhibited
proliferation, migration and invasion in the transfected HeLa
cells. As expected, the levels of proteins including p85β, p110α,
p-PDK1, p-AKT, p-p70S6K, p-S6, p-GSK3β, cyclin D1, MRCKα, ROCK1,
p-PAK1 and p-PLCγ1 were decreased after transfection of the
miR-126-3p mimic. Notably, p85β was significantly downregulated
both at 48 and 72 h, whereas, p110a downregulation was noted at 48
h only. Thus, miR-126-3p mainly exerts its effects via inhibition
of p85b.
Akt is one of the most frequently hyperactivated
kinases in human cancers and plays an important role in cell
migration (37). S6K1 is a
serine-threonine kinase that is implicated in the regulation of
several cellular processes, such as growth, survival and metabolism
(38). The main target of S6K is
ribosomal protein S6, a component of the 40S ribosomal subunit. S6
is essential for protein synthesis and cell cycle progression. S6K
shows increased activity in several cancer cells and has been
associated with resistance to drugs and chemotherapeutic treatment,
emphasizing the role of these proteins in carcinogenesis. S6K1
inhibition prevents the migration of breast tumor cells, suggesting
its important role in metastatic breast cancer. The S6K1 inhibitor,
PF-4708671, was found to markedly reduce cell migration and
invasion, suggesting that this drug may be suitable for
anti-metastatic therapy for breast cancer (39). GSK-3 is associated with various
signaling pathways, including Wnt/β-catenin, PI3K/PTEN/Akt/mTORC,
Ras/Raf/MEK/ERK, Ηedgehog, Notch and TP53, all related to
epithelial-mesenchymal transition and to cancer stem cells
(40). Phosphorylated AKT induces
phosphorylated GSK3β (inactive form). Dephosphorylated GSK3β
(active form) inhibits cyclin D1 (41,42)
and the inactive form of phosphorylated GSK3β induces cyclin D1.
Therefore, miR-126-3p-downregulated phosphorylated AKT is likely to
inhibit cell turnover through downregulation of cyclin D1.
Furthermore, inhibition of the PI3K/Akt pathway can lead to the
apoptosis of cervical cancer cells (43). Our results demonstrated that
inhibition of the PI3K/Akt pathway by miR-126-3p promoted caspase
3/7 activity through the control of anti-apoptotic proteins such as
Bcl-xL and pro-apoptotic proteins including Bax and Bad. Activated
AKT and mTOR are indicators of poor prognosis in cervical cancer
(31,44). In clinical trials, PI3K/AKT/mTOR
inhibitors have been used as molecular targeted therapies in
cervical cancer, and the PI3K/AKT inhibitor, LY294002 was found to
increase radiation sensitivity in cervical cancer cell lines
(45). The PI3K/mTOR inhibitor,
NVP-BEZ235, was also found to exert an effect on cervical cancer
cells (46). Recent research has
revealed that Akt/mTOR signaling promotes neuroendocrine cervical
cancer, which has an extremely poor prognosis, suggesting that this
pathway may be utilized for novel anticancer treatment strategies
(47).
We found that miR-126-3p suppresses PDK1 and its
downstream proteins. PDK1 regulates cell migration via AKT, PAK1,
ROCK1, MRCKα and PLCγ1 (48) and is
centrally involved in cell signaling via regulation of PI3K
signaling and activation of multiple downstream effectors. PAK1 can
be activated by phosphorylation of threonine 423 by phosphorylated
PDK1. Both PDK1 and PAK1 have been reported to regulate the
migration of vascular smooth muscle cells. PDK1 regulates cell
migration and invasion by activating ROCK1 and MRCKα and PLCγ1.
Overexpression of PDK1 correlates with a more aggressive cancer
phenotype and poorer patient prognosis (44). Thus, PDK1 targeting could also
provide an effective therapeutic intervention in cancer where
migration and invasion play a crucial role in prognosis. Here, we
presented data on the kinetics of effects on protein expression
after transfection that are consistent with the hierarchy of this
pathway (Fig. 7).
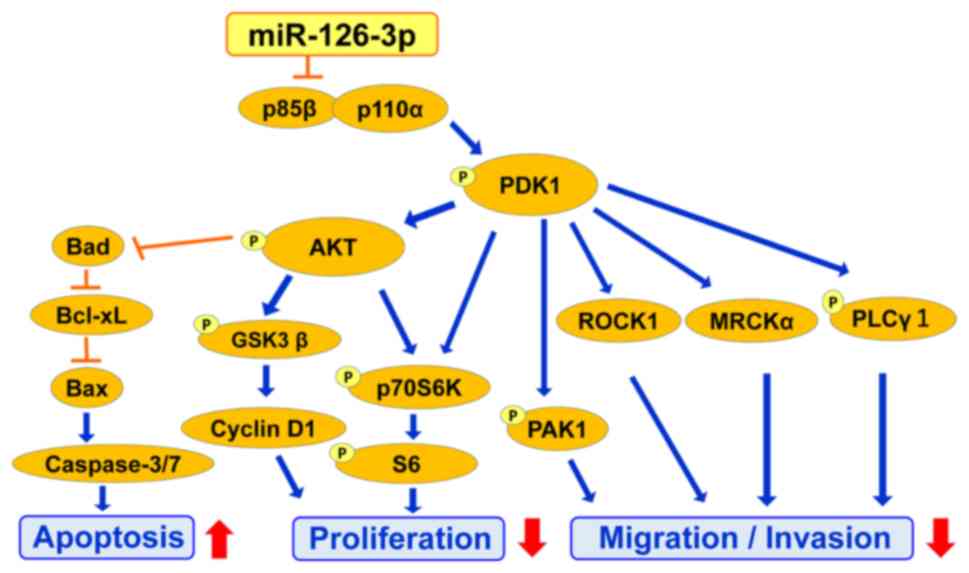 | Figure 7.A schematic model of the underlying
mechanisms of miR-126-3p-mediated suppression of proliferation,
migration and invasion through the PI3K/PDK1/AKT pathway.
miR-126-3p suppresses progression of cervical cancer cells through
the PI3K/PDK1/AKT signaling pathway. The AKT pathway is associated
with proliferation of cells through its downstream signaling
pathway including GSK3β, cyclin D1, p70S6K and S6 activation. The
PDK1 pathway is associated with migration and invasion of cells
through mediators of its downstream signaling pathway including
p70S6K, MRCKα, ROCK1, PLCγ-1 and PAK1. PI3K, phosphoinositide 3
kinase; AKT, protein kinase B; PDK1, 3-phosphoinositide-dependent
protein kinase-1; GSK3β, glycogen synthase kinase 3β; MRCKα,
myotonic dystrophy-related CDC42-binding kinases α; ROCK1, Rho
associated coiled-coil containing protein kinase 1; PAK1,
p21-activated kinase 1; PLCγ1, phospholipase C γ1; Bad,
Bcl-2-associated agonist of cell death; Bcl-xL, B-cell
lymphoma-extra-large; Bax, Bcl-2-associated X. |
There are some limitations to the present study.
Although many cervical cancer cells are available for in
vitro experiments, we used HeLa cells. This was because the
combination of HeLa cells, transfection reagents, and miRNA mimics
used in our experiments produced the highest transfection rates
(data not shown). If transfection efficiency is improved, other
cell lines may also become amenable for such a study. We could not
estimate the expression level compared with normal cells as normal
cells display heterogeneity including epithelia, stroma and
hematopoietic cells. In addition, we did not investigate whether
p85b could reverse the effect of miR-126-3p. This experiment is
technically very challenging as the enforced expression of
miR-126-3p and p85b is not stable due to the transient transfection
technique employed. The situation would be much clearer if we could
show a direct link between miR-126-3p target genes and suppression
of the PI3K/PDK1/Akt pathway as alternative pathways and target
proteins associated with miR-126-3p are possible. In fact, Xu et
al reported that miR-126-3p inhibited cell proliferation and
migration via the target protein ZEB1, which is associated with
mesenchymal-related protein (49).
We did not conduct knockdown or inhibitor experiments of endogenous
miR-126-3p, downstream of the PI3K/AKT pathway or a xenograft model
to demonstrate the potential application of miR-126-3p into human
patients. Despite this limitation, the strength of our study is
that enforced expression of miR-126-3p affected the levels of 15
proteins related to the PI3K/AKT pathway in the cell. We showed
that miR-126-3p affects the PI3K/PDK1/AKT signaling pathway cascade
(Fig. 7).
In cervical cancer, HPV E6 and E7 are known to play
vital roles in carcinogenesis. Therefore, in this cancer, the
PI3K/PDK1/AKT signaling pathway may have escaped control by
endogenous miR-126-3p. Taken together with in vivo and in
vitro studies, we hypothesize that miR-126-3p opposes some of
the effects of E6 and E7. If so, it is not unexpected that strong
expression of miR-126-3p is observed in patients with cervical
cancer or precursor lesions. We now report that enforced expression
of a miR-126-3p mimic negatively influenced cellular functions
including proliferation, migration, invasion and apoptosis, and
suggest that increased endogenous miR-126-3p expression may be a
compensatory mechanism attempting to control carcinogenesis.
Exogenous miR-126-3p and its target molecules may thus be
candidates for miRNA-based cervical cancer therapy even in cases
where the PI3K gene is not mutated or amplified. For further
research, an animal model is necessary in the event we are able to
develop drugs using miR-126-3p.
Supplementary Material
Supporting Data
Acknowledgements
We thank Yuko Nakagawa and Yumiko Usui for typing
the manuscript. We thank Dr Takashi Iwata, Department of Obstetrics
and Gynecology, School of Medicine, Keio University, Tokyo, Japan,
for providing the HeLa cells.
Funding
This study was partly supported by JSPS KAKENHI
(grant no. JP 26462540), and a Fujita Health University Research
Grant-in-Aid.
Availability of data and materials
The analyzed datasets generated during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
TF and RI designed the experiments. RK and AI
conducted the experiments. TF, RK, AI, HN, SO, EN and RI analyzed
the data. RI, TF, RK, SO, EN and AI contributed to the writing of
the manuscript. All authors read and approved the final manuscript
and agree to be accountable for all aspects of the research in
ensuring that the accuracy or integrity of any part of the work are
appropriately investigated and resolved.
Ethics approval and consent to
participate
This study was approved by the Institutional
Biosafety Committee of Fujita Health University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare no potential competing
interests.
References
|
1
|
International agency for research on
cancer, GLOBOCAN 2012: Estimated cancer incidence, mortality and
prevalence. http://gco.iarc.fr/Worldwidein 2012.
|
|
2
|
Gupta S, Kumar P and Das BC: HPV:
Molecular pathways and targets. Curr Probl Cancer. 42:161–174.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hou MM, Liu X, Wheler J, Naing A, Hong D,
Coleman RL, Tsimberidou A, Janku F, Zinner R, Lu K, et al: Targeted
PI3K/AKT/mTOR therapy for metastatic carcinomas of the cervix: A
phase I clinical experience. Oncotarget. 5:11168–11179. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bregar AJ and Growdon WB: Emerging
strategies for targeting PI3K in gynecologic cancer. Gynecol Oncol.
140:333–344. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Tinker AV, Ellard S, Welch S, Moens F,
Allo G, Tsao MS, Squire J, Tu D, Eisenhauer EA and MacKay H: Phase
II study of temsirolimus (CCI-779) in women with recurrent,
unresectable, locally advanced or metastatic carcinoma of the
cervix. A trial of the NCIC Clinical Trials Group (NCIC CTG IND
199). Gynecol Oncol. 130:269–274. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Rashmi R, DeSelm C, Helms C, Bowcock A,
Rogers BE, Rader JL, Grigsby PW and Schwarz JK: AKT inhibitors
promote cell death in cervical cancer through disruption of mTOR
signaling and glucose uptake. PLoS One. 9:e929482014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Esteller M: Non-coding RNAs in human
disease. Nat Rev Genetics. 12:861–874. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Granados López AJ and López JA: Multistep
model of cervical cancer: Participation of miRNAs and coding genes.
Int J Mol Sci. 15:15700–15733. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ma D, Zhang YY, Guo YL, Li ZJ and Geng L:
Profiling of microRNA-mRNA reveals roles of microRNAs in cervical
cancer. Chin Med J (Engl). 125:4270–4276. 2012.PubMed/NCBI
|
|
10
|
Cheung TH, Man KN, Yu MY, Yim SF, Siu NS,
Lo KW, Doran G, Wong RR, Wang VW, Smith DI, et al: Dysregulated
microRNAs in the pathogenesis and progression of cervical neoplasm.
Cell Cycle. 11:2876–2884. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Liu D, Liu C, Wang X, Ingvarsson S and
Chen H: MicroRNA-451 suppresses tumor cell growth by
down-regulating IL6R gene expression. Cancer Epidemiol. 38:85–92.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ding H, Wu YL, Wang YX and Zhu FF:
Characterization of the microRNA expression profile of cervical
squamous cell carcinoma metastases. Asian Pac J Cancer Prev.
15:1675–1679. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yu Q, Tong S, Yan J, Hong C, Zhai W and Li
Y: Preparative separation of quaternary ammonium alkaloids from
Corydalis yanhusuo W. T. Wang by pH-zone-refining counter-current
chromatography. J Sep Sci. 34:278–285. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yang Y, Song KL, Chang H and Chen L:
Decreased expression of microRNA-126 is associated with poor
prognosis in patients with cervical cancer. Diagn Pathol.
9:2202014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Huang TH and Chu TY: Repression of miR-126
and upregulation of adrenomedullin in the stromal endothelium by
cancer-stromal cross talks confers angiogenesis of cervical cancer.
Oncogene. 33:3636–3647. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Liu R, Zhang YS, Zhang S, Cheng ZM, Yu JL,
Zhou S and Song J: MiR-126-3p suppresses the growth, migration and
invasion of NSCLC via targeting CCR1. Eur Rev Med Pharmacol Sci.
23:679–689. 2019.PubMed/NCBI
|
|
17
|
Luo W, Yan D, Song Z, Zhu X, Liu X, Li X
and Zhao S: miR-126-3p sensitizes glioblastoma cells to
temozolomide by inactivating Wnt/β-catenin signaling via targeting
SOX2. Life Sci. 226:98–106. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Fujii T, Shimada K, Tatsumi Y, Fujimoto K
and Konishi N: Syndecan-1 responsive microRNA-126 and 149 regulate
cell proliferation in prostate cancer. Biochem Biophys Res Commun.
456:183–189. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Otsubo T, Akiyama Y, Hashimoto Y, Shimada
S, Goto K and Yuasa Y: MicroRNA-126 inhibits SOX2 expression and
contributes to gastric carcinogenesis. PLoS One. 6:e166172011.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang X, Tang S, Le SY, Lu R, Rader JS,
Meyers C and Zheng ZM: Aberrant expression of oncogenic and
tumor-suppressive microRNAs in cervical cancer is required for
cancer cell growth. PLoS One. 3:e25572008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yu Q, Liu SL, Wang H, Shi G, Yang P and
Chen XL: miR-126 suppresses the proliferation of cervical cancer
cells and alters cell sensitivity to the chemotherapeutic drug
bleomycin. Asian Pac J Cancer Prev. 14:6569–6572. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kawai S, Fujii T, Kukimoto I, Yamada H,
Yamamoto N, Kuroda M, Otani S, Ichikawa R, Nishio E, Torii Y, et
al: Identification of miRNAs in cervical mucus as a novel
diagnostic marker for cervical neoplasia. Sci Rep. 8:70702018.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Du C, Lv Z, Cao L, Ding C, Gyabaah OA, Xie
H, Zhou L, Wu J and Zheng S: MiR-126-3p suppresses tumor metastasis
and angiogenesis of hepatocellular carcinoma by targeting LRP6 and
PIK3R2. J Transl Med. 12:2592014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Guo C, Sah JF, Beard L, Willson JK,
Markowitz SD and Guda K: The noncoding RNA, miR-126, suppresses the
growth of neoplastic cells by targeting phosphatidylinositol
3-kinase signaling and is frequently lost in colon cancers. Genes
Chromosomes Cancer. 47:939–946. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Xiao J, Lin HY, Zhu YY, Zhu YP and Chen
LW: MiR-126 regulates proliferation and invasion in the bladder
cancer BLS cell line by targeting the PIK3R2-mediated PI3K/Akt
signaling pathway. Onco Targets Ther. 9:5181–5193. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Liu LY, Wang W, Zhao LY, Guo B, Yang J,
Zhao XG, Hou N, Ni L, Wang AY, Song TS, et al: Mir-126 inhibits
growth of SGC-7901 cells by synergistically targeting the oncogenes
PI3KR2 and Crk, and the tumor suppressor PLK2. Int J Oncol.
45:1257–1265. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Qu Y, Wu J, Deng JX, Zhang YP, Liang WY,
Jiang ZL, Yu QH and Li J: MicroRNA-126 affects rheumatoid arthritis
synovial fibroblast proliferation and apoptosis by targeting PIK3R2
and regulating PI3K-AKT signal pathway. Oncotarget. 7:74217–74226.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Meng Q, Wang W, Yu X, Li W, Kong L, Qian
A, Li C and Li X: Upregulation of MicroRNA-126 contributes to
endothelial progenitor cell function in deep vein thrombosis via
its target PIK3R2. J Cell Biochem. 116:1613–1623. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Vlachos IS, Paraskevopoulou MD, Karagkouni
D, Georgakilas G, Vergoulis T, Kanellos I, Anastasopoulos IL,
Maniou S, Karathanou K, Kalfakakou D, et al: DIANA-TarBase v7.0:
Indexing more than half a million experimentally supported
miRNA:mRNA interactions. Nucleic Acids Res. 43((Database Issue)):
D153–D159. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Temkin SM, Yamada SD and Fleming GF: A
phase I study of weekly temsirolimus and topotecan in the treatment
of advanced and/or recurrent gynecologic malignancies. Gynecol
Oncol. 117:473–476. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Faried LS, Faried A, Kanuma T, Aoki H,
Sano T, Nakazato T, Tamura T, Kuwano H and Minegishi T: Expression
of an activated mammalian target of rapamycin in adenocarcinoma of
the cervix: A potential biomarker and molecular target therapy. Mol
Carcinog. 47:446–457. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
32
|
Tomao F, Di Tucci C, Imperiale L, Boccia
SM, Marchetti C, Palaia I, Muzii L and Panici PB: Cervical cancer:
Are there potential new targets? An update on preclinical and
clinical results. Curr Drug Targets. 15:1107–1120. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wright AA, Howitt BE, Myers AP, Dahlberg
SE, Palescandolo E, Van Hummelen P, MacConaill LE, Shoni M, Wagle
N, Jones RT, et al: Oncogenic mutations in cervical cancer: Genomic
differences between adenocarcinomas and squamous cell carcinomas of
the cervix. Cancer. 119:3776–3783. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wilting SM and Steenbergen RDM: Molecular
events leading to HPV-induced high grade neoplasia. Papillomavirus
Res. 2:85–88. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Macville M, Schrock E, Padilla-Nash H,
Keck C, Ghadimi BM, Zimonjic D, Popescu N and Ried T: Comprehensive
and definitive molecular cytogenetic characterization of HeLa cells
by spectral karyotyping. Cancer Res. 59:141–150. 1999.PubMed/NCBI
|
|
36
|
Vazquez-Mena O, Medina-Martinez I,
Juárez-Torres E, Barrón V, Espinosa A, Villegas-Sepulveda N,
Gómez-Laguna L, Nieto-Martinez K, Orozco L, Roman-Basaure E, et al:
Amplified genes may be overexpressed, unchanged, or downregulated
in cervical cancer cell lines. PLoS One. 7:e326672012. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Gonzalez E and McGraw TE: The Akt kinases:
Isoform specificity in metabolism and cancer. Cell Cycle.
8:2502–2508. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Tavares MR, Pavan IC, Amaral CL,
Meneguello L, Luchessi AD and Simabuco FM: The S6K protein family
in health and disease. Life Sci. 131:1–10. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Pearce LR, Alton GR, Richter DT, Kath JC,
Lingardo L, Chapman J, Hwang C and Alessi DR: Characterization of
PF-4708671, a novel and highly specific inhibitor of p70 ribosomal
S6 kinase (S6K1). Biochem J. 431:245–255. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
McCubrey JA, Fitzgerald TL, Yang LV,
Lertpiriyapong K, Steelman LS, Abrams SL, Montalto G, Cervello M,
Neri LM, Cocco L, et al: Roles of GSK-3 and microRNAs on epithelial
mesenchymal transition and cancer stem cells. Oncotarget.
8:14221–14250. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Suprynowicz FA, Kamonjoh CM, Krawczyk E,
Agarwal S, Wellstein A, Agboke FA, Choudhury S, Liu X and Schlegel
R: Conditional cell reprogramming involves non-canonical β-catenin
activation and mTOR-mediated inactivation of Akt. PLoS One.
12:e01808972017. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Engelman JA: Targeting PI3K signalling in
cancer: Opportunities, challenges and limitations. Nat Rev Cancer.
9:550–562. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Kim MS, Kim JH, Bak Y, Park YS, Lee DH,
Kang JW, Shim JH, Jeong HS, Hong JT and Yoon DY: 2,4-bis
(p-hydroxyphenyl)-2-butenal (HPB242) induces apoptosis via
modulating E7 expression and inhibition of PI3K/Akt pathway in SiHa
human cervical cancer cells. Nutr Cancer. 64:1236–1244. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Bahrami A, Hasanzadeh M, Hassanian SM,
ShahidSales S, Ghayour-Mobarhan M, Ferns GA and Avan A: The
potential value of the PI3K/Akt/mTOR signaling pathway for
assessing prognosis in cervical cancer and as a target for therapy.
J Cell Biochem. 118:4163–4169. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Lee CM, Fuhrman CB, Planelles V, Peltier
MR, Gaffney DK, Soisson AP, Dodson MK, Tolley HD, Green CL and
Zempolich KA: Phosphatidylinositol 3-kinase inhibition by LY294002
radiosensitizes human cervical cancer cell lines. Clin Cancer Res.
12:250–256. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Xie G, Wang Z, Chen Y, Zhang S, Feng L,
Meng F and Yu Z: Dual blocking of PI3K and mTOR signaling by
NVP-BEZ235 inhibits proliferation in cervical carcinoma cells and
enhances therapeutic response. Cancer Lett. 388:12–20. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Cho SY, Choi M, Ban HJ, Lee CH, Park S,
Kim H, Kim YS, Lee YS and Lee JY: Cervical small cell
neuroendocrine tumor mutation profiles via whole exome sequencing.
Oncotarget. 8:8095–8104. 2017.PubMed/NCBI
|
|
48
|
Gagliardi PA, di Blasio L and Primo L:
PDK1: A signaling hub for cell migration and tumor invasion.
Biochim Biophys Acta. 1856:178–188. 2015.PubMed/NCBI
|
|
49
|
Xu J, Wang H, Wang H, Chen Q, Zhang L,
Song C, Zhou Q and Hong Y: The inhibition of miR-126 in cell
migration and invasion of cervical cancer through regulating ZEB1.
Hereditas. 156:112019. View Article : Google Scholar : PubMed/NCBI
|