Introduction
Hepatocellular carcinoma (HCC) is one of the most
notorious solid tumors and has become a rapidly increasing cause of
cancer-associated death in the USA (1–3). Based on
the 2018 Global Cancer Statistics, liver cancer has an incidence of
4.7% and a mortality rate of 8.2% (4). Although surgery, local ablation,
trans-arterial chemoembolization and systemic therapy have greatly
advanced treatment of patients with HCC (5), distant and intrahepatic metastasis and
postoperative recurrence remain a concern and result in poor
outcomes in these patients (6,7).
Therefore, identifying or discovering novel early diagnostic
biomarkers and elucidating the underlying mechanisms of cancer
metastasis has received much focus for improving treatment efficacy
and prognosis in patients with liver cancer.
HCC is an inflammation-associated cancer (8,9), with
chronic inflammation closely associated with disease progression.
Tumor-associated macrophages (TAMs) are predominantly inflammatory
cells in the tumor microenvironment (TME) that serve a pivotal role
in tumor initiation, metastasis, angiogenesis and suppression of
adaptive immunity (10). In the TME,
TAMs exhibit high plasticity and heterogeneity in response to
different stimuli, existing as classically activated macrophages
(M1 macrophages) and alternatively activated macrophages (M2
macrophages) (11). In response to
inflammatory mediators, M1 macrophages express inducible nitric
oxide synthase (iNOS), which uses L-arginine as a substrate to
produce nitric oxide (12), and
exhibit pro-inflammatory and anti-tumorigenic properties.
Conversely, M2 macrophages constitutively express the enzyme
arginase-1 (Arg-1), which hydrolyzes L-arginine to L-ornithine
(12) and demonstrates
anti-inflammatory and pro-tumor characteristics. TAMs predominantly
exhibit M2 properties with a pro-tumor phenotype (13), promote tumor cell proliferation,
invasion, metastasis, neovascularization and suppression of the
adaptive immune response in the TME (14,15).
Therefore, identifying mechanisms through which macrophages are
polarized in the TME is crucial for understanding their roles in
tumor progression.
Triggering receptor expressed on myeloid cells
(TREM) proteins are a family of immunoglobulin cell surface
receptors expressed on myeloid cells that consist of a single
extracellular immunoglobulin-like domain, a trans-membrane region
and a short cytoplasmic tail (16).
The TREM gene cluster is located on human chromosome 6p21, and
includes genes that encode TREM1 and TREM2; on mouse chromosome
17c3, it encodes for TREM3 (16).
TREM1 activation is associated with downstream signaling adaptor
DNAX activation protein of 12 kDa (17) and peptidoglycan recognition protein-1
(18). TREM1 acts as a potent
amplifier of inflammatory responses and leads to the increased
secretion of pro-inflammatory cytokines, including monocyte
chemotactic protein-1, TNFα, IL-1α, IL-1β, IL-6, IL-8 and
macrophage colony-stimulating factor (19). In human non-small cell lung cancer,
high expression levels of TREM1 on TAMs are associated with tumor
recurrence and poor prognosis (20),
suggesting that TREM1 may play an important role in cancer
progression. In experimental xenograft mouse models of pancreatic
cancer, inhibiting TREM1 attenuates tumor growth and prolongs
survival (21). Similarly, in a
chemically induced model of HCC, TREM1 serves a role in the
connection between the activation of Kupffer cells and
tumorigenesis (22). However, the
role of TREM1 in the regulation of macrophage polarization in
tumor-associated inflammation and its microenvironment has not been
established. The present study aimed to investigate the influence
and possible signaling pathway of TREM1 on macrophage polarization.
Additionally, the effects of TREM1 downregulation in macrophages on
the migration and invasion of liver cancer cells were explored.
Materials and methods
Patients and specimens
Incisional biopsy samples (tumor and adjacent normal
tissues) were collected from 20 clinically diagnosed patients with
HCC between August 2014 and April 2016, and were sent for
histological confirmation according to classical histopathological
features. The distance of the adjacent tissue from the tumor was
>5 cm. Patients with other systemic tumors, infections or
incomplete clinical data were excluded. All samples were obtained
from the Department of Hepatobiliary Surgery of The First
Affiliated Hospital of Fujian Medical University (Fuzhou, China).
The mean age of the patients was 53 years (age range, 41–65 years).
Clinical and laboratory parameters of patients such as disease
severity were evaluated using the Child-Pugh score (23). Tumor staging was based on the
Barcelona-Clinic Liver Cancer system (24). All patients signed informed consent
forms in accordance with the Declaration of Helsinki. The study
protocol was approved by the Ethical Committee of The First
Affiliated Hospital of Fujian Medical University [approval no.
(2015) 132].
Immunohistochemistry
For immunohistochemistry, three areas of tumor
specimens and adjacent normal tissues from 20 patients with HCC
were fixed with 10% formalin for 24 h at 37°C and embedded in a
paraffin block. Subsequently, the samples were de-paraffinized in
two changes of xylene for 5 min each at 37°C and rehydrated in 100,
95, 70 and 50% alcohol for 5 min each at 37°C, and finally
sectioned into 5-µm-thick slices. For antigen retrieval, samples
were treated with 10 mM citrate buffer (pH 6.0) at 100°C for 10
min. After quenching endogenous peroxidase with 3% hydrogen
peroxide for 30 min at 37°C, 0.5% bovine serum albumin (BSA; cat.
no. ST025-5 g; Beyotime Institute of Biotechnology) was used for 1
h at 37°C to block non-specific binding. The sections were then
incubated overnight at 4°C with polyclonal rabbit anti-human TREM1
antibody (1:200; cat. no. YT5133; ImmunoWay Biotechnology Company).
Subsequently, the sections were incubated with the appropriate goat
anti-rabbit IgG secondary antibody (1:1,000; cat. no. ab6721;
Abcam) for 60 min at 37°C according to the manufacturer's
instructions. Images of the labeled specimens were captured with an
Olympus CX41 light microscope (Olympus Corporation; magnification,
×400) and analyzed using ImageJ software (v1.4; National Institutes
of Health). All experiments were performed in triplicate.
Immunohistochemical double
staining
Immunohistochemical double staining (CD206+Arg-1,
CD16+iNOS and CD206+TREM1) was performed using the DouMaxVision•
immunohistochemical double staining test kit (cat. no. KIT-9998;
Fuzhou Maixin Biotech Co., Ltd.) according to the manufacturer's
protocol, and the number of positively stained cells in five
high-power fields was counted under an Olympus CX41 light
microscope (Olympus Corporation; magnification, ×400). Samples were
incubated with anti-CD16 (1:200; cat. no. ab46629), anti-iNOS
(1:200; cat. no. ab53769), anti-CD206 (1:200; cat. no. ab64693) and
anti-Arg-1 primary antibodies (1:200; cat. no. ab133543) for 2 h at
37°C. Biotinylated goat anti-rabbit/mouse IgG secondary antibodies
(1:1,000; cat. no. KIT-9998; Fuzhou Maixin Biotech Co., Ltd) were
added for 60 min at 37°C.
Cell culture and chemicals
The human leukemia monocytic cell line (THP-1), the
293T cell line and the HepG2 and MHCC97H cell lines were purchased
from The Cell Bank of Type Culture Collection of The Chinese
Academy of Sciences. THP-1 cells were cultured in RPMI-1640 medium
(Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% fetal
calf serum (Gibco; Thermo Fisher Scientific, Inc.) and 0.05 mM
β-mercaptoethanol (Sigma-Aldrich; Merck KGaA). 293T, HepG2 and
MHCC97H cell lines were cultured in high-glucose DMEM (Gibco;
Thermo Fisher Scientific, Inc.) supplemented with 10% fetal calf
serum. All cell lines were cultured with 5% CO2 at
37°C.
Cell transfection
The third generation of lentiviral vector
(hU6-MCS-Ubiquitin-EGFP-IRES-puromycin) containing short hairpin
(sh)RNAs against TREM1 (shTREM1) used in the present study were
produced by Shanghai GeneChem Co., Ltd., and were used for TREM1
gene knockdown. The target sequences of shRNAs were as follows:
shTREM1-1, 5′-AGCCAGAAAGCTTGGCAGATA-3′; shTREM1-2,
5′-GAGGATCATACTAGAAGACTA-3′; and shTREM1-3,
5′-GTGGAAGATTCTGGACTGTAT-3′. Three plasmids (Shanghai GeneChem Co.,
Ltd.), namely 20 µg GV248 carrying the target sequence, 15 µg
pHelper1.0 (carrying gag, pol and rev genes) and 10 µg pHelper2.0
(carrying the VSV-G gene), were mixed with
Lipofectamine® 2000 (cat. no. 11668030; Invitrogen;
Thermo Fisher Scientific, Inc.) for 15 min at room temperature.
Subsequently, the mixture was transfected into 293T cells for 6 h
at 37°C to produce lentivirus. Lentiviral particles were collected
and the virus titer were detected to be 2×109 TU/ml.
Transfections of shRNA into THP-1 cells were performed using
polybrene (cat. no. REVG0001; Shanghai GeneChem Co., Ltd.)
according to the manufacturer's instructions, and lentivirus at a
multiplicity of infection of 50 was chosen for further experiments.
After 12 h at 37°C, the medium was changed into RPMI-1640 medium
supplemented with 10% fetal calf serum and 0.05 mM
β-mercaptoethanol containing no virus. The fluorescence intensity
was observed under a fluorescence microscope at ×100 and ×400
magnification. Puromycin (5 µg/ml) was used to screen the
shRNA-transfected THP-1 cells after 7 days at 37°C. The knockdown
efficiency of shTREM1 was tested by quantitative PCR assay and
western blot analysis. A scrambled non-specific shRNA
(5′-TTCTCCGAACGTGTCACGT-3′) was used as a negative control
(sh-ctrl).
Induced M2 macrophages from THP-1
cells
M2 macrophages were derived from THP-1 cells.
shTREM1-treated macrophages were derived from shTREM1 THP-1 cells
stimulated with 100 ng/ml phorbol 12-myristate 13-acetate (PMA;
Sigma-Aldrich; Merck KGaA) for 24 h at 37°C. After PMA treatment,
20 ng/ml IL-4 and 20 ng/ml IL-13 (PeproTech, Inc.) were added for
24 h at 37°C (25).
Conditioned medium preparation
After incubating M2 and shTREM1 macrophages with
fresh RPMI-1640 medium for an additional 24 h at 37°C, the cells'
conditioned media (CM) was harvested, centrifuged at 3,000 × g for
15 min at 37°C, filter-sterilized through a 0.22-µm nylon filter
(Nalgene; Thermo Fisher Scientific, Inc.) and finally stored at
−80°C for further experiments.
Double-labeling cells
immunofluorescence
For immunofluorescence double labeling, cells were
fixed in 4% paraformaldehyde for 20 min at 37°C and permeabilized
with 0.1% Triton X-100 for 10 min at 37°C. After blocking
non-specific binding with 0.1% BSA for 30 min at 37°C, cells were
incubated with primary antibodies against CD163 (cat. no. ab156769;
Abcam) and TREM1 antibody (1:200; cat. no. YT5133; ImmunoWay
Biotechnology Company) at a 1:500 dilution overnight at 4°C.
Subsequently, the samples were incubated with Alexa
Fluor® 488-labeled donkey anti-rabbit IgG antibody
(1:1,000; cat. no. A-21206; Thermo Fisher Scientific, Inc.) and
Alexa Fluor® 546-labeled donkey anti-mouse IgG antibody
(1:1,000; cat. no. A-10036; Thermo Fisher Scientific, Inc.) for 30
min at room temperature, and 1 µg/ml DAPI (Invitrogen; Thermo
Fisher Scientific, Inc.) was used for nuclear labeling for 5 min at
room temperature. Images of the labeled specimens were captured
with a Zeiss confocal laser scanning microscope (Carl Zeiss AG) at
×2,000 magnification and analyzed using ImageJ software. All
treatments were performed in triplicate.
Reverse transcription-quantitative
(RT-q)PCR
Total cellular RNA was extracted using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.) and was reverse transcribed into cDNA using PrimeScript™ 1st
strand cDNA synthesis kit (cat. no. 6110A; Takara Bio, Inc.)
according to the manufacturer's protocol. Subsequently, qPCR was
performed using a SYBR Green kit (Takara Bio, Inc.) with specific
primers for TREM1, iNOS, CD16, IL-12, CD163, Arg-1, IL-10 and GAPDH
(used as the reference gene). PCR settings were adjusted according
to the manufacturer's instructions: Initial denaturation at 95°C
for 30 sec and 40 cycles at 95°C for 5 sec and 60°C for 30 sec. All
samples were assayed in triplicate and relative mRNA expression was
quantified using the 2−∆∆Cq method (26). The primer sequences used were as
follows: TREM1 forward, 5′-GGCAGATAATAAGGGACGGAGAG-3′ and reverse,
5′-CATTCGGACGCGCAGTAAA-3′; iNOS forward, 5′-GAGCCAGGCCACCTCTATGT-3′
and reverse, 5′-GTCCTCGACCTGCTCCTCAT-3′; CD16 forward,
5′-CCAGTGTGGCATCATGTGG-3′ and reverse, 5′-ATTGAGGCTCCAGGAACACC-3′;
IL-12 forward, 5′-CTGAAGAAGATGGTATCACCTGGAC-3′ and reverse,
5′-TTAGAACCTCGCCTCCTTTGTG-3′; CD163 forward,
5′-TTCCTGTTCTGGACGTGTGG-3′ and reverse, 5′-AGCTGGACCACAGCCAAGTT-3′;
Arg-1 forward, 5′-GGTGTTGCCTGCTGCCTTCC-3′ and reverse,
5′-GTTCTGAAGAGGTGAGTGGCTGTC-3′; IL-10 forward,
5′-TCAGAGAGGGGGTTAGACCTG-3′ and reverse,
5′-GAGTTGGTCCTGCCAGACTT-3′; GAPDH forward,
5′-CCCTTCATTGACCTCAACTACATG-3′ and reverse,
5′-TGGGATTTCCATTGATGACAAGC-3′.
Western blotting
Cells were washed with cold PBS, and then lysed with
radioimmunoprecipitation assay buffer with phenylmethylsulfonyl
fluoride (Beyotime Institute of Biotechnology). Cell lysates were
collected and then centrifuged at 12,000 × g for 15 min at 4°C. The
bicinchoninic acid method was used to detect protein sample
concentrations. A total of 20 µg protein/lane was separated via 10%
SDS-PAGE, and the proteins were transferred to polyvinylidene
fluoride membranes. The membranes were blocked against non-specific
binding with 5% non-fat milk for 30 min at 37°C, followed by
overnight incubation at 4°C with primary antibodies (all 1:1,000)
against TREM1 (cat. no. YT5133; ImmunoWay Biotechnology Company),
iNOS (cat. no. ab53769), CD16 (cat. no. ab46629), IL-12 (cat. no.
ab106270), CD163 (cat. no. ab156769), Arg-1 (cat. no. ab133543),
IL-10 (cat. no. ab34843), phosphorylated (p)-PI3K (cat. no.
ab138364), p-AKT (ab183758), p-mTOR (cat. no. ab84400), PI3K (cat.
no. ab151549), AKT (cat. no. ab179463), mTOR (cat. no. ab2732),
snail (cat. no. ab229701), E-cadherin (cat. no. ab15148) and GAPDH
(cat. no. ab181602) (all Abcam). After three washes with TBS with
0.3% Triton X-100, the membranes were incubated with secondary
HRP-conjugated goat anti-rabbit IgG (cat. no. ab6721) or goat
anti-mouse IgG (cat. no. ab6789) secondary antibodies (both
1:2,000; Abcam) for 1 h at room temperature. The membranes were
then washed three times and exposed via enhanced chemiluminescence
reagents (BeyoECL Moon kit; cat. no. P0018FS; Beyotime Institute of
Biotechnology). The protein bands were captured using Invitrogen
iBright FL1500 Imaging System (Thermo Fisher Scientific, Inc.) and
analyzed using ImageJ software.
Cell migration/invasion co-culture
assay
Transwell chambers (8-µm; EMD Millipore) were used
to evaluate cancer cell migration and invasion as previously
described (27). A total of 80 µl
high-glucose DMEM (Gibco; Thermo Fisher Scientific, Inc.)
supplemented with 2% fetal calf serum (Gibco; Thermo Fisher
Scientific, Inc.) containing 2×105 HepG2 and MHCC97H
cells was added into the upper chamber inserts. Serum-free
RPMI-1640 medium, M2 macrophage-CM, shTREM1 macrophage-CM and
sh-ctrl macrophage-CM were added into the bottom chambers to
evaluate their influence on cancer cell migration. Before seeding
HepG2 and MHCC97H cells, Matrigel (100 µl; Becton, Dickinson and
Company) was placed into the upper chamber for invasion assays for
4 h at 37°C, and the remaining steps were the same as for
migration. After a 24-h incubation at 37°C, the upper inserts were
washed three times in PBS, fixed with 4% formaldehyde for 20 min at
37°C and stained with crystal violet for 2 h at 37°C. Five fields
of view were randomly photographed with an Olympus CX41 light
microscope (Olympus Corporation; magnification, ×100) and the
numbers of migrated and invaded cells per field were determined
using ImageJ software.
Gap closure assay
A total of 2×105 HepG2 and MHCC97H cells
were seeded into culture inserts (Ibidi GmbH); the culture inserts
were then removed, and a 500-µm wide gap was generated. Cells were
incubated with serum-free RPMI-1640 medium, M2 macrophage-CM,
shTREM1 macrophage-CM and sh-ctrl macrophage-CM for 24 h at 37°C.
Five fields of view were randomly photographed with an Olympus CX41
light microscope (Olympus Corporation; magnification, ×100) and the
gap closure rate was estimated by calculating the gap area with
ImageJ software.
Cell proliferation assay
HepG2 and MHCC97H cells were seeded into 96-well
plates at a density of 1×104 cells/well. Cells were
incubated with serum-free RPMI-1640 medium, M2 macrophage-CM,
shTREM1 macrophage-CM and sh-ctrl macrophage-CM for 24 h at 37°C.
According to the instructions of a BrdU Cell Proliferation Assay
kit (cat. no. 6813; Cell Signaling Technology, Inc.), BrdU was
added to wells and incubated for 12 h at 37°C. Subsequently, cells
were fixed and denatured using 100 µl Fix/Denature solution (part
of the aforementioned kit) for 30 min at 37°C. Cells were washed
with cold PBS three times. Cells were incubated with 100 µl
anti-BrdU antibody (part of the aforementioned kit) for 1 h at 37°C
and were then washed three times with cold PBS. Subsequently,
HRP-conjugated secondary antibody (part of the aforementioned kit)
was added for 1 h at 37°C. A total of 100 µl Substrate Solution was
then added and incubated for 5 min at 37°C. Finally, 100 µl Stop
Solution was added to stop the enzyme reaction, and the levels of
BrdU incorporation was determined using an ELISA reader at a
wavelength of 450 nm (Bio-Rad Laboratories, Inc.).
Statistical analysis
All continuous variables were presented as the mean
± SD. The paired Student's t-test was used for comparative analysis
of paired cancerous and adjacent normal tissues. The unpaired
Student's t-test was applied for comparisons between 2 groups for
the other assays, and one-way ANOVA followed by Tukey's multiple
comparison test was used when >2 groups were evaluated.
Two-sided P<0.05 was considered to indicate a statistically
significant difference. All statistical analysis was performed
using SPSS version 19.0 (IBM Corp.).
Results
High TREM1 expression on M2
macrophages in HCC
The clinical characteristics of the study population
are described in Table SI.
Immunohistochemistry was performed to determine the expression
levels of TREM1 in HCC tissues and adjacent normal tissues. The
percentage of TREM1+ cells per field in HCC tissues was
significantly higher than that in adjacent normal tissues
(11.23±0.8840 vs. 2.253±0.8662%; P<0.01; Fig. 1A-C). Immunohistochemical double
staining was used to detect the tumor-associated macrophage
phenotype in HCC by analyzing CD206 and Arg-1 expression. The
results revealed that the infiltration of M2 macrophages in liver
cancer tissues was significantly increased compared with that in
adjacent normal tissues (3.50±0.5000 vs. 0.5100±0.1813%; P<0.01;
Fig. 1D-F). No double-positive cells
of CD16 and iNOS (M1-associated markers) were observed, indicating
no M1 macrophage infiltration in liver cancer tissues and adjacent
normal tissues (Fig. 1G-I). In
addition, immunohistochemical double staining with the M2
macrophage surface markers CD206 and TREM1 revealed that TREM1 was
highly expressed on the surface of M2 macrophages in HCC tissues
(16.51±0.9327 vs. 6.644±1.221%; P<0.01; Fig. 1J-L). The current results suggested
that M2 macrophages mainly infiltrate HCC tissues, while no
infiltration of M1 macrophages was observed in both HCC and
adjacent normal tissues. M2 macrophages in HCC tissues highly
express TREM1. These results indicated that TREM1 expression on M2
macrophages may serve an important role in the progression of
HCC.
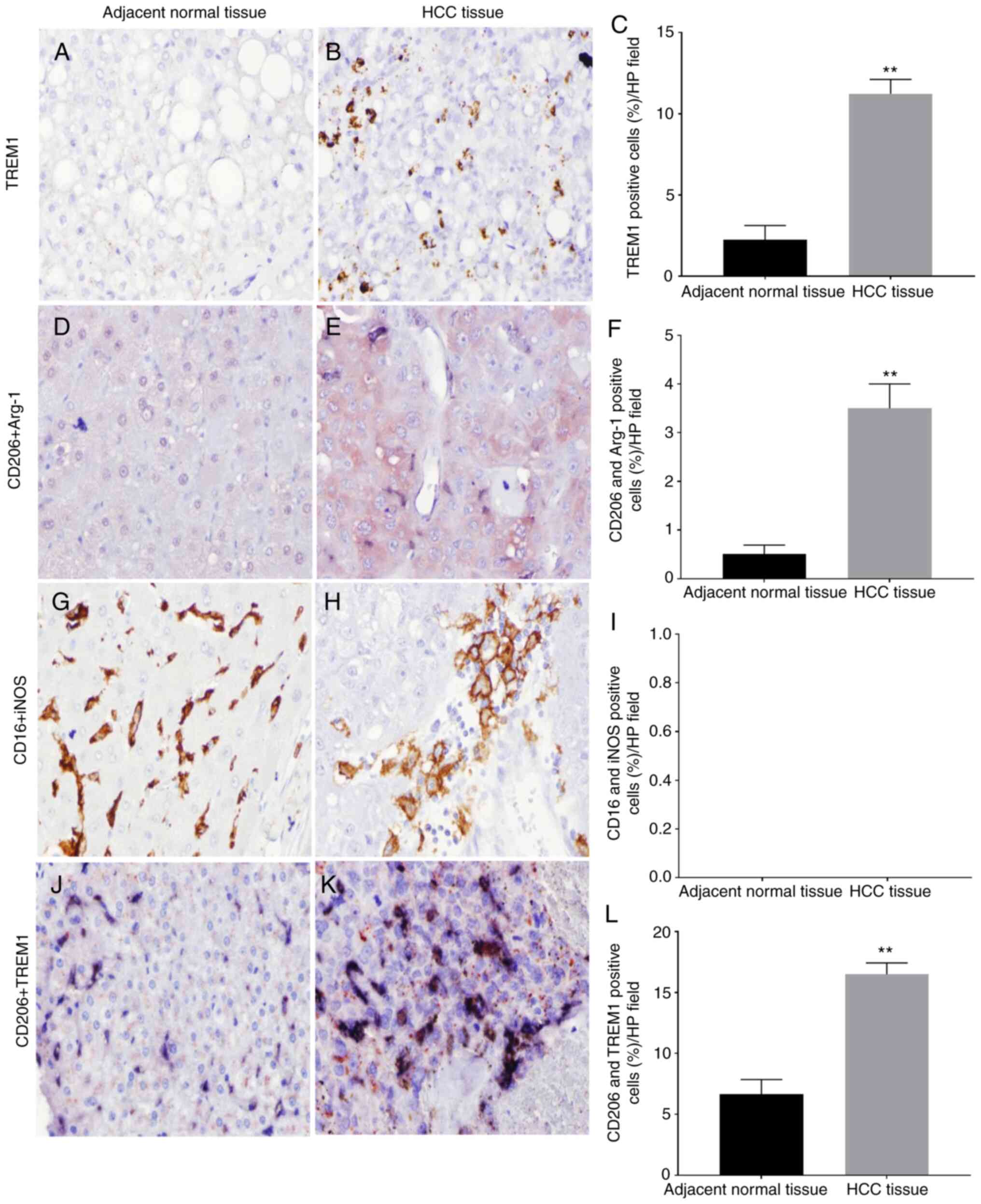 | Figure 1.TREM1/macrophage-associated marker
expression in HCC and adjacent normal tissues. Images were captured
at ×400 magnification. TREM1 expression in (A) normal and (B) HCC
tissues. Some pale-stained brown precipitate was observed outside
the cell membrane in adjacent tissues, but a large number of
brown-black precipitates were observed on the cell membrane in HCC
tissues. (C) Quantification of TREM1 expression. M2
macrophage-associated markers (CD206 and Arg-1) in (D) normal and
(E) HCC tissues. The blue-black precipitate on the membrane was
positive for CD206, and the red precipitate in the cytoplasm was
positive for Arg-1. Cells labeled with two colors (both markers)
were M2 macrophages. (F) Quantification of CD206+ and
Arg-1+ cells. M1 macrophage-associated markers (CD16 and
iNOS) in (G) normal and (H) HCC tissues. The brown and black
precipitates on the cell membrane were positive for CD16, no red
precipitates for iNOS were found in the cytoplasm and nuclei,
indicating that there were no double-positive cells for CD16 and
iNOS. (I) Quantification of CD16+ and iNOS+
cells. TREM1 expression on the surface of M2 macrophages in (J)
normal and (K) HCC tissues. The blue-black precipitate on the
membrane was positive for CD206, and the red precipitate was
positive for TREM1. (L) Quantification of CD206+ and
TREM1+ cells. **P<0.01. HCC, hepatocellular
carcinoma; TREM1, triggering receptor expressed on myeloid cells-1;
iNOS, inducible nitric oxide synthase; Arg-1, arginase-1; HP, high
power. |
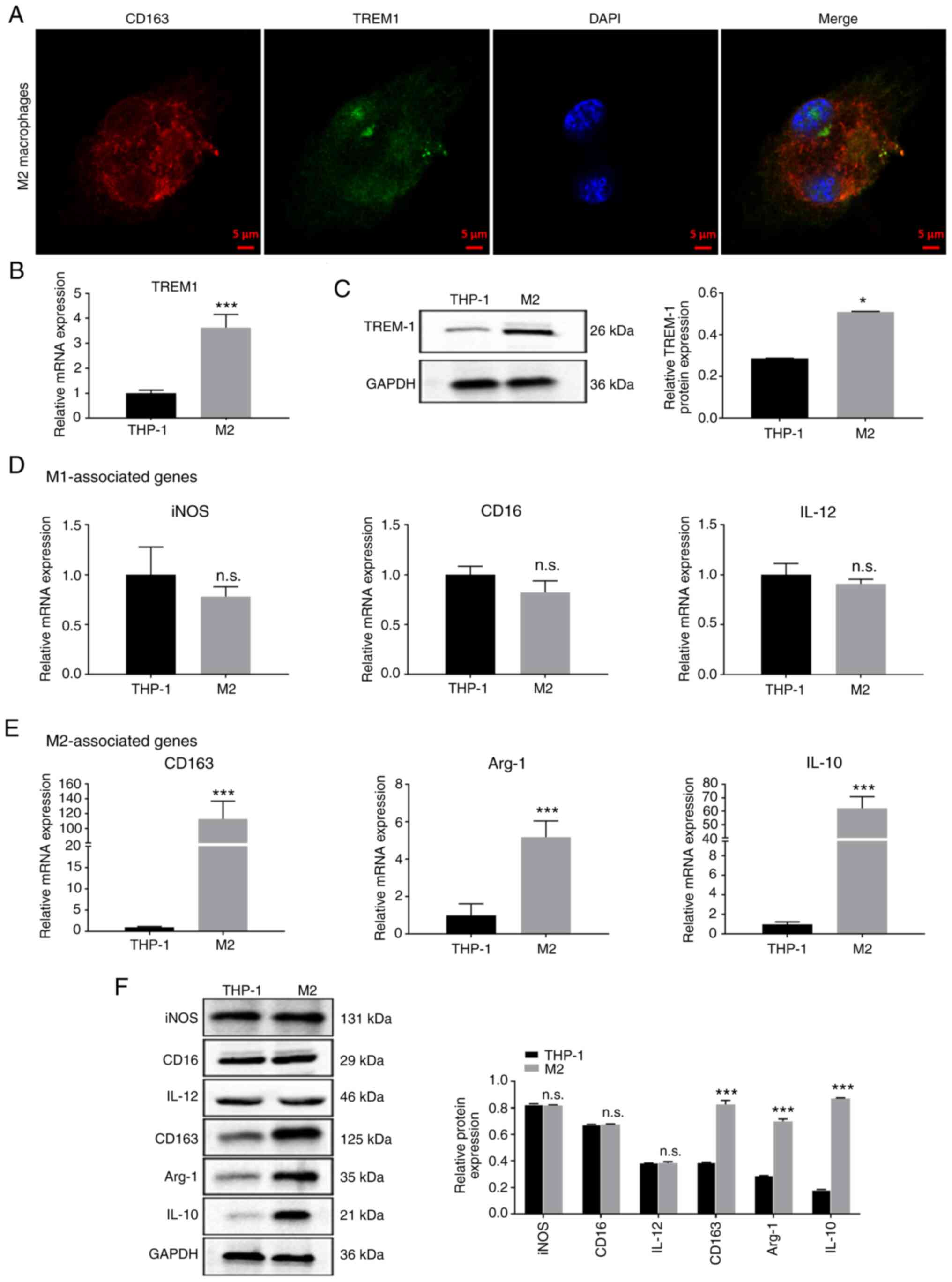
M2 macrophages induced from human
leukemia monocytic cells
To establish a model of human M2 macrophages, human
leukemia monocytic cells (THP-1) were stimulated using PMA, IL-4
and IL-13. Double-immunofluorescence labeling was used to identify
co-expression of CD163 and TREM1 in M2 macrophages (Fig. 2A). RT-qPCR and western blot analysis
revealed that TREM1 expression in M2 macrophages was significantly
higher than in THP-1 cells (Fig. 2B and
C). The mRNA expression levels of iNOS, CD16 and IL-12 (M1
macrophage-associated markers) were not significantly different
between THP-1 cells and M2 macrophages, while the expression levels
of CD163, Arg-1 and IL-10 (M2 macrophage-associated markers) were
significantly increased in M2 macrophages compared with in THP-1
cells (Fig. 2D and E). Similarly,
protein expression levels of iNOS, CD16 and IL-12 exhibited no
significant difference between THP-1 cells and M2 macrophages,
while protein expression levels of CD163, Arg-1 and IL-10 were
significantly upregulated in M2 macrophages compared with in THP-1
cells (Fig. 2F). Overall, the present
data indicated that M2 macrophages were successfully induced from
the THP-1 monocytic cell line.
Downregulation of TREM1 inhibits M2
macrophage polarization and promotes a shift towards the M1
phenotype
To discover possible functions of TREM1 in
macrophage polarization, shRNAs were used to knock down TREM1
expression. As shown in Fig. 3A,
shTREM1-2 was the most effective at TREM1 silencing and stable
regulation of TREM1 in THP-1 cells and was therefore used for
subsequent experiments. Western blot analysis revealed that
transfection with shTREM1-2 resulted in a significant
downregulation of TREM1 expression compared with
sh-ctrl-transfected cells (Fig. 3B).
RT-qPCR and western blot analysis demonstrated that iNOS, CD16 and
IL-12 expression was significantly upregulated in shTREM1
macrophages, while CD163, Arg-1 and IL-10 expression was
significantly downregulated in shTREM1 macrophages compared with in
sh-ctrl macrophages (Fig. 3C-E).
Overall, the present results indicated that TREM1 downregulation
inhibited M2 macrophage polarization and promoted a shift towards
the M1 phenotype.
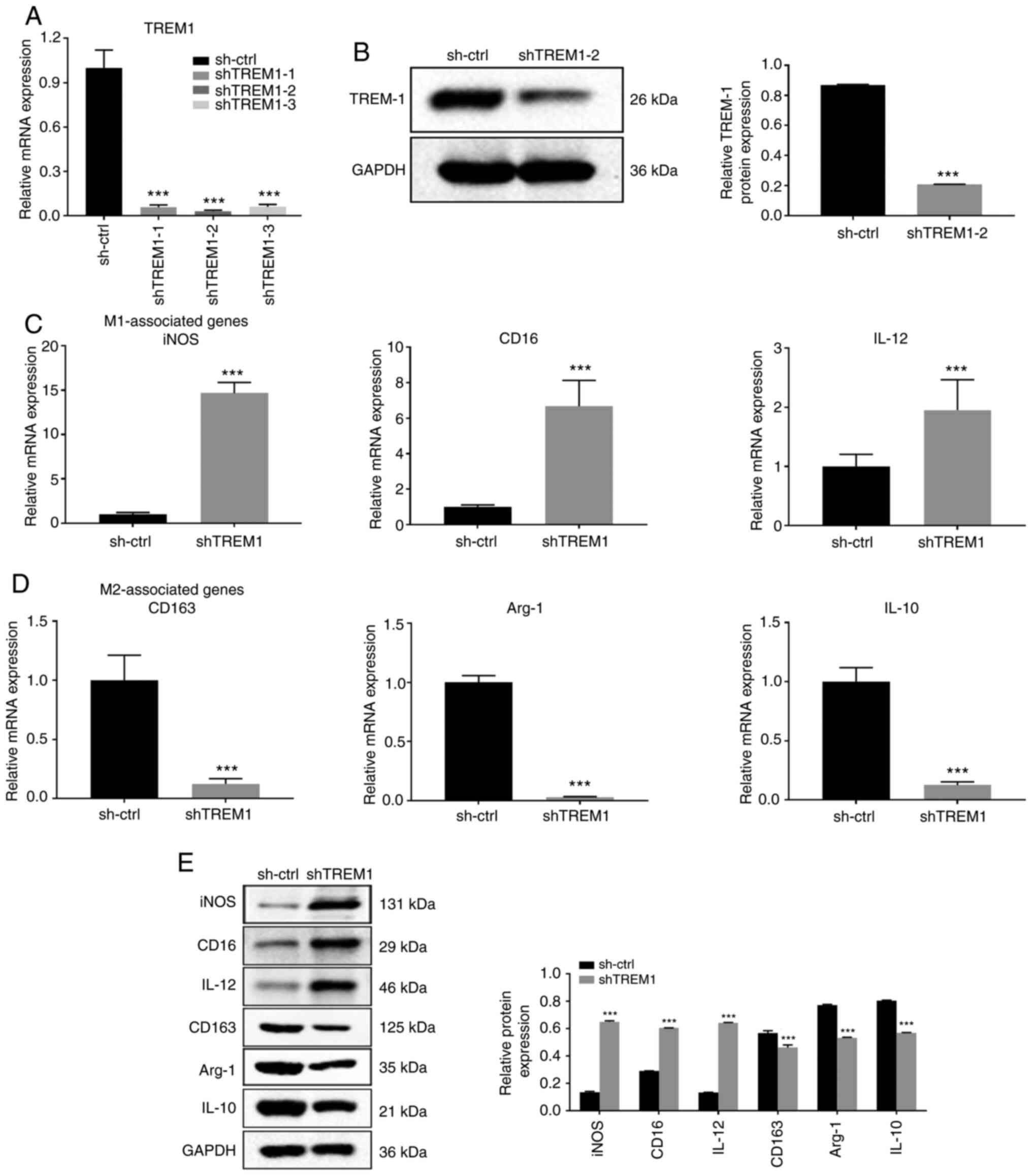 | Figure 3.Downregulation of TREM1 in
macrophages inhibits M2 polarization and shifts macrophages into an
M1 phenotype. shRNA against TREM1 was used in THP-1 cells. M2
macrophages were derived from THP-1 cells, and shTREM1 macrophages
were derived from shTREM1 THP-1 cells with stimulation using
phorbol 12-myristate 13-acetate, IL-4 and IL-13 for 24 h. (A)
shTREM1-1, shTREM1-2 and shTREM1-3 were used for TREM1-knockdown.
TREM1 mRNA expression was assessed via RT-qPCR. (B) TREM1 protein
expression was assessed by western blotting. (C) mRNA expression
levels of the M1 macrophage-associated markers (iNOS, CD16 and
IL-12) and (D) M2 macrophage-associated markers (CD163, Arg-1 and
IL-10) were measured by RT-qPCR. (E) Protein expression levels of
M1 and M2 macrophage-associated markers were determined by western
blotting. Data are shown as the mean ± SD of three independent
experiments. ***P<0.001 vs. sh-ctrl. shRNA, short hairpin RNA;
ctrl, control; iNOS, inducible nitric oxide synthase; Arg-1,
arginase-1; TREM1, triggering receptor expressed on myeloid
cells-1; RT-qPCR, reverse transcription-quantitative PCR. |
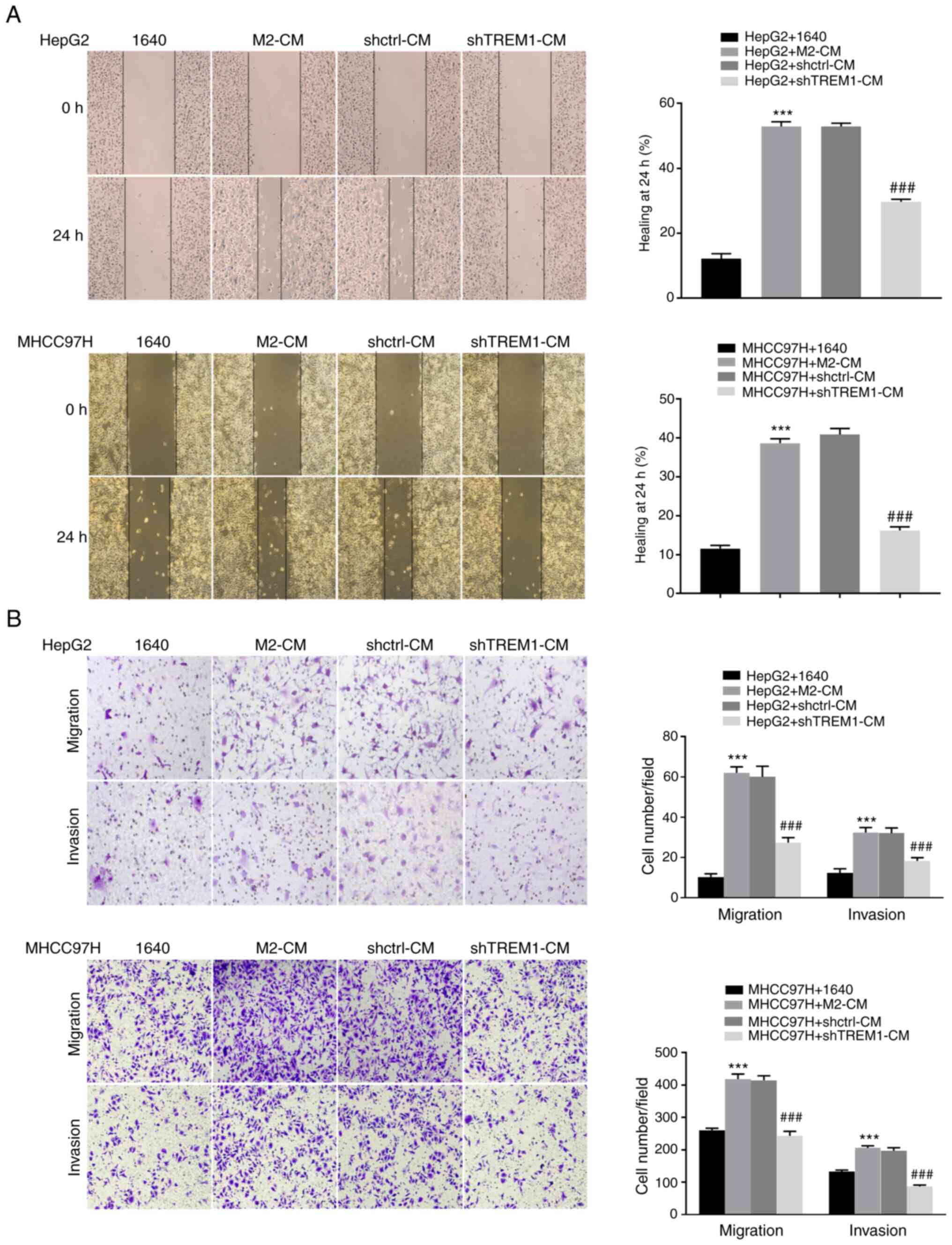
Downregulation of TREM1 in macrophages
inhibits the migration and invasion of tumor cells
A co-culture system was used to investigate the
effect of shTREM1 macrophages on the migration and invasion of
cancer cells. The results revealed that M2 macrophage-CM (M2-CM)
significantly increased cell gap closure (Fig. 4A), as well as migration and invasion
(Fig. 4B), in HepG2 and MHCC97H cell
lines compared with RPMI-1640 medium (1640); meanwhile, shTREM1
macrophage-CM (shTREM1-CM) significantly decreased this effect. The
present results indicated that downregulation of TREM1 can inhibit
the migration and invasion of tumor cells.
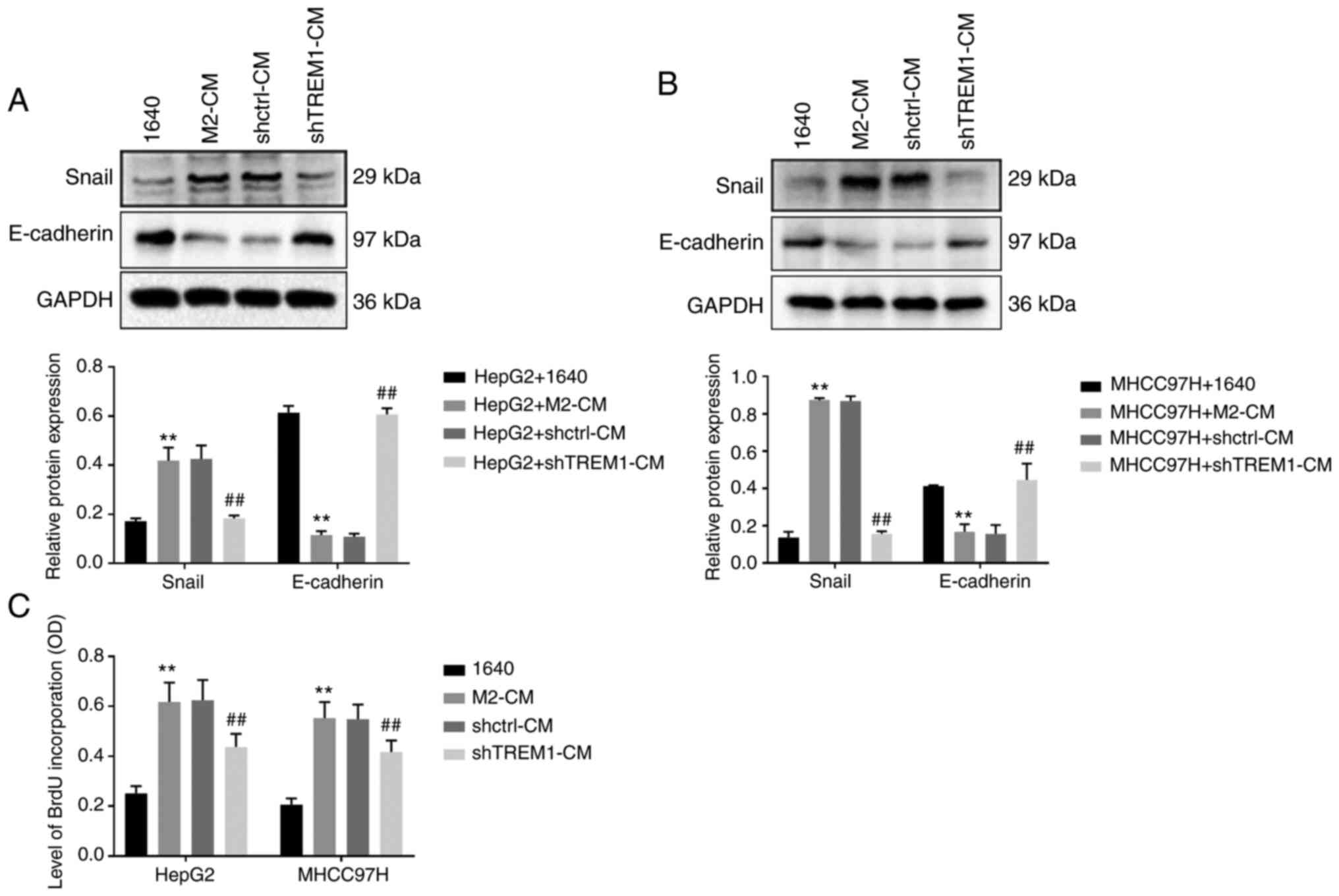
Downregulation of TREM1 in macrophages
inhibits epithelial-mesenchymal transition (EMT) and the
proliferation of tumor cells
A co-culture system was used to investigate the
effect of shTREM1 macrophages on EMT of cancer cells. The results
revealed that M2-CM significantly increased the protein expression
levels of snail and significantly decreased those of E-cadherin in
HepG2 (Fig. 5A) and MHCC97H (Fig. 5B) cells compared with RPMI-1640
medium; meanwhile, shTREM1-CM reversed this effect. These results
indicated that downregulation of TREM1 inhibited the EMT of tumor
cells. A BrdU kit was used to investigate the effect of shTREM1
macrophages on the proliferation of cancer cells. The results
demonstrated that M2-CM significantly increased proliferation of
HepG2 and MHCC97H cells compared with RPMI-1640 medium; meanwhile,
shTREM1-CM reversed this effect (Fig.
5C). The present results indicated that downregulation of TREM1
inhibited the proliferation of tumor cells.
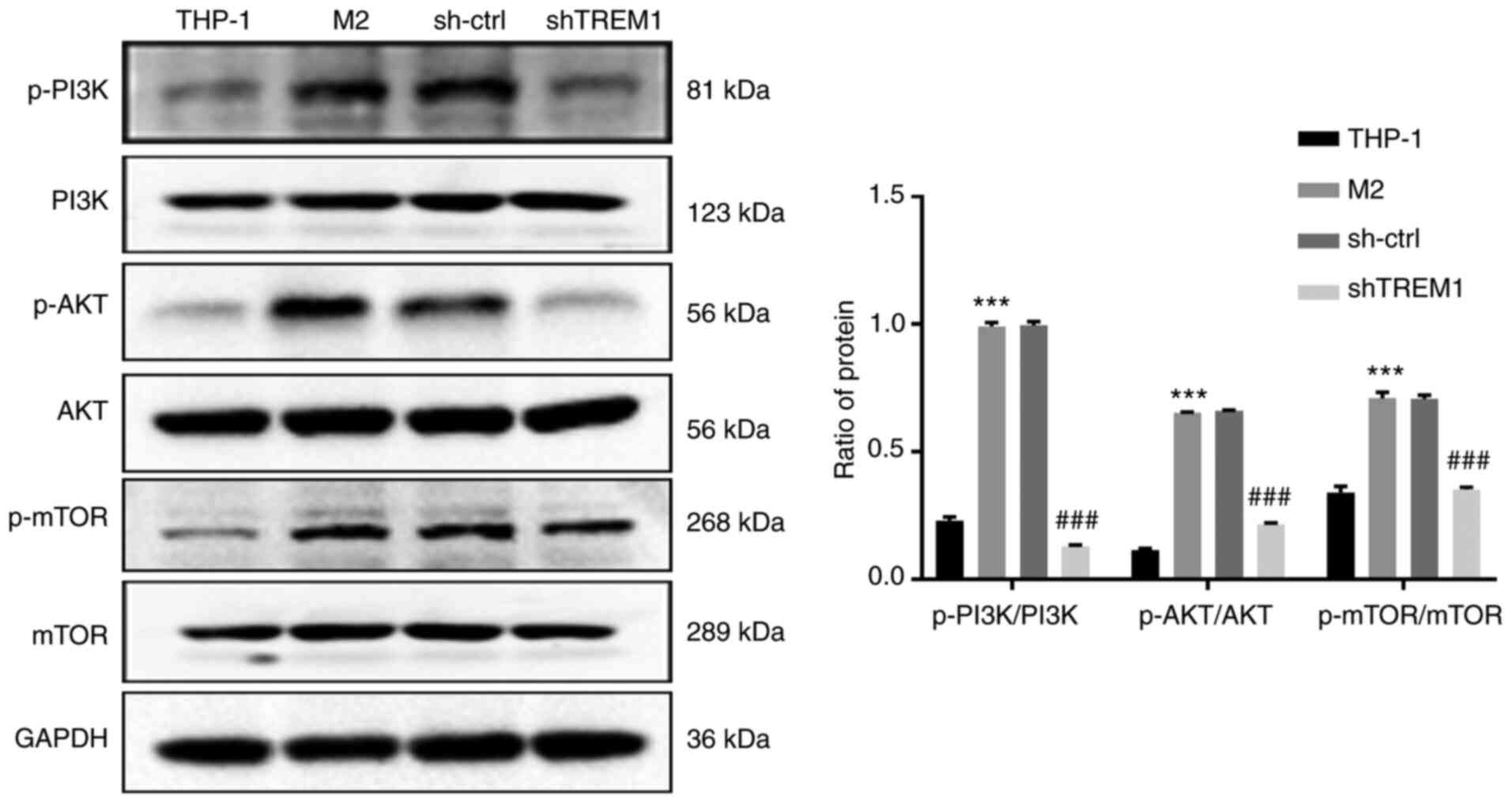
Downregulation of TREM1 inhibits
PI3K/AKT signaling in the M2 polarization of macrophages
To discover the molecular mechanisms of the effects
of silencing TREM1 in macrophages and inhibition of M2 polarization
of macrophages, western blotting was used to determine the protein
expression levels of p-PI3K (p85), PI3K, p-AKT, AKT, p-mTOR and
mTOR. As illustrated in Fig. 6, the
ratios of p-PI3K/PI3K, p-AKT/AKT and p-mTOR/mTOR were significantly
upregulated in M2-polarized macrophages, while downregulation of
TREM1 in macrophages reversed this effect. These results indicated
that the PI3K/AKT/mTOR signaling pathway may be involved in the
regulation of macrophage polarization by TREM1.
Discussion
The TME is a complex and dynamic cellular system
that is associated with cancer occurrence, progression and response
to therapy (28). TAMs are
predominant inflammatory components in the TME and can exist as
classically activated macrophages (M1 macrophages) and
alternatively activated macrophages (M2 macrophages) in response to
different stimuli within the TME (10). In numerous types of cancer, such as
ovarian, breast and pancreatic cancer, TAMs tend to exhibit M2
polarization characteristics (10,13). The
functions of M2 macrophages in cancer are complex, including
promotion of angiogenesis, enhancing cell proliferation, migration
and invasion, and suppression of the immune response in the TME
(29). Yeung et al (14) revealed that M2 macrophages promote
tumor growth and invasiveness in HCC through CCL2-induced EMT.
HCC is a well-known and extensively investigated
inflammation-associated cancer (8,9). TREM1 is
regarded as an effective amplifier of inflammatory responses in the
immunoglobulin superfamily (30).
Previous studies have investigated TREM1 expression in HCC cells
and hepatic stellate cells (31,32). Duan
and Wang (31) reported that the
influence of TREM1 expression on HCC cell proliferation, invasion
and apoptosis was not mediated by monocytes or macrophages,
indicating that TREM1 had a direct pro-tumor effect in cancer
cells. The present study confirmed that TREM1 expression was
elevated in tumor tissues of patients with HCC. Furthermore, the
current results revealed that TREM1 was highly expressed in M2
macrophages in HCC tissues through immunohistochemical
double-staining with the surface markers CD206 and TREM1. This
result indicated that TREM1 may have an indirect effect on HCC
mediated by macrophages. The activation of TREM1 expression in
Kupffer cells (resident macrophages in liver tissue) stimulates
tumorigenesis during diethylnitrosamine-induced
hepatocarcinogenesis (22).
Therefore, TREM1 expression on macrophages may serve an important
role in HCC formation.
Recruited monocyte-derived macrophages are thought
to be more plastic and prone to switch from an antitumor to a
pro-tumor phenotype under the influence of the growing tumor cells
(33). These phenotypes are usually
defined by the M1/M2 polarization classification and different
functional patterns. As a pro-inflammatory receptor, TREM1 protein
expression on macrophages may serve as a regulator to switch the
phenotype of macrophages and change their function (34). The cross-talk between TREM1 and
macrophages in the TME during liver cancer development remains
poorly understand. To explore the mechanisms by which TREM1
influences macrophage polarization, the present study knocked down
TREM1 expression in macrophages by shRNA, revealing that M2
polarization markers were inhibited and M1 macrophage-associated
markers were upregulated in shTREM1 macrophages. The present
results suggested that downregulation of TREM1 may promote a shift
in shTREM1 macrophages towards the M1 phenotype, presenting as iNOS
upregulation and Arg-1 downregulation. Additionally, the CM from
TREM1-knockdown macrophages was co-cultured with liver cancer
cells, as well as the CM from M2 macrophages used as a control
group. Blocking TREM1 shifted the polarization of macrophages to
inhibit migration, invasion, EMT and proliferation of liver cancer
cells. Similarly, a recent study has found that lncRNA cox-2 small
interfering RNA decreases the ability of M1 macrophages to inhibit
tumor cell proliferation, invasion, migration and EMT, while
strengthening the ability of M2 macrophages to promote tumor cell
proliferation and inhibit apoptosis (35). Therefore, the present results provided
evidence for macrophage polarization in the TME, which increases
the current understanding of the role of TAMs in cancer
progression. The current study further defined phenotypic
transition as one of the major mechanisms by which TREM1 signaling
may act on activated M2 macrophages to drive hepatocarcinogenesis.
Notably, TAMs are being identified as increasingly complex, with
several studies suggesting that distinct populations may have
different roles in tumorigenesis (22,31).
The PI3K signaling pathway is a classical signaling
pathway that converges inflammatory and metabolic signals during
phagocytosis, autophagy and cell metabolism (36,37). Class
IA PI3K includes a p110 catalytic subunit, which interacts with an
SH2 domain-containing p85 regulatory subunit (36,37). The
p85 regulatory subunit representative of PI3K phosphorylation was
assessed in the present study. Phosphorylation of PI3K results in
the activation of downstream signal effectors, including AKT and
mTOR; PI3K/AKT and its downstream effectors are regarded as central
mediators of the inflammatory response and polarization of
macrophages (38). In bone
marrow-derived macrophages, the PI3K/AKT signaling pathway promotes
M2 polarization (39), while
inhibition of either PI3K or AKT attenuates the expression levels
of M2-associated genes (39,40), illustrating the importance of the
PI3K/AKT signaling pathway in M2 polarization. The present findings
suggested that TREM1 regulated macrophage polarization through the
PI3K/AKT signaling pathway. Upregulated expression levels of
markers of PI3K/AKT signaling pathway activation were detected in
the current study, suggesting that the M2 macrophage polarization
depended on this activation. In addition, downregulation of TREM1
in macrophages blocked the activation of PI3K/AKT signaling in M2
polarization. Although the present results do not formally prove a
cause-effect association, they clearly support that the PI3K/AKT
signaling pathway may partially mediate an M1/M2 phenotypic switch
in the TME. A limitation of the present study is that it was only
demonstrated that TREM1 inhibited liver cancer cells by regulating
macrophage polarization. The function of TREM-1+ TAMs
and their relevant mechanisms of immunosuppression in the tumor
microenvironment require further investigation.
In conclusion, the present study provided novel
evidence that knocking down TREM1 expression may shift M2
macrophages towards a M1 phenotype, likely through PI3K/AKT
signaling. These findings enhance the understanding of TREM1
expression on M2 macrophages and its role in the pathogenesis of
liver cancer. Finally, the present study supports targeting TREM1
and/or TAMs as a therapeutic strategy against cancer to improve the
efficacy of current systemic therapy.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by grants from the
Middle-Aged Backbone Training Project in the Health System of
Fujian province (grant no. 2016-ZQN-40) and the Startup Fund for
scientific research of Fujian Medical University (grant no.
2017XQ1077).
Availability of data and materials
The datasets generated and/or analyzed during the
current study are not publicly available due to restrictions of the
First Affiliated Hospital of Fujian Medical University, but are
available from the corresponding author on reasonable request.
Authors' contributions
MC completed most of the experiments and wrote the
manuscript. QZ designed the study and reviewed/edited the drafts.
RL analyzed the data, completed part of the experiments and edited
the drafts. XL and WC performed follow-up of patients and collected
data. HW completed the immunohistochemical analysis and co-wrote
the final draft. MC, RL and QZ were responsible for the
authenticity of the data. QZ is the guarantor of this work and, as
such, had full access to all the data in the study and takes
responsibility for the integrity of the data and the accuracy of
the data analysis. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
All patients signed informed consent forms in
accordance with the Declaration of Helsinki. The study protocol was
approved by the Ethical Committee of The First Affiliated Hospital
of Fujian Medical University [Fuzhou, China; approval no. (2015)
132].
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
Arg-1
|
arginase-1
|
|
CM
|
conditioned media
|
|
EMT
|
epithelial-mesenchymal transition
|
|
HCC
|
hepatocellular carcinoma
|
|
iNOS
|
inducible nitric oxide synthase
|
|
PMA
|
phorbol 12-myristate 13-acetate
|
|
shRNA
|
short hairpin RNA
|
|
TAMs
|
tumor-associated macrophages
|
|
TME
|
Tumor microenvironment
|
|
TREM1
|
triggering receptor expressed on
myeloid cells-1
|
References
|
1
|
Dawkins J and Webster RM: The
hepatocellular carcinoma market. Nat Rev Drug Discov. 18:13–14.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ryerson AB, Eheman CR, Altekruse SF, Ward
JW, Jemal A, Sherman RL, Henley SJ, Holtzman D, Lake A, Noone AM,
et al: Annual report to the nation on the status of cancer,
1975–2012, featuring the increasing incidence of liver cancer.
Cancer. 122:1312–1337. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Forner A, Reig M and Bruix J:
Hepatocellular carcinoma. Lancet. 391:1301–1314. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bruix J, Qin S, Merle P, Granito A, Huang
YH, Bodoky G, Pracht M, Yokosuka O, Rosmorduc O, Breder V, et al:
Regorafenib for patients with hepatocellular carcinoma who
progressed on sorafenib treatment (RESORCE): A randomised,
double-blind, placebo-controlled, phase 3 trial. Lancet. 389:56–66.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
El-Khoueiry AB, Sangro B, Yau T, Crocenzi
TS, Kudo M, Hsu C, Kim TY, Choo SP, Trojan J, Welling TH Rd, et al:
Nivolumab in patients with advanced hepatocellular carcinoma
(CheckMate 040): An open-label, non-comparative, phase 1/2 dose
escalation and expansion trial. Lancet. 389:2492–2502. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Nikolaou K, Sarris M and Talianidis I:
Molecular pathways: The complex roles of inflammation pathways in
the development and treatment of liver cancer. Clin Cancer Res.
19:2810–2816. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Arzumanyan A, Reis HM and Feitelson MA:
Pathogenic mechanisms in HBV- and HCV-associated hepatocellular
carcinoma. Nat Rev Cancer. 13:123–135. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Solinas G, Germano G, Mantovani A and
Allavena P: Tumor-associated macrophages (TAM) as major players of
the cancer-related inflammation. J Leukoc Biol. 86:1065–1073. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Boniakowski AE, Kimball AS, Jacobs BN,
Kunkel SL and Gallagher KA: Macrophage-mediated inflammation in
normal and diabetic wound healing. J Immunol. 199:17–24. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Glass CK and Natoli G: Molecular control
of activation and priming in macrophages. Nat Immunol. 17:26–33.
2016. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
Binnemars-Postma K, Storm G and Prakash J:
Nanomedicine strategies to target tumor-associated macrophages. Int
J Mol Sci. 18:9792017. View Article : Google Scholar
|
|
14
|
Yeung OW, Lo CM, Ling CC, Qi X, Geng W, Li
CX, Ng KT, Forbes SJ, Guan XY, Poon RT, et al: Alternatively
activated (M2) macrophages promote tumour growth and invasiveness
in hepatocellular carcinoma. J Hepatol. 62:607–616. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Capece D, Fischietti M, Verzella D,
Gaggiano A, Cicciarelli G, Tessitore A, Zazzeroni F and Alesse E:
The inflammatory microenvironment in hepatocellular carcinoma: A
pivotal role for tumor-associated macrophages. Biomed Res Int.
2013:1872042013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Sharif O and Knapp S: From expression to
signaling: Roles of TREM-1 and TREM-2 in innate immunity and
bacterial infection. Immunobiology. 213:701–713. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Tessarz AS and Cerwenka A: The
TREM-1/DAP12 pathway. Immunol Lett. 116:111–116. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Read CB, Kuijper JL, Hjorth SA, Heipel MD,
Tang X, Fleetwood AJ, Dantzler JL, Grell SN, Kastrup J, Wang C, et
al: Cutting edge: Identification of neutrophil PGLYRP1 as a ligand
for TREM-1. J Immunol. 194:1417–1421. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
van Bremen T, Dromann D, Luitjens K, Dodt
C, Dalhoff K, Goldmann T and Schaaf B: Triggering receptor
expressed on myeloid cells-1 (Trem-1) on blood neutrophils is
associated with cytokine inducibility in human E. coli
sepsis. Diagn Pathol. 8:242013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ho CC, Liao WY, Wang CY, Lu YH, Huang HY,
Chen HY, Chan WK, Chen HW and Yang PC: TREM-1 expression in
tumor-associated macrophages and clinical outcome in lung cancer.
Am J Respir Crit Care Med. 177:763–770. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Shen ZT and Sigalov AB: Novel TREM-1
inhibitors attenuate tumor growth and prolong survival in
experimental pancreatic cancer. Mol Pharm. 14:4572–4582. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wu J, Li J, Salcedo R, Mivechi NF,
Trinchieri G and Horuzsko A: The proinflammatory myeloid cell
receptor TREM-1 controls Kupffer cell activation and development of
hepatocellular carcinoma. Cancer Res. 72:3977–3986. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Tandon P, Abraldes JG, Keough A,
Bastiampillai R, Jayakumar S, Carbonneau M, Wong E, Kao D, Bain VG
and Ma M: Risk of bacterial infection in patients with cirrhosis
and acute variceal hemorrhage, based on Child-Pugh class, and
effects of antibiotics. Clin Gastroenterol Hepatol.
13:1189–1196.e2. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Llovet JM, Brú C and Bruix J: Prognosis of
hepatocellular carcinoma: The BCLC staging classification. Semin
Liver Dis. 19:329–338. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Genin M, Clement F, Fattaccioli A, Raes M
and Michiels C: M1 and M2 macrophages derived from THP-1 cells
differentially modulate the response of cancer cells to etoposide.
BMC Cancer. 15:5772015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T). Methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhang L, Wang JN, Tang JM, Kong X, Yang
JY, Zheng F, Guo LY, Huang YZ, Zhang L, Tian L, et al: VEGF is
essential for the growth and migration of human hepatocellular
carcinoma cells. Mol Biol Rep. 39:5085–5093. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Polyak K, Haviv I and Campbell IG:
Co-evolution of tumor cells and their microenvironment. Trends
Genet. 25:30–38. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Condeelis J and Pollard JW: Macrophages:
Obligate partners for tumor cell migration, invasion, and
metastasis. Cell. 124:263–266. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Bouchon A, Dietrich J and Colonna M:
Cutting edge: Inflammatory responses can be triggered by TREM-1, a
novel receptor expressed on neutrophils and monocytes. J Immunol.
164:4991–4995. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Duan M, Wang ZC, Wang XY, Shi JY, Yang LX,
Ding ZB, Gao Q, Zhou J and Fan J: TREM-1, an inflammatory
modulator, is expressed in hepatocellular carcinoma cells and
significantly promotes tumor progression. Ann Surg Oncol.
22:3121–3129. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Liao R, Sun TW, Yi Y, Wu H, Li YW, Wang
JX, Zhou J, Shi YH, Cheng YF, Qiu SJ and Fan J: Expression of
TREM-1 in hepatic stellate cells and prognostic value in hepatitis
B-related hepatocellular carcinoma. Cancer Sci. 103:984–992. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Colegio OR, Chu NQ, Szabo AL, Chu T,
Rhebergen AM, Jairam V, Cyrus N, Brokowski CE, Eisenbarth SC,
Phillips GM, et al: Functional polarization of tumour-associated
macrophages by tumour-derived lactic acid. Nature. 513:559–563.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Lo TH, Tseng KY, Tsao WS, Yang CY, Hsieh
SL, Chiu AW, Takai T, Mak TW, Tarng DC and Chen NJ: TREM-1
regulates macrophage polarization in ureteral obstruction. Kidney
Int. 86:1174–1186. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ye Y, Xu Y, Lai Y, He W, Li Y, Wang R, Luo
X, Chen R and Chen T: Long non-coding RNA cox-2 prevents immune
evasion and metastasis of hepatocellular carcinoma by altering
M1/M2 macrophage polarization. J Cell Biochem. 119:2951–2963. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Guillermet-Guibert J, Bjorklof K, Salpekar
A, Gonella C, Ramadani F, Bilancio A, Meek S, Smith AJ, Okkenhaug K
and Vanhaesebroeck B: The p110beta isoform of phosphoinositide
3-kinase signals downstream of G protein-coupled receptors and is
functionally redundant with p110gamma. Proc Natl Acad Sci USA.
105:8292–8297. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Vergadi E, Ieronymaki E, Lyroni K,
Vaporidi K and Tsatsanis C: Akt signaling pathway in macrophage
activation and M1/M2 polarization. J Immunol. 198:1006–1014. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Byles V, Covarrubias AJ, Ben-Sahra I,
Lamming DW, Sabatini DM, Manning BD and Horng T: The TSC-mTOR
pathway regulates macrophage polarization. Nat Commun. 4:28342013.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Rauh MJ, Ho V, Pereira C, Sham A, Sly LM,
Lam V, Huxham L, Minchinton AI, Mui A and Krystal G: SHIP represses
the generation of alternatively activated macrophages. Immunity.
23:361–374. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Ruckerl D, Jenkins SJ, Laqtom NN,
Gallagher IJ, Sutherland TE, Duncan S, Buck AH and Allen JE:
Induction of IL-4Rα-dependent microRNAs identifies PI3K/Akt
signaling as essential for IL-4-driven murine macrophage
proliferation in vivo. Blood. 120:2307–2316. 2012. View Article : Google Scholar : PubMed/NCBI
|