Long non-coding RNAs (lncRNAs) are defined as
transcripts >200 nucleotides in length that display limited
protein coding potential (1). Certain
lncRNAs with open reading frames can be translated into peptides
(2), and a previous study reported
that certain non-coding RNA (ncRNA) genes could undergo active
protein translation in mouse macrophages (3). The human genome project confirmed that
<2% of genomes encode proteins, whereas >98% are transcribed
into ncRNAs, of which 76% are lncRNAs (4,5). lncRNAs
were initially considered to be the ‘noise’ of genomic
transcription, lacking biological functions (6,7); however,
with the development of sequencing technology, rapid progress has
been made in the field of lncRNA research.
lncRNAs can be placed into one or more of the
following categories: Sense, antisense, bidirectional, intronic and
intergenic (8). Previous studies on
well-characterized lncRNAs have demonstrated that lncRNAs can
regulate gene expression at epigenetic, transcriptional and
post-transcriptional levels by functioning as enhancers of their
neighbouring genes, scaffolds for protein-protein interactions,
guides for protein-DNA interactions, or decoys for proteins or
microRNAs (miRs) to participate in various biological processes,
including cell proliferation, differentiation, survival and
migration (9–11). Moreover, dysregulation of lncRNAs
impacts different human diseases, including cancer (12). lncRNAs are also involved in DNA damage
repair, cell cycle regulation, metabolism and other physiological
or pathological processes that contribute to regulating the
occurrence and development of cancer (1,13–15). Therefore, lncRNAs have become a
research hotspot in the field of life science.
Metabolism is defined as biochemical processes that
produce or consume energy in living organisms (16). Otto Warburg suggested that cancer is a
metabolic disease (17). Increasing
evidence has demonstrated that the process of tumorigenesis is
typically accompanied by alterations in metabolic patterns,
including glycolysis, the tricarboxylic acid (TCA) cycle,
mitochondrial oxidative phosphorylation (OXPHOS), the pentose
phosphate pathway (PPP), glutamine metabolism and lipid metabolism,
which is also known as metabolic reprogramming (18–23). At
present, metabolic reprogramming is considered as an emerging
hallmark of cancer cells (24). Under
aerobic conditions, the majority of differentiated cells metabolize
glucose to carbon dioxide and GTP via the oxidation of pyruvate
produced by glycolysis. Pyruvate is converted into acetyl Coenzyme
A (CoA), which then enters the TCA cycle of aerobic oxidation,
which is also known as the citric acid cycle or Krebs cycle. It is
only when oxygen is unavailable that differentiated cells produce
ATP via glycolysis. By contrast, cancer cells typically obtain
energy from glycolysis rather than relying on mitochondrial OXPHOS,
even in the presence of oxygen, a phenomenon known as the Warburg
effect or aerobic glycolysis, which was first reported by Otto
Warburg in the 1920s (25).
Another important aspect of altered energy
metabolism in cancer cells is glutamine addiction, which is
characterized by enhanced uptake and utilization of glutamine. In
addition to glucose, glutamine is another major source of energy
for tumor cells, as it can function as a substrate for the TCA
cycle, but can also serve as a nitrogen donor for nucleic acid
synthesis and as a precursor for protein and glutathione
biosynthesis (26). Abnormal lipid
metabolism is also a hallmark of tumor metabolic reprogramming,
which is characterized by abnormal fatty acid (FA) metabolism and
lipolysis (26). As important
regulators, lncRNAs have attracted increasing attention for their
role in regulating tumor metabolic reprogramming and their
potential clinical application. The present review discussed the
current understanding of the roles of lncRNAs in the metabolic
reprogramming of cancer cells and summarized the mechanisms
underlying lncRNA-mediated regulation of chemoresistance via
metabolic pathways. In addition, the present review discussed the
drugs that are currently in clinical use or clinical trials, as
well as a number of potential challenges associated with targeting
metabolic reprogramming.
As an important energy substance, glucose is also
essential for the occurrence and development of tumors (27). Glucose is taken up by cells via
glucose transporters (GLUTs) and then enters three pathways for
further oxidative decomposition, including the glycolysis pathway
to produce lactic acid, the PPP to produce nicotinamide adenine
dinucleotide phosphate (NADPH) and nucleic acids, and the TCA cycle
and the OXPHOS pathway to produce oxygen and ATP (28). lncRNAs primarily regulate the process
of glucose metabolism by directly regulating the enzymes and
transporters involved in the aforementioned pathways or by
indirectly regulating the associated transcription factors
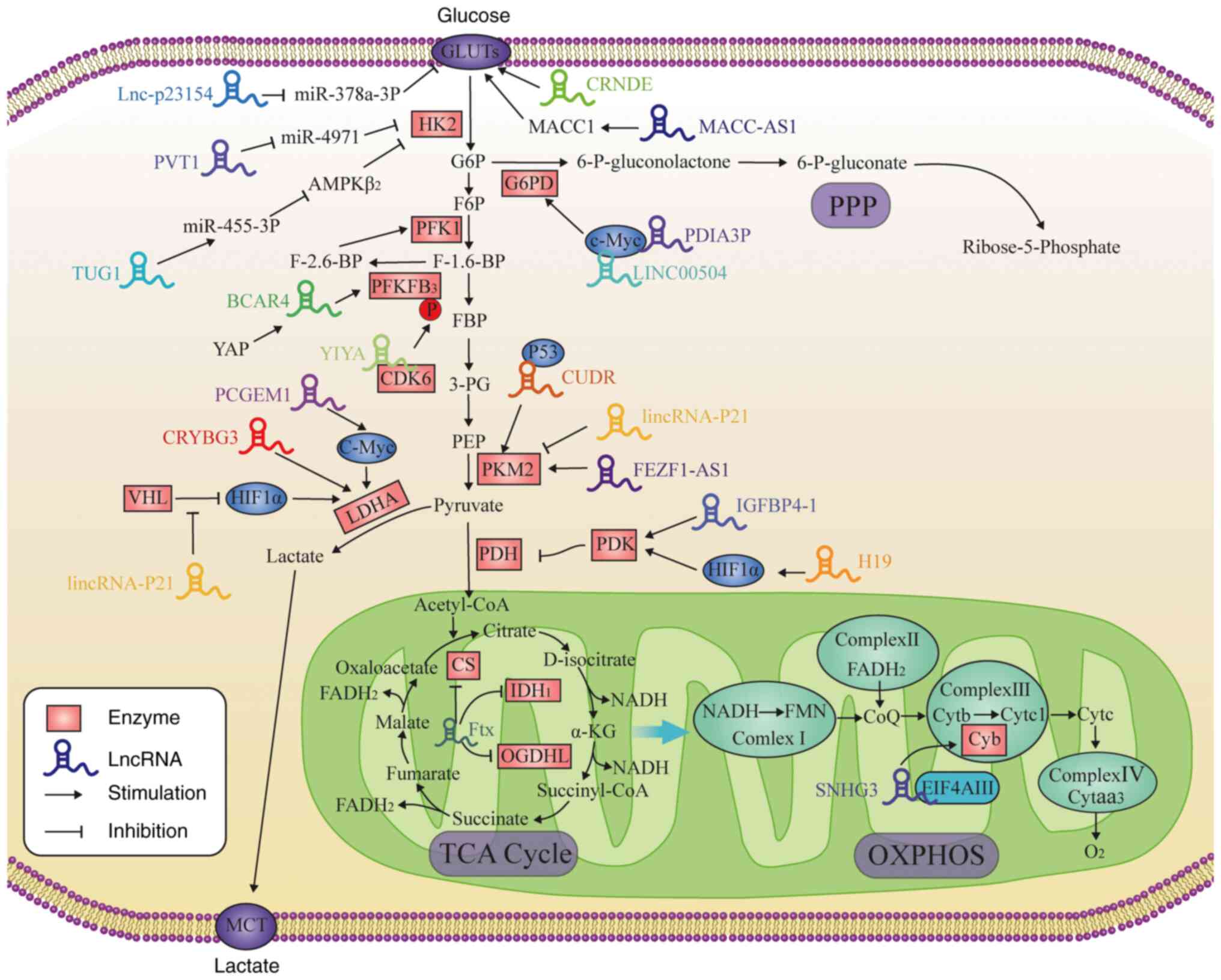
(Fig. 1 and Table I) (20,29–33).
In aerobic glycolysis, there is an excessive
conversion of pyruvate to lactate rather than acetyl CoA, which
only produces 2 ATPs per molecule of glucose. By contrast, the
complete OXPHOS of one molecule of glucose produces up to 36 ATPs
(28). Therefore, an interesting
question is why cancer cells utilize a less efficient metabolic
pathway, at least for the production of ATP. One possible
explanation is that aerobic glycolysis can produce intermediate
metabolites for further de novo nucleotide and lipid
synthesis. In addition, compared with mitochondrial OXPHOS, aerobic
glycolysis reduces reactive oxygen species (ROS) production, as ROS
are a by-product of electron transport (34). Moreover, the acidic microenvironment
caused by aerobic glycolysis is also beneficial for cancer cell
migration and invasion (35,36). For example, the bloodstream has a
higher oxygen concentration compared with most other tissues, yet
leukemic cells are highly dependent on aerobic glycolysis, which is
beneficial to cell migration (37,38).
Furthermore, aerobic glycolysis requires fewer steps, thus it
provides energy at a faster rate for tumor proliferation.
Therefore, cancer cells tend to obtain energy from aerobic
glycolysis.
GLUTs are membrane proteins that serve a pivotal
role in glucose metabolism in cancer cells by transporting glucose
into cells. At present, 14 members of the GLUT family have been
identified, among which GLUT1 GLUT3 and GLUT4 are closely
associated with glucose metabolism in cancer (39,40).
lncRNA-p23154 can interact with the promoter region of miR-378a-3p,
which inhibits miR-378a-3p transcription. miR-378a-3p targets the
3′-untranslated region (UTR) of GLUT1, leading to downregulation of
GLUT1 expression. Thus, the ability of lncRNA-p23154 to promote
oral squamous metastasis is mediated by GLUT1 (41). lncRNA HOX transcript antisense
intergenic RNA (HOTAIR) also regulates GLUT1 expression by
activating mTOR signaling in hepatocellular carcinoma (HCC)
(23). Similarly, lncRNA colorectal
neoplasia differentially expressed (CRNDE) positively modulates
GLUT4 expression and glucose uptake in colorectal cancer (CRC).
High CRNDE expression promotes glucose uptake and leads to an
enhanced Warburg effect (42).
Furthermore, the dysregulation of lncRNAs that regulate GLUT
expression promotes the proliferation and metastasis of other
tumors, including osteosarcoma and lung cancer (LC) (19,43,44). In
addition to directly regulating the expression of GLUTs, lncRNAs
can also modulate the distribution of GLUTs, thereby regulating
their capacity to transport glucose. MACC1 antisense RNA 1
(MACC1-AS1) is the cognate antisense lncRNA of the oncogene MET
transcriptional regulator MACC1 (MACC1). Under metabolic stress,
lncRNA MACC1-AS1 is induced, which promotes MACC1 mRNA stability
via the 5′AMP-activated protein kinase (AMPK)/lin-28 homolog A
signaling pathway, further enhancing the distribution of GLUT1 to
the cell membrane and promoting glucose absorption (45).
Hexokinase 2 (HK2) is a key enzyme in glycolysis
that catalyses the conversion of glucose to glucose-6-phosphate
(G6P) (46). In osteosarcoma, lncRNA
Pvt1 oncogene (PVT1) can promote glycolysis and tumor progression
by serving as a molecular sponge to suppress miR-497, and miR-497
can downregulate HK2 expression. Therefore, PVT1 regulates tumor
progression via the miR-497/HK2 axis (47). Another study demonstrated that lncRNA
can affect the stability of HK2. lncRNA taurine-upregulated gene 1
(TUG1) can upregulate the expression of miR-455-3p, and miR-455-3p
can regulate the expression of AMPKβ2 via binding to AMPKβ2 3′UTR
at positions 54–61. TUG1 affects HK2 stability via modulating the
p21/miR-455-3p axis, promoting HCC cell migration (48). Phosphofructo-1-kinase (PFK1) catalyzes
the conversion of G6P to fructose-1,6-bisphosphate, which is the
rate-limiting step of glycolysis.
6-Phosphofructo-2-kinase/fructose-2,6-bisphosphatase 3 (PFKFB3)
catalyzes the conversion of G6P to fructose-2,6-bisphosphate, which
can increase the activity of PFK1. Thus, PFKFB3 can indirectly
promote glycolysis (29). A previous
study suggested that lncRNA long intergenic non-protein coding RNA
(LINC)00538 can associate with CDK6 and F-box and WD repeat domain
containing 7 (FBXW7) to promote the association between CDK6 and
cyclin D3, further enhancing the phosphorylation of PFKFB3, and
ultimately promoting glucose consumption and lactate production
(29). In addition to direct
involvement in the modulation of glycolytic enzymes, lncRNAs can
also be involved in various cancer-related signaling pathways to
indirectly regulate glycolytic enzymes. A previous study reported
that lncRNA breast cancer anti-estrogen resistance 4 (BCAR4)
participates in the regulation of glycolysis via the
Hippo/Yes-associated protein (YAP) signaling pathway. YAP can
promote the transcription of BCAR4 via directly binding to the
BCAR4 promoter region. BCAR4 can activate HK2 and PFKFB3
transcription by recruiting p300, which promotes the acetylation of
histones marked by H3K27ac (49).
To date, four members of the pyruvate kinase (PK)
family have been identified: L, R, M1 and M2 types, among which the
abnormal expression of pyruvate kinase M2 (PKM2) is the most common
in tumor cells (50–52). LINC-p21 can function as a
tumor-suppressive lncRNA, as LINC-p21 knockdown upregulates the
expression of PKM2 via the PTEN/AKT/mTOR cascade (30). In addition, lncRNAs can regulate not
only the expression of enzymes, but also the stability of enzymes
via modulating the degradation pathway of enzymes. FEZF1 antisense
RNA 1 can bind to PKM2 protein to inhibit the ubiquitin-proteasome
signaling pathway and increase its stability, promoting activation
of STAT3 signaling and the Warburg effect (53).
Pyruvate dehydrogenase kinase (PDK) phosphorylates
and negatively modulates pyruvate dehydrogenase, inhibiting
pyruvate entry into the TCA cycle. In human cells, four PDK
isoenzymes (1–4) have been identified. In LC cells, a novel
lncRNA, insulin-like growth factor binding protein 4-1 (IGFBP4-1),
has been reported to upregulate the expression of HK2, PDK1 and
lactate dehydrogenase (LDH)A (54).
LDH catalyzes the last step of aerobic glycolysis,
the conversion of pyruvate to lactate. To date, four members of the
LDH family have been identified: LDHA, LDHB, LDHC and LDHD
(55). Increasing evidence has
demonstrated that LDHA is overexpressed and correlated with the
poor prognosis of several types of tumors (56,57). The
novel lncRNA crystalline βγ domain containing 3 has been reported
to promote glycolysis via directly interacting with LDHA and
further promote the LC cell proliferation (58).
In addition to the aforementioned enzymes, other
enzymes can also be regulated by lncRNAs (59–61).
Although numerous studies on these enzymes have been conducted, the
regulatory mechanisms underlying lncRNA-mediated regulation of
these enzymes are not completely understood. Therefore, further
investigations should be conducted to explore additional specific
mechanisms for the molecular targeted treatment of tumors.
In addition to regulating glycolytic enzymes,
lncRNAs can also regulate the expression and stability of
transcription factors associated with glycolysis, or be regulated
by these transcription factors, thereby regulating the occurrence
and development of tumors (32,62). The
primary transcription factors associated with lncRNAs in glycolysis
are c-Myc, hypoxia-inducible factor 1α (HIF1α), transcription
factor 7 like 2 (TCF7L2) and p53. MYC is a potent oncogene that can
promote tumorigenesis in multiple tissues (63). Under normoxic conditions, aerobic
glycolysis is commonly driven by c-Myc, a protein product of MYC.
lncRNA-Myc inhibitory factor (MIF) can serve as a molecular sponge
for miR-586 to attenuate the inhibitory effect of miR-586 on FBXW7,
thereby upregulating FBXW7 expression. FBXW7 is a E3 ubiquitin
ligase, and c-Myc and c-Jun are the substrates of FBXW7. Thus,
lncRNA-MIF can decrease c-Myc and c-Jun expression to inhibit
aerobic glycolysis. Furthermore, c-Myc can activate lncRNA-MIF
transcription via binding to the promoter and intronic region of
the lncRNA-MIF gene, thus forming a positive feedback loop between
c-Myc and lncRNA-MIF (31). In
addition, lncRNA prostate cancer gene expression marker 1 (PCGEM1)
can increase the transactivation activity of c-Myc, promoting
aerobic glycolysis. Mechanistically, PCGEM1 can recruit c-Myc to
metabolic gene promoters and enhance histone hyper-acetylation on
the majority metabolic genes, thereby upregulating the expression
of these target genes (62).
The transcription factor HIF1α is another important
regulator of glycolysis that is upregulated in hypoxia (64,65). In
breast carcinoma, lncRNA H19 imprinted maternally expressed
transcript can increase HIF1α expression by serving as an
endogenous sponge of let7, which further enhances PDK1 expression
and leads to increased glycolysis in hypoxia (66). In nasopharyngeal carcinoma (NPC),
lncRNA cancer susceptibility candidate 9 interacts with HIF1α to
increase its stability, promoting NPC cell glycolysis and
tumorigenesis (67). Prolyl
hydroxylases (PHDs) are oxygen sensors that hydroxylate HIF1α under
normoxia, leading to the degradation of HIF1α via the
von-Hippel-Lindau (VHL)-mediated ubiquitin-proteasome signaling
pathway. However, PHD activity is inhibited under hypoxia, causing
HIF1α accumulation (68). LINC-p21 is
a hypoxia-responsive lncRNA. In hypoxia, HIF1α associates with the
chromatin fragments corresponding to hypoxia response elements
within the LINC-p21 gene to promote transcription of LINC-p21.
Conversely, LINC-p21 can then increase HIF1α stability by
disrupting the interaction between VHL and HIF1α, which protects
HIF1α from being degraded via the VHL-mediated ubiquitin-proteasome
signaling pathway (18). However, a
previous study demonstrated that lncRNAs can increase HIF1α
stability under normoxic conditions. lncRNA LINC for kinase
activation is a cytoplasmic lncRNA that can interact with protein
tyrosine kinase 6 and leucine rich repeat kinase 2, subsequently
phosphorylating Tyr565 and Ser797 of HIF1α, respectively. Tyr565
phosphorylation represses pro 564 hydroxylation, which stabilizes
HIF1α in normoxia, whereas Ser797 phosphorylation enhances its
transcriptional activity, leading to increased glycolysis and
breast cancer progression (32). In
line with the aforementioned studies, a recent study also confirmed
that a novel hypoxia-induced lncRNA G077640 can bind to H2A.X
variant histone to enhance the stability of HIF1α, leading to
enhanced hypoxia-related glycolysis, which ultimately promotes
esophageal squamous cell carcinoma cell proliferation and migration
(unpublished data).
Similar to HIF1α and c-Myc, TCF7L2 is also a
metabolic transcription factor. TCF7L2 is a member of the TCF
family that is involved in the formation of β-catenin/TCF, a key
regulator of the canonical Wnt signaling pathway (69). Additionally, the Wnt signaling pathway
has been reported to serve a crucial role in metabolic
reprogramming (70,71). lncRNA metastasis associated lung
adenocarcinoma transcript 1 (MALAT1) can regulate the expression of
TCF7L2 at the post-transcriptional level via splicing oncoprotein
serine and arginine rich splicing factor 1 in HCC cells. In
addition, MALAT1 can promote glycolysis and inhibit gluconeogenesis
by modulating the translation of TCF7L2 (72).
p53, a tumor suppressor, can downregulate GLUT1 and
GLUT4 expression and regulate multiple metabolism-related enzymes
(73,74). Increasing evidence has demonstrated
that p53 mutations can promote tumorigenesis (75,76).
Mutant p53 (N340Q/L344R) can bind to lncRNA urothelial cancer
associated 1 (UCA1) to form a complex, which then binds to the
promoter regions of PKM2 to promote the expression and
phosphorylation of PKM2, accelerating liver cancer cell
proliferation. Thus, UCA1 serves a crucial role in the p53-mediated
regulation of glycolysis (33).
Alterations in the PPP often occur during tumor
metabolic reprogramming, and large amounts of NADPH and
ribose-5-phosphate can be produced via this pathway, which can
maintain the reduction-oxidation status of cells and provide the
raw material for the synthesis of various substances, respectively
(77). The conversion of G6P to
6-phosphogluconate is catalyzed by G6P dehydrogenase (G6PD), which
is the first key enzyme in the PPP (62). Using nuclear magnetic resonance
spectroscopy, a previous study reported that lncRNA protein
disulfide isomerase family A member 3 pseudogene 1 (PDIA3P) can
increase the PPP flux and NADPH level via enhancing the expression
and activity of G6PD. Mechanistically, PDIA3P can bind to c-Myc to
increase its transactivation activity and binding to the G6PD
promoter (20). Similarly, lncRNA
LINC00504 was also reported to modulate G6PD activity by
interacting with c-Myc and increasing its transactivation activity
(78). In addition, lncRNA PCGEM1 can
also regulate the PPP via regulating the expression and activity of
G6PD (62).
The abnormal expression of lncRNAs is often
accompanied by alterations in OXPHOS during the regulation of
glycolysis. Increased glycolysis is sometimes accompanied by either
decreased or increased OXPHOS (19,21,79).
Although the reasons for this phenomenon are unclear, these
adaptive alterations are ultimately intended to promote the
tumorigenesis and progression of cancer. A recent study
demonstrated that lncRNA LINC00184 can recruit DNA
methyltransferase 1 to promote promoter methylation of PTEN,
leading to increased Akt phosphorylation and promoting glycolysis,
as well as reducing the expression of mitochondrial COMPLEX II,
III, IV and V subunits and the ATP level (21). lncRNA FTX transcript, XIST regulator
(Ftx) was also reported to increase the expression of GLUT1 and
GLUT4, and decrease isocitrate dehydrogenase (IDH)1, citrate
synthase and oxoglutarate dehydrogenase expression by activating
the peroxisome proliferator-activated receptor (PPAR)γ signaling
pathway, indicating that lncRNA Ftx might promote glycolysis and
inhibit the TCA cycle in HCC cells (14). By contrast, lncRNA small nucleolar RNA
host gene 3 can simultaneously upregulate the expression of PKM,
IDH2 and cytochrome b reductase 1 via binding eukaryotic
translation initiation factor 4A3 (79).
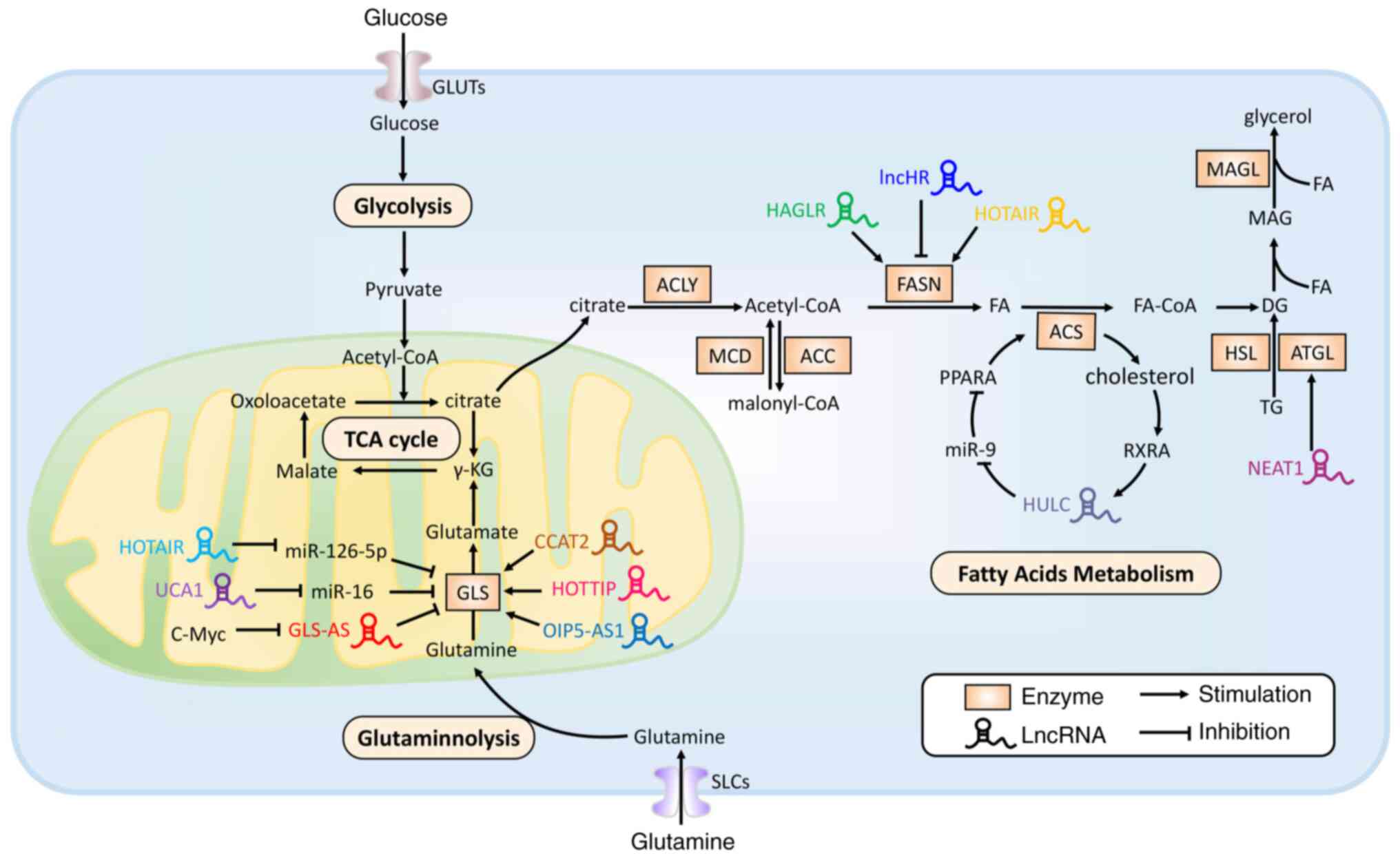
Although glutamine is a nonessential amino acid that
can be synthesized from glucose, cancer cells also display an
increased dependence on glutamine to feed the TCA cycle. Moreover,
certain cancer cells display a sensitivity to glutamine starvation,
which is known as glutamine addiction (80,81). In
1950, Harry Eagle (71) reported that
HeLa cells require excess glutamine compared with other amino acids
present in the culture medium. In addition, it has been reported
that tumors consume glutamine faster than surrounding healthy
tissues (82) and the level of blood
glutamine is increased in patients with advanced cancer (83). Glutamine is transferred into cells via
the solute carrier (SLC) family members, including SLC1, SLC6, SLC7
and SLC38 members. When in the cell, glutamine is deaminated by
glutaminase (GLS)1 and GLS2 to produce glutamate. Glutamine
dehydrogenase, glutamate oxaloacetate transaminase or glutamine
pyruvate transaminase can further catalyze glutamate to
α-ketoglutarate to fuel the TCA cycle (84). Alternatively, glutamate can be further
oxidized to glutathione by glutathione cysteine ligase, which can
neutralize mitochondrial ROS (85).
Under certain circumstances, reduced acetyl CoA entering the TCA
cycle may also promote the compensatory oxidation of glutamine to
further fuel the TCA cycle and OXPHOS in mitochondria (86).
GLSs are the rate-limiting enzymes in glutamine
metabolism, which are encoded by the human genes GLS1 and GLS2, and
have been reported to be involved in the development of numerous
malignant tumors (87–90). GLS-antisense (AS) can repress GLS
expression at the post-transcriptional level via adenosine
deaminases acting on RNA/dicer-dependent RNA interference. Under
nutrient stress circumstances, Myc can bind to the promoter region
of GLS-AS to inhibit the transcriptional activity of GLS-AS.
Furthermore, GLS and GLS-AS can also regulate Myc protein
stability. Therefore, there is a feedback loop between GLS-AS and
Myc, leading to reduced expression, which increases GLS expression
and promotes pancreatic cancer progression (91). UCA1 expression is positively
correlated with GLS2. UCA1 can bind to miR-16, and miR-16 can
further regulate the expression of GLS2 via directly binding to the
3′UTR of GLS2 mRNA. Thus, UCA1 can bind to miR-16 to repress the
inhibitory effect of miR-16 on GLS2, leading to increased GLS2
expression, glutamine uptake and reduced ROS production (88). Similarly, lncRNAs HOTAIR, HOXA distal
transcript antisense RNA and OIP5 antisense RNA 1 have also been
reported to affect glutaminolysis by regulating GLS (92–94).
In addition to the aforementioned effects, lncRNAs
have been reported to regulate the alternative splicing of GLS.
Single nucleotide polymorphisms (SNPs) in lncRNAs have been
identified to be associated with susceptibility to numerous types
of cancer (95–97). lncRNA colon cancer-associated
transcript 2 (CCAT2) harboring the rs6983267 SNP could modulate
cancer metabolism via binding the cleavage factor I complex, which
can regulate the alternative splicing of GLS. Compared with control
cells, CCAT2 T allele- or G allele-overexpression HCT116 cells both
display enhanced GLS activity. However, compared with T
allele-overexpression HCT116 cells, G allele-overexpression HCT116
cells display higher enzymatic activity. Therefore, the
aforementioned results revealed an allele-specific regulatory
mechanism underlying glutamine metabolism in colon cancer (Fig. 2 and Table
I) (22).
Abnormal lipid metabolism is characterized by
abnormal FA metabolism and abnormal lipolysis. The majority of
healthy cells obtain FA primarily via the exogenous pathway.
However, 90% of FA obtained by tumor cells is derived via de
novo synthesis. FA β oxidation can provide energy to meet the
needs of rapid tumor cell proliferation and growth, and lipids are
the major components of cell membranes (98).
With respect to the modulation of lipid metabolism
in tumor cells, three regulatory mechanisms should be taken into
consideration: i) Regulation of the expression of the enzymes in
the lipid synthesis pathway, including acetyl CoA carboxylase,
fatty acid synthase (FASN), acetyl CoA synthase (ACS), malonyl-CoA
decarboxylase and ATP citrate lyase (ACLY); ii) regulation of the
expression of the enzymes in lipolysis, including adipose
triacylglyceride lipase (ATGL), hormone-sensitive lipase and
monoacylglycerol lipase; and iii) regulation of related
transcription factors, including sterol regulatory element-binding
protein 1 (SREBP-1), liver X-activated receptor, retinoid X
receptors (RXRs) and PPAR-α (Fig. 2)
(99,100). ACS long chain family member 1
(ASCL1) is a member of the ACSL family that can catalyze the first
step in FA metabolism and can be activated by the transcription
factor PPAR-α in the liver (99). A
study has demonstrated that highly upregulated lncRNA in liver
cancer (HULC) can downregulate miR-9 expression levels by inducing
methylation of CpG islands in its promoter, and miR-9 can
downregulate PPAR-α expression by directly binding to the 3′UTR of
PPAR-α. Therefore, HULC can upregulate the level of ACSL1. By
contrast, the cholesterol product of ACSL1 can upregulate HULC
expression by activating RXRA, a member of the RXR family, which
serves a crucial role in the control of various physiologic
processes and can be activated by sterol (101), thereby forming a positive feedback
loop between HULC/miR-9/PPAR-α/ACSL1/cholesterol/RXRA/HULC in
hepatoma cells (102). snoRNA host
gene 16 (SNHG16) is highly expressed and regulated by the Wnt
signaling pathway and c-Myc in CRC. SNHG16 regulates the genes
involved in lipid metabolism potentially via a competitive
endogenous (ce)RNA-related mechanism and serves an oncogenic role
in CRC (103).
Nuclear paraspeckle assembly transcript 1 (NEAT1) is
an lncRNA that is primarily located in cytoplasm and can also
promote the formation of nuclear paraspeckles by serving as a
scaffolding factor. ATGL is a primary enzyme of lipolysis that can
hydrolyze triglyceride (TAG) into diacylglycerol (DAG) and free
fatty acid (FFA) during TAG metabolism (109). A previous report demonstrated that
NEAT1 can regulate ATGL expression via binding to miR-124-3p to
disrupt the lipolysis of hepatoma cells, leading to elevated levels
of DAG and FFA. In addition, NEAT1 can also regulate PPAR-α
expression. Thus, NEAT1 can modulate HCC cell proliferation via the
miR-124-3p/ATGL/DAG+FFA/PPAR-α signaling pathway (110).
Lymph node (LN) metastasis indicates poor prognosis
of patients with cancer, but the mechanism underlying this process
is not completely understood. lncRNA associated with LN metastasis
in cervical cancer (LNMICC) upregulation was observed in patients
with cervical cancer with LN metastasis. LNMICC can recruit the
nuclear factor nucleophosmin 1 to the promoter of fatty acid
binding protein 5, a fatty acid binding protein that is
indispensable for FA uptake and transport, to activate fatty acid
metabolism, further promoting LN metastasis and accelerating
lipogenesis in vitro and in vivo. However, this
effect can be suppressed by miR190, which can directly bind to
LNMICC (111).
SREBP-1c is a transcriptional factor that can
regulate hepatic lipid homeostasis. The novel human specific lncRNA
HCV regulated 1 can negatively modulate SREBP-1c and FASN
expression, thereby inhibiting oleic acid-induced triglyceride and
lipid droplet accumulation in Huh7 cells (112). Thus, lncRNAs can affect cellular
lipid metabolism by regulating the associated enzymes and
transcription factors (Fig. 2 and
Table I).
Chemotherapy is one of the primary treatment
strategies for multiple types of tumors, and the major hurdle of
chemotherapy is chemoresistance. A number of different mechanisms
lead to chemoresistance, including cancer stem cells (113), mitochondrial alteration (114), epithelial-mesenchymal transition
(115), DNA repair (116) and autophagy (117). Recent studies have confirmed that
metabolic reprogramming is involved in the regulation of lncRNAs in
the chemoresistance of multiple tumors, including glycolysis, lipid
synthesis and FA oxidation (FAO) (118–120).
Elevated glycolysis rates were observed in cisplatin-resistant
colon cancer cells. lncRNA differentiation antagonizing non-coding
RNA (DANCR) is upregulated in colon cancer tissues, cells and
cisplatin-resistant colon cancer cells. DANCR can regulate
miR-125b-5p expression via a ceRNA mechanism, and miR-125b-5p can
bind to HK2 to modulate glycolysis, further leading to enhanced
cisplatin resistance. Thus, DANCR can modulate cisplatin resistance
via the miR-125b-5p/HK2 axis (118).
Similarly, lncRNA-suppressing androgen receptor in renal cell
carcinoma can improve the sensitivity of osteosarcoma cells to
cisplatin via targeting HK2 to regulate miR-143-mediated glycolysis
(121). In addition to glycolysis,
lipid metabolism is also involved in the regulation of lncRNAs on
chemoresistance. lncRNA TINCR ubiquitin domain containing (TINCR)
can bind to ACLY to protect it from ubiquitin degradation, leading
to increased cellular acetyl CoA levels and further promoting NPC
cisplatin resistance. Mechanistically, TINCR regulates NPC
cisplatin resistance via the peptidyl arginine deiminase
1/MAPK/matrix metallopeptidase 2/9 signaling pathway (119). FAO regulates drug resistance in
breast cancer stem cell (122). In
gastric cancer cells, lncRNA MACC1-AS1, induced by mesenchymal stem
cells-derived TGF-β1, can promote FAO-dependent chemoresistance by
antagonizing miR-145-5p (120).
Therefore, targeting these lncRNAs and associated signaling
pathways may serve as an effective strategy to increase the
sensitivity of tumors to chemotherapy.
The occurrence of tumors is due to abnormal cell
growth and proliferation, which requires the materials for cell
construction, including nucleic acids, proteins and lipids, as well
as the energy for tumor cell proliferation. As aforementioned,
glucose, glutamine and lipid metabolism are not independent in
tumor cells; they cooperate with one another to allow tumor cells
to rapidly proliferate. lncRNAs, as important regulators of
metabolic reprogramming, do not serve singular roles. Certain
lncRNAs have only been reported to regulate glycolysis, such as the
aforementioned lncRNA IGFBP4-1 and LINC00538 (29,54).
However, it is possible that lncRNAs can also regulate other
metabolic processes because metabolic processes are not independent
and may display feedback mechanisms. Certain lncRNAs have been
reported to simultaneously regulate multiple metabolic pathways.
For example, CCAT2 can enhance glutamine metabolism and glycolysis
(22), lncRNA Ftx can promote
glycolysis and inhibit the TCA cycle (19), and PCGEM1 can regulate multiple
metabolic pathways, including glucose metabolism, glutamine
metabolism, and nucleotide and fatty acid biosynthesis (62). Similarly, lncRNA HOTAIR has been
reported to regulate glutaminolysis, the expression of FASN and
glucose metabolism in different experiments (23,92,108).
Metabolic pathways are not independent, which makes it difficult to
target metabolic pathways for tumor therapy due to the possible
compensatory effects of other pathways.
Tumor-associated alterations in metabolism involve
numerous molecules and pathways that are potential targets of tumor
therapy. Therefore, targeting metabolism is an effective strategy
as the heterogeneity in the genetic landscape of tumors is greater
than the metabolic heterogeneity. The first small-interfering RNA
(siRNA) drug, Patisiran, was approved by the Food and Drug
Administration in 2018, which might aid in the development of siRNA
drugs targeting lncRNAs or other genes involved in metabolic
reprogramming (123). Notably, some
inhibitors or RNA interference-mediated inhibition of enzymes and
transcription factors involved in metabolic reprogramming have been
demonstrated to inhibit the proliferation of tumor cells, some of
which are entering clinical evaluation or currently undergoing
clinical evaluation. PKM2 and GLS inhibitors have been reported to
inhibit gastric cancer cell proliferation (89). Similarly, a report demonstrated that a
GLS inhibitor, Bis-2 (5-p henylacetamido-1,2,4-thiadiazol-2-yl)
ethyl sulfide, displayed antitumor efficacy in several tumor models
both in vitro and in vivo (124,125).
Moreover, antisense oligonucleotide-targeting HIF-1a, EZN-2968, can
bind to HIF-1a mRNA and reduce HIF-1a protein expression levels. In
addition, the study demonstrated that EZN-2968 can be safely used
in patients with advanced solid tumors (126). However, whether these inhibitors are
toxic to healthy cells requires further investigation. For example,
the glycolysis pathway is also widely used by immune cells to
provide energy. In addition, the compensatory expression of other
genes after the administration of inhibitors is also unclear.
Therefore, further studies of metabolic reprogramming should
provide additional options for targeted therapy in patients with
tumors.
Although lncRNAs were previously believed to not
encode proteins, a previous study demonstrated that certain ncRNA
genes can undergo active protein translation in mouse macrophages
(3). lncRNAs perform complex
biological functions in various tumors, and further studies should
be conducted to understand how lncRNAs regulate tumorigenesis for
the identification of novel therapeutic targets for cancer
treatment and biomarkers for prognostic assessment.
Not applicable.
The present study was supported by the Science and
Technology Commission Foundation of Luzhou city (grant nos.
2019-RCM-97 and 2016LZXNYD-T06), the Science and Technology
Commission Foundation of Sichuan Province (grant no. 2018JY0285),
the National Natural Science Foundations of China (grant nos.
81672458 and 81972296), the Talent Development Project of The
Affiliated Hospital of Southwest Medical University (grant no.
20079)and the Affiliated Hospital of Southwest Medical University
(grant no. 15039).
Not applicable.
CL and HL confirm the authenticity of all the raw
data. CL and HL drafted the review. FC and XZ assisted with
developing the tables and graphs. RX and QW revised the review with
regard to grammar, text format and framework, revised the
manuscript for important intellectual content and collated the
references. SY and TL revised the manuscript. SL and ML provided
the direction and ideas for the writing, made repeated revisions to
the review and provided guidance throughout the process. All
authors read and approved the final version of the manuscript.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
|
1
|
Rinn JL and Chang HY: Genome regulation by
long noncoding RNAs. Annu Rev Biochem. 81:145–166. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Huang JZ, Chen M, Chen D, Gao XC, Zhu S,
Huang H, Hu M, Zhu H and Yan GR: A peptide encoded by a putative
lncRNA HOXB-AS3 suppresses colon cancer growth. Mol Cell.
68:171–184.e6. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Jackson R, Kroehling L, Khitun A, Bailis
W, Jarret A, York AG, Khan OM, Brewer JR, Skadow MH, Duizer C, et
al: The translation of non-canonical open reading frames controls
mucosal immunity. Nature. 564:434–438. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Stein LD: Human genome: End of the
beginning. Nature. 431:915–916. 2004. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Mercer TR, Dinger ME and Mattick JS: Long
non-coding RNAs: Insights into functions. Nat Rev Genet.
10:155–159. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Struhl K: Transcriptional noise and the
fidelity of initiation by RNA polymerase II. Nat Struct Mol Biol.
14:103–105. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ebisuya M, Yamamoto T, Nakajima M and
Nishida E: Ripples from neighbouring transcription. Nat Cell Biol.
10:1106–1113. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ponting CP, Oliver PL and Reik W:
Evolution and functions of long noncoding RNAs. Cell. 136:629–641.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bian Z, Jin L, Zhang J, Yin Y, Quan C, Hu
Y, Feng Y, Liu H, Fei B, Mao Y, et al: LncRNA-UCA1 enhances cell
proliferation and 5-fluorouracil resistance in colorectal cancer by
inhibiting miR-204-5p. Sci Rep. 6:238922016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Pauli A, Rinn JL and Schier AF: Non-coding
RNAs as regulators of embryogenesis. Nat Rev Genet. 12:136–149.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yang SZ, Xu F, Zhou T, Zhao X, McDonald JM
and Chen Y: The long non-coding RNA HOTAIR enhances pancreatic
cancer resistance to TNF-related apoptosis-inducing ligand. J Biol
Chem. 292:10390–10397. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Schmitt AM and Chang HY: Long noncoding
RNAs in cancer pathways. Cancer Cell. 29:452–463. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Batista PJ and Chang HY: Long noncoding
RNAs: Cellular address codes in development and disease. Cell.
152:1298–1307. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Guttman M and Rinn JL: Modular regulatory
principles of large non-coding RNAs. Nature. 482:339–346. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Khaitan D, Dinger ME, Mazar J, Crawford J,
Smith MA, Mattick JS and Perera RJ: The melanoma-upregulated long
noncoding RNA SPRY4-IT1 modulates apoptosis and invasion. Cancer
Res. 71:3852–3862. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang Y, Xia Y and Lu Z: Metabolic features
of cancer cells. Cancer Commun (Lond). 38:652018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Warburg O, Wind F and Negelein E: The
metabolism of tumors in the body. J Gen Physiol. 8:519–530. 1927.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yang F, Zhang H, Mei Y and Wu M:
Reciprocal regulation of HIF-1α and lincRNA-p21 modulates the
Warburg effect. Mol Cell. 53:88–100. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li X, Zhao Q, Qi J, Wang W, Zhang D, Li Z
and Qin C: lncRNA Ftx promotes aerobic glycolysis and tumor
progression through the PPARγ pathway in hepatocellular carcinoma.
Int J Oncol. 53:551–566. 2018.PubMed/NCBI
|
|
20
|
Yang X, Ye H, He M, Zhou X, Sun N, Guo W,
Lin X, Huang H, Lin Y, Yao R and Wang H: LncRNA PDIA3P interacts
with c-Myc to regulate cell proliferation via induction of pentose
phosphate pathway in multiple myeloma. Biochem Biophys Res Commun.
498:207–213. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Li W, Huang K, Wen F, Cui G, Guo H, He Z
and Zhao S: LINC00184 silencing inhibits glycolysis and restores
mitochondrial oxidative phosphorylation in esophageal cancer
through demethylation of PTEN. EBioMedicine. 44:298–310. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Redis RS, Vela LE, Lu W, Ferreira de
Oliveira J, Ivan C, Rodriguez-Aguayo C, Adamoski D, Pasculli B,
Taguchi A, Chen Y, et al: Allele-specific reprogramming of cancer
metabolism by the long non-coding RNA CCAT2. Mol Cell. 61:520–534.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wei S, Fan Q, Yang L, Zhang X, Ma Y, Zong
Z, Hua X, Su D, Sun H, Li H and Liu Z: Promotion of glycolysis by
HOTAIR through GLUT1 upregulation via mTOR signaling. Oncol Rep.
38:1902–1908. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Warburg O: On the origin of cancer cells.
Science. 123:309–314. 1956. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wang C, Wu J, Wang Z, Yang Z, Li Z, Deng
H, Li L, Peng X and Feng M: Glutamine addiction activates
polyglutamine-based nanocarriers delivering therapeutic siRNAs to
orthotopic lung tumor mediated by glutamine transporter SLC1A5.
Biomaterials. 183:77–92. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Vander Heiden MG, Cantley LC and Thompson
CB: Understanding the warburg effect: The metabolic requirements of
cell proliferation. Science. 324:1029–1033. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
DeBerardinis RJ, Lum JJ, Hatzivassiliou G
and Thompson CB: The biology of cancer: Metabolic reprogramming
fuels cell growth and proliferation. Cell Metab. 7:11–20. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Xing Z, Zhang Y, Liang K, Yan L, Xiang Y,
Li C, Hu Q, Jin F, Putluri V, Putluri N, et al: Expression of long
noncoding RNA YIYA promotes glycolysis in breast cancer. Cancer
Res. 78:4524–4532. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wang X, Xu Y, Wang X, Jiang C, Han S, Dong
K, Shen M and Xu D: LincRNA-p21 suppresses development of human
prostate cancer through inhibition of PKM2. Cell Prolif.
50:e123952017. View Article : Google Scholar
|
|
31
|
Zhang P, Cao L, Fan P, Mei Y and Wu M:
LncRNA-MIF, a c-Myc-activated long non-coding RNA, suppresses
glycolysis by promoting Fbxw7-mediated c-Myc degradation. EMBO Rep.
17:1204–1220. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Lin A, Li C, Xing Z, Hu Q, Liang K, Han L,
Wang C, Hawke DH, Wang S, Zhang Y, et al: The LINK-A lncRNA
activates normoxic HIF1α signalling in triple-negative breast
cancer. Nat Cell Biol. 18:213–224. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wu M, An J, Zheng Q, Xin X, Lin Z, Li X,
Li H and Lu D: Double mutant P53 (N340Q/L344R) promotes
hepatocarcinogenesis through upregulation of Pim1 mediated by PKM2
and LncRNA CUDR. Oncotarget. 7:66525–66539. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Dröge W: Free radicals in the
physiological control of cell function. Physiol Rev. 82:47–95.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Shiraishi T, Verdone JE, Huang J, Kahlert
UD, Hernandez JR, Torga G, Zarif JC, Epstein T, Gatenby R,
McCartney A, et al: Glycolysis is the primary bioenergetic pathway
for cell motility and cytoskeletal remodeling in human prostate and
breast cancer cells. Oncotarget. 6:130–143. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Peppicelli S, Bianchini F and Calorini L:
Extracellular acidity, a ‘reappreciated’ trait of tumor environment
driving malignancy: Perspectives in diagnosis and therapy. Cancer
Metastasis Rev. 33:823–832. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Gottschalk S, Anderson N, Hainz C,
Eckhardt SG and Serkova NJ: Imatinib (STI571)-mediated changes in
glucose metabolism in human leukemia BCR-ABL-positive cells. Clin
Cancer Res. 10:6661–6668. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Elstrom RL, Bauer DE, Buzzai M, Karnauskas
R, Harris MH, Plas DR, Zhuang H, Cinalli RM, Alavi A, Rudin CM and
Thompson CB: Akt stimulates aerobic glycolysis in cancer cells.
Cancer Res. 64:3892–3899. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Joost HG, Bell GI, Best JD, Birnbaum MJ,
Charron MJ, Chen YT, Doege H, James DE, Lodish HF, Moley KH, et al:
Nomenclature of the GLUT/SLC2A family of sugar/polyol transport
facilitators. Am J Physiol Endocrinol Metab. 282:E974–E976. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Mueckler M and Thorens B: The SLC2 (GLUT)
family of membrane transporters. Mol Aspects Med. 34:121–138. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Wang Y, Zhang X, Wang Z, Hu Q, Wu J, Li Y,
Ren X, Wu T, Tao X, Chen X, et al: LncRNA-p23154 promotes the
invasion-metastasis potential of oral squamous cell carcinoma by
regulating Glut1-mediated glycolysis. Cancer Lett. 434:172–183.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Ellis BC, Graham LD and Molloy PL: CRNDE,
a long non-coding RNA responsive to insulin/IGF signaling,
regulates genes involved in central metabolism. Biochim Biophys
Acta. 1843:372–386. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Kang Y, Zhu X, Xu Y, Tang Q, Huang Z, Zhao
Z, Lu J, Song G, Xu H, Deng C and Wang J: Energy stress-induced
lncRNA HAND2-AS1 represses HIF1α-mediated energy metabolism and
inhibits osteosarcoma progression. Am J Cancer Res. 8:526–537.
2018.PubMed/NCBI
|
|
44
|
Chang L, Xu W, Zhang Y and Gong F: Long
non-coding RNA-NEF targets glucose transportation to inhibit the
proliferation of non-small-cell lung cancer cells. Oncol Lett.
17:2795–2801. 2019.PubMed/NCBI
|
|
45
|
Zhao Y, Liu Y, Lin L, Huang Q, He W, Zhang
S, Dong S, Wen Z, Rao J, Liao W and Shi M: The lncRNA MACC1-AS1
promotes gastric cancer cell metabolic plasticity via AMPK/Lin28
mediated mRNA stability of MACC1. Mol Cancer. 17:692018. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Wolf A, Agnihotri S, Micallef J, Mukherjee
J, Sabha N, Cairns R, Hawkins C and Guha A: Hexokinase 2 is a key
mediator of aerobic glycolysis and promotes tumor growth in human
glioblastoma multiforme. J Exp Med. 208:313–326. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Song J, Wu X, Liu F, Li M, Sun Y, Wang Y,
Wang C, Zhu K, Jia X, Wang B and Ma X: Long non-coding RNA PVT1
promotes glycolysis and tumor progression by regulating miR-497/HK2
axis in osteosarcoma. Biochem Biophys Res Commun. 490:217–224.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Lin YH, Wu MH, Huang YH, Yeh CT, Cheng ML,
Chi HC, Tsai CY, Chung IH, Chen CY and Lin KH: Taurine up-regulated
gene 1 functions as a master regulator to coordinate glycolysis and
metastasis in hepatocellular carcinoma. Hepatology. 67:188–203.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Zheng X, Han H, Liu GP, Ma YX, Pan RL,
Sang LJ, Li RH, Yang LJ, Marks JR, Wang W and Lin A: LncRNA wires
up Hippo and Hedgehog signaling to reprogramme glucose metabolism.
EMBO J. 36:3325–3335. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Noguchi T, Yamada K, Inoue H, Matsuda T
and Tanaka T: The L- and R-type isozymes of rat pyruvate kinase are
produced from a single gene by use of different promoters. J Biol
Chem. 262:14366–14371. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Noguchi T, Inoue H and Tanaka T: The M1-
and M2-type isozymes of rat pyruvate kinase are produced from the
same gene by alternative RNA splicing. J Biol Chem.
261:13807–13812. 1986. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Mazurek S, Boschek CB, Hugo F and
Eigenbrodt E: Pyruvate kinase type M2 and its role in tumor growth
and spreading. Semin Cancer Biol. 15:300–308. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Bian Z, Zhang J, Li M, Feng Y, Wang X,
Zhang J, Yao S, Jin G, Du J, Han W, et al: LncRNA-FEZF1-AS1
promotes tumor proliferation and metastasis in colorectal cancer by
regulating PKM2 signaling. Clin Cancer Res. 24:4808–4819. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Yang B, Zhang L, Cao Y, Chen S, Cao J, Wu
D, Chen J, Xiong H, Pan Z, Qiu F, et al: Overexpression of lncRNA
IGFBP4-1 reprograms energy metabolism to promote lung cancer
progression. Mol Cancer. 16:1542017. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Feng Y, Xiong Y, Qiao T, Li X, Jia L and
Han Y: Lactate dehydrogenase A: A key player in carcinogenesis and
potential target in cancer therapy. Cancer Med. 7:6124–6136. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Ping W, Senyan H, Li G, Yan C and Long L:
Increased lactate in gastric cancer tumor-infiltrating lymphocytes
is related to impaired T cell function due to miR-34a deregulated
lactate dehydrogenase A. Cell Physiol Biochem. 49:828–836. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Girgis H, Masui O, White NM, Scorilas A,
Rotondo F, Seivwright A, Gabril M, Filter ER, Girgis AH, Bjarnason
GA, et al: Lactate dehydrogenase A is a potential prognostic marker
in clear cell renal cell carcinoma. Mol Cancer. 13:1012014.
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Chen H, Pei H, Hu W, Ma J, Zhang J, Mao W,
Nie J, Xu C, Li B, Hei TK, et al: Long non-coding RNA CRYBG3
regulates glycolysis of lung cancer cells by interacting with
lactate dehydrogenase A. J Cancer. 9:2580–2588. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Rupaimoole R, Lee J, Haemmerle M, Ling H,
Previs RA, Pradeep S, Wu SY, Ivan C, Ferracin M, Dennison JB, et
al: Long noncoding RNA ceruloplasmin promotes cancer growth by
altering glycolysis. Cell Rep. 13:2395–2402. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Xiang S, Gu H, Jin L, Thorne RF, Zhang XD
and Wu M: LncRNA IDH1-AS1 links the functions of c-Myc and HIF1α
via IDH1 to regulate the Warburg effect. Proc Natl Acad Sci USA.
115:E1465–E1474. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Zhao L, Ji G, Le X, Wang C, Xu L, Feng M,
Zhang Y, Yang H, Xuan Y, Yang Y, et al: Long noncoding RNA
LINC00092 acts in cancer-associated fibroblasts to drive glycolysis
and progression of ovarian cancer. Cancer Res. 77:1369–1382. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Hung CL, Wang LY, Yu YL, Chen HW,
Srivastava S, Petrovics G and Kung HJ: A long noncoding RNA
connects c-Myc to tumor metabolism. Proc Natl Acad Sci USA.
111:18697–18702. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Sodir NM, Swigart LB, Karnezis AN, Hanahan
D, Evan GI and Soucek L: Endogenous Myc maintains the tumor
microenvironment. Genes Dev. 25:907–916. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Semenza GL, Rue EA, Iyer NV, Pang MG and
Kearns WG: Assignment of the hypoxia-inducible factor 1alpha gene
to a region of conserved synteny on mouse chromosome 12 and human
chromosome 14q. Genomics. 34:437–439. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Wang GL, Jiang BH, Rue EA and Semenza GL:
Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS
heterodimer regulated by cellular O2 tension. Proc Natl Acad Sci
USA. 92:5510–5514. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Peng F, Wang JH, Fan WJ, Meng YT, Li MM,
Li TT, Cui B, Wang HF, Zhao Y, An F, et al: Glycolysis gatekeeper
PDK1 reprograms breast cancer stem cells under hypoxia. Oncogene.
37:1062–1074. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Su X, Li G and Liu W: The Long noncoding
RNA cancer susceptibility candidate 9 promotes nasopharyngeal
carcinogenesis via stabilizing HIF1α. DNA Cell Biol. 36:394–400.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Jaakkola P, Mole DR, Tian YM, Wilson MI,
Gielbert J, Gaskell SJ, von Kriegsheim A, Hebestreit HF, Mukherji
M, Schofield CJ, et al: Targeting of HIF-alpha to the von
Hippel-Lindau ubiquitylation complex by O2-regulated prolyl
hydroxylation. Science. 292:468–472. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Shao W, Wang D, Chiang YT, Ip W, Zhu L, Xu
F, Columbus J, Belsham DD, Irwin DM, Zhang H, et al: The Wnt
signaling pathway effector TCF7L2 controls gut and brain
proglucagon gene expression and glucose homeostasis. Diabetes.
62:789–800. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Sethi JK and Vidal-Puig A: Wnt signalling
and the control of cellular metabolism. Biochem J. 427:1–17. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Laplante M and Sabatini DM: mTOR signaling
in growth control and disease. Cell. 149:274–293. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Malakar P, Stein I, Saragovi A, Winkler R,
Stern-Ginossar N, Berger M, Pikarsky E and Karni R: Long noncoding
RNA MALAT1 regulates cancer glucose metabolism by enhancing
mTOR-mediated translation of TCF7L2. Cancer Res. 79:2480–2493.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Aquilano K, Baldelli S, Pagliei B, Cannata
SM, Rotilio G and Ciriolo MR: p53 orchestrates the PGC-1α-mediated
antioxidant response upon mild redox and metabolic imbalance.
Antioxid Redox Signal. 18:386–399. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Rajeshkumar NV, Dutta P, Yabuuchi S, de
Wilde RF, Martinez GV, Le A, Kamphorst JJ, Rabinowitz JD, Jain SK,
Hidalgo M, et al: Therapeutic targeting of the Warburg effect in
pancreatic cancer relies on an absence of p53 function. Cancer Res.
75:3355–3364. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Greenblatt MS, Feitelson MA, Zhu M,
Bennett WP, Welsh JA, Jones R, Borkowski A and Harris CC: Integrity
of p53 in hepatitis B × antigen-positive and -negative
hepatocellular carcinomas. Cancer Res. 57:426–432. 1997.PubMed/NCBI
|
|
76
|
Feitelson MA, Zhu M, Duan LX and London
WT: Hepatitis B × antigen and p53 are associated in vitro and in
liver tissues from patients with primary hepatocellular carcinoma.
Oncogene. 8:1109–1117. 1993.PubMed/NCBI
|
|
77
|
Patra KC and Hay N: The pentose phosphate
pathway and cancer. Trends Biochem Sci. 39:347–354. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Feng J, Ma J, Liu S, Wang J and Chen Y: A
noncoding RNA LINC00504 interacts with c-Myc to regulate tumor
metabolism in colon cancer. J Cell Biochem. 120:14725–14734. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Li N and Zhan X and Zhan X: The lncRNA
SNHG3 regulates energy metabolism of ovarian cancer by an analysis
of mitochondrial proteomes. Gynecol Oncol. 150:343–354. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Wise DR and Thompson CB: Glutamine
addiction: A new therapeutic target in cancer. Trends Biochem Sci.
35:427–433. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Wu MC, Arimura GK and Yunis AA: Mechanism
of sensitivity of cultured pancreatic carcinoma to asparaginase.
Int J Cancer. 22:728–733. 1978. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Eagle H: The minimum vitamin requirements
of the L and HeLa cells in tissue culture, the production of
specific vitamin deficiencies, and their cure. J Exp Med.
102:595–600. 1955. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Lai HS, Lee JC, Lee PH, Wang ST and Chen
WJ: Plasma free amino acid profile in cancer patients. Semin Cancer
Biol. 15:267–276. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Hsieh AL, Walton ZE, Altman BJ, Stine ZE
and Dang CV: MYC and metabolism on the path to cancer. Semin Cell
Dev Biol. 43:11–21. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Wu G, Fang YZ, Yang S, Lupton JR and
Turner ND: Glutathione metabolism and its implications for health.
J Nutr. 134:489–492. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Yang C, Ko B, Hensley CT, Jiang L, Wasti
AT, Kim J, Sudderth J, Calvaruso MA, Lumata L, Mitsche M, et al:
Glutamine oxidation maintains the TCA cycle and cell survival
during impaired mitochondrial pyruvate transport. Mol Cell.
56:414–424. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Yu Y, Yu X, Fan C, Wang H, Wang R, Feng C
and Guan H: Targeting glutaminase-mediated glutamine dependence in
papillary thyroid cancer. J Mol Med (Berl). 96:777–790. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Li HJ, Li X, Pang H, Pan JJ, Xie XJ and
Chen W: Long non-coding RNA UCA1 promotes glutamine metabolism by
targeting miR-16 in human bladder cancer. Jpn J Clin Oncol.
45:1055–1063. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Kitayama K, Yashiro M, Morisaki T, Miki Y,
Okuno T, Kinoshita H, Fukuoka T, Kasashima H, Masuda G, Hasegawa T,
et al: Pyruvate kinase isozyme M2 and glutaminase might be
promising molecular targets for the treatment of gastric cancer.
Cancer Sci. 108:2462–2469. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Lampa M, Arlt H, He T, Ospina B, Reeves J,
Zhang B, Murtie J, Deng G, Barberis C, Hoffmann D, et al:
Glutaminase is essential for the growth of triple-negative breast
cancer cells with a deregulated glutamine metabolism pathway and
its suppression synergizes with mTOR inhibition. PLoS One.
12:e01850922017. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Deng SJ, Chen HY, Zeng Z, Deng S, Zhu S,
Ye Z, He C, Liu ML, Huang K, Zhong JX, et al: Nutrient
stress-dysregulated antisense lncRNA GLS-AS impairs GLS-mediated
metabolism and represses pancreatic cancer progression. Cancer Res.
79:1398–1412. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Liu L, Cui S, Wan T, Li X, Tian W, Zhang
R, Luo L and Shi Y: Long non-coding RNA HOTAIR acts as a competing
endogenous RNA to promote glioma progression by sponging
miR-126-5p. J Cell Physiol. 233:6822–6831. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Ge Y, Yan X, Jin Y, Yang X, Yu X, Zhou L,
Han S, Yuan Q and Yang M: MiRNA-192 (corrected) and miRNA-204
Directly Suppress lncRNA HOTTIP and Interrupt GLS1-mediated
glutaminolysis in hepatocellular carcinoma. PLoS Genet.
11:e10057262015. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Luan W, Zhang X, Ruan H, Wang J and Bu X:
Long noncoding RNA OIP5-AS1 acts as a competing endogenous RNA to
promote glutamine catabolism and malignant melanoma growth by
sponging miR-217. J Cell Physiol. Feb 18–2019.(Epub ahead of
print). doi: 10.1002/jcp.28335. View Article : Google Scholar
|
|
95
|
Zhuo ZJ, Zhang R, Zhang J, Zhu J, Yang T,
Zou Y, He J and Xia H: Associations between lncRNA MEG3
polymorphisms and neuroblastoma risk in Chinese children. Aging
(Albany NY). 10:481–491. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Li S, Lin A, Han D, Zhou H, Cheng J, Zhang
J, Fu W, Zhuo Z and He J: LINC00673 rs11655237 C>T and
susceptibility to Wilms tumor: A five-center case-control study. J
Gene Med. 21:e31332019. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Li Y, Zhuo ZJ, Zhou H, Liu J, Liu Z, Zhang
J, Cheng J, Li S, Zhou H, Zhou R, et al: Additional data support
the role of LINC00673 rs11655237 C>T in the development of
neuroblastoma. Aging (Albany NY). 11:2369–2377. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Hapala I, Marza E and Ferreira T: Is fat
so bad? Modulation of endoplasmic reticulum stress by lipid droplet
formation. Biol Cell. 103:271–285. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Phillips CM, Goumidi L, Bertrais S, Field
MR, Cupples LA, Ordovas JM, Defoort C, Lovegrove JA, Drevon CA,
Gibney MJ, et al: Gene-nutrient interactions with dietary fat
modulate the association between genetic variation of the ACSL1
gene and metabolic syndrome. J Lipid Res. 51:1793–1800. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Zhou W, Tu Y, Simpson PJ and Kuhajda FP:
Malonyl-CoA decarboxylase inhibition is selectively cytotoxic to
human breast cancer cells. Oncogene. 28:2979–2987. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Núñez V, Alameda D, Rico D, Mota R,
Gonzalo P, Cedenilla M, Fischer T, Boscá L, Glass CK, Arroyo AG and
Ricote M: Retinoid X receptor alpha controls innate inflammatory
responses through the up-regulation of chemokine expression. Proc
Natl Acad Sci USA. 107:10626–10631. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Cui M, Xiao Z, Wang Y, Zheng M, Song T,
Cai X, Sun B, Ye L and Zhang X: Long noncoding RNA HULC modulates
abnormal lipid metabolism in hepatoma cells through an
miR-9-mediated RXRA signaling pathway. Cancer Res. 75:846–857.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Christensen LL, True K, Hamilton MP,
Nielsen MM, Damas ND, Damgaard CK, Ongen H, Dermitzakis E, Bramsen
JB, Pedersen JS, et al: SNHG16 is regulated by the Wnt pathway in
colorectal cancer and affects genes involved in lipid metabolism.
Mol Oncol. 10:1266–1282. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Gong J, Shen S, Yang Y, Qin S, Huang L,
Zhang H, Chen L, Chen Y, Li S, She S, et al: Inhibition of FASN
suppresses migration, invasion and growth in hepatoma carcinoma
cells by deregulating the HIF-1α/IGFBP1 pathway. Int J Oncol.
50:883–892. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Jiang Y, Yin X, Wu L, Qin Q and Xu J:
MAPK/P53-mediated FASN expression in bone tumors. Oncol Lett.
13:4035–4038. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Tao BB, He H, Shi XH, Wang CL, Li WQ, Li
B, Dong Y, Hu GH, Hou LJ, Luo C, et al: Up-regulation of USP2a and
FASN in gliomas correlates strongly with glioma grade. J Clin
Neurosci. 20:717–720. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Lu C, Ma J and Cai D: Increased HAGLR
expression promotes non-small cell lung cancer proliferation and
invasion via enhanced de novo lipogenesis. Tumour Biol. Apr
26–2017.(Epub ahead of print). doi:
org/10.1177/1010428317697574.
|
|
108
|
Ma DD, Yuan LL and Lin LQ: LncRNA HOTAIR
contributes to the tumorigenesis of nasopharyngeal carcinoma via
up-regulating FASN. Eur Rev Med Pharmacol Sci. 21:5143–5152.
2017.PubMed/NCBI
|
|
109
|
Bolsoni-Lopes A and Alonso-Vale MI:
Lipolysis and lipases in white adipose tissue-an update. Arch
Endocrinol Metab. 59:335–342. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Liu X, Liang Y, Song R, Yang G, Han J, Lan
Y, Pan S, Zhu M, Liu Y, Wang Y, et al: Long non-coding RNA
NEAT1-modulated abnormal lipolysis via ATGL drives hepatocellular
carcinoma proliferation. Mol Cancer. 17:902018. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Shang C, Wang W, Liao Y, Chen Y, Liu T, Du
Q, Huang J, Liang Y, Liu J, Zhao Y, et al: LNMICC promotes nodal
metastasis of cervical cancer by reprogramming fatty acid
metabolism. Cancer Res. 78:877–890. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Li D, Cheng M, Niu Y, Chi X, Liu X, Fan J,
Fan H, Chang Y and Yang W: Identification of a novel human long
non-coding RNA that regulates hepatic lipid metabolism by
inhibiting SREBP-1c. Int J Biol Sci. 13:349–357. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Kulsum S, Sudheendra HV, Pandian R,
Ravindra DR, Siddappa G, R N, Chevour P, Ramachandran B, Sagar M,
Jayaprakash A, et al: Cancer stem cell mediated acquired
chemoresistance in head and neck cancer can be abrogated by
aldehyde dehydrogenase 1 A1 inhibition. Mol Carcinog. 56:694–711.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Yao Z, Jones AW, Fassone E, Sweeney MG,
Lebiedzinska M, Suski JM, Wieckowski MR, Tajeddine N, Hargreaves
IP, Yasukawa T, et al: PGC-1β mediates adaptive chemoresistance
associated with mitochondrial DNA mutations. Oncogene.
32:2592–2600. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Zhang Y, Zeng S, Ma J, Deng G, Qu Y, Guo C
and Shen H: Nestin overexpression in hepatocellular carcinoma
associates with epithelial-mesenchymal transition and
chemoresistance. J Exp Clin Cancer Res. 35:1112016. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
McNeil EM and Melton DW: DNA repair
endonuclease ERCC1-XPF as a novel therapeutic target to overcome
chemoresistance in cancer therapy. Nucleic Acids Res.
40:9990–10004. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Kim M, Jung JY, Choi S, Lee H, Morales LD,
Koh JT, Kim SH, Choi YD, Choi C, Slaga TJ, et al: GFRA1 promotes
cisplatin-induced chemoresistance in osteosarcoma by inducing
autophagy. Autophagy. 13:149–168. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Shi H, Li K, Feng J, Liu G, Feng Y and
Zhang X: LncRNA-DANCR Interferes with miR-125b-5p/HK2 axis to
desensitize colon cancer cells to cisplatin vis activating
anaerobic glycolysis. Front Oncol. 10:10342020. View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Zheng ZQ, Li ZX, Guan JL, Liu X, Li JY,
Chen Y, Lin L, Kou J, Lv JW, Zhang LL, et al: Long noncoding RNA
TINCR-mediated regulation of acetyl-coa metabolism promotes
nasopharyngeal carcinoma progression and chemoresistance. Cancer
Res. 80:5174–5188. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
He W, Liang B, Wang C, Li S, Zhao Y, Huang
Q, Liu Z, Yao Z, Wu Q, Liao W, et al: MSC-regulated lncRNA
MACC1-AS1 promotes stemness and chemoresistance through fatty acid
oxidation in gastric cancer. Oncogene. 38:4637–4654. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Wen JF, Jiang YQ, Li C, Dai XK, Wu T and
Yin WZ: LncRNA-SARCC sensitizes osteosarcoma to cisplatin through
the miR-143-mediated glycolysis inhibition by targeting Hexokinase
2. Cancer Biomark. 28:231–246. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Wang T, Fahrmann JF, Lee H, Li YJ,
Tripathi SC, Yue C, Zhang C, Lifshitz V, Song J, Yuan Y, et al:
JAK/STAT3-Regulated fatty acid beta-oxidation is critical for
breast cancer stem cell self-renewal and chemoresistance. Cell
Metab. 27:136–150.e5. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Hoy SM: Patisiran: First global approval.
Drugs. 78:1625–1631. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Yang J, Guo Y, Seo W, Zhang R, Lu C, Wang
Y, Luo L, Paul B, Yan W, Saxena D and Li X: Targeting cellular
metabolism to reduce head and neck cancer growth. Sci Rep.
9:49952019. View Article : Google Scholar : PubMed/NCBI
|
|
125
|
Elgogary A, Xu Q, Poore B, Alt J,
Zimmermann SC, Zhao L, Fu J, Chen B, Xia S, Liu Y, et al:
Combination therapy with BPTES nanoparticles and metformin targets
the metabolic heterogeneity of pancreatic cancer. Proc Natl Acad
Sci USA. 113:E5328–E5336. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
126
|
Qing L and Qing W: Hypoxia inducible
factor 1 inhibitors for cancer therapy. Minerva Chir. 74:442–444.
2019. View Article : Google Scholar : PubMed/NCBI
|