Introduction
Esophageal cancer (EC) is the sixth leading cause of
cancer-related mortality worldwide and the 5-year survival rate of
patients is only 15–25% (1). Despite
the significant advancements made in the treatment of EC, patients
with EC have a poor prognosis, as EC cells can metastasize to lymph
nodes, even at an early stage, and migrate to distant sites
(2). Thus, possible biological
targets for the treatment of EC require further investigation.
Partner of NOB1 homolog (PNO1) is a highly conserved
protein with a K homology (KH) domain at its C-terminal and two
putative nuclear localization signals at its N-terminal (3,4). In mice,
PNO1 was discovered to be involved in immune responses and
proteasome activities (5). Currently,
the oncogenic role of PNO1 in hepatocellular carcinoma and
colorectal cancer has been determined (6,7). However,
the expression levels, biological effects and mechanisms of action
of PNO1 in EC remain to be elucidated.
The present study first analyzed the expression
levels of PNO1 in EC tissues using data obtained from The Cancer
Genome Atlas (TCGA) database. Subsequently, the biological effects
of PNO1 in EC were determined. Finally, potential downstream
targets of PNO1 in EC were investigated.
Materials and methods
TCGA database
RNA-sequencing profiles of PNO1 expression in 162 EC
and 11 normal samples were downloaded from the TCGA database
(https://tcga-data.nci.nih.gov/tcga).
In addition, RNA-sequencing profiles of PNO1 expression in 653
normal samples were obtained from the Genotype-Tissue Expression
(GTEx) database (http://xena.ucsc.edu), which is a
database that provides information on normal samples from healthy
participants. Both the 653 samples in GTEx database and 11 normal
samples in TCGA database were used as normal samples (8,9). The
differentially expressed genes (DEGs) between control and cancerous
samples were identified using the Limma package of R software
(http://bioconductor.org/packages/release/bioc/html/limma.html)
(10). The cut-off values for DEGs
were |logfold change| >1 and P<0.05.
Cell lines and culture
EC cell lines (EC9706, Eca-109 and TE-1), the normal
esophageal epithelial cell line HEEC, and the 293T cell line
(Procell Life Technology Co., Ltd., Wuhan, China) were cultured in
DMEM (HyClone; Cytiva) supplemented with 10% FBS (Gibco; Thermo
Fisher Scientific, Inc.) at 37°C with 5% CO2.
Construction of stable PNO1-knockdown
(KD) and β-catenin (CTNNB1)-overexpressing cells
PNO1 was stably knocked down in Eca-109 and TE-1
cells using short hairpin RNA (sh), and CTNNB1 was stably
overexpressed in Eca-109 cells only. sh-PNO1-KD, sh-PNO1-negative
control (NC), sh-CTNNB1-OE and sh-CTNNB1-negative control (NC) were
used for transfection. Lentiviral vectors were used for Eca-109 and
TE-1 cell transfection as previously described (11). Images of the cells were obtained under
a fluorescence microscope following transfection for 72 h. Reverse
transcription-quantitative PCR (RT-qPCR) and western blotting were
used to analyze the transfection efficiency.
RT-qPCR
Total RNA was extracted and reversed transcribed
into cDNA using TRIzol® reagent (Invitrogen; Thermo
Fisher Scientific, Inc.) and Promega's Universal Riboclone cDNA
synthesis system (Promega Corp.), respectively, according to the
manufacturers' protocols. qPCR was subsequently performed using a
SYBR Green Master mix (Takara Biotechnology Co., Ltd.), using GAPDH
as the endogenous control. The following forward and reverse
primers sequences were used for the qPCR: GAPDH forward,
5′-TGACTTCAACAGCGACACCCA-3′ and reverse,
5′-CACCCTGTTGCTGTAGCCAAA-3′; and PNO1 forward,
5′-TGTTAAACCCCTAAAGGGAGACC-3′ and reverse,
5′-CCTTGTCCGTGTCACATTCTCT-3′. Expression levels were quantified
using the 2−ΔΔCq method (12).
Western blotting
Total protein was extracted from cells using
radioimmunoprecipitation lysis buffer (RIPA, Solarbio Technology
Co., Beijing, China). The extracted protein was separated by
SDS-PAGE and transferred onto PVDF membranes. The membranes were
subsequently incubated with primary antibodies (Table I). Following the primary antibody
incubation, the membranes were incubated with anti-mouse IgG
(1:5,000; cat. no. sc-2005; Santa Cruz Biotechnology, Inc.) and
anti-rabbit IgG (1:5,000; cat. no. sc-2004; Santa Cruz
Biotechnology, Inc.) secondary antibodies. Protein bands were
visualized using a Pierce™ ECL Western Blotting substrate (Thermo
Fisher Scientific, Inc.).
 | Table I.List of the primary antibodies used
in the western blot analysis. |
Table I.
List of the primary antibodies used
in the western blot analysis.
| Gene | Abbreviation | Host | Company | Cat. no. | Weight (kDa) | Dilution |
|---|
| Cadherin 1 | CDH1 | Mouse | Cell Signaling
Technology, Inc. (CST) | 14472s | 135 | 1:100 |
| Mitogen-activated
protein kinase 14 | P38 | Rabbit | CST | 8690 | 40 | 1:300 |
|
Phosphorylated-nuclear factor κB | p-NFKB | Rabbit | CST | 3033s | 65 | 1:200 |
| Matrix
metallopeptidase 9 | MMP9 | Rabbit | CST | 13667s | 84 | 1:300 |
| Mechanistic target
of rapamycin kinase | mTOR | Rabbit | CST | 2983 | 289 | 1:300 |
| Catenin β1 | CTNNB1 | Rabbit | CST | 9562 | 92 | 1:300 |
| Phosphorylated-AKT
serine/threonine kinase | p-AKT | Rabbit | CST | 4060 | 60 | 1:1,000 |
| Cadherin 2 | CDH2 | Rabbit | CST | 13116 | 140 | 1:100 |
|
Phosphorylated-mitogen-activated protein
kinase 14 | p-P38 | Rabbit | CST | 4631 | 43 | 1:300 |
|
Phosphorylated-catenin β1 |
p-CTNNB1 | Rabbit | CST | 2009s | 92 | 1:300 |
| Nuclear factor κB
p65 | NF-κB
p65 | Rabbit | CST | 8242 | 65 | 1:500 |
| MYC proto-oncogene,
bHLH transcription factor | myc | Rabbit | CST | ab32072 | 57 | 1:100 |
| NFKB inhibitor
α | NFKBIA | Rabbit | CST | ab7217 | 35.6 | 1:300 |
| Matrix
metallopeptidase 2 | MMP2 | Rabbit | CST | ab37150 | 72 | 1:300 |
| Twist family bHLH
transcription factor | Twist | Rabbit | CST | ab50581 | 21 | 1:300 |
| Fibronectin 1 | FN1 | Mouse | CST | ab6328 | >250 | 1:100 |
| AKT
serine/threonine kinase 1 | AKT1 | Rabbit | CST | ab183758 | 56 | 1:300 |
|
Phosphorylated-mechanistic target of
rapamycin kinase | p-mTOR | Rabbit | CST | ab109268 | 289 | 1:300 |
| Fartner of NOB1
homolog | PNO1 | Rabbit | Santa Cruz
Biotechnology, Inc. | sc-133263 | 31 | 1:100 |
|
Glyceraldehyde-3-phosphate
dehydrogenase | GAPDH | Mouse | Santa Cruz
Biotechnology, Inc. | sc-32233 | 36 | 1:500 |
Celigo cell counting assay
A Celigo cell counting assay was performed as
previously described (13). Briefly,
Eca-109 and TE-1 cells were seeded into 96-well plates at a density
of 2×103 cells/well. Cells were cultured for a total of
120 h, and cells were counted with a Celigo® Cell
Imaging cytometer (Nexcelom Bioscience) every 24 h.
Colony formation assay
A cell colony formation assay was performed as
previously described (14). Briefly,
Eca-109 and TE-1 cells were seeded into 6-well plates at a density
of 800 cells/well. Following the culture of the cells for 2 weeks,
cells were fixed with 1 ml paraformaldehyde (4%) for 40 min and
stained with 1 ml crystal violet dye solution (0.1%) for 15 min.
Stained cells were visualized under a microscope (Olympus) and
colonies with more than 10 cells were counted.
MTT assay
The MTT assay was performed as previously described
(15). Briefly, Eca-109 and TE-1
cells were seeded into 96-well plates at a density of
2×103 cells/well and cultured for 5 days. Subsequently,
20 µl MTT solution (5 mg/ml; Gen-view Scientific, Inc.) was added
into each well and incubated for 4 h at 37°C. Following the
incubation, 100 µl DMSO was added to each well to dissolve the
purple formazan crystals. The absorbance was measured at a
wavelength of 490 nm to detect the optical density (OD) value.
Cell apoptosis assay
Cell apoptosis assay was performed using an Annexin
V Apoptosis Detection kit (eBioscience; Thermo Fisher Scientific,
Inc.) as previously described (16).
Briefly, Eca-109 and TE-1 cells (1×106 cells/tube) were
stained with 10 µl Annexin V-allophycocyanin, and apoptotic cells
were analyzed by flow cytometry (FACSCalibur, Beckman Coulter).
Wound healing assay
The wound healing assay was performed as previously
described (17) using a Celigo
cytometer. Briefly, Eca-109 and TE-1 cells expressing GFP were
seeded into 96-well plates at a density of 5×104
cells/well and a scratch was made in the cell monolayer. The
fluorescence indicates the efficiency of transfection. After
scratching, the cells were cultured in serum-free DMEM for 24 h.
Images of the scratch were obtained at 0 and 24 h using Celigo
which can identify cells with green fluorescence and images were
captured.
Cell Transwell assay
Cell Transwell assay was performed with Transwell
kits (Corning, Inc.) as previously described (18). Briefly, 1×105 cells
suspended in 100 µl serum-free DMEM were seeded into the upper
chambers. The lower chambers were filled with 600 µl DMEM
supplemented with 30% FBS. Following culture for 8 h, cells were
fixed with 4% paraformaldehyde for 30 min and stained with crystal
violet aqueous solution (0.5%). Cells were subsequently visualized
under a microscope (Olympus).
Cell invasion assay
Cell invasion assay was performed using BioCoat™
Matrigel® Invasion chambers (Corning, Inc.) as
previously described (19). Briefly,
500 µl serum-free medium was plated into both the upper and lower
chambers for 2 h at 37°C to rehydrate the Matrigel matrix.
Subsequently, 1×105 cells in 500 µl serum-free DMEM were
seeded into the upper chamber and 750 µl DMEM supplemented with 30%
FBS was added into the lower chamber. Following incubation for 8 h,
Giemsa staining solution was added, and images were captured using
a microscope (Olympus).
Protein-protein interaction
analysis
The BioGRID database (https://thebiogrid.org) (20) was used to identify proteins that
interacted with PNO1 in humans.
Gene set enrichment analysis
(GSEA)
GSEA version 3.0 software (software.broadinstitute.org/gsea/index.jsp) was used
for GSEA (21). A false discovery
rate (FDR q-val) of ≤25% and nominal P<0.05 were set as the
cut-off values. The ggplot2 package (https://cran.r-project.org/web/packages/ggplot2/index.html)
was used to merge the selected images.
Xenograft experiments
Twenty female BALB/c nude mice (age, 4 weeks,
Shanghai Slake Experimental Animal Co., Ltd.) were subcutaneously
inoculated with sh-PNO1-NC- or sh-PNO1-KD-transfected Eca-109 cells
(4×106 cells suspended in 200 µl PBS) to form tumors.
All mice were euthanized by intraperitoneal injection of an
overdose of 2% sodium pentobarbital (100 mg/kg), and the death was
confirmed by cervical dislocation. The tumor volume of each mice
was measured every three days for 14 consecutive days two weeks
after inoculation using the following equation: 3.14/6 × (length ×
width × width). All animal experiments were conducted in accordance
with the Guide for the Care and Use of Laboratory Animals of the
National Institutes of Health [National Research Council (US)
Institute for Laboratory Animal Research, 1996] and were approved
by the Ethics Committee of The First Affiliated Hospital of Bengbu
Medical College.
Statistical analysis
Statistical analysis was performed using SPSS 22.0
software (IBM Corp.) and GraphPad Prism 7.0 software (GraphPad
Software, Inc.). For western blot analysis, only one repeat was
performed. However, for all other experiments, three repeats were
conducted and experiments were further repeated if discordant
results were obtained. Data are presented as the mean ± SD.
Statistical differences between groups were determined using
unpaired Student's t-test or one-way ANOVA with Tukey test. The
expression data of PNO1 mRNA in the 8 paired samples obtained from
the TCGA database met the requirement for a parametric analysis,
and paired Student's t-test was utilized. P<0.05 was indicative
of statistically significant differences.
Results
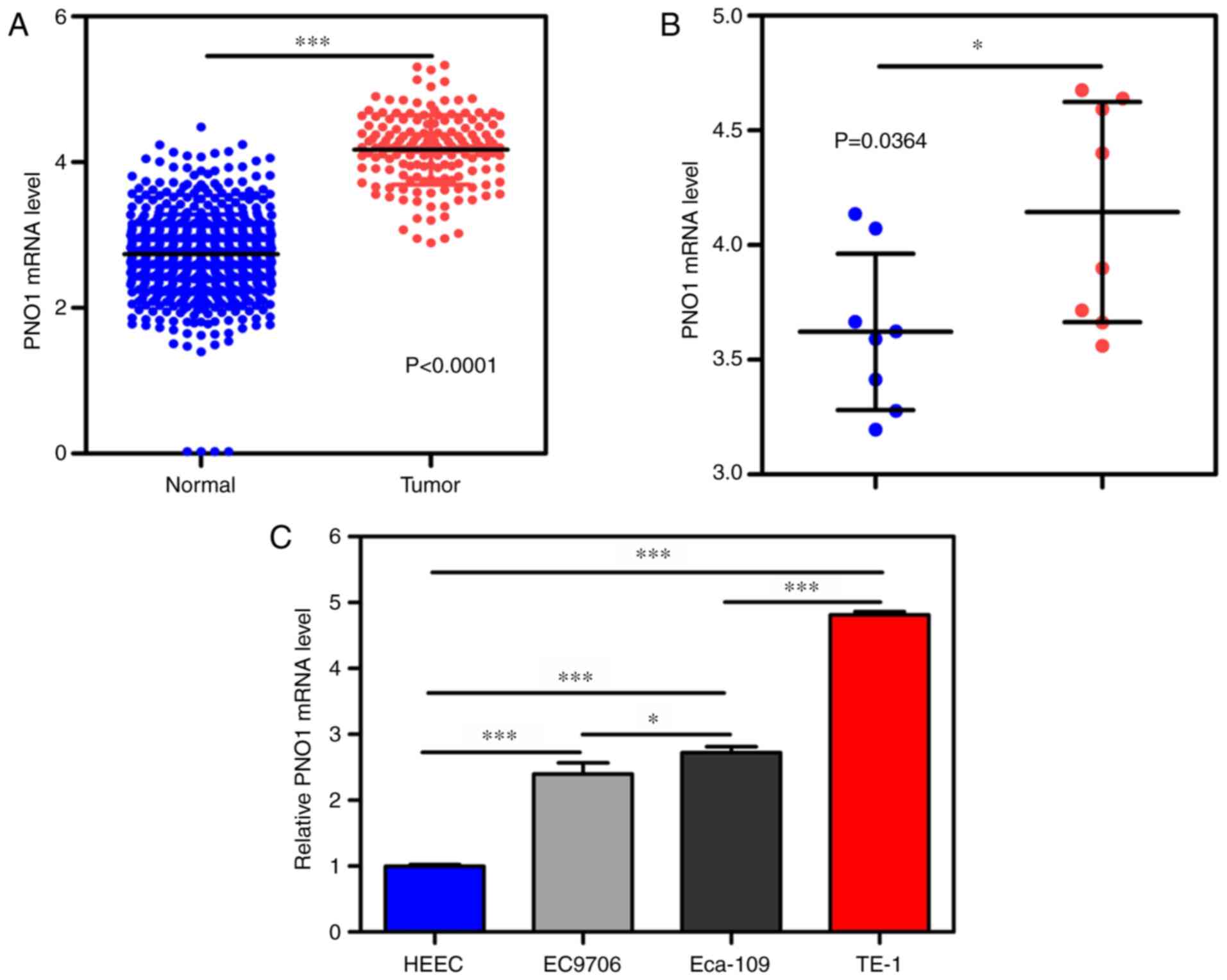
Expression levels of PNO1 in EC
To determine the expression levels of PNO1 in EC,
PNO1 expression levels in 162 EC and 664 normal samples from
databases were analyzed. As shown in Fig.
1A, PNO1 expression levels were upregulated in the tumor
tissues compared with that in the 664 normal tissues (P<0.0001).
Paired tumor and non-tumor samples from the dataset were selected,
and similar results were obtained (P=0.0364; Fig. 1B). In addition, PNO1 mRNA expression
levels in EC cell lines (EC9706, Eca-109 and TE-1) and the normal
esophageal epithelial cell line, HEEC, were analyzed. As shown in
Fig. 1C, PNO1 expression levels were
upregulated in EC cells compared with that noted in the HEEC cells
(P<0.0001). The PNO1 expression levels were highest in TE-1
cells, then Eca-109 cells, then EC9706 cells, and the lowest in
HEEC cells. These data suggested that PNO1 mRNA expression levels
may be upregulated in tumor samples compared with the levels in
normal samples.
Successful establishment of stable
PNO1 KD Eca-109 and TE-1 cells
To determine the function and mechanism of PNO1 in
EC, PNO1 expression was knocked down in Eca-109 and TE-1 cells. The
green fluorescence intensity was used to evaluate whether the
virus-carried plasmid was successfully transfected into the cells.
As shown in the fluorescence images in Fig. S1A and B, PNO1 expression was
successfully knocked down in the Eca-109 and TE-1 cells, with a
transduction efficiency in both cells of >80%. In addition, PNO1
expression was successfully knocked down in Eca-109 and TE-1 cells,
with transduction efficiencies of >90 and 70%, respectively.
Furthermore, RT-qPCR (Fig. S1C and
D) and western blotting (Fig. S1E
and F) were conducted to verify the transduction efficiency,
and the results were consistent with the fluorescence microscopy
results (P<0.0001). These results suggested that PNO1 expression
was successfully silenced in both Eca-109 and TE-1 cells.
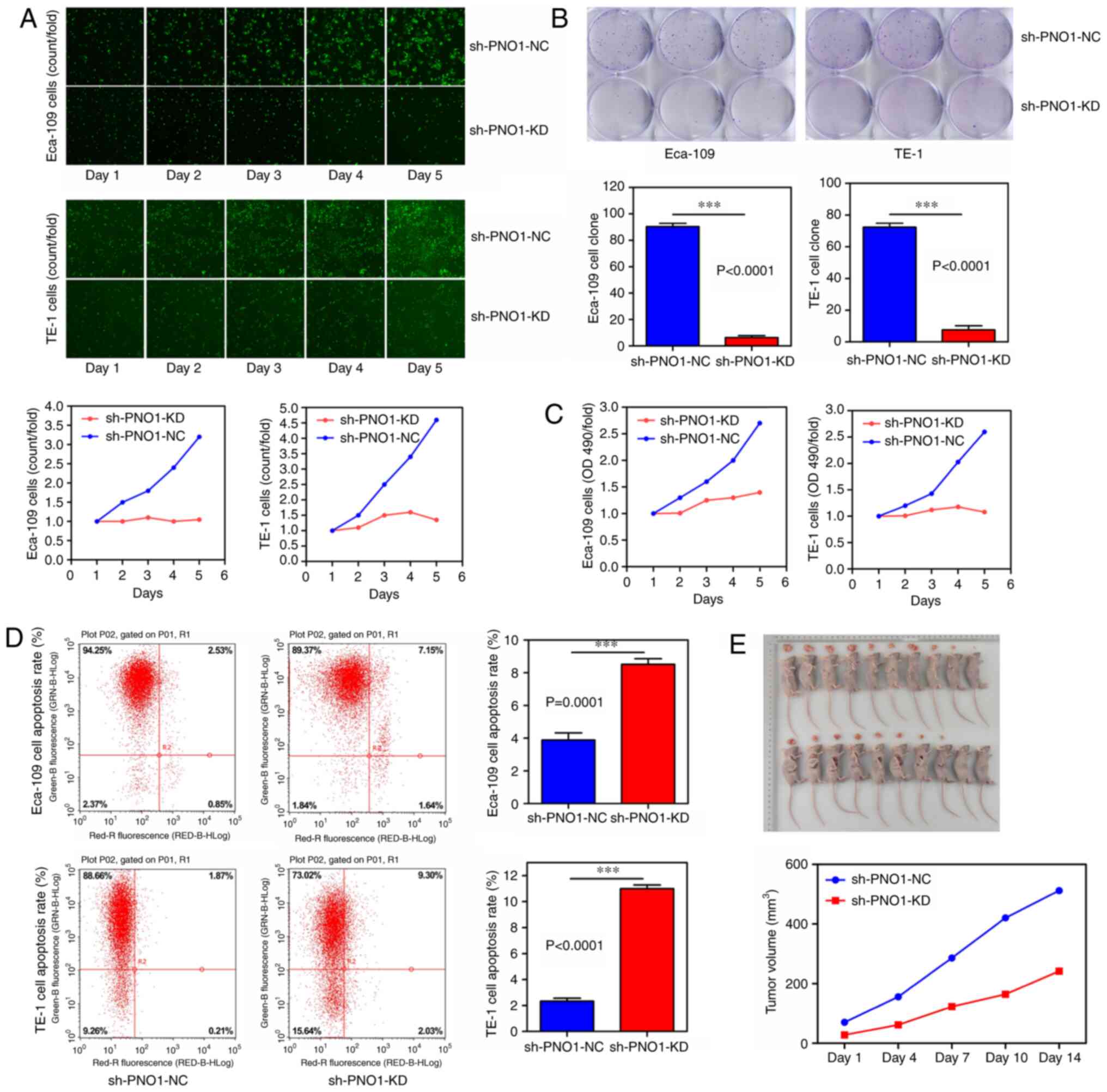
KD of PNO1 expression inhibits EC cell
tumorigenesis in vitro and in vivo
To determine the effects of PNO1 on the
proliferation of Eca-109 and TE-1 cells, Celigo cell counting,
colony formation and MTT assays were performed. The results of the
cell counting assay revealed that the cell amount/fold value was
decreased by 3-fold in the sh-PNO1-KD-transfected cells compared
with sh-PNO1-NC-transfected cells (Fig.
2A). Furthermore, the results of the colony formation assay
demonstrated that the colony formation in sh-PNO1-KD-transfected
Eca-109 cells was decreased by ~15-fold compared with
sh-PNO1-NC-transfected Eca-109 cells, while the colony forming
ability of sh-PNO1-KD-transfected TE-1 cells was ~9-fold decreased
compared with sh-PNO1-NC-transfected TE-1 cells (both P<0.0001;
Fig. 2B). In addition, the results of
the MTT assay found that the OD490/fold value of
sh-PNO1-KD-transfected Eca-109 cells was decreased by ~2-fold
compared with sh-PNO1-NC-transfected Eca-109 cells, while the
OD490/fold value of sh-PNO1-KD-transfected TE-1 cells was decreased
by ~2.5-fold compared with sh-PNO1-NC-transfected TE-1 cells
(Fig. 2C).
To determine the effects of PNO1 on the apoptosis of
Eca-109 and TE-1 cells, the apoptotic rate of cells was
investigated using flow cytometry. As shown in Fig. 2D, the apoptotic rate of
sh-PNO1-KD-transfected Eca-109 cells was increased by 2-fold
compared with sh-PNO1-NC-trasfected Eca-109 cells (P<0.0001),
while that of sh-PNO1-KD-transfected TE-1 cells was increased by
5-fold compared with sh-PNO1-NC-transfected TE-1 cells
(P<0.0001).
To verify the effects of PNO1 on EC growth, in
vivo xenograft experiments were performed and the tumor volume
was calculated for ~2 weeks. As shown in Fig. 2E, the tumor volume was significantly
decreased in the sh-PNO1-KD group compared with the sh-PNO1-NC
group. Overall, these results suggested that the KD of PNO1
inhibited tumorigenesis in vitro and in vivo.
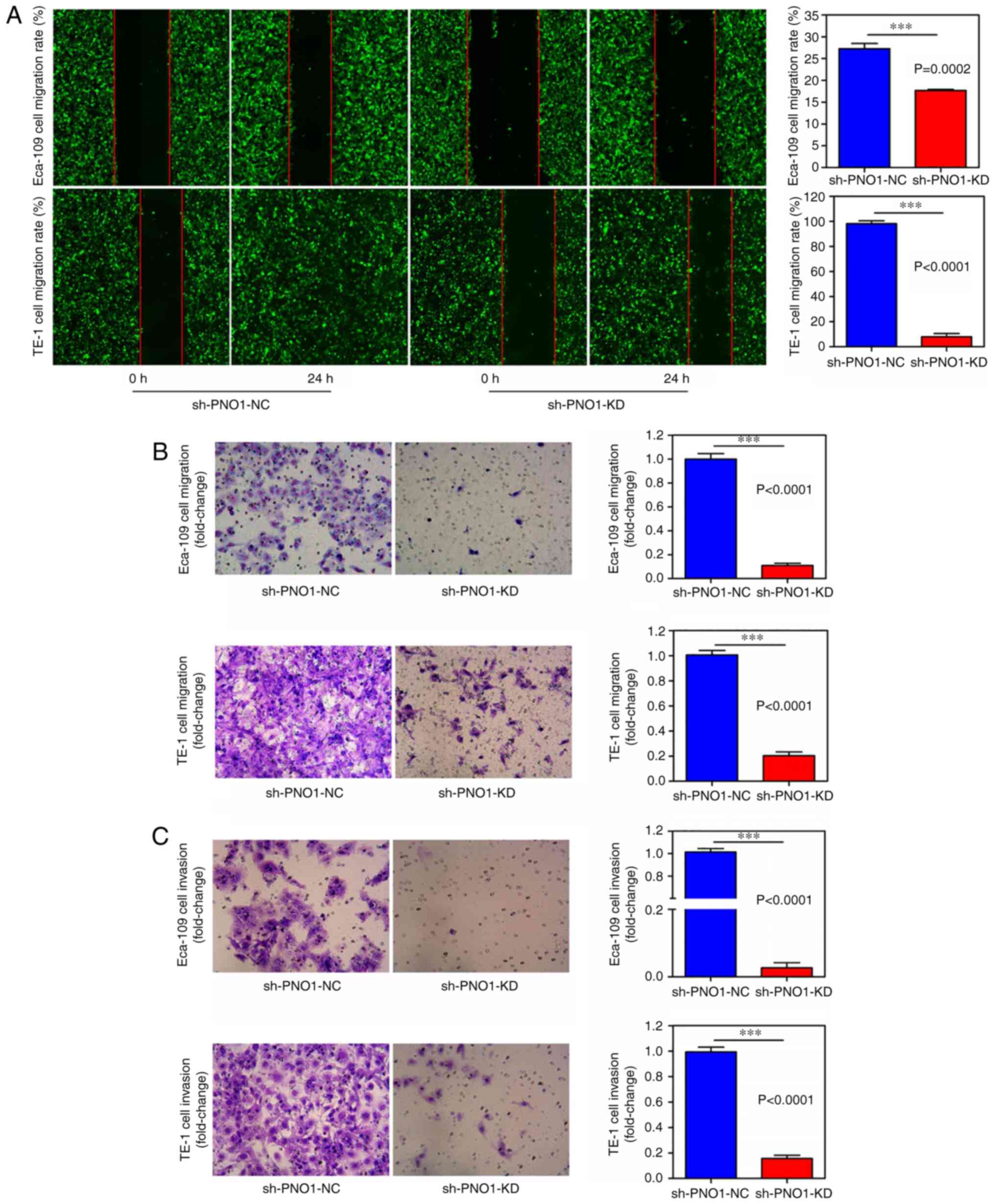
KD of PNO1 suppresses cell migration
and invasion
To determine the effects of PNO1 on the migration
and invasion of Eca-109 and TE-1 cells, wound healing, and
Transwell migration and invasion assays were performed. The wound
healing assay revealed that the migratory rates of the
sh-PNO1-KD-transfected Eca-109 cells (P<0.0001) and
sh-PNO1-KD-transfected TE-1 cells (P<0.001) were significantly
decreased compared with sh-PNO1-NC-transfected Eca-109 and
sh-PNO1-NC-transfected TE-1 cells (Fig.
3A). In particular, the migration rate of
sh-PNO1-KD-transfected TE-1 cells was decreased by ~12-fold
compared with sh-PNO1-NC-transfected TE-1 cells (Fig. 3A). Furthermore, the number of
migratory sh-PNO1-KD-transfected Eca-109 and sh-PNO1-KD-transfected
TE-1 cells was decreased compared with the sh-PNO1-NC-transfected
Eca-109 and sh-PNO1-NC-transfected TE-1 cells (P<0.0001;
Fig. 3B). In more detail, the number
of migratory sh-PNO1-KD-transfected Eca-109 cells was decreased by
~10-fold compared with the number of migratory
sh-PNO1-NC-transfected Eca-109 cells, while the number of migratory
sh-PNO1-KD-transfected TE-1 cells was decreased by ~5-fold compared
with sh-PNO1-NC-transfected TE-1 cells. In addition, the number of
invasive sh-PNO1-KD-transfected Eca-109 and sh-PNO1-KD-transfected
TE-1 cells was significantly decreased compared with the number of
sh-PNO1-NC-transfected Eca-109 and sh-PNO1-NC-transfected TE-1
cells. In fact, very few sh-PNO1-KD-transfected Eca-109 cells
underwent invasion (P<0.0001; Fig.
3C).
Biological mechanisms of PNO1 in
EC
To determine the biological mechanisms of PNO1 in
EC, GSEA was performed using data obtained from TCGA database. A
total of 178 gene sets were enrolled for analysis, in which 21 and
25 gene sets met the criterion for the PNO1 high and low expression
phenotypes, respectively. In samples with PNO1 high expression
phenotypes, pathways related to the cell cycle and DNA replication
were upregulated, while pathways associated with chemokine
signaling pathways and cytokine-cytokine receptor interaction were
downregulated in the PNO1 low expression phenotype (Fig. 4A), which indicated that PNO1 may
promote EC growth via regulating genes related to the cell cycle
and DNA replication. Thus, the expression levels of molecules
related to the cell cycle were investigated in Eca-109 cells. As
shown in Fig. 4B and C, the protein
expression levels of AKT1, Twist, Myc, mTOR, matrix
metalloproteinase (MMP)2, NF-κB p65 and CTNNB1 were downregulated
in sh-PNO1-KD-transfected Eca-109 cells compared with in
sh-PNO1-NC-transfected Eca-109 cells. However, the protein
expression levels of cadherin (CDH)1, CDH2, p38, phosphorylated
(p)-p38, fibronectin 1, MMP9, NF-κB inhibitor α, p-AKT, p-mTOR,
p-CTNNB1 and p-NF-κB were similar in both sh-PNO1-KD-transfected
Eca-109 and sh-PNO1-NC-transfected Eca-109 cells (Fig. 4B and C).
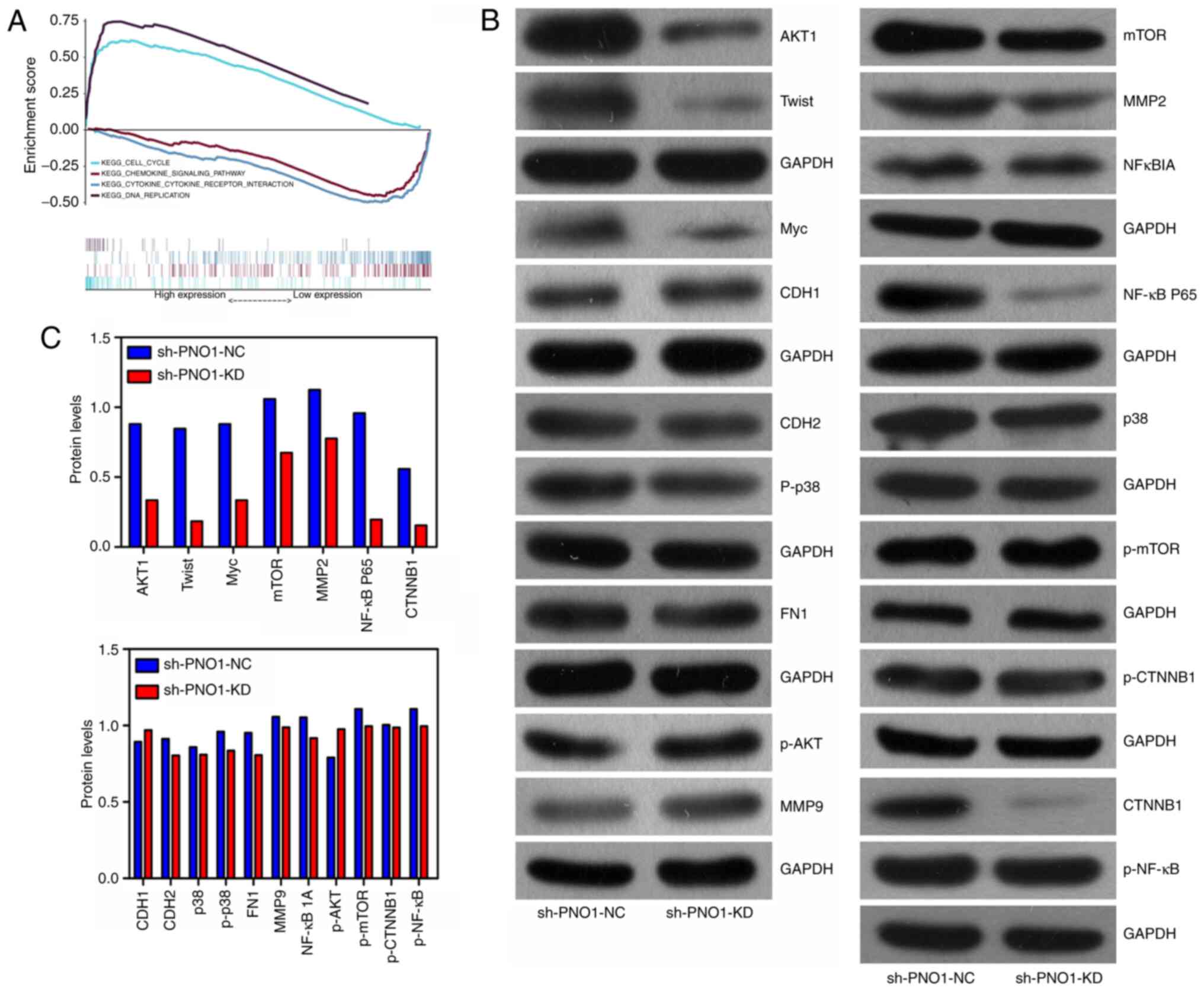 | Figure 4.Biological mechanisms of PNO1 in
esophageal cancer. (A) Gene Set Enrichment Analysis based on data
from The Cancer Genome Atlas database found that pathways related
to the cell cycle and DNA replication were upregulated in the PNO1
high expression phenotype, while pathways associated with chemokine
signaling pathways and cytokine-cytokine receptor interaction were
downregulated in the PNO1 low expression phenotype. (B and C)
Protein expression levels of AKT1, Twist, Myc, mTOR, MMP2, NF-κB
p65 and CTNNB1 were downregulated in sh-PNO1-KD-transfected Eca-109
cells compared with sh-PNO1-NC-transfected Eca-109 cells. Protein
expression levels of CDH1, CDH2, p38, p-p38, FN1, MMP9, NFKBIA,
p-AKT, p-mTOR, p-CTNNB1, p-NF-κB and Slug were similar in the
sh-PNO1-KD-transfected Eca-109 and sh-PNO1-NC-transfected Eca-109
cells. CDH1, cadherin 1; p-, phosphorylated; FN1, fibronectin 1;
MMP9, matrix metalloproteinase 9; NFKBIA, NF-κB inhibitor α; PNO1,
partner of NOB1 homolog; MMP2, matrix metalloproteinase 2; CTNNB1,
β-catenin 1; KD, knockdown; sh, short hairpin RNA; NC, negative
control. |
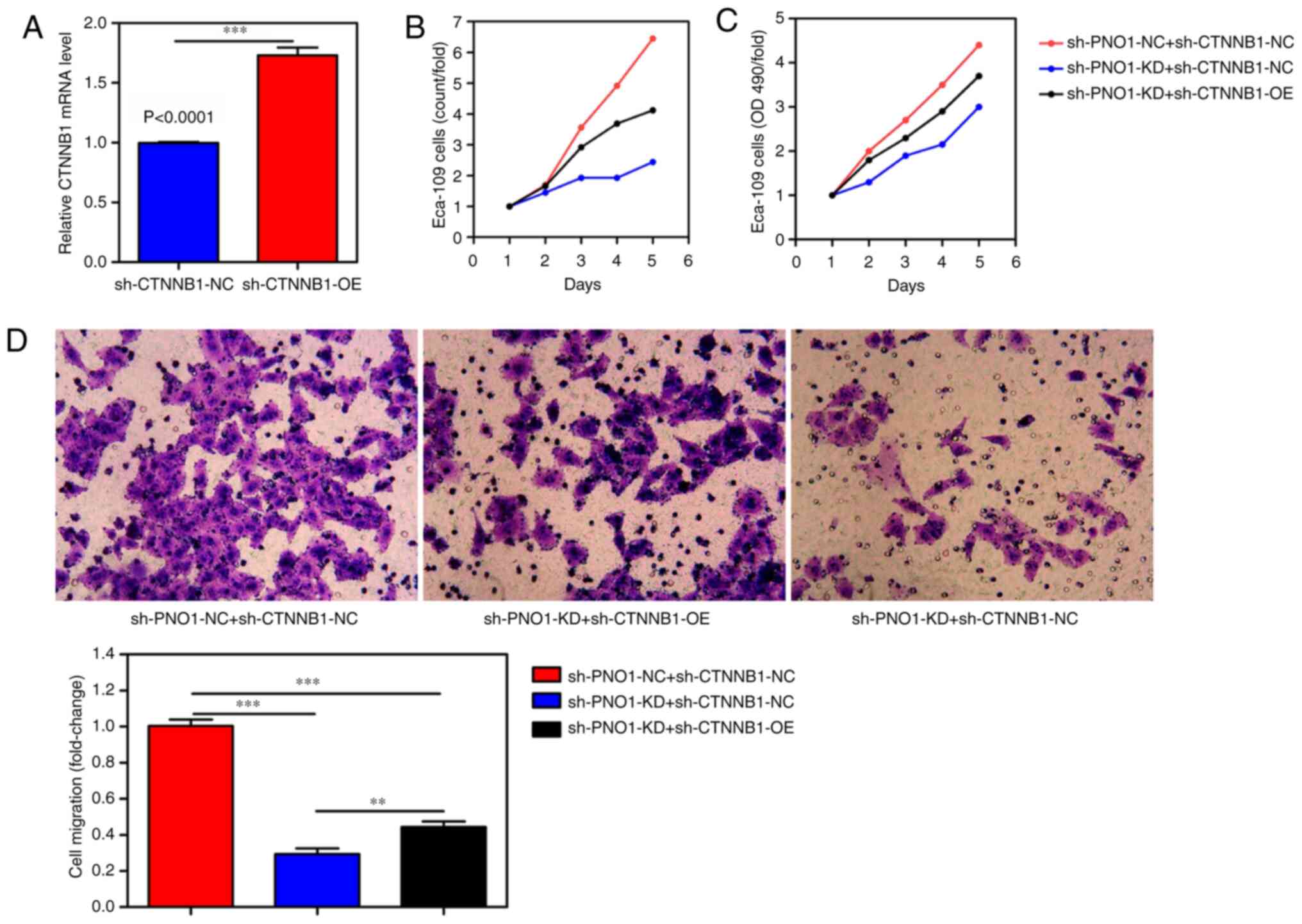
CTNNB1 may be a potential direct downstream target
of PNO1 in EC. As shown in Fig. 4B and
C, CTNNB1 was found to be regulated by PNO1. In addition, the
results obtained from BioGRID identified that CTNNB1 interacted
with PNO1 (data not shown). Hence, a rescue experiment was designed
to verify the relationship between CTNNB1 and PNO1 in EC. We here
overexpressed CTNNB1 in Eca-109 cells (Fig. 5A) and split Eca-109 cells into three
groups: i) sh-PNO1-NC+sh-CTNNB1-NC-transfected Eca-109 cells; ii)
sh-PNO1-KD+sh-CTNNB1-NC-transfected Eca-109 cells; and iii)
sh-PNO1-KD + sh-CTNNB1-overexpression (OE)-transfected Eca-109
cells. Cell counting (Fig. 5B), MTT
(Fig. 5C) and Transwell assays
(Fig. 5D) were performed with the
three groups. Compared with the sh-PNO1-NC+sh-CTNNB1-NC group, the
proliferation and invasion of the
sh-PNO1-KD+sh-CTNNB1-NC-transfected Eca-109 cells was decreased.
Conversely, compared with the sh-PNO1-KD+sh-CTNNB1-NC group, the
proliferation and invasion of the
sh-PNO1-KD+sh-CTNNB1-OE-transfected Eca-109 cells was increased.
Altogether, these results suggested that the OE of CTNNB1 may
abolish the effects of the KD of PNO1 in Eca-109 cells. Thus,
CTNNB1 may be a potential direct downstream target of PNO1 in
EC.
Discussion
Partner of NOB1 homolog (PNO1) is located on human
chromosome 2q14 and consists of five introns and seven exons
(4). The length of the full cDNA
sequence of human PNO1 is 1,637 base pairs, which includes an open
reading frame of 759 base pairs in length, and the weight of the
PNO1 protein is 35 kDa (3,4). In addition, PNO1 contains a KH domain,
which is responsible for binding RNA and NIN1 (RPN12) binding
protein 1 homolog (NOB1) at amino acids 157–230 in the human PNO1
C-terminal, and two nuclear localization signals at the PNO1
C-terminal from amino acids 23–29 and amino acids 53–58 (4). The biological function of PNO1 has been
investigated over the past few years. In yeast, the interaction
between PNO1 and NOB1 was found to be related to ribosome
biogenesis (5,7). The KD of PNO1 led to assembly defects of
26S and 40S ribosomal RNA (rRNA), decreased the levels of 18S rRNA
and led to an accumulation of 32S, 33S and 35S rRNA (7,22–24). In addition, previous studies reported
that PNO1 was associated with the immune response and proteasome
activities. These aforementioned findings indicated the potential
crucial role of PNO1 in the physiological state. Furthermore, PNO1
has also been also identified as an oncogene in hepatocellular
carcinoma and colorectal cancer (5,7). However,
to the best of our knowledge, the role of PNO1 in esophageal cancer
(EC) remains unclear.
To investigate the role of PNO1 in EC, the present
study analyzed the expression levels of PNO1 in EC tissues using
data from TCGA database. Subsequently, the biological effects of
PNO1 in EC were determined. Finally, potential downstream targets
of PNO1 in Eca-109 cells were identified.
Shen et al analyzed microarray data performed
with 14 pairs of colorectal cancer and corresponding tissues and
preliminary identified PNO1, NUF2 component of NDC80 kinetochore
complex, cell division cycle associated 5 and dachshund family
transcription factor 1 as oncogenes in colorectal cancer (7). In addition, the study reported that PNO1
was a negative factor for predicting the overall survival of
patients with colorectal cancer. The present study analyzed PNO1
expression levels in EC tissues from TCGA and GTEx databases and
verified its expression in EC cells. The data demonstrated that
PNO1 expression levels were upregulated in EC tissues compared with
normal tissues. The differential expression of PNO1 indicated that
PNO1 may play a role in EC progression. To verify this hypothesis,
PNO1 expression was knocked down in Eca-109 and TE-1 cells, and the
results revealed that the cell proliferation, migration and
invasion abilities decreased, while the cell apoptosis ability was
increased following the KD. In addition, in nude mice, a smaller
tumor volume was observed following the KD of PNO1 expression.
These results indicated that PNO1 may promote EC progression.
Previous studies have also identified PNO1 as a tumor-promoting
factor in hepatocellular carcinoma and colorectal cancer. For
example, Dai et al reported that the growth and metastasis
of hepatocellular carcinoma was inhibited following the silencing
of PNO1 (6). Wang et al and
Shen et al knocked down PNO1 expression in colorectal cell
lines (PKO and HCT116) and found that the cell viability and colony
formation rate were decreased and that the percentage of cells in
the G0/G1 phase and undergoing apoptosis
increased (5,7). These findings suggested that PNO1 may be
an oncogene in EC.
As described above, PNO1 was identified as an
oncogene in EC. Thus, the mechanism underlying the effects of PNO1
in EC was determined. Through GSEA, gene sets related to the cell
cycle and DNA replication were found to be upregulated in the PNO1
high expression phenotype. These results indicated that PNO1 may
promote EC growth via regulating genes related to the cell cycle
and DNA replication. To verify this hypothesis, the expression
levels of molecules related to the cell cycle were analyzed in
Eca-109 cells with or without PNO1 KD. The NF-κB and Wnt signaling
pathways are involved in tumor proliferation, and NF-κB and CTNNB1
are key genes in the NF-κB and Wnt signaling pathways, respectively
(25,26). Thus, the expression levels of NF-κB
and CTNNB1 were analyzed following the KD of PNO1. The results
revealed that the expression levels of both NF-κB and CTNNB1 were
downregulated in the sh-PNO1-KD-transfected Eca-109 cells. In
addition, the OE of CTNNB1 in sh-PNO1-KD-transfected Eca-109 cells
reversed the decreased proliferation of Eca-109 cells. However,
knockdown of PNO1 failed to change the level of phosphorylated
CTNNB1. Two reasons may cause this phenomenon: Firstly, the level
of phosphorylation is a process of modification of proteins after
translation, and the level of total protein also represent its
level before translation. So, CTNNB1 may be phosphorylated after
translation. Secondly, protein level is regulated by upstream
molecules, while phosphorylated protein is affected by kinases.
Thus, it is possible that PNO1 affected the expression of some
kinases and then the kinases regulated the phosphorylation of
CTNNB1. Although, the level of phosphorylated CTNNB1 did not
change, we can still draw a conclusion that CTNNB1 is a downstream
target of PNO1 in EC.
PNO1 was observed to promote EC metastasis in
previous studies, and the expression levels of Twist, Myc and MMP2,
which are genes known to participate in the tumor metastasis
process (27–29), were also analyzed following the KD of
PNO1. The results suggested that PNO1 may promote EC metastasis via
upregulating Twist, Myc and MMP2 expression levels. Altogether
these data suggest that PNO1 may promote EC progression via
upregulating NF-κB p65, CTNNB1, Twist, Myc and MMP2 expression. Our
study was consistent with previous studies (27–29).
There were limitations to the present research.
Experiments to determine the direct association between PNO1 and
CTNNB1 were not performed. Also, we failed to detect the expression
of PNO1 in fresh samples to verify the results obtained from public
databases. Finally, we failed to detect the transfection efficiency
of PNO1 in vivo.
In conclusion, the findings of the present study
suggested that PNO1 may promote EC progression by regulating the
expression of AKT1, Twist, Myc, mTOR, MMP2, NF-κB p65 and
CTNNB1.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The Natural Science Key Projects of Bengbu Medical
College (no. BYKY2019087ZD) funded this research.
Availability of data and materials
The datasets used during the present study are
available from the corresponding author upon reasonable
request.
Authors' contributions
TT and GW both made substantial contributions to the
conception and design of this study. GW, QL, CL, GD, HS, HD, YY and
CM made substantial contributions to the analysis and
interpretation of the data. TT and GW both made substantial
contributions to writing the manuscript. All authors approved the
final version to be published and are accountable for all aspects
of the work.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of the First Affiliated Hospital of Bengbu Medical
College (Bengbu, Anhui, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Domper Arnal MJ, Ferrández Arenas Á and
Lanas Arbeloa Á: Esophageal cancer: Risk factors, screening and
endoscopic treatment in western and eastern countries. World J
Gastroenterol. 21:7933–7943. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Udagawa H and Akiyama H: Surgical
treatment of esophageal cancer: Tokyo experience of the three-field
technique. Dis Esophagus. 14:110–114. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Gibson TJ, Thompson JD and Heringa J: The
KH domain occurs in a diverse set of RNA-binding proteins that
include the antiterminator NusA and is probably involved in binding
to nucleic acid. FEBS Lett. 324:361–366. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhou GJ, Zhang Y, Wang J, Guo JH, Ni J,
Zhong ZM, Wang LQ, Dang YJ, Dai JF and Yu L: Cloning and
characterization of a novel human RNA binding protein gene PNO1.
DNA Seq. 15:219–224. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wang X, Wu T, Hu Y, Marcinkiewicz M, Qi S,
Valderrama-Carvajal H, Luo H and Wu J: Pno1 tissue-specific
expression and its functions related to the immune responses and
proteasome activities. PLoS One. 7:e460932012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Dai H, Zhang S, Ma R and Pan L: Celecoxib
inhibits hepatocellular carcinoma cell growth and migration by
targeting PNO1. Med Sci Monit. 25:7351–7360. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Shen A, Chen Y, Liu L, Huang Y, Chen H, Qi
F, Lin J, Shen Z, Wu X, Wu M, et al: EBF1-mediated upregulation of
ribosome assembly factor PNO1 contributes to cancer progression by
negatively regulating the p53 signaling pathway. Cancer Res.
79:2257–2270. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Gao L, Li X, Nie X, Guo Q, Liu Q, Qi Y,
Liu J and Lin B: Integrated analysis of lymphocyte
infiltration-associated lncRNA for ovarian cancer via TCGA, GTEx
and GEO datasets. PeerJ. 8:e89612020. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhang X, Wen X, Feng N, Chen A, Yao S,
Ding X and Zhang L: Increased expression of T-box transcription
factor protein 21 (TBX21) in skin cutaneous melanoma predicts
better prognosis: A study based on the cancer genome atlas (TCGA)
and genotype-tissue expression (GTEx) databases. Med Sci Monit.
26:e9230872020. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW,
Shi W and Smyth GK: limma powers differential expression analyses
for RNA-sequencing and microarray studies. Nucleic Acids Res.
43:e472015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Fang KP, Dai W, Ren YH, Xu YC, Zhang SM
and Qian YB: Both Talin-1 and Talin-2 correlate with malignancy
potential of the human hepatocellular carcinoma MHCC-97 L cell. BMC
Cancer. 16:452016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Vinci M, Gowan S, Boxall F, Patterson L,
Zimmerman M, Court W, Lomas C, Mendiola M, Hardisson D and Eccles
SA: Advances in establishment and analysis of three-dimensional
tumor spheroid-based functional assays for target validation and
drug evaluation. BMC Biol. 10:292012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sun R, Wu J, Chen Y, Lu M, Zhang S, Lu D
and Li Y: Down regulation of Thrombospondin2 predicts poor
prognosis in patients with gastric cancer. Mol Cancer. 13:2252014.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Moodley S, Koorbanally NA, Moodley T,
Ramjugernath D and Pillay M: The
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT)
assay is a rapid, cheap, screening test for the in vitro
anti-tuberculous activity of chalcones. J Microbiol Methods.
104:72–78. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Fadok VA, Voelker DR, Campbell PA, Cohen
JJ, Bratton DL and Henson PM: Exposure of phosphatidylserine on the
surface of apoptotic lymphocytes triggers specific recognition and
removal by macrophages. J Immunol. 148:2207–2216. 1992.PubMed/NCBI
|
|
17
|
Arsic N, Bendris N, Peter M, Begon-Pescia
C, Rebouissou C, Gadéa G, Bouquier N, Bibeau F, Lemmers B and
Blanchard JM: A novel function for cyclin A2: Control of cell
invasion via RhoA signaling. J Cell Biol. 196:147–162. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Song Y, Dong MM and Yang HF: Effects of
RNA interference targeting four different genes on the growth and
proliferation of nasopharyngeal carcinoma CNE-2Z cells. Cancer Gene
Ther. 18:297–304. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zengel P, Ramp D, Mack B, Zahler S,
Berghaus A, Muehlenweg B, Gires O and Schmitz S: Multimodal therapy
for synergic inhibition of tumour cell invasion and tumour-induced
angiogenesis. BMC Cancer. 10:922010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chatr-Aryamontri A, Oughtred R, Boucher L,
Rust J, Chang C, Kolas NK, O'Donnell L, Oster S, Theesfeld C,
Sellam A, et al: The BioGRID interaction database: 2017 update.
Nucleic Acids Res. 45D:D369–D379. 2017. View Article : Google Scholar
|
|
21
|
Subramanian A, Tamayo P, Mootha VK,
Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub
TR, Lander ES and Mesirov JP: Gene set enrichment analysis: A
knowledge-based approach for interpreting genome-wide expression
profiles. Proc Natl Acad Sci USA. 102:15545–15550. 2015. View Article : Google Scholar
|
|
22
|
Senapin S, Clark-Walker GD, Chen XJ,
Séraphin B and Daugeron MC: RRP20, a component of the 90S
preribosome, is required for pre-18S rRNA processing in
Saccharomyces cerevisiae. Nucleic Acids Res. 31:2524–2533. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Tone Y and Toh-EA: Nob1p is required for
biogenesis of the 26S proteasome and degraded upon its maturation
in Saccharomyces cerevisiae. Genes Dev. 16:3142–3157. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Vanrobays E, Leplus A, Osheim YN, Beyer
AL, Wacheul L and Lafontaine DL: TOR regulates the subcellular
distribution of DIM2, a KH domain protein required for
cotranscriptional ribosome assembly and pre-40S ribosome export.
RNA. 14:2061–2073. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Dolcet X, Llobet D, Pallares J and
Matias-Guiu X: NF-κB in development and progression of human
cancer. Virchows Arch. 446:475–482. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Tanabe S, Aoyagi K, Yokozaki H and Sasaki
H: Regulation of CTNNB1 signaling in gastric cancer and stem cells.
World J Gastrointest Oncol. 8:592–598. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Khan MA, Chen HC, Zhang D and Fu J: Twist:
A molecular target in cancer therapeutics. Tumour Biol.
34:2497–2506. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wang XN, Su XX, Cheng SQ, Sun ZY, Huang ZS
and Qu TM: MYC modulators in cancer: A patent review. Expert Opin
Ther Pat. 29:353–367. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wang HL, Zhou PY, Zhang Y and Liu P:
Relationships between abnormal MMP2 expression and prognosis in
gastric cancer: A meta-analysis of cohort studies. Cancer Biother
Radiopharm. 29:166–172. 2014. View Article : Google Scholar : PubMed/NCBI
|