Introduction
Endometrial carcinoma (EC) is the sixth most common
malignancy of the female genital tract worldwide, with 380,000 new
cases in 2018 (1–3). Despite improvements in EC diagnosis and
treatment, the prognosis and survival rate of patients with EC is
still unsatisfactory due to invasion and recurrence, with
>89,000 deaths in 2018 (2,3). Therefore, it is necessary to discover
the underlying mechanism of the tumorigenesis of EC to develop
novel biomarkers and improve therapy for patients with EC.
Long non-coding (lnc)RNAs are a group of ncRNAs
>200 nucleotides in length that affect key biological processes,
including transcriptional regulation, protein translation and
degradation (4,5). Novel lncRNAs have been discovered and
identified as crucial regulators involved in neoplastic
tumorigenesis and development, such as tumor promoter lncRNA Xist,
lncRNA HOTAIR and tumor suppressor lncRNA MEG3, TDRG1 (6–10),
offering the possibility of lncRNAs as effective biomarkers and
therapeutic targets for cancer. However, the association between
the deregulation of lncRNAs and carcinogenesis remains unknown
(5,11). Thus, identification of new lncRNAs not
only provides insight into their functional roles but may also
elucidate the potential mechanism of lncRNAs in human cancer,
including EC.
Recently, LINC01224, a cancer-associated lncRNA, has
been identified to be involved in carcinogenesis and to promote
tumor proliferation, invasion, migration and apoptosis; its
upregulation is associated with tumor grade and poor prognosis in
epithelial ovarian cancer and hepatocellular carcinoma (12,13).
However, the role and underlying molecular mechanism of LINC01224
in EC remains unclear. MicroRNAs (miRNAs/miRs) are short
non-protein-coding RNAs ~22 nucleotides in length that inhibit gene
expression by targeting the 3′-untranslated region (UTR) of mRNA.
Accumulating evidence has demonstrated that miRNAs serve crucial
roles in multiple biological processes, including cell growth,
differentiation and apoptosis (14,15).
lncRNAs serve as competing endogenous RNAs by sponging miRNAs, thus
restoring miRNA-induced functions of target genes (6,16).
Moreover, it has been reported that LINC01224 promotes epithelial
ovarian cancer progression via sponging certain miRNAs and
LINC01224 inhibits hepatocellular carcinoma progression via
sponging miR-330-5p (12,13). However, whether LINC01224 interacts
with miRNA in EC remains to be determined. Therefore, discovering
the novel lncRNA-miRNA axis in EC is urgent for the identification
of promising diagnostic biomarkers and therapeutic targets for EC
treatment.
LINC01224 expression levels in EC tumor tissue and
cell lines, and its association with survival of patients with EC,
were assessed. The phenotypes of LINC01244 knockdown both in
vitro and in vivo were also determined. The present
study aimed to investigate the functional mechanism and clinical
significance of LINC01224 in EC.
Materials and methods
Clinical patient samples
A total of 50 pairs of EC and adjacent (distance, ≥3
cm) para-carcinoma tissue samples were obtained from the Affiliated
Yixing Hospital of Jiangsu University (Yixing, China). A total of
50 female patients (age, 42–68 years) were enrolled from January
2017 to January 2019. Patients were diagnosed with primary EC by
two pathologists and had not received any therapy before sample
collection. The tissues were instantly placed into liquid nitrogen
to protect RNA integrity and stored at −80°C for further use. All
patient sample collections and experiments were approved by the
ethics committee of the Affiliated Yixing Hospital of Jiangsu
University and performed in accordance with the guidelines of the
Declaration of Helsinki. Written informed consent was obtained from
all patients.
Cell lines
The human endometrial stromal cell (ESC) line,
CL0453, was provided by Cell Bank of Chinese Academy of Sciences
(cat. no. BNCC267006; Shanghai, China). Three EC cell lines (HEC1A,
HEC1B and Ishikawa) and 293T cells were purchased from American
Type Culture Collection (ATCC). Human ESC cells were cultured
according to the protocol of the Cell Bank of Chinese Academy of
Sciences [1:1 DMEM and Ham's F-12 medium (both Gibco; Thermo Fisher
Scientific, Inc.) containing 1.2 g/l sodium bicarbonate, 2.5 mM
L-glutamine, 15 mM HEPES and 0.5 mM sodium pyruvate supplemented
with 2.0 mM L-Alanyl-L-Glutamine, 0.1 mM non-essential amino acids,
0.1 mM 2-mercaptoethanol and 4 ng/ml basic fibroblastic growth
factor; 5% knockout serum replacement (Gibco; Thermo Fisher
Scientific, Inc.) and 15% fetal bovine serum]; HEC1A cell line was
cultured in McCoy's 5A medium (Gibco; Thermo Fisher Scientific,
Inc.); 293T, HEC1B and Ishikawa cell lines were cultured in DMEM
(Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% (v/v)
FBS (Gibco; Thermo Fisher Scientific, Inc.). All cell lines were
cultured in a humidified 37°C incubator with 5% CO2.
Cell infection
To knock down the expression of LINC01244 in EC
cells, a lentiviral short hairpin (sh)RNA vector targeting
shLINC01244 was generated by inserting double-stranded
oligonucleotides (shLINC01244#1, 5′-GGTTGTTGCTTCCTAGTCTGG-3′;
shLINC01244#2, 5′-GCTTCCTAGTCTGGTGGTGAA-3′) into pLKO-puro Vector
(Thermo Fisher Scientific, Inc.) plasmids (pLKO.1-shRNA, 1 µg;
pCMV-ΔR8.2, 1 µg; pM2D.G, 0.5 µg/per well in 6-well plates).
Specific LINC01244 shRNA lentivirus was infected with HEC1A and
Ishikawa cell lines for 48 h (Shanghai GenePharma Co., Ltd.). A
total of 1×105 HEC1A and Ishikawa cells were seeded in
6-well plates and incubated for 24 h at 37°C and 5% CO2.
For the negative control (con) group, cells were added with
lentivirus containing scramble shRNA (sh-con); for the LINC01244
knockdown group, cells were added with lentivirus containing
shLINC01244#1 and shLINC01244#2. The stably infected cells were
screened using puromycin (2 µg/ml) for 10 days.
Reverse transcription-quantitative
(RT-q)PCR
Total RNA from HEC1A, HEC1B and Ishikawa cells was
extracted following transfection using a miRNeasy Mini kit (cat.
no. 217004; Qiagen GmbH). The first-strand cDNA was reverse
transcribed using PrimeScript™ RT Master Mix (cat. no. RR036Q,
Takara Bio, Inc.) according to manufacturer's protocol. RT-qPCR was
performed using SYBR Green SuperMix (Roche Diagnostics) following
the manufacturer's protocols. GAPDH and U6 were used as the
internal control for lncRNA/mRNA and miRNAs, respectively. Each
sample was run in triplicate using samples from independent
experiments. The relative mRNA expression levels were expressed as
a function of threshold cycle (Cq) and analyzed by the
2−ΔΔCq method (17).
The primer sequences were as follows: LINC01224
forward, 5′-AGAGCTTGGGATCGCTTTCTG-3′ and reverse,
5′-TTACTCAGGTGCCTTTCCCAC-3′; miR-485-5p forward,
5′-AGAGGCTGGCCGTGAT-3′ and reverse, 5′-ATGTGTTGCTGTGTTTGTCG-3′;
GAPDH forward, 5′-TATCGTGATGCTAGTCCGATG-3′ and reverse,
5′-TGCAGCTAGCTGCATCGATCGG-3′ and U6 forward,
5′-CTCGCTTCGGCAGCACA-3′ and reverse, 5′-AACGCTTCACGAATTTGCGT-3′.
The thermocycling conditions were as follows: 95°C for 15 min,
followed by 95°C for 30 sec, 65°C for 30 sec and 72°C for 30 sec
(35–45 cycles) and 72°C for 5 min.
Cell counting kit (CCK)-8 assay
Cell proliferation was evaluated by CCK-8 assay
(Dojindo Molecular Technologies, Inc.) following the manufacturer's
protocol. The transfected HEC1A and Ishikawa cells were seeded at a
density of 2×103/well in 96-well plates. Cells were
cultured for 24, 48, 72 or 96 h at 37°C with 5% CO2.
Then, 10 µl CCK-8 solution was added to each well and incubated for
1 h at 37°C. The absorbance was measured at 450 nm with a
microplate reader (Synergy H4 Hybrid Reader; BioTek Instruments,
Inc.). Data are presented as the mean ± SD of three independent
experiments.
Colony formation
A total of 1,000-1,500 cells were seeded into 6-well
plates. After culturing for 14 days in a humidified 37°C incubator
with 5% CO2, the colonies were fixed with 10%
formaldehyde for 30 min at room temperature and then stained with
0.5% crystal violet (Beyotime Institute of Biotechnology) for 30
min at room temperature. The colonies were photographed under a
light microscope at ×20 magnification (Olympus Corporation).
Apoptosis assay
Transfected HEC1A and Ishikawa cells were seeded in
a 6-well plate (1×106 cells/well). Following
transfection, cells were harvested by centrifugation at 200 × g for
5 min at room temperature and washed with 1X PBS three times, then
incubated with 5 µl FITC-conjugated Annexin V and 5 µl PI (25
µg/ml) (both BD Biosciences) for 15 min at room temperature in the
dark. The stained cells were detected using a BD FACSAria II flow
cytometer (BD Biosciences). FlowJo V10.5.2 (BD Biosciences)
software was used to analyze data.
Western blot analysis
Transfected HEC1A and Ishikawa cells were lysed in
RIPA buffer (Beyotime Institute of Biotechnology). Total protein
concentration was qualified with a BCA Protein Assay kit (Sangon
Biotech Co., Ltd.). Next, protein samples (~1,000 ng/lane) were
separated by 12% SDS-PAGE and transferred to PVDF membranes
(MilliporeSigma). After blocking with 5% non-fat milk for 1 h at
room temperature, the membranes were incubated with primary
antibodies [cleaved caspase-3, 1:500, cat. no. ab32042; Bcl-2,
1:1,000, cat. no. ab32124; proliferating cell nuclear antigen
(PCNA), 1:1,000, cat. no. ab29; AKT3, 1:2,000, cat. no. ab152157;
β-actin, 1:5,000, cat. no. ab6276] at 4°C overnight. Anti-rabbit
(cat. no. ab150077, for cleaved caspase-3, Bcl-2, AKT3, β-actin)
and anti-mouse IgG (cat. no. ab150113, 1:10,000, for PCNA) were
used as the secondary antibody at room temperature for 2 h. All the
antibodies was purchased from Abcam (Shanghai, China). ECL kit
(Sangon Biotech Co., Ltd.) was used for chemiluminescent detection
of immobilized proteins according to the manufacturer' protocol.
All antibodies were obtained from Abcam. The protein bands were
quantified with ImageJ V1.8.0 software (National Institutes of
Health).
Tumor xenograft experiments
A total of 12 female BALB/c nude mice (age, 4 weeks;
weight, 13–15 g; Shanghai Model Organisms Center, Inc.) were used
for xenograft experiments. All mice were housed in 25°C, 50%
humidity and specific-pathogen-free conditions with a 12/12-h
light/dark cycle. Sterile food and water were provided daily. All
animal protocols were approved by the Institutional Animal Care and
Use Committee at the Affiliated Yixing Hospital of Jiangsu
University. Briefly, lentivirus-transfected HEC1A cells
(1×106) were resuspended in 100 µl DMEM (Gibco; Thermo
Fisher Scientific, Inc.) without FBS and subcutaneously injected
into the flank of mice. After ~1 week, the tumor was visible (100
mm3) and injected with sh-con, shLINC01224#1 and
shLINC01224#2 (10 nmol/20 g) twice per week (n=6/group). Tumor
volumes were measured every 7 days. Tumor volume was calculated as
follows: Tumor volume (mm3) = (height) ×
(width)2/2. After 28 days, mice were sacrificed for
analysis and tumor weight was measured. An intraperitoneal
injection of 150 mg/kg pentobarbital was used for euthanasia. The
dose of pentobarbital used for anesthesia was 10% in saline, 50
mg/kg, via intraperitoneal injection. Cessation of movement,
breathing, and heartbeat were considered to indicate death. Mouse
death was verified by cutting the chest cavity to confirm no
heartbeat and touching the eyeball, which produced no reflex
activity. Based on Institutional Animal Care and Use Committee
guidelines, tumor size >20 mm at the largest diameter was
considered to be the humane endpoint (18). After 28 days, the maximum tumor weight
observed was 7.8% of mouse body weight. The maximum tumor diameter
observed was 17.5 mm.
Luciferase reporter assay
The day before transfection, 293T cells were plated
in 24-well plates (4×104 cells/well). Full-length
LINC01224 gene and 3′-UTR of the AKT3 gene were cloned into pmirGLO
plasmids containing luciferase (Promega Corporation) according to
the manufacturer's protocol and transfected into 293T cells.
GeneArt™ Site-Directed Mutagenesis system (Thermo Fisher
Scientific, Inc.) was used to produce the mutant (MUT) LINC01224
and AKT3 gene reporter. LINC01224 wild-type (WT)/MUT or AKT3 WT/MUT
plasmids were co-transfected with miR-negative control (NC) or
miR-485-5p mimics into 293T cells using Lipofectamine®
2000 (Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's instructions for 48 h. Following transfection for 48
h, luciferase activity was measured immediately by Dual-Luciferase
Reporter Assay system (Promega Corporation) following the
manufacturer's protocol, and firefly luciferase activity was
normalized against Renilla luciferase activity.
Bioinformatics analysis
Gene Expression Profiling Interactive Analysis
(GEPIA2; gepia2.cancer-pku.cn/#index) was used to analyze LINC01224
expression levels in EC. StarBase 3.0 (starbase.sysu.edu.cn/) was used to predict the
interaction between LINC01224 and miRNAs and the targets of
miR-485-5p.
RNA pull-down assay
LINC01224 and NC sequence (nonsense sequence) were
biotin-labeled using Biotin RNA Labeling Mix and T7/SP6 RNA
polymerase (Roche Diagnostics) to create bio-LINC01224 and bio-NC
by Shanghai GenePharma Co., Ltd. by using pcDNA 3.1 (GenePharma
Co., Ltd., China) with T7 promoter. according to the manufacturer's
protocol. Following transfection, HEC1A and Ishikawa cell lines
were lysed by lysis buffer (Invitrogen; Thermo Fisher Scientific,
Inc.) according to the manufacturer's protocol and incubated with
biotinylated RNAs for 48 h at room temperature under moderate
agitation on a tube rotator. Next, 200 µl lysed cells were
collected and incubated with 400 µl Streptavidin agarose beads
(Invitrogen; Thermo Fisher Scientific, Inc.) for 1 h at 37°C under
moderate agitation on a tube rotator at room temperature. After
beads were washed with wash buffer (Invitrogen; Thermo Fisher
Scientific, Inc.), the bound RNAs were analyzed by RT-qPCR
assay.
Statistical analysis
Data are presented as the mean ± SD of three
independent biological experiments. Statistical analysis was
performed using Microsoft Office Excel 2016 (Microsoft
Corporation). Significance of differences between two groups was
calculated with Student's unpaired t-test. One-way ANOVA followed
by Tukey's post hoc test was used to determine the significance
among multiple groups. Overall survival was calculated by
Kaplan-Meier and log-rank test. Pearson's correlation analysis was
performed to assess correlation between RNA expression levels.
P<0.05 was considered to indicate a statistically significant
difference.
Results
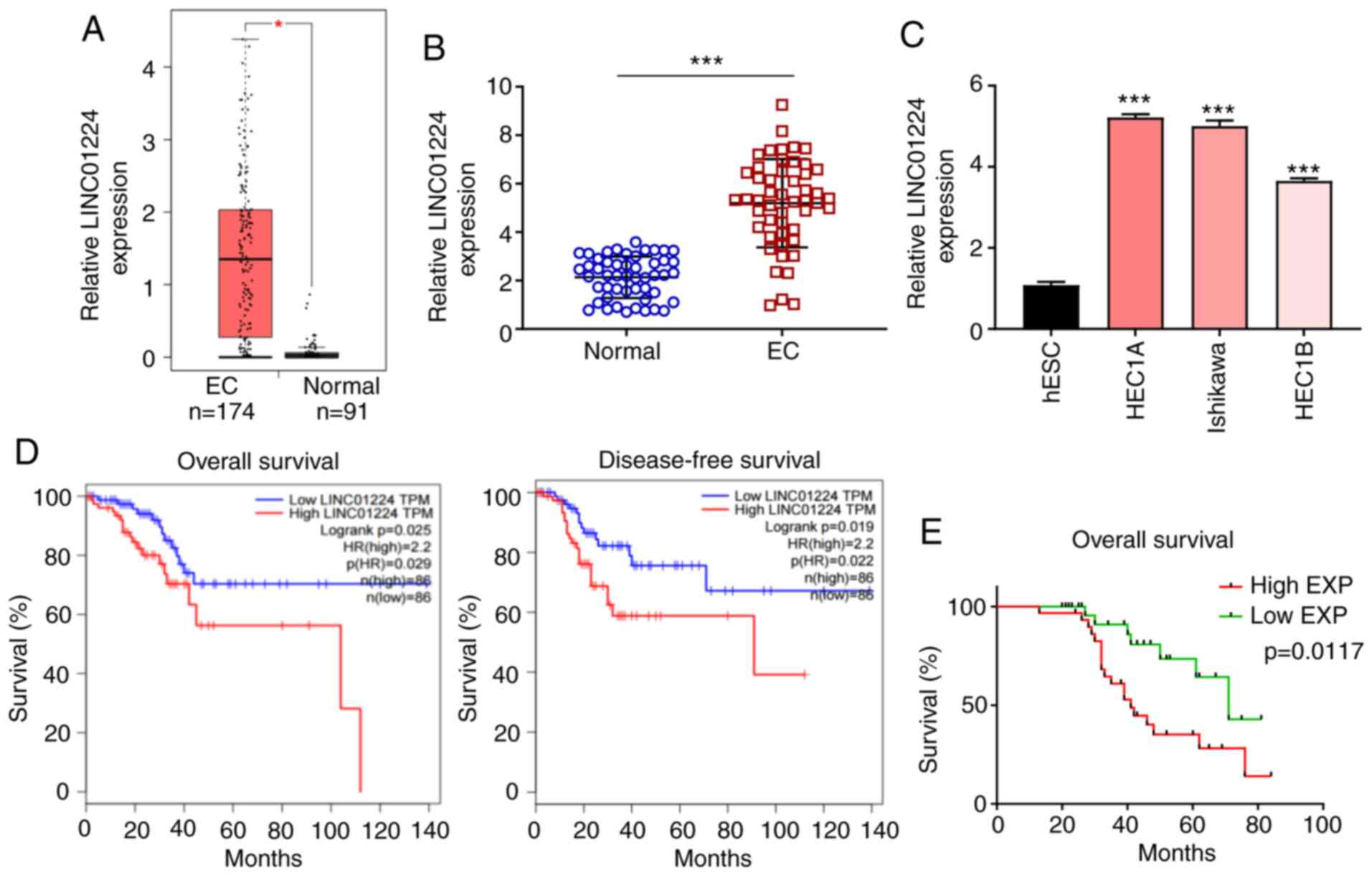
LINC01224 is significantly increased
in EC and associated with poor survival rate of patients with
EC
RNA sequencing data of LINC01224 expression levels
from The Cancer Genome Atlas (TCGA) and the Genotype Tissue
Expression (GTEx) projects (gepia.cancer-pku.cn/) was used to
evaluate LINC01224 expression in EC using GEPIA2. The data showed
that LINC01224 expression was higher in EC tissue (n=174) than in
normal endometrial tissue (n=91; Fig.
1A). LINC01224 expression was assessed in 50 pairs of EC
clinical samples by RT-qPCR. LINC01224 expression was significantly
upregulated in EC tissue compared with matched adjacent normal
tissue (Fig. 1B), which was
consistent with expression levels in three EC cell lines (HEC1A,
HEC1B and Ishikawa) compared with human normal ESCs (Fig. 1C). In the TCGA dataset, patients with
EC with high LINC01224 expression had shorter overall and
disease-free survival compared with patients with low LINC01224
expression (Fig. 1D). The median
LINC01224 expression in EC tissue (Fig.
1B) was selected as the cut-off value (4.813) to divide
patients into LINC01224 low-(n=25) and high-expression groups
(n=25). Kaplan-Meier curve of overall survival for patients with EC
demonstrated that high expression levels of LINC01224 was
associated with poor prognosis (Fig.
1E). These results indicated that LINC01224 exerted a
tumor-promoting effect on EC progression.
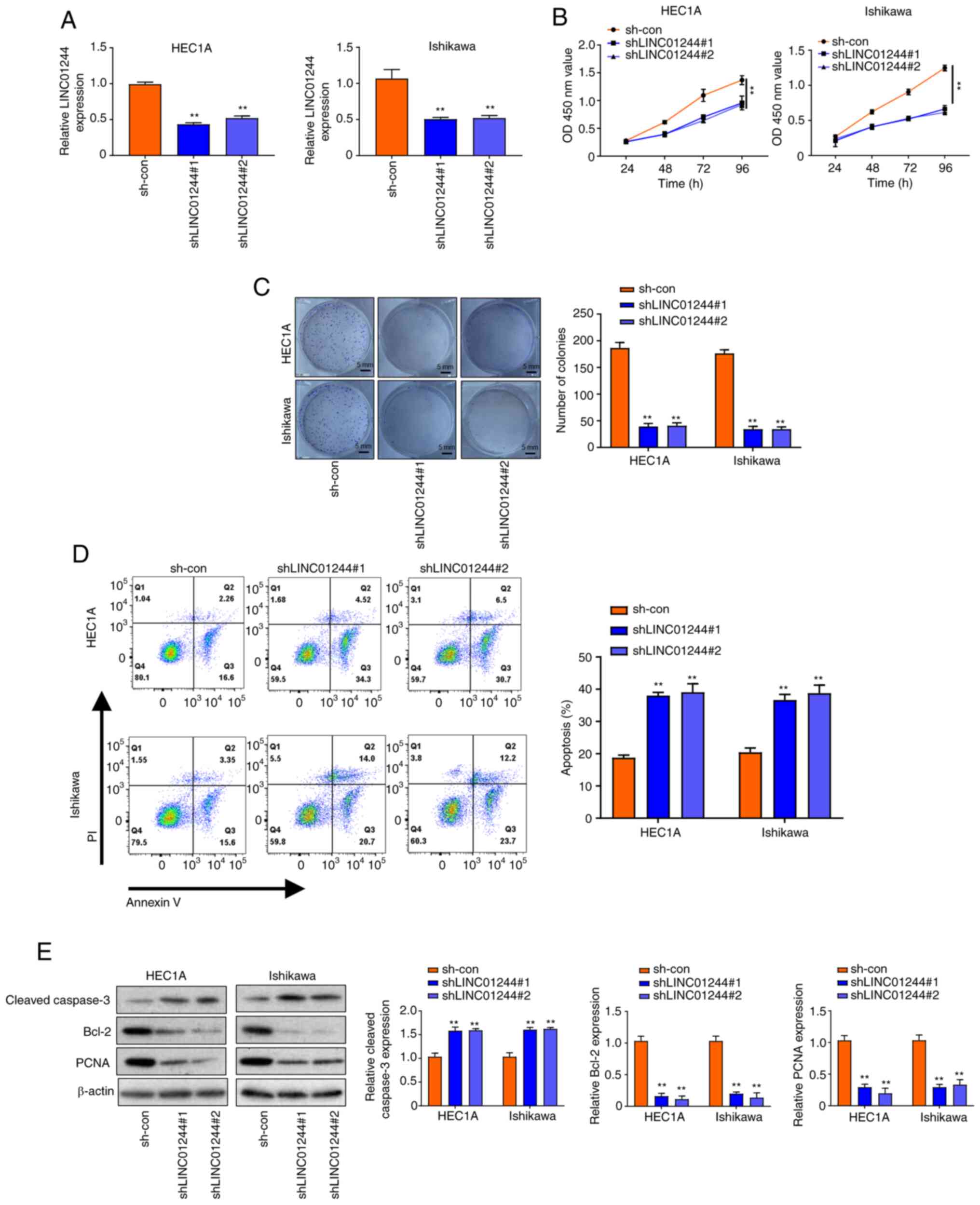
LINC01224 promotes proliferation and
inhibits apoptosis of EC cells
To investigate the functional role of LINC01224 in
EC, loss of function experiments were performed using HEC1A and
Ishikawa cell lines with high LINC01244 expression. Firstly,
LINC01224 was knocked down in HEC1A and Ishikawa cell lines using
shRNA (Fig. 2A). CCK-8 assay results
demonstrated that knockdown of LINC01224 (shLINC01224#1 and
shLINC01224#2) decreased proliferation of HEC1A and Ishikawa cells
in a time-dependent manner (Fig. 2B);
this was only significant at 96 h. Colony formation assay indicated
that LINC01224 knockdown decreased the number of colonies of HEC1A
and Ishikawa cells (Fig. 2C).
Moreover, flow cytometry assay showed that apoptosis of HEC1A and
Ishikawa cells transfected with shLINC01224 significantly increased
compared with the sh-con group (Fig.
2D). Western blotting results showed that knockdown of
LINC01224 increased the protein levels of cleaved caspase-3 but
decreased those of Bcl-2 and PCNA (Fig.
2E). Moreover, overexpression of LINC01224 promoted
proliferation and colony formation of HEC1A and Ishikawa cells
(Fig. S1A-C). To validate the in
vivo effect of LINC01224 in EC progression, a xenograft model
of EC was established by subcutaneously injecting
LINC01224-knockdown (shLINC01224#1 and shLINC01224#2) and sh-con
lentiviral-transfected stable HEC1A cells into mice (Fig. S1D). Tumor sizes were measured every 7
days. The results showed that knockdown of LINC01224 decreased both
tumor size and weight (Fig. S2A-C).
Consistent with cellular assay results, LINC01224 downregulation
decreased Bcl-2 and PCNA expression and increased cleaved caspase-3
expression in the HEC1A xenograft tumors (Fig. S2A-D). Collectively, these data
suggested that knockdown of LINC01224 inhibited proliferation and
apoptosis of EC cells.
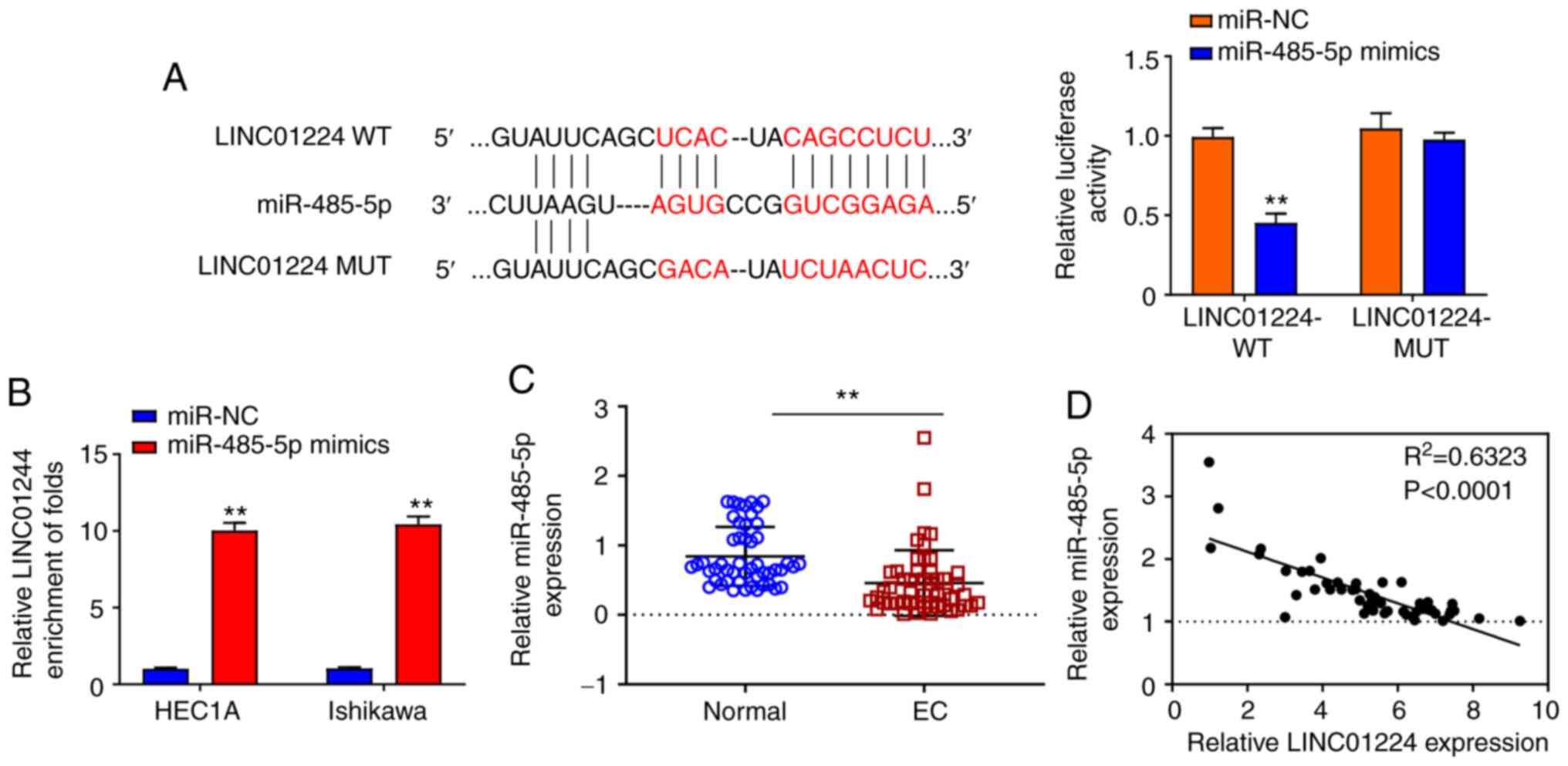
LINC01224 directly binds to and
negatively regulates expression of miR-485-5p
Accumulating evidence suggested that lncRNAs serve
as miRNA sponges to inhibit expression and regulate the biofunction
of miRNAs (6,16). Thus, the potential binding miRNA
partner of LINC01224 was analyzed using Starbase (data not shown).
Results showed that LINC01224 potentially bound to miR-485-5p.
Luciferase reporter assay demonstrated that overexpression of
miR-485-5p significantly repressed the luciferase activity of
LINC01224 WT but not LINC01224 MT (Fig.
3A). RNA pull-down assay indicated that the LINC01224
expression was more enriched on the miR-485-5p probe (Fig. 3B). In 50 pairs of samples from
patients with EC, miR-485-5p was significantly downregulated in EC
tumor tissue (Fig. 3C), and there was
a strong negative correlation between LINC01224 and miR-485-5p
(Fig. 3D; P<0.0001). These results
demonstrated that LINC01224 directly interacted with, and inhibited
expression of, miR-485-5p.
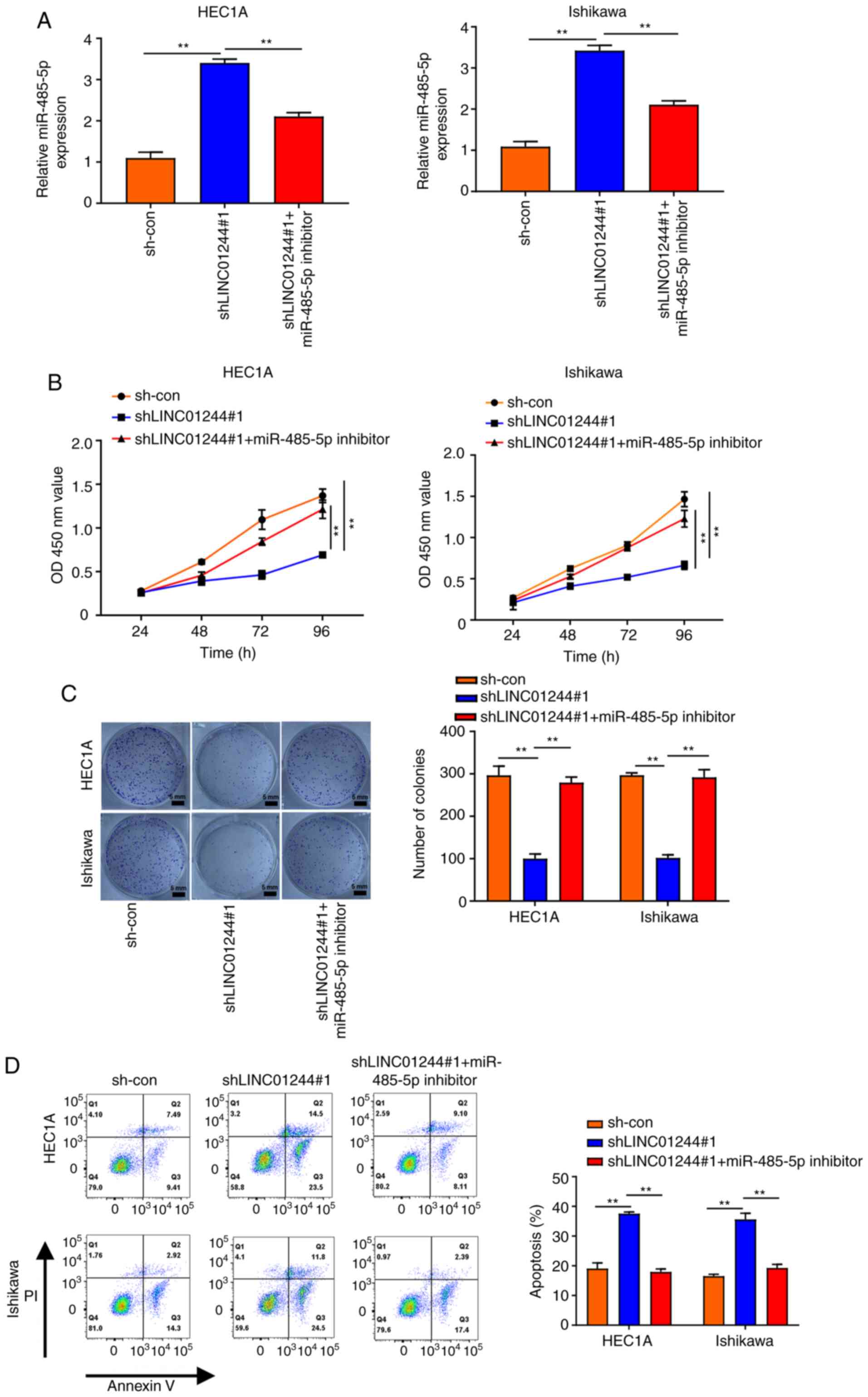
LINC01224 promotes proliferation and
inhibits apoptosis of EC cells via sponging miR-485-5p
To investigate the role of LINC01224 in EC
progression, LINC01224 was knocked down in HEC1A and Ishikawa cells
(Fig. 4A) and miR-485-5p expression
levels were assessed. Knockdown of LINC01224 significantly
increased the expression of miR-485-5p; this was partially rescued
by treatment with miR-485-5p inhibitor (Figs. 4A and S1F). Based on the aforementioned results,
it was hypothesized that the LINC01224/miR-485-5p axis is linked to
EC progression. CCK-8 and colony formation assays showed that
LINC01224 knockdown inhibited the proliferation of HEC1A and
Ishikawa cells but miR-485-5p inhibitor treatment partially
reversed this effect (Fig. 4B and C).
Flow cytometry assay displayed a similar result for EC cell
apoptosis (Fig. 4D), consistent with
western blotting results demonstrating that miR-485-5p inhibition
restored the increased protein levels of cleaved caspase-3 and
decreased protein levels of Bcl-2 and PCNA in EC cells transfected
with shLINC01224 (Fig. S3). Taken
together, these results demonstrated that LINC01224 promoted
proliferation and inhibited apoptosis of EC cells via sponging
miR-485-5p.
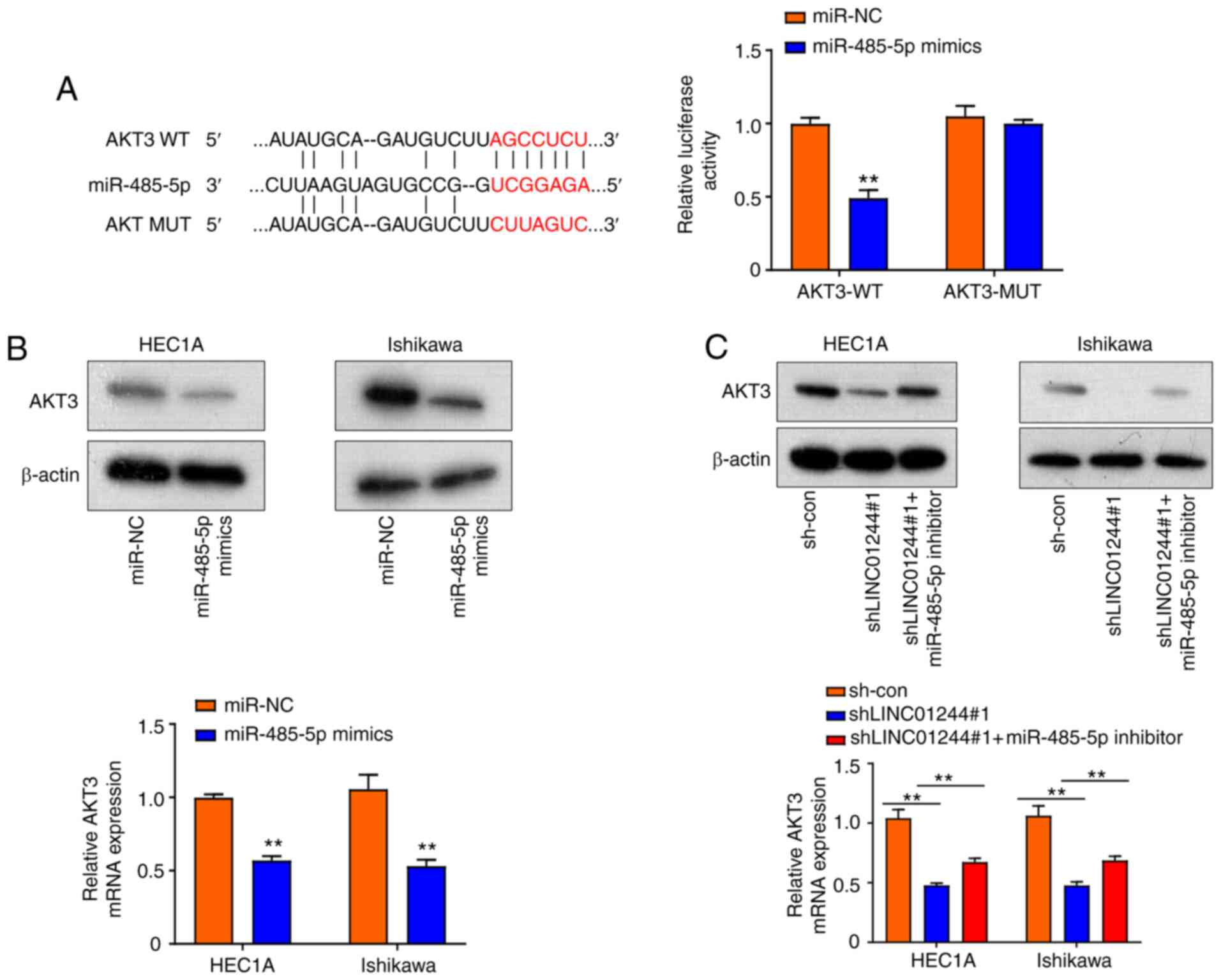
LINC01224 elevates AKT3 expression via
sponging miR-485-5p in EC cells
Previous research has demonstrated that AKT3 is a
potential target of miRNA (19,20). It
has been reported that lncRNAs serve as competing endogenous RNAs
that interact with miRNAs to regulate the expression of key
oncogenes (21), involved in
tumorigenesis (21). The present
study demonstrated that miR-485-5p bound to LINC01224. Starbase 3.0
predicted that miR-485-5p targets the 3′-UTR of the AKT3 gene
(Fig. 5A). Thus, AKT3 was selected
for subsequent analysis. Luciferase reporter assay demonstrated
that miR-485-5p mimics significantly inhibited luciferase activity
of AKT3 WT; this effect was diminished following mutation of the
predicted binding site for miR-485-5p (Fig. 5A), confirming the physical interaction
of AKT3 with miR-485-5p. Both the mRNA and protein levels of AKT3
in HEC1A and Ishikawa cells were downregulated by overexpression of
miR-485-5p mimics (Fig. 5B).
Furthermore, knockdown of LINC01244 significantly decreased AKT3
protein and mRNA levels, which were effectively restored by
miR-485-5p inhibitor (Fig. 5C).
Collectively, these results indicated that LINC01224 elevated AKT3
expression via sponging miR-485-5p in EC cells.
Discussion
EC is the most common cancer in women, but overall
clinical outcomes are still unsatisfactory due to the lack of
effective therapeutic targets (1–3).
Increasing evidence has demonstrated that lncRNAs function as
endogenous miRNA sponges by binding to miRNAs and regulating their
function, thus participating in tumor initiation and progression,
thus presenting potential therapeutic targets and prognostic
biomarkers for cancer (6,9,10,16,22). A
novel lncRNA, LINC01224, has been discovered to exhibit a
tumor-promoting effect in cancer (including hepatocellular
carcinoma and ovarian cancer) via miRNA-mediated target gene
expression (12,13). However, its clinical significance and
underlying functional role of LINC01224 in EC remain unclear. The
present study demonstrated that expression of LINC01224 was
significantly upregulated in EC tissue and cell lines; this was
associated with shorter survival time of patients with EC.
Knockdown of LINC01224 impaired EC cell proliferation but promoted
apoptosis. The nude mouse xenograft model of LINC01224 knockdown
further revealed that LINC01224 increased EC tumor growth in
vivo. These results suggested that LINC01224 functioned as an
oncogene to promote EC progression.
The involvement of miRNAs in cancer and their
significance as clinical biomarkers is increasingly appreciated
(14,23). miR-485-5p has been recently found to
be downregulated in cancer, including cholangiocarcinoma,
hepatocellular carcinoma and breast and lung cancer, and its low
expression is positively associated with risk and poor prognosis of
cancer, implying that miR-485-5p serves as a tumor suppressor
(24–28). AKT3 has been found to mediate
resistance to apoptosis in B-RAF-targeted melanoma cells (29). In the present study, bioinformatics
analysis (Starbase 3.0) predicted that miR-485-5p was the only
downstream miRNA target of LINC01224. RNA pull-down and luciferase
assays further verified their direct interaction in EC.
Additionally, AKT3 was shown to be a direct target of miR-485-5p in
EC cells. Functionally, LINC01224 inhibition significantly
suppressed EC cell proliferation but increased apoptosis via
sponging miR-485-5p; these effects were partially abrogated by
miR-485-5p inhibition. Collectively, the present study determined
the expression levels, clinical implication and functional
mechanism of LINC01224 in EC and revealed that LINC01224
facilitated EC progression via mediating the miR-485-5p/AKT3 axis,
thus offering a potential diagnostic biomarker and therapeutic
target for EC treatment.
In summary, LINC01224 served as an oncogenic lncRNA
to promote EC cell proliferation and inhibit apoptosis via sponging
miR-485-5p to elevate AKT3 expression levels, thus providing a
promising therapeutic target for EC treatment.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by the Affiliated
Yixing Hospital of Jiangsu University.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
XZ, WL, XY and HZ conceived and supervised the
project. XZ, WL, XY, TM, YR, MH, HY, HW and HZ performed the
biological experiments. XZ, WL, XY and HZ analyzed data and wrote
the manuscript. XY and HZ confirm the authenticity of all the raw
data. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the ethics
committee of the Affiliated Yixing Hospital of Jiangsu University
(approval no. 2017-00238). All patients provided written informed
consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
CCK-8
|
Cell Counting Kit-8
|
|
EC
|
Endometrial carcinoma
|
|
miRNA
|
microRNA
|
|
MT
|
mutant
|
|
Con
|
Control
|
|
RT-q
|
reverse transcription-quantitative
|
|
GTEx
|
Genotype Tissue Expression
|
|
NC
|
negative control
|
|
shRNA
|
short hairpin RNA
|
|
TCGA
|
The Cancer Genome Atlas
|
|
UTR
|
untranslated region
|
|
WT
|
wild-type
|
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
D'Andrilli G, Bovicelli A, Paggi MG and
Giordano A: New insights in endometrial carcinogenesis. J Cell
Physiol. 227:2842–2846. 2012. View Article : Google Scholar
|
|
3
|
Buhtoiarova TN, Brenner CA and Singh M:
Endometrial carcinoma: Role of current and emerging biomarkers in
resolving persistent clinical dilemmas. Am J Clin Pathol. 145:8–21.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lorenzi L, Avila Cobos F, Decock A,
Everaert C, Helsmoortel H, Lefever S, Verboom K, Volders PJ,
Speleman F, Vandesompele J and Mestdagh P: Long noncoding RNA
expression profiling in cancer: Challenges and opportunities. Genes
Chromosomes Cancer. 58:191–199. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Derrien T, Johnson R, Bussotti G, Tanzer
A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG,
et al: The GENCODE v7 catalog of human long noncoding RNAs:
Analysis of their gene structure, evolution, and expression. Genome
Res. 22:1775–1789. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Geisler S and Coller J: RNA in unexpected
places: Long non-coding RNA functions in diverse cellular contexts.
Nat Rev Mol Cell Biol. 14:699–712. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Khalil AM, Guttman M, Huarte M, Garber M,
Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van
Oudenaarden A, et al: Many human large intergenic noncoding RNAs
associate with chromatin-modifying complexes and affect gene
expression. Proc Natl Acad Sci USA. 106:11667–11672. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
DiStefano JK: The emerging role of long
noncoding RNAs in human disease. Disease Gene Identification:
Methods and Protocols. DiStefano JK: Springer; New York, NY: pp.
91–110. 2018, View Article : Google Scholar
|
|
9
|
Hu G, Niu F, Humburg BA, Liao K, Bendi S,
Callen S, Fox HS and Buch S: Molecular mechanisms of long noncoding
RNAs and their role in disease pathogenesis. Oncotarget.
9:18648–18663. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kondo Y, Shinjo K and Katsushima K: Long
non-coding RNAs as an epigenetic regulator in human cancers. Cancer
Sci. 108:1927–1933. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Haemmerle M and Gutschner T: Long
non-coding RNAs in cancer and development: Where do we go from
here? Int J Mol Sci. 16:1395–1405. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gong D, Feng PC, Ke XF, Kuang HL, Pan LL,
Ye Q and Wu JB: Silencing long non-coding RNA LINC01224 inhibits
hepatocellular carcinoma progression via microRNA-330-5p-induced
inhibition of CHEK1. Mol Ther Nucleic Acids. 19:482–497. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Xing S, Zhang Y and Zhang J: LINC01224
exhibits cancer-promoting activity in epithelial ovarian cancer
through microRNA-485-5p-mediated PAK4 upregulation. Onco Targets
Ther. 13:5643–5655. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Mishra S, Yadav T and Rani V: Exploring
miRNA based approaches in cancer diagnostics and therapeutics. Crit
Rev Oncol Hematol. 98:12–23. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hansen TB, Jensen TI, Clausen BH, Bramsen
JB, Finsen B, Damgaard CK and Kjems J: Natural RNA circles function
as efficient microRNA sponges. Nature. 495:384–388. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Smith CJ and Osborn AM: Advantages and
limitations of quantitative PCR (Q-PCR)-based approaches in
microbial ecology. FEMS Microbiol Ecol. 67:6–20. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Pritt SL and Smith TM: Institutional
animal care and use committee postapproval monitoring programs: A
proposed comprehensive classification scheme. J Am Assoc Lab Anim
Sci. 59:127–131. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li L and Ma L: Upregulation of miR-582-5p
regulates cell proliferation and apoptosis by targeting AKT3 in
human endometrial carcinoma. Saudi J Biol Sci. 25:965–970. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Fang Y, Liang X, Xu J and Cai X: miR-424
targets AKT3 and PSAT1 and has a tumor-suppressive role in human
colorectal cancer. Cancer Manag Res. 10:6537–6547. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wang Y, Hou J, He D, Sun M, Zhang P, Yu Y
and Chen Y: The emerging function and mechanism of ceRNAs in
cancer. Trends Genet. 32:211–224. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wang KC and Chang HY: Molecular mechanisms
of long noncoding RNAs. Mol Cell. 43:904–914. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Adams BD, Kasinski AL and Slack FJ:
Aberrant regulation and function of microRNAs in cancer. Curr Biol.
24:R762–R776. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Bao W, Cao F, Ni S, Yang J, Li H, Su Z and
Zhao B: lncRNA FLVCR1-AS1 regulates cell proliferation, migration
and invasion by sponging miR-485-5p in human cholangiocarcinoma.
Oncol Lett. 18:2240–2247. 2019.PubMed/NCBI
|
|
25
|
Tu J, Zhao Z, Xu M, Chen M, Weng Q and Ji
J: LINC00460 promotes hepatocellular carcinoma development through
sponging miR-485-5p to up-regulate PAK1. Biomed Pharmacother.
118:1092132019. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Peng Y, Leng W, Duan S and Hong M: Long
noncoding RNA BLACAT1 is overexpressed in hepatocellular carcinoma
and its downregulation suppressed cancer cell development through
endogenously competing against hsa-miR-485-5p. Biomed Pharmacother.
116:1090272019. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lu H, Wang C, Xue L, Zhang Q, Luh F, Wang
J, Lin TG, Yen Y and Liu X: Human mitotic centromere-associated
kinesin is targeted by MicroRNA 485-5p/181c and prognosticates poor
survivability of breast cancer. J Oncol. 2019:23162372019.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Gao F, Wu H, Wang R, Guo Y, Zhang Z, Wang
T, Zhang G, Liu C and Liu J: MicroRNA-485-5p suppresses the
proliferation, migration and invasion of small cell lung cancer
cells by targeting flotillin-2. Bioengineered. 10:1–12. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Shao Y and Aplin AE: Akt3-mediated
resistance to apoptosis in B-RAF-targeted melanoma cells. Cancer
Res. 70:6670–6681. 2010. View Article : Google Scholar : PubMed/NCBI
|