Introduction
Oesophageal carcinoma (EC) is the ninth most common
malignancy worldwide, and its mortality rate is the sixth highest
among all tumour types (1). In China,
EC incidence and mortality are especially high, accounting for ~50%
of worldwide morbidity and mortality rates (2). As the predominant form of EC in China,
oesophageal squamous cell carcinoma (ESCC) is generally associated
with a poor prognosis due to inadequate effective clinical
approaches for early detection (3).
With or without neoadjuvant therapy or adjuvant therapy, the
five-year survival rate for patients with ESCC undergoing surgery
is 25–45% (4), and the infiltration
and metastasis of ESCC are the primary causes of ESCC-related
deaths (5). Therefore, the
identification of molecular markers for predicting ESCC prognosis
is crucial.
Exosomes range between 30–100 nm in diameter, are
composed of a lipid bilayer, and are widely distributed in serum,
urine, plasma and malignant ascites (6). To sustain their survival and
reproduction, tumour-originated exosomes harbour oncogenic
biomolecules, including RNA, DNA and proteins, that maintain
internal cancer cell homeostasis. Numerous studies have documented
that exosomes may also be involved in tumour progression (7). Therefore, investigation of the
biological features of patients with ESCC-derived exosomes may be
of great importance for the early diagnosis of ESCC, monitoring of
treatment efficacy, and evaluation of their functions in tumour
progression and metastasis. Pyruvate kinase isoenzyme type M2
(PKM2) is a metabolic enzyme and signalling modulator in the
cytoplasm, and a transcriptional regulator in the nucleus (8). In our previous proteomics study,
screening for exosomal differential proteins revealed that PKM2 was
upregulated in patients with stage III compared with stage II ESCC.
In addition, several studies indicated that PKM2 was packaged into
exosomes (9–14). However, whether PKM2 can be assembled
into exosomes in ESCC, and the function of exosomal PKM2 in
patients with ESCC, remain unknown.
Therefore, the aim of the present study was to
investigate the protein level of plasma-derived exosomal PKM2. The
impacts of plasma-derived exosomes from ESCC patients on the
proliferation, migration and invasion abilities of oesophageal
squamous cell lines was also determined. To clarify the possible
mechanism of exosomal PKM2 in ESCC, the differences in PKM2 and
STAT3 mRNA expression between EC and non-malignant oesophageal
tissues were analysed using the Gene Expression Profiling
Interactive Analysis (GEPIA) database. In addition, the levels of
PKM2 and pSTAT3Tyr705 in ESCC clinical samples were
assessed and verified by immunohistochemistry.
Materials and methods
Patients
The present study was approved by the local ethics
committee of the Affiliated Tumor Hospital of Xinjiang Medical
University (approval no. K-2019054). From December 2013 to November
2014, patients with ESCC at various stages, who were hospitalized
in the Cancer Hospital Affiliated with Xinjiang Medical University,
were enrolled in the study. All subjects were treated by surgical
dissection. The study subjects had not received surgical resection,
chemotherapy or radiotherapy prior to recruitment. The TNM staging
system of the American Joint Commission on Cancer (8th edition,
published in 2017) was used for tumour staging (15). A cohort of 76 candidates [52 patients
with ESCC, 18 healthy volunteers and 6 patients with oesophageal
intraepithelial neoplasia (EIN)] who were seen in the Department of
Thoracic Surgery of the Cancer Hospital Affiliated with Xinjiang
Medical University (Urumqi, China) were recruited for ELISA.
Detailed information on all participants is summarized in Table SI and SII; two tissue microarrays
(HEso-Squ180Sur-01 and HEso-Squ180Sur-04; Shanghai Outdo Biotech
Co., Ltd.) including 95 ESCC and 85 matched non-malignant control
tissues, as well as a group of surgical specimens from 52 patients
with ESCC (administered from the Department of Thoracic Surgery of
the Cancer Hospital Affiliated with Xinjiang Medical University)
were immunostained for analysis of the protein level of PKM2 and
pSTAT3Tyr705.
Extraction of exosomes from
plasma
Blood samples (4 ml) were obtained from individuals
with ESCC and healthy controls prior to surgery, and were
centrifuged at 4,000 × g for 20 min to remove cells and debris. The
plasma was stored at −80°C until required. Isolation of exosomes
was performed using the exoEasy Maxi Kit (Qiagen GmbH), per the
manufacturers protocol. Specifically, the plasma was mixed with an
equivalent volume of XBP buffer and loaded onto exoEasy spin
columns. The mixture was centrifuged at 500 × g for 1 min at room
temperature (RT). The flow-through was discarded and 10 ml XWP
buffer was added to each column, followed by centrifugation at
5,000 × g (5 min at RT) to remove any residual buffer. After
washing with XWP, the columns were transferred into new collection
tubes. Buffer XE (1 ml) was added to the membrane and incubated for
1 min. The exosomes were collected following centrifugation at 500
and 5,000 × g (5 min each at 4°C).
Transmission electron microscopy
(TEM)
TEM was used to examine and photograph the exosomes.
First, the exosomes were isolated and coated onto a carbon grid as
previously described (16). Briefly,
exosome solution was dropped onto 100-mesh sample-loaded copper
mesh and fixed with 2% glutaraldehyde and 2% paraformaldehyde in
0.1 mol/l sodium cacodylate buffer at pH 7.3 for 3 h at RT.
Afterwards, the grids were air dried and the exosome morphology
photographed using a transmission electron microscope (Tecnai™ G2
spirit Bio-Twin; FEI; Thermo Fisher Scientific, Inc.) at an
acceleration voltage of 100 kV. Digital images were captured with a
charge-coupled device camera (Veleta; Olympus Soft Imaging
Solutions GmbH).
Exosome size and concentration
analysis
A volume of 5 µl exosomes was diluted in PBS to 30
µl. After the standard sample was tested, the exosome sample was
loaded. Information regarding the size and concentration of
exosomes was detected and analysed using Nano flow cytometry
(NanoFCM) (instrumentation, Flow NanoAnalyzer; NanoFCM) as
previously described (17).
NanoFCM analysis of exosomes-bound
beads
A volume of 10 µl exosomes was diluted to 0.2 µg/µl.
Then, 20 µl FITC mouse anti-human CD9 (cat. no. 555371; BD
Biosciences), FITC mouse anti-human CD81 (cat. no. 551108; BD
Biosciences) and FITC mouse IgG (cat. no. 400108, BioLegend, Inc.)
was added. The sample was incubated at 37°C for 30 min in the dark,
and rinsed twice with PBS by ultracentrifugation at 100,000 × g (2
h at 4°C). The pellet was subsequently resuspended in 100 µl PBS
for detection and analysis using NanoFCM. Instrumentation; Flow
NanoAnalyzer; NanoFCM (Flow NanoAnalyzer, Model No: N30E, S/N:
FNAN30E2007151114; Software:V1.08).
Cell culture
Eca109, KYSE30, KYSE150, TE-1 and KYSE510 cells were
purchased from Wuhan University, China, and maintained in RPMI-1640
medium (HyClone; Cytiva) enriched with 10% FBS (Shanghai VivaCell
Biosciences, Ltd.) and penicillin-streptomycin (HyClone; Cytiva) in
a humidified atmosphere containing 5% CO2 at 37°C.
Uptake of exosomes by Eca109
cells
To determine whether Eca109 cells could take up
exosomes from patients with ESCC, the PKH67 Green Fluorescent Cell
Linker Kit (Sigma-Aldrich; Merck KGaA) was used to label exosomes,
according to the manufacturers protocol. Briefly, ESCC patient
exosomes were diluted and resuspended in sterile PBS to a final
concentration of 50 µg/ml. Diluent C (250 µl; PKH67 solution) was
added to 1.6 µl PKH67 dye, and 200 µl exosomes were mixed with 250
µl PKH67 solution in a 1.5-ml microfuge tube. The samples were
gently mixed for 4 min at 37°C, and 500 µl 1% BSA was added to bind
the excess PKH67 dye. Next, the ExoEasy Maxi Kit was used to
collect PKH67-labelled exosomes, after which the PKH67-labelled
exosomes were resuspended in RPMI-1640. Eca109 cells were seeded
onto culture dishes (35-mm diameter) at a density of
3×105 cells/dish, and incubated in complete medium for
24 h at 37°C (5% CO2). Subsequently, the plates were
rinsed three times with PBS to remove the effect of serum exosomes,
and medium containing 100 µl PKH67-labelled exosomes and an
equivalent volume of the PKH67-PBS or PBS control, was added to the
appropriate wells. After that, cells and ESCC exosomes were
co-cultured for 1, 2, 4 and 24 h, after which the dishes were
gently rinsed in PBS and fixed with 4% paraformaldehyde solution
for 30 min at RT. The dishes were then rinsed again three times
using PBS. After nuclear staining was performed using ProLong Gold
Antifade Reagent with DAPI (Beijing Solarbio Science &
Technology Co., Ltd.), the dishes were viewed under a fluorescence
microscope.
ELISA
The PKM2 ELISA kit (cat. no. tw041272; Shanghai
Tongwei Biological Technology Co., Ltd.) was equilibrated at RT for
60 min. Plasma was completely lysed in a water bath set at 37° for
30 min. After standing at RT for 30 min, the supernatant was
obtained. For sample addition, 50 µl standard or 10 µl sample mixed
with 40 µl sample diluent was added to each ELISA plate well, and
100 µl biotin-conjugated antibody working solution was added to all
wells, followed by incubation for 1 h at 37°C. The supernatant was
discarded, and the plate was rinsed five times with wash solution
(in the kit). Then, 50 µl each of substrate solution A and B was
added to each well, followed by incubation at 37°C in the dark for
15 min. Then, 50 µl stop solution was added to terminate the
reaction, and 5 min later, the OD value of each well was determined
using a microplate reader set at 450 nm. A standard curve of the OD
values was plotted by using GraphPad Prism 5.0 software (GraphPad
Software, Inc.), and the concentration of plasma-derived PKM2 in
each sample was determined.
Cellular proliferation assay
Eca109 cells were washed with PBS and seeded into
96-well plates (3,000 cells/well) in RPMI-1640 medium enriched with
10% exosome-free FBS, with or without exosomes (50 µg/ml). After
incubation for 2 h, the Cell Counting Kit 8 (APExBIO Technology
LLC) was used to assess proliferative capacity after 0, 24, 48, 72
and 96 h according to the product instructions.
Transwell invasion assay
Diluted Matrigel (60 µl) was added to the upper
compartment of the Transwell plates (Corning, Inc.) and incubated
at 37°C for 2 h. Subsequently, 5×103 Eca109 cells were
added into the upper compartment in medium without or with exosomes
(50 µg/ml); 500 µl medium with 10% exosome-free FBS was added to
the lower compartment and incubated, and the plate was incubated
for 24 h. The cells were then fixed with 4% paraformaldehyde at 4°C
for 30 min, followed by staining with crystal violet (Beijing
Solarbio Science & Technology Co., Ltd.) at RT for 5 min. The
migrated cells were counted and photographed using an inverted
phase-contrast microscope.
Wound-healing assay
Eca109 Cells were seeded into a 6-well flat-bottomed
plate (1×105/well) in RPMI 1640 medium enriched with 10%
exosome-free FBS as previously described (18). Subsequently, a pipette tip was used to
create a wound in the cell monolayer, the floating cells were
removed, and medium without or with 50 µg/ml exosomes was added to
each well. Thereafter, an inverted microscope was used to capture
images at 0 and 24 h post-scratching. The motility of cells was
determined by comparing the closure distance between the two time
points.
Colony formation assay
Eca109 cells were plated into 6-well plates
(1×103 per well) in RPMI 1640 medium with 30%
exosome-free FBS, with or without exosomes (50 µg/ml). The plates
were incubated at 37°C with 5% CO2 for 10–14 days.
Formed colonies were fixed with 4% paraformaldehyde at 4°C for 30
min, stained with crystal violet for 15 min at RT and washed with
distilled water. Colonies containing >50 cells were counted by
eye.
Western blotting
Total protein from the exosomal, cell or co-culture
cell samples was extracted using RIPA lysis buffer (Beijing
Solarbio Science & Technology Co., Ltd.), and the concentration
was calculated using a BCA kit. The proteins were boiled at 100°C
for 5 min, fractionated on 10% SDS-PAGE gels for 120 min, and then
blotted onto a PVDF membrane for 100 min. Thereafter, 5% skimmed
milk was used to block the membranes for 2 h at RT, followed by
inoculation with antibodies against CD9 (1:1,000; cat. no.
20597-1-AP; ProteinTech Group, Inc.), CD63 (1:1,000; cat. no.
25682-1-AP; ProteinTech Group, Inc.), CD81 (1:1,000; cat. no.
27855-1-AP; ProteinTech Group, Inc.), TSG101 (1:1,000; cat. no.
14497-1-AP; ProteinTech Group, Inc.), PKM2 (1:1,000; cat. no.
15822-1-AP; ProteinTech Group, Inc.), STAT3 (1:1,1000; cat. no.
10253-2-AP; ProteinTech Group, Inc.), pSTAT3Tyr705
(1:1,000; cat. no. 9145S; Cell Signaling Technology, Inc.) and
GAPDH (1:10,000; cat. no. ab8245; Abcam) at 4°C overnight. Then,
the membranes were incubated with secondary goat anti-mouse IgG
antibody (1:1,000; cat. no. BA1010) and goat anti-rabbit IgG
antibody (1:1,000, cat. no. BA1011) (both Wuhan Boster Biological
Technology, Ltd.) at RT for 60 min. Finally, the membranes were
rinsed three times with PBST (PBS + 0.05% Tween 20), followed by
soaking in AP Chromogenic Substrate (Invitrogen; Thermo Fisher
Scientific, Inc.) for signal development. Grayscale quantification
was performed using ImageJ software (1.51j8, National Institutes of
Health).
Immunofluorescence analysis
A total of 4×105 Eca109 cells were
inoculated into a 35-mm petri dish with RPMI 1640 medium
supplemented with 10% exosome-free FBS, with or without exosomes
(50 µg/ml), and incubated for 24 h. The cells were then fixed with
4% paraformaldehyde at RT for 30 min, and then treated with 0.5%
Triton X-100 diluted with PBS at RT for 15 min. Cells were
incubated with antibodies specific for PKM2 (1:40; cat. no. 15822-1
AP; ProteinTech Group, Inc.) and pSTAT3Tyr705 (1:150,
cat. no. 9145S, Cell Signaling Technology) overnight at 4°C, and
then with Alexa Fluor-labelled secondary antibodies (1:100; cat.
no. BA1142; Wuhan Boster Biological Technology, Ltd.) in the dark
for 45 min at RT. DAPI was used to counterstain the nuclei for 20
min at RT, and images were captured using a laser scanning confocal
microscope (magnification, ×400; LSM710; Zeiss AG).
GEPIA
GEPIA (http://gepia.cancer-pku.cn/) is a widely utilized
interactive web resource for plotting the expression patterns of
specified genes. GEPIA contains 9,736 tumours and 8,587
non-malignant tissue samples from The Cancer Genome Atlas, as well
as GTEx data resources. It is employed to conduct survival analysis
on the basis of levels of specified gene expression, per the
user-specified sample selections and approaches (19). Herein, GEPIA was used to determine PKM
and STAT3 mRNA levels in EC, as well as subsequent correlation
analyses.
Immunohistochemistry
A total of 147 ESCC and 85 non-malignant control
tissues were examined. The tissue microarray and tissue samples
were cut at 4-µm thickness, dewaxed, hydrated in a descending
alcohol series, treated with EDTA antigen repair solution
(Sigma-Aldrich; Merck KGaA) for 98°C for 13 min, and then blocked
in serum (OriGene Technologies, Inc.) for 1 h at RT. Endogenous
peroxidase activity was blocked with 3% H2O2.
The sections were incubated overnight with rabbit anti-PKM2 (cat.
no. 15822-1 AP; ProteinTech Group, Inc.) at a dilution of 1:200,
and rabbit anti-pSTAT3Tyr705 (cat. no. 9145S, Cell
Signaling Technology) at a dilution of 1:150. Then, the sections
were incubated with goat anti-rabbit IgG (1:800; cat. no. 31926;
Cell Signaling Technology) for 60 min at 37°C. A semiquantitative
scoring technique was used to determine the PKM2 and
pSTAT3Tyr705 expression levels. An evaluation of the
staining intensity and proportion of positive cells was used to
assess the expression of PKM2 and pSTAT3Tyr705. Staining
intensity was scored using a four-point scale as follows: 3, strong
(tan); 2, medium (brown-yellow); 1, weak (light yellow); and 0,
none. The proportion of positive cells was scored as 0, 0%; 1,
≤10%; 2, 11–50%; 3, 51–75%; and 4, >75% (20). The staining index (SI) was determined
as the sum of the staining intensity score and the number of
positive cells. Patients with ESCC were stratified into two groups
on the basis of SI: i) Patients with a total score of 0–3 were
clustered into the low-expression group (Fig. S1A-D); and ii) those with a total
score of 3–7 were stratified into the high-expression group
(21).
Statistical analysis
Numerical data are presented as the mean ± standard
deviation. Student's t-test was used to compare statistical
significance between two groups, while Bonferroni's post hoc
analysis was used following ANOVA. For categorical data, Fisher's
exact test was used to analyse significant differences when dealing
with expected values <5. Spearman's correlation analysis was
used to analyse the correlation between the protein levels of PKM2
and pSTAT3Tyr705. All in vitro experiments were
performed 2–4 times. Survival analyses for patients with ESCC were
performed using the Kaplan-Meier method, and the log-rank test was
used to determine the statistical significance of the difference
between the two groups. Analyses were conducted using GraphPad
Prism 8.0 software (GraphPad Software) or STATA 15.0 software
(StataCorp LP) at the 95% confidence level. P<0.05 was
considered to indicate a statistically significant difference.
Results
Characterization of exosomes extracted
from plasma
Exosomes were successfully derived from the plasma
of patients and healthy controls, and exosomes isolated from
patients with ESCC were validated in terms of morphology, size and
specific markers. Fig. 1A indicates
that extracted exosomes from patients with ESCC appeared as discs
in the TEM images. In addition, ~2.84×109 vesicles were
detected in most of the 1-ml plasma samples (Fig. 1B). Exosome diameter ranged from 30 to
150 nm based on NanoFCM analysis, and the mean diameter was ~81.97
nm (Fig. 1C). In addition, NanoFCM
displayed that the proportions of CD9 and CD81 were 7.3 and 14.5%,
respectively (Fig. 1D). In order to
further verify exosome extraction, the expression of other exosomal
proteins was confirmed by western blot analyses. The results showed
that CD9, CD63, CD81 and TSG101 were expressed in exosomes
(Fig. 1E). Altogether, this isolation
approach was suitable for the following experiments. As shown in
Fig. 1F, quantitative analysis of the
western blotting data showed that the expression of exosomal PKM2
was lower in healthy subjects (Exo-H), than in individuals with
ESCC (Exo-P) (P<0.001). To determine whether host cells could
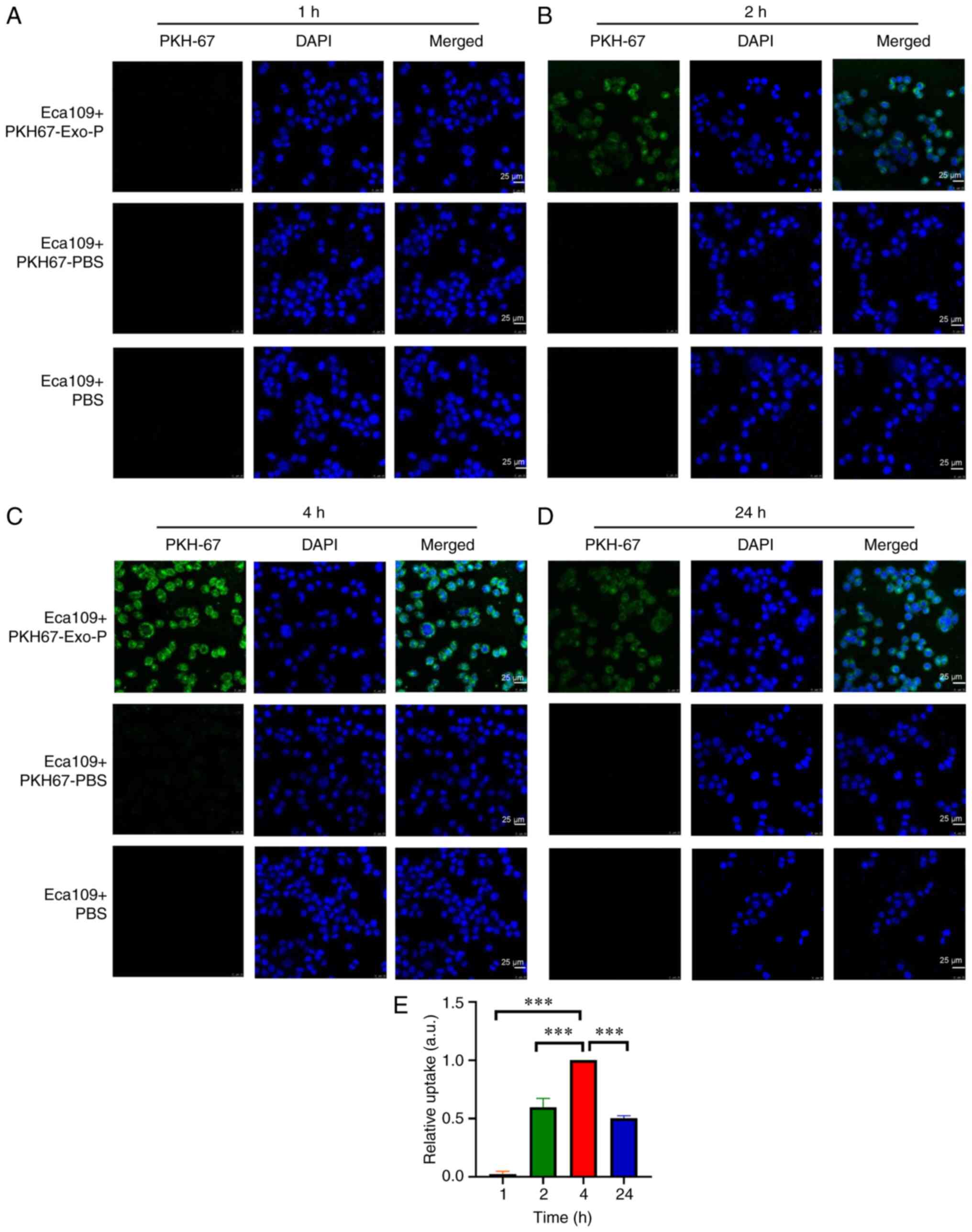
take up ESCC patient exosomes, PKH67 dye (green fluorescence) was
used to label ESCC patient exosomes, which were then incubated with
Eca109 ESCC cells in vitro. As indicated in Fig. 2A, there was no obvious phagocytosis
following co-culture for 1 h. However, the cytoplasm and nucleus
exhibited green fluorescence at 2 (Fig.
2B), 4 (Fig. 2C) and 24 h
(Fig. 2D) in exosomes from patients
with ESCC, implying that Eca109 cells took up a significant number
of exosomes compared with the PKH67-PBS control, or the PBS
control. Quantification of internalized exosomes (green) showed
that the average optical densities at 2 and 24 h were 59.67 and
50.33% of that at 4 h, respectively. Notably, phagocytosis was the
strongest at 4 h (Fig. 2E).
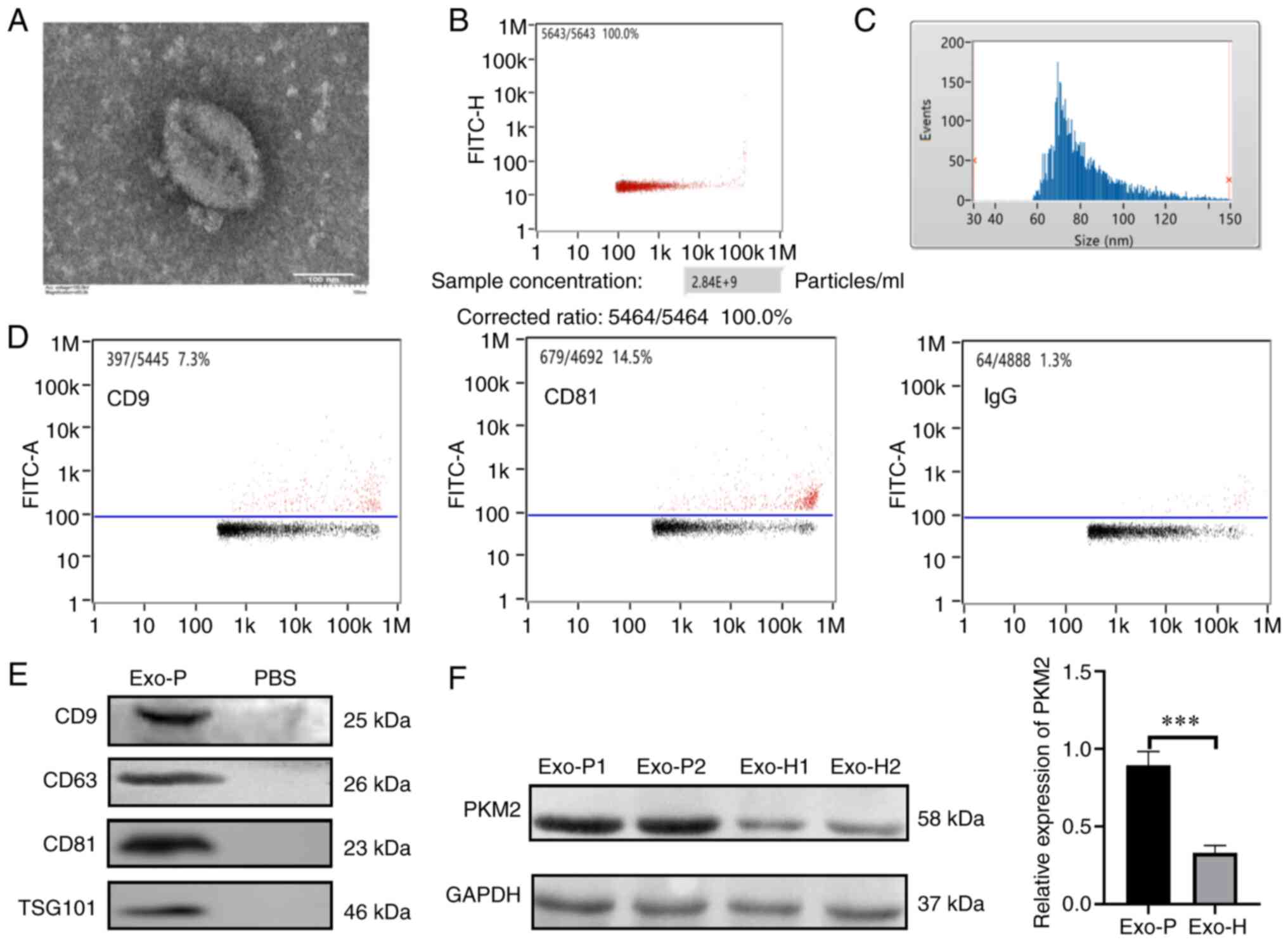 | Figure 1.Characterization of plasma-derived
exosomes of ESCC. (A) Exosomes isolated from patients with ESCC
were observed under a transmission electron microscope to have a
diameter of 50–150 nm (scale bar, 100 nm). (B) Vesicles from 1- ml
plasma detected by NanoFCM. (C) Size distribution of exosomes
measured by NanoFCM (mean value, 81.97 nm). (D) Exosome-enriched
proteins, CD9 and CD81, were analysed by FCM. (E) Exosome-enriched
CD9, CD63, CD81 and TSG101 were analysed by western blotting. (F)
PKM2 levels in exosomes were analysed by western blotting in Exo-P
and Exo-H cells. ***P<0.001. ESCC, oesophageal squamous cell
carcinoma; Exo-P, exosomes from patients with ESCC; Exo-H, exosomes
from healthy controls; FCM, flow cytometry; PKM2, pyruvate kinase
isoenzyme type M2. |
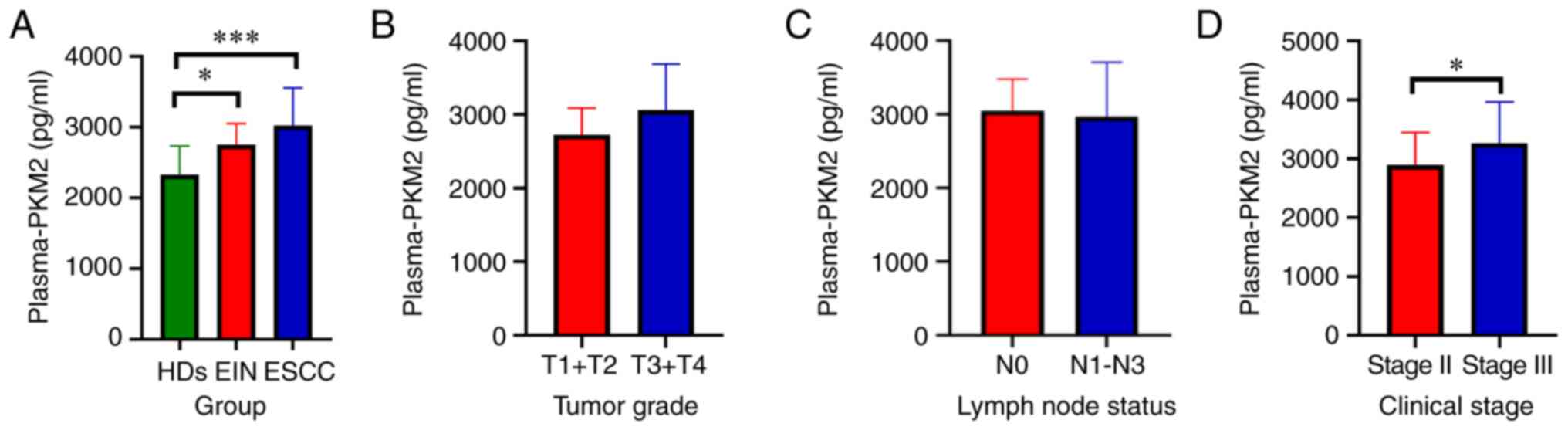
Concentration of plasma-derived PKM2
in ESCC
The aforementioned results prompted investigation
into the role of exosomal PKM2 expression in patients with ESCC,
which was assessed by ELISA. The clinicopathological
characteristics of the plasma samples of 76 patients were acquired
via diagnostic procedures, and the characteristics of ESCC, EIN and
healthy donors (HDs) are summarised in Tables SI and SII. ESCC, EIN and HDs accounted for 68.4,
7.9 and 23.7%, respectively. The data from patients with ESCC were
extracted; 27 (51.9%) of the 52 patients had stage II ESCC, and 25
(41.9%) had stage III ESCC. In addition, most patients presented
with T3 or T4 (78.8%) and a positive nodal status (61.5%).
The profile of plasma-derived PKM2 was assayed via
ELISA, and the data showed that the PKM2 content was markedly
higher in patients with ESCC (3022±528.0 pg/ml plasma) and EIN
patients (2748±300.6 pg/ml plasma) than in healthy controls
(2327±409.7 pg/ml serum) (P<0.001 and P<0.05, respectively)
(Fig. 3A). No significant difference
was identified in tumour grade (Fig.
3B) or lymph node stage (Fig.
3C). However, the PKM2 level of patients with stage III disease
(3265±697.9 pg/ml plasma) was markedly higher than that of patients
with stage II disease (2893±552.8 pg/ml plasma) (P<0.05,
Fig. 3D).
Exosomes from patients with ESCC
enhance the proliferation and motility of ESCC cells, and PKM2 can
be transferred by exosomes
To assess the oncogenic potential of ESCC
patient-derived exosomes, the basal expression of PKM2 in a panel
of ESCC cell lines was assessed by western blotting (Fig. 4A). Among these, the expression of PKM2
was the lowest in Eca109 cells. Next, Eca109 cells were treated
with or without exosomes from patients with ESCC, and the colony
formation assay showed that patient exosomes promoted cellular
proliferation (Fig. 4B, P<0.001).
The CCK-8 assay results showed that exosomes derived from patients
with ESCC promoted the proliferation of ESCC cells at 24 and 48 h,
though this effect was not observed at 72 and 96 h (Fig. S1E). Although the CCK-8 assay results
suggested that exosomes had limited ability to promote
proliferation, the clone formation assay revealed that exosomes
promoted colony formation significantly. Thus collectively, these
two experiments indicated that exosomes promoted cellular
proliferation. When the mobility of recipient cells was
investigated via Transwell migration assay, exosome-treated cells
also exhibited elevated infiltration compared with the control
cells (Fig. 4C, P<0.05).
Furthermore, in the wound-healing assay, patient-derived exosomes
exerted increased cellular motility. At 24 h, exosomes facilitated
a higher rate of wound closure than PBS control (Fig. 4D, P<0.001). Moreover, to validate
that exosomes mediate PKM2 delivery and activate STAT3, Eca109
cells were exposed to exosomes from patients with ESCC and HDs, and
the protein expression level of PKM2 and pSTAT3Tyr705
was evaluated by immunofluorescence analysis and western blotting.
Quantitative analyses showed that in the ESCC-derived exosome
group, PKM2 and pSTAT3Tyr705 expression in ESCC cells
was significantly increased compared with that in the PBS group; by
contrast, no significant changes in total STAT3 were observed
between ESCC-derived exosome group and HDs group (Fig. 4E-I).
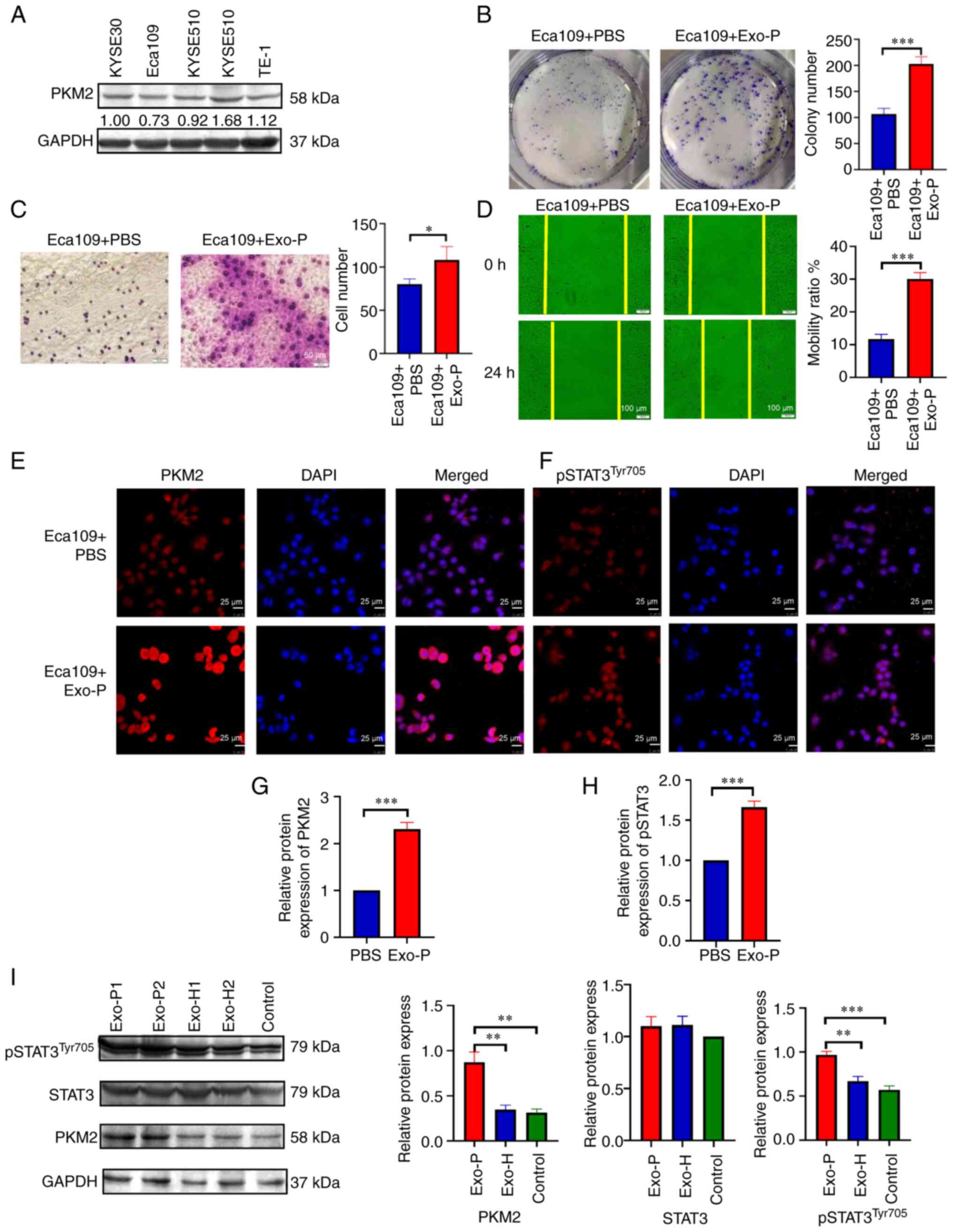 | Figure 4.Exosomes affect the proliferation,
migration and invasiveness of recipient Eca109 cells after
co-cultivation for 24 h. (A) Endogenous expression of PKM2 in ESCC
cell lines. (B) Proliferation of recipient Eca109 cells with PBS or
Exo-P was assessed using the colony formation method. (C)
Invasiveness of Eca109 cells with PBS or Exo-P was assessed by
Transwell assay. (D) Migration of Eca109 cells treated with PBS or
Exo-P was assessed using the wound-healing method. (E) PKM2 protein
level of recipient Eca109 cells treated with PBS or Exo-P was
analysed by immunofluorescence analysis. Magnification, ×400; scale
bar, 25 µm. (F) pSTAT3Tyr705 protein of recipient Eca109
cells with PBS or Exo-P was analysed by immunofluorescence
analysis. Magnification, ×400; scale bar, 25 µm. Quantification of
immunofluorescence of (G) PKM2 and (H) pSTAT3Tyr705
levels in Eca109 cells. (I) PKM2, total STAT3 and
pSTAT3Tyr705 protein levels of recipient Eca109 cells
treated with Exo-P and Exo-H were analysed by western blotting.
*P<0.05, **P<0.01 and ***P<0.001. ESCC, oesophageal
squamous cell carcinoma; Exo-P, Exosomes from patients with ESCC.
PKM2, pyruvate kinase isoenzyme type M2; Exo-H, Exosomes from
healthy controls. |
PKM2 mRNA expression correlates with
that of STAT3 in EC
To verify whether PKM2 exerts its role through
phosphorylation of STAT3, the mRNA levels in EC and non-malignant
oesophageal tissues were examined. The findings revealed elevated
mRNA levels of PKM2 and STAT3 in EC samples compared with
non-malignant oesophageal tissue (Fig.
S1F and G); however, no significant difference was observed.
Nevertheless, there was a remarkable correlation between PKM2 and
STAT3 expression in EC (P<0.05, Fig.
S1H).
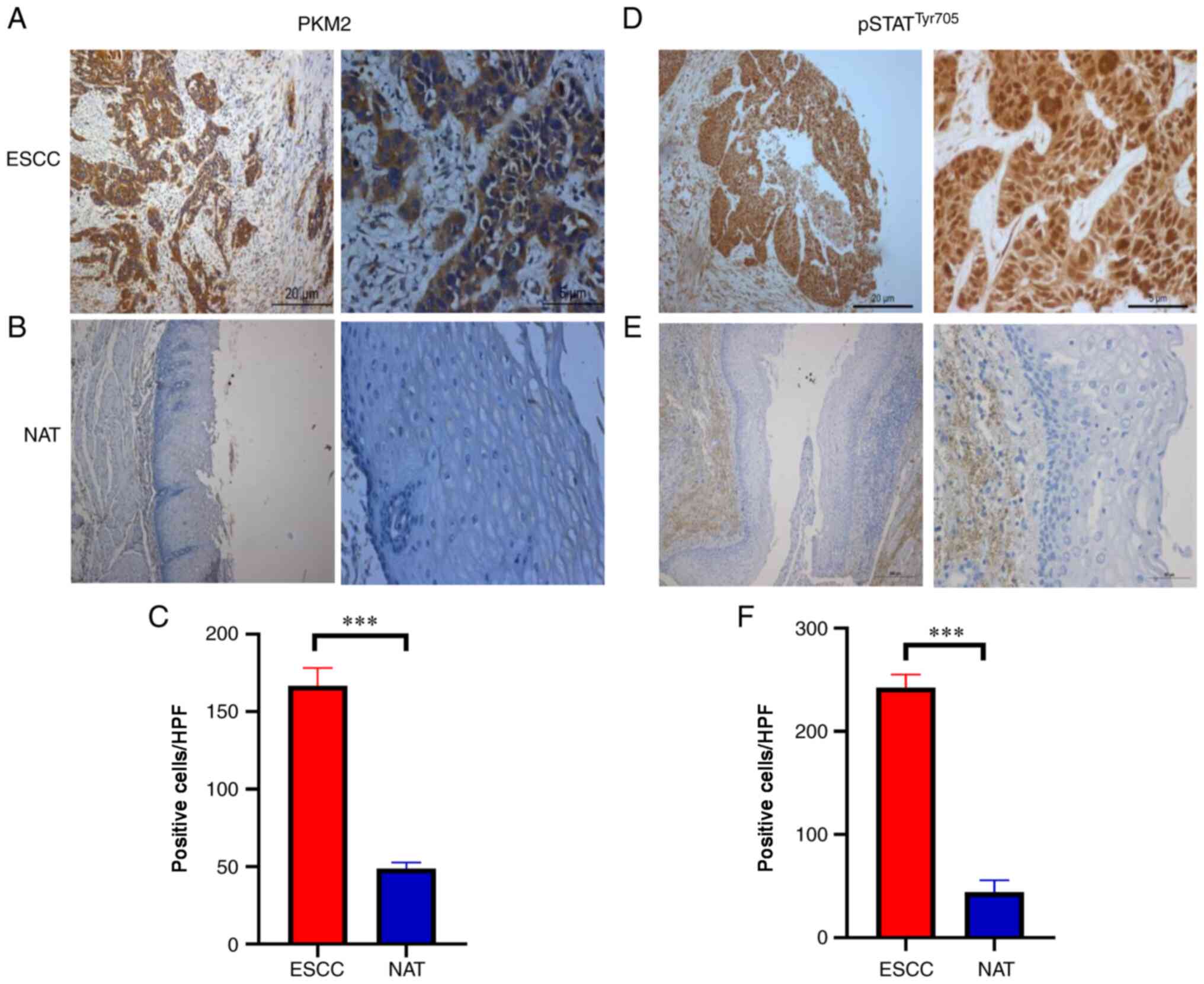
PKM2 and pSTAT3Tyr705 are
associated with clinico-pathological features and prognosis in
patients with ESCC
Tissue microarrays and 52 ESCC tumour tissues
obtained from diagnostic procedures were immunostained for PKM2 and
pSTAT3Tyr705. To validate the correlation between PKM2
and pSTAT3Tyr705, their protein levels were detected by
immunohistochemical analysis. As ESCC showed increased PKM2
expression and pSTAT3Tyr705 compared with
normal-adjacent tissues (Fig. 5A-F),
further assays to assess the clinicopathological potential of PKM2
and pSTAT3Tyr705 in ESCC were performed. The PKM2
expression level was positively linked to metastasis to the lymph
nodes (χ2=8.200; P=0.004; Table I), TNM stage (χ2=7.718;
P=0.022; Table I), and the
upregulation of pSTAT3Tyr705 was associated with TNM
stage (χ2=7.408; P=0.006; Table II).
 | Table I.Association between PKM2 expression
and clinical characteristics of patients with ESCC. |
Table I.
Association between PKM2 expression
and clinical characteristics of patients with ESCC.
|
|
| PKM2
expression |
|
|
|---|
|
|
|
|
|
|
|---|
| Characteristic | Patients, N | Negative | Positive |
χ2-value | P-value |
|---|
| Tissue type |
|
|
|
|
|
|
ESCC | 147 | 56 | 91 | 29.864 | 0.000a |
|
Adjacent normal | 85 | 64 | 21 |
|
|
| Age |
|
|
|
|
|
|
<60 | 50 | 16 | 34 | 1.194 | 0.275 |
|
≥60 | 97 | 40 | 57 |
|
|
| Sex |
|
|
|
|
|
|
Male | 116 | 43 | 73 | 0.246 | 0.620 |
|
Female | 31 | 13 | 18 |
|
|
| T stage |
|
|
|
|
|
|
T1-T2 | 33 | 10 | 23 | 1.096 | 0.295 |
|
T3-T4 | 114 | 46 | 68 |
|
|
| N stage |
|
|
|
|
|
|
N0-N1 | 75 | 37 | 38 | 8.200 | 0.004a |
|
N2-N3 | 72 | 19 | 53 |
|
|
| TNM stage |
|
|
|
|
|
| I | 8 | 3 | 5 | 7.718 | 0.022a |
| II | 71 | 35 | 36 |
|
|
|
III | 68 | 18 | 50 |
|
|
| Differentiation
degree |
|
|
|
|
|
|
Well | 10 | 4 | 6 | 1.992 | 0.681 |
|
Moderate | 99 | 34 | 65 |
|
|
|
Poor | 38 | 18 | 20 |
|
|
| Gross
classification |
|
|
|
|
|
|
Ulcerative type | 85 | 34 | 51 | 1.127 | 0.749 |
|
Medullary type | 46 | 15 | 31 |
|
|
|
Protrude type | 7 | 4 | 3 |
|
|
|
Fungating type | 9 | 4 | 5 |
|
|
 | Table II.Association between
pSTAT3Tyr705 expression and clinical characteristics of
patients with ESCC. |
Table II.
Association between
pSTAT3Tyr705 expression and clinical characteristics of
patients with ESCC.
|
|
|
pSTAT3Tyr705 |
|
|
|---|
|
|
|
|
|
|
|---|
| Characteristic | Patients, N | Negative | Positive |
χ2-value | P-value |
|---|
| Tissue type |
|
|
|
|
|
| ESCC | 147 | 57 | 90 | 21.804 | 0.000a |
| Adjacent
normal | 85 | 60 | 25 |
|
|
| Age |
|
|
|
|
|
| ≤60 | 97 | 36 | 61 | 0.332 | 0.565 |
| >60 | 50 | 21 | 29 |
|
|
| Sex |
|
|
|
|
|
| Male | 116 | 45 | 71 | 0.001 | 0.993 |
| Female | 31 | 12 | 19 |
|
|
| T stage |
|
|
|
|
|
| T1-T2 | 33 | 12 | 21 | 0.104 | 0.747 |
| T3-T4 | 114 | 45 | 69 |
|
|
| N stage |
|
|
|
|
|
| N0-N1 | 75 | 34 | 41 | 2.774 | 0.096 |
| N2-N3 | 71 | 23 | 49 |
|
|
| TNM stage |
|
|
|
|
|
| I | 8 | 6 | 2 | 7.408 | 0.006a |
| II | 71 | 28 | 43 |
|
|
| III | 68 | 23 | 45 |
|
|
| Differentiation
degree |
|
|
|
|
|
| Well | 10 | 5 | 5 | 0.597 | 0.540 |
| Moderate | 99 | 38 | 61 |
|
|
| Poor | 38 | 14 | 24 |
|
|
| Gross
classification |
|
|
|
|
|
| Ulcerative
type | 85 | 33 | 52 | 0.163 | 0.983 |
| Medullary type | 46 | 18 | 28 |
|
|
| Protrude type | 7 | 3 | 4 |
|
|
| Fungating type | 9 | 3 | 6 |
|
|
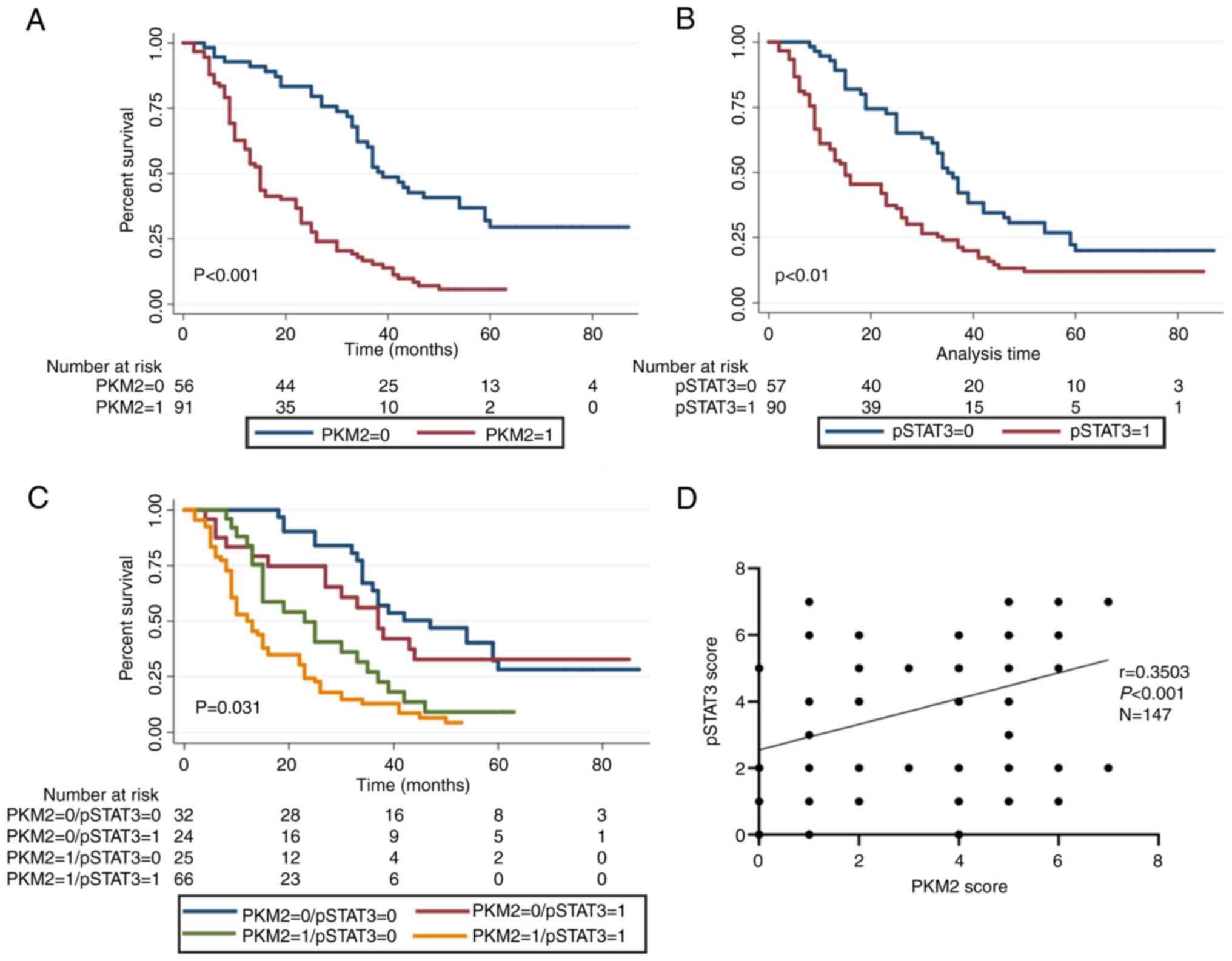
The Kaplan-Meier survival curve showed that the
median OS of ESCC patients with high and low PKM2 expression was
20.3 and 49.1 months, respectively, exhibiting a marked difference
(P<0.001; Fig. 6A). Similarly, the
median OS in ESCC patients with high and low
pSTAT3Tyr705 expression was 25.7 and 42.2 months,
respectively, displaying a statistically significant difference
(P<0.01; Fig. 6B). After
determining the risks associated with PKM2 expression and
pSTAT3Tyr705, the 147 ESCC patients were stratified into
4 groups on the basis of pSTAT3Tyr705 and PKM2 level:
pSTAT3Tyr705-/PKM2-(S-/P-);
pSTAT3Tyr705-/PKM2+ (S-/P+);
pSTAT3Tyr705+/PKM2-(S+/P-) and
pSTAT3Tyr705+/PKM2+(S+/P+). Using the Kaplan-Meier
approach, patients with the S+/P+expression had the shortest OS
(17.5±1.7 months), while patients with the S-/P-expression trend
had the longest OS (52.3±4.5 months) (Fig. 6C). In addition, the relationship
between PKM2 and STAT3 expression was assessed in 147 patients with
ESCC. The data illustrated that PKM2 expression was markedly linked
to pSTAT3Tyr705 (r=0.3503; P<0.001; Fig. 6D). To determine whether PKM2 or
pSTAT3Tyr705 expression levels were independent of other
predictive factors, univariate and multivariate analyses were
conducted using a Cox multivariate proportional hazard regression
model. The univariate assessment data demonstrated that PKM2
upregulation, pSTAT3Tyr705 upregulation, male sex, lymph
node metastasis, clinical classification and gross classification
were predictors of poor prognosis for ESCC (Table III). Multivariate analysis showed
that PKM2 expression, pSTAT3Tyr705 and TNM stage were
independent prognostic factors of ESCC survival (Table III).
 | Table III.Univariate and multivariate analysis
of oesophageal squamous cell carcinoma for overall survival. |
Table III.
Univariate and multivariate analysis
of oesophageal squamous cell carcinoma for overall survival.
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|---|
| Clinical
feature | P-value | 95% CI for HR | P-value | 95% CI for HR |
|---|
| PKM2 |
<0.001a | 2.198–5.000 |
<0.001a | 2.272–5.732 |
|
pSTAT3Tyr705 | 0.001a | 1.341–2.874 | 0.005a | 1.190–2.646 |
| Sex | 0.017a | 0.235–0.894 | 0.251 | 0.414–1.258 |
| Age | 0.306 | 0.976–1.007 |
|
|
| T stage | 0.110 | 0.918–2.330 |
|
|
| N stage |
<0.001a | 1.465–4.074 | 0.673 | 0.556–2.577 |
| TNM stage |
<0.001a | 1.874–3.713 | 0.003a | 1.447–2.851 |
| Differentiation
degree | 0.842 | 0.749–1.425 |
|
|
| Gross
classification | 0.002a | 0.535–0.864 | 0.126 | 0.860–3.408 |
Discussion
The present study illustrated that PKM2 was
assembled into exosomes; in vitro, exosomes from patients
with ESCC promoted the proliferation, invasion and migration of
ESCC cells. Furthermore, the results indicated that PKM2 could be
transferred by exosomes, and that exosomal PKM2 may function by
activating STAT3.
To the best of our knowledge, the present study was
the first to investigate the biological function of exosomes in
ESCC, and to establish that PKM2 can be transferred by exosomes and
acts by activating STAT3. In this respect, the results have
important implications for understanding the progression of
ESCC.
Exosomes are extracellular vesicles found in the
blood (22,23), urine (24) and other bodily fluids (25). Exosomes are secreted in excess by
tumour cells under oxidative stress conditions (26), and are involved in their interaction
with the cancer microenvironment (14). As core communication centres, exosomes
are rich in bioactive molecules (13), including RNA, DNA and proteins. Cancer
cells have been documented to produce exosomes harbouring PKM2
(14,26–28).
However, whether PKM2 could be packaged and transferred in exosomes
from patients with ESCC was unknown. Other studies (29,30) and
our previous study (31–33) have shown that the expression of PKM2
in tumour tissues and cells was higher than that in normal-adjacent
tissues and oesophageal epithelial cells, and that upregulation of
the PKM2 isoform was associated with the increased Warburg effect
of tumour cells (34,35). Moreover, exosomes express
blastocyte-derived surface markers (36). Cancer cells, which secrete more
exosomes than normal tissue cells (37), secrete elevated levels of PKM2. In
addition, seminal studies from prostate cancer (14) and liver cancer (38) have demonstrated that the elevated PKM2
in plasma exosomes of tumour patients was secreted by tumour cells.
The present data were consistent with previous investigations, and
verified that PKM2 was expressed by circulating exosomes of cancer
patients (12). PKM2 in circulating
exosomes possesses clinical significance and modulates biological
roles in tumours (8). In the present
study, ELISA also indicated that the plasma level of PKM2 in
individuals with ESCC differed markedly to that in the healthy
controls, which was in accordance with the results of a study on
prostate cancer (14).
It was further confirmed that the invasion rate of
ESCC cells inoculated with ESCC patient exosomes was higher than
that from ESCC cells without exosomes. In the present study,
although the expression level of PKM2 in plasma was not correlated
with tumour grade or lymph node metastasis, it was closely
associated with clinical stage. Compared with tumour grade and
lymph node metastasis, clinical stage combined with these
parameters may be more value in predicting the prognosis and
outcome of patients. Therefore, exosomal PKM2 derived from patients
with ESCC may promote proliferation and motility. This finding may
provide novel insights and strategies for the averting the distant
metastasis of tumours in clinical practice.
The results of the proliferation experiment in the
present study were consistent with published results (39), where a blank control was used. Thus in
the present study, PBS served as the mock control. Exosomes
promoted cellular proliferation at 24 and 48 h, though this effect
was not observed at 72 and 96 h. This was attributed to the
following possible reasons. On one hand, the cell activity detected
by CCK-8 assays is the joint result of proliferation and apoptosis,
and is the overall result from a large number of cells. Therefore,
the ability to assesses cellular proliferation alone is limited. In
addition, exosomes were added to the cell culture medium when the
cells adhered to the well. Therefore, the result may be due to the
weakened proliferation-promoting ability of exosomes after they
were metabolized by cells. Although the CCK-8 assay results
suggested that exosomes had limited ability to promote
proliferation, the clone formation assay revealed that exosomes
significantly colony formation, which focuses on the proliferation
ability of single cells, crucial for subsequent colony formation.
The combined results of these two experiments indicated that
exosomes promoted cellular proliferation.
When examining the basal expression of PKM2, the
levels were comparatively lowest in Eca109 cells. Western blotting
and immunofluorescence analysis indicated that PKM2 could be
packaged into and transferred by plasma-derived exosomes.
Originally reported in 1934, pyruvate kinase was shown to exist as
two different isoforms, PKM1 and PKM2 (40). Increaesd expression of PKM2 promotes
multiple cancer cell characteristics, including extracellular
signal transduction and metabolism, and is closely associated with
tumorigenesis. Ma et al (27)
demonstrated that exosomal PKM2 triggers a tumour-like phenotype in
mesenchymal stem cells by activating glycolysis in glioma. In the
current study, PKM2 was primarily expressed in the cytoplasm, and
occasionally in the nucleus in the high-expression group, while in
patients in the low-expression group, PKM2 was almost exclusively
expressed in the cytoplasm. According to the follow-up results, the
prognosis of the low-expression group was more favourable than that
of the high-expression group, which indirectly confirms the
prognostic value of PKM2. Furthermore, 91 patients had high PKM2
expression, and 56 patients had low PKM2 expression. According to
previous studies, the positive expression rate of PKM2 in solid
tumours is ~20-70% (29,41,42), which
is consistent with the present study. However, it was slightly
higher than in our previous study (31). The possible reasons are as follows: i)
the results of immunohistochemical staining were quantitatively
analysed; and ii) scores ≤3 were included in the low-expression
group.
Western blotting and immunofluorescence analysis
suggested that pSTAT3Tyr705 was increased in co-cultured
cells. STAT3 is widely expressed in various tissues and cell types,
where it participates in the regulation of physiological functions
including cellular differentiation, proliferation, malignant
transformation and apoptosis inhibition. Overexpression of STAT3
can result in abnormal cellular proliferation and inhibition of
apoptosis (43), and several studies
have revealed that PKM2 regulation of STAT3 expression is
associated with cancer cell migration and invasiveness (9,10). PKM2
plays a vital role in the phosphorylation of STAT3 at Tyr705,
resulting in cancer cell proliferation (31). Another study using colon cancer cells
demonstrated that PKM2 enhanced cellular migration via increased
transcription of the STAT3 gene and the phosphorylation of STAT3 at
Ser727. However, the association between PKM2 and
pSTAT3Tyr705 and their prognostic value in ESCC remains
unclear.
By searching the GEPIA database, it was established
that the PKM mRNA expression in EC was associated with the
expression level of STAT3. Tumour PKM2 and pSTAT3Tyr705
immunohistochemical profiles were also analysed. Similarly, the
expression level of PKM2 was positively correlated with the level
of pSTAT3Tyr705. High expression of PKM2 and
pSTAT3Tyr705, or co-expression of PKM2 and
pSTAT3Tyr705, were all associated with OS. These results
suggested that STAT3 may be a critical downstream regulator of the
exosomal PKM2 signalling pathway. In future studies, the exact
mechanism by which exosomal PKM2 promotes tumour invasion and
metastasis by activating STAT3 will be investigated.
In the current study, cells treated with PBS served
as the control group. It seems somewhat far-fetched, but the
selection of the control group was based on currently published
literature (39). The findings of the
present study suggest that patient-derived exosomes promote the
malignant phenotype of ESCC cells. In future studies exploring the
mechanism by which exosomal PKM2 promotes the migration and
invasiveness of oesophageal squamous cells, we aim to use healthy
donors as the control group. In addition, plasma levels of PKM2 in
stage I patients was not included in the results, as no blood
samples were collected from such patients in the sampling process.
Instead, blood samples from patients with precancerous lesions were
collected and analysed, namely, high-grade intraepithelial
neoplasia. EIN is a necessary stage of transformation from normal
oesophagus or esophagitis to ESCC. Therefore, it is reasonable to
presume that the 6 cases of EIN included in the current analysis
were of representative significance for the determination of PKM2
in plasma exosomes of patients with early-stage ESCC. The results
suggested that exosomal PKM2 functions through STAT3. Nevertheless,
the exact mechanisms remain largely unknown. The aim of further
studies will be to explore the mechanism by which exosomal PKM2
promotes the migration and invasiveness of oesophageal squamous
cells.
Collectively, exosomes were successfully isolated
from the plasma of patients with ESCC, and were characterized for
quality confirmation. ELISA indicated that exosomal PKM2 was
associated with tumour stage in ESCC. In vitro cellular
functional experiments also confirmed that exosomes from ESCC
patient plasma promoted and accelerated cellular proliferation,
invasiveness and migration. Subsequently, immunohistochemical
analysis suggested that PKM2 expression was associated with the
level of pSTAT3Tyr705. In addition, PKM2 can be
transferred by exosomes. However, the identification of its
function in ESCC should be further investigated in future.
Supplementary Material
Supporting Data
Acknowledgements
The authors would like to thank Dr Adili Salai and
Dr Xiaohong Sun of the Department of Thoracic Surgery of Affiliated
Tumour Hospital of Xinjiang Medical University for their help with
data collection.
Funding
The study was funded by the Natural Science
Foundation of China (grant no. 81960527, U1603284 and 81860511),
the National Innovation Research Group Cultivation Program (grant
no. xyd2021C001), the Key Research and Development Project of the
Xinjiang Uygur Autonomous Region (grant no. 2020B03003-1), the
Science and Technology Projects of Xinjiang Uygur Autonomous Region
(grant no. 2018E02067), the Tianshan Xuesong Project of the
Xinjiang Uygur Autonomous Region (grant no. 2018XS19), the General
Project from the State Key Laboratory of Pathogenesis, Prevention,
Treatment of High Incidence Diseases in Central Asia, Urumqi,
Xinjiang Uygur Autonomous Region (grant no. SKL-HIDCA-2020-11,12)
and the Natural Science Foundation of Xinjiang Uygur Autonomous
Region (grant no. 2020D01C218).
Availability of data and materials
The datasets used and/or analysed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
LY contributed to the study design, performed the
experiments, and prepared the manuscript. AT performed the cell
culture, and QZ collected the clinical samples. XH carried out the
ELISA. SZ interpreted the data and contributed to critical
discussion. QL and TL analysed the experimental results and
confirmed the authenticity of all the raw data. XL conceived and
oversaw the whole study. All authors have read and approved the
final manuscript.
Ethics approval and consent to
participate
The present study was approved by the local ethics
committee of the Affiliated Tumor Hospital of Xinjiang Medical
University (approval no. K-2019054). All patients provided written
informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Zhao A, Guo L, Xu J, Zheng L, Guo Z, Ling
Z, Wang L and Mao W: Identification and validation of circulating
exosomes-based liquid biopsy for esophageal cancer. Cancer Med.
8:3566–3574. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Qin HD, Liao XY, Chen YB, Huang SY, Xue
WQ, Li FF, Ge XS, Liu DQ, Cai Q, Long J, et al: Genomic
characterization of esophageal squamous cell carcinoma reveals
critical genes underlying tumorigenesis and poor prognosis. Am J
Hum Genet. 98:709–727. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhong R, Chen Z, Mo T, Li Z and Zhang P:
Potential role of circPVT1 as a proliferative factor and treatment
target in esophageal carcinoma. Cancer Cell Int. 19:2672019.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhang W, Hong R, Li L, Wang Y, Du P, Ou Y,
Zhao Z, Liu X, Xiao W, Dong D, et al: The chromosome 11q13. 3
amplification associated lymph node metastasis is driven by
miR-548k through modulating tumor microenvironment. Mol Cancer.
17:1252018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Tkach M and Théry C: Communication by
extracellular vesicles: Where we are and where we need to go. Cell.
164:1226–1232. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ludwig S, Floros T, Theodoraki MN, Hong
CS, Jackson EK, Lang S and Whiteside TL: Suppression of lymphocyte
functions by plasma exosomes correlates with disease activity in
patients with head and neck cancer. Clin Cancer Res. 23:4843–4854.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hsu MC and Hung WC: Pyruvate kinase M2
fuels multiple aspects of cancer cells: From cellular metabolism,
transcriptional regulation to extracellular signaling. Mol Cancer.
17:352018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Gao X, Wang H, Yang JJ, Liu X and Liu ZR:
Pyruvate kinase M2 regulates gene transcription by acting as a
protein kinase. Mol Cell. 45:598–609. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yang P and Li Z, Fu R, Wu H and Li Z:
Pyruvate kinase M2 facilitates colon cancer cell migration via the
modulation of STAT3 signalling. Cell Signal. 26:1853–1862. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Buschow SI, Van Balkom BW, Aalberts M,
Heck AJ, Wauben M and Stoorvogel W: MHC class II-associated
proteins in B-cell exosomes and potential functional implications
for exosome biogenesis. Immunol Cell Biol. 88:851–856. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Li L, Zhang Y, Qiao J, Yang JJ and Liu ZR:
Pyruvate kinase M2 in blood circulation facilitates tumor growth by
promoting angiogenesis. J Biol Chem. 289:25812–25821. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yang P and Li Z, Wang Y, Zhang L, Wu H and
Li Z: Secreted pyruvate kinase M2 facilitates cell migration via
PI3K/Akt and Wnt/β-catenin pathway in colon cancer cells. Biochem
Biophys Res Commun. 459:327–332. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Dai J, Escara-Wilke J, Keller JM, Jung Y,
Taichman RS, Pienta KJ and Keller ET: Primary prostate cancer
educates bone stroma through exosomal pyruvate kinase M2 to promote
bone metastasis. J Exp Med. 216:2883–2899. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Sudo N, Ichikawa H, Muneoka Y, Hanyu T,
Kano Y, Ishikawa T, Hirose Y, Miura K, Shimada Y, Nagahashi M, et
al: Clinical utility of ypTNM stage grouping in the 8th edition of
the American joint committee on cancer TNM staging system for
esophageal squamous cell carcinoma. Ann Surg Oncol. 28:650–660.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Muller L, Hong CS, Stolz DB, Watkins SC
and Whiteside TL: Isolation of biologically-active exosomes from
human plasma. J Immunol Methods. 411:55–65. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wei P, Wu F, Kang B, Sun X, Heskia F,
Pachot A, Liang J and Li D: Plasma extracellular vesicles detected
by single molecule array technology as a liquid biopsy for
colorectal cancer. J Extracell Vesicles. 9:18097652020. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Luo J, Liu L, Shen J, Zhou N, Feng Y,
Zhang N, Sun Q and Zhu Y: MiR-576-5p promotes
epithelial-to-mesenchymal transition in colorectal cancer by
targeting the Wnt5a-mediated Wnt/β-catenin signaling pathway. Mol
Med Rep. 23:942021. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Tang Z, Li C, Kang B, Gao G, Li C and
Zhang Z: GEPIA: A web server for cancer and normal gene expression
profiling and interactive analyses. Nucleic Acids Res. 45:W98–W102.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yan Y, Xu Z, Huang J, Guo G, Gao M, Kim W,
Zeng X, Kloeber JA, Zhu Q, Zhao F, et al: The deubiquitinase USP36
Regulates DNA replication stress and confers therapeutic resistance
through PrimPol stabilization. Nucleic Acids Res. 48:12711–12726.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yu S, Zhang Y, Li Q, Zhang Z, Zhao G and
Xu J: CLDN6 promotes tumor progression through the YAP1-snail1 axis
in gastric cancer. Cell Death Dis. 10:9492019. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhang W, Ou X and Wu X: Proteomics
profiling of plasma exosomes in epithelial ovarian cancer: A
potential role in the coagulation cascade, diagnosis and prognosis.
Int J Oncol. 54:1719–1733. 2019.PubMed/NCBI
|
|
23
|
Matsumoto Y, Kano M, Akutsu Y, Hanari N,
Hoshino I, Murakami K, Usui A, Suito H, Takahashi M, Otsuka R, et
al: Quantification of plasma exosome is a potential prognostic
marker for esophageal squamous cell carcinoma. Oncol Rep.
36:2535–2543. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
McKiernan J, Donovan MJ, O'Neill V,
Bentink S, Noerholm M, Belzer S, Skog J, Kattan MW, Partin A,
Andriole G, et al: A novel urine exosome gene expression assay to
predict high-grade prostate cancer at initial biopsy. JAMA Oncol.
2:882–889. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Severino V, Dumonceau JM, Delhaye M, Moll
S, Annessi-Ramseyer I, Robin X, Frossard JL and Farina A:
Extracellular vesicles in bile as markers of malignant biliary
stenoses. Gastroenterology. 153:495–504. e8. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Dorayappan KDP, Wanner R, Wallbillich JJ,
Saini U, Zingarelli R, Suarez AA, Cohn DE and Selvendiran K:
Hypoxia-induced exosomes contribute to a more aggressive and
chemoresistant ovarian cancer phenotype: A novel mechanism linking
STAT3/Rab proteins. Oncogene. 37:3806–3821. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ma Z, Cui X, Lu L, Chen G, Yang Y, Hu Y,
Lu Y, Cao Z, Wang Y and Wang X: Exosomes from glioma cells induce a
tumor-like phenotype in mesenchymal stem cells by activating
glycolysis. Stem Cell Res Ther. 10:602019. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wei Y, Wang D, Jin F, Bian Z, Li L, Liang
H, Li M, Shi L, Pan C, Zhu D, et al: Pyruvate kinase type M2
promotes tumour cell exosome release via phosphorylating
synaptosome-associated protein 23. Nat Commun. 8:140412017.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wang C, Wang J, Chen Z, Gao Y and He J:
Immunohistochemical prognostic markers of esophageal squamous cell
carcinoma: A systematic review. Chin J Cancer. 36:652017.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Xiaoyu H, Yiru Y, Shuisheng S, Keyan C,
Zixing Y, Shanglin C, Yuan W, Dongming C, Wangliang Z, Xudong B and
Jie M: The mTOR pathway regulates PKM2 to affect glycolysis in
esophageal squamous cell carcinoma. Technol Cancer Res Treat.
17:15330338187800632018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ma R, Liu Q, Zheng S, Liu T, Tan D and Lu
X: PKM2-regulated STAT3 promotes esophageal squamous cell carcinoma
progression via TGF-β1-induced EMT. J Cell Biochem. 2019.(Epub
ahead of print). View Article : Google Scholar
|
|
32
|
Liu Q, Liang M, Liu T, Vuitton L, Zheng S,
Gao X, Lu M, Li X, Sheyhidin I and Lu X: M2 isoform of pyruvate
kinase (PKM2) is upregulated in Kazakh's ESCC and promotes
proliferation and migration of ESCC cells. Tumour Biol.
37:2665–2672. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhang X, Liu T, Zheng S, Liu Q, Shen T,
Han X, Zhang Q, Yang L and Lu X: SUMOylation of HSP27 regulates
PKM2 to promote esophageal squamous cell carcinoma progression.
Oncol Rep. 44:1355–1364. 2020.PubMed/NCBI
|
|
34
|
Zhang Z, Deng X, Liu Y, Liu Y, Sun L and
Chen F: PKM2, function and expression and regulation. Cell Biosci.
9:522019. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Li S, Huang P, Gan J, Ling X, Du X, Liao
Y, Li L, Meng Y, Li Y and Bai Y: Dihydroartemisinin represses
esophageal cancer glycolysis by down-regulating pyruvate kinase M2.
Eur J Pharmacol. 854:232–239. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Song J, Sun T, Tang Z, Ruan Y, Liu K, Rao
K, Lan R, Wang S, Wang T and Liu J: Exosomes derived from smooth
muscle cells ameliorate diabetes-induced erectile dysfunction by
inhibiting fibrosis and modulating the NO/cGMP pathway. J Cell Mol
Med. 24:13289–13302. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Hrustincova A, Krejcik Z, Kundrat D,
Szikszai K, Belickova M, Pecherkova P, Klema J, Vesela J, Hruba M,
Cermak J, et al: Circulating small noncoding RNAs have specific
expression patterns in plasma and extracellular vesicles in
myelodysplastic syndromes and are predictive of patient outcome.
Cells. 9:7942020. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Hou PP, Luo LJ, Chen HZ, Chen QT, Bian XL,
Wu SF, Zhou JX, Zhao WX, Liu JM, Wang XM, et al: Ectosomal PKM2
promotes HCC by inducing macrophage differentiation and remodeling
the tumor microenvironment. Mol Cell. 78:1192–1206. e10. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Tanaka Y, Kamohara H, Kinoshita K,
Kurashige J, Ishimoto T, Iwatsuki M, Watanabe M and Baba H:
Clinical impact of serum exosomal microRNA-21 as a clinical
biomarker in human esophageal squamous cell carcinoma. Cancer.
119:1159–1167. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Noguchi T, Inoue H and Tanaka T: The
M1-and M2-type isozymes of rat pyruvate kinase are produced from
the same gene by alternative RNA splicing. J Biol Chem.
261:13807–13812. 1986. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Wu J, Hu L, Chen M, Cao W, Chen H and He
T: Pyruvate kinase M2 overexpression and poor prognosis in solid
tumors of digestive system: Evidence from 16 cohort studies. Onco
Targets Ther. 9:4277–4288. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Wang Y, Li Y, Jiang L, Ren X, Cheng B and
Xia J: Prognostic value of glycolysis markers in head and neck
squamous cell carcinoma: A meta-analysis. Aging (Albany NY).
13:7284–7299. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Haura EB, Turkson J and Jove R: Mechanisms
of disease: Insights into the emerging role of signal transducers
and activators of transcription in cancer. Nat Clin Pract Oncol.
2:315–324. 2005. View Article : Google Scholar : PubMed/NCBI
|