Introduction
Human ovarian cancer (HOC) is a major
life-threatening gynecological malignancy that accounts for ~3% of
all cancer incidences in females, with 240,000 newly diagnosed
cases and 150,000 deaths from HOC annually worldwide (1). Owing to the absence of early symptoms,
HOC is usually diagnosed at advanced stages that are difficult to
cure by simple surgical resection, and relapse commonly occurs
during the clinical course of HOC (2). Most patients with HOC exhibit peritoneal
metastases and suffer from perennial malignant ascites leading to
advanced stages, a massive progression and poor prognosis of HOC
(3,4).
Despite surgical interventions and the administration of anticancer
drug combinations, the 5-year survival rate of patients with
advanced-stage HOC remains low (5);
owing to the distinct biology of HOC, the effective treatment for
this disease remains limited (1).
Thus, it is urgent to develop novel and more effective treatments
for this disease.
A previous study revealed that several members of
the sirtuin (SIRT) family (SIRT1-SIRT7), the
mammalian homologues of the yeast silent information regulator
(Sir2) gene, serve crucial roles in the development of
cancers (6). The SIRT family includes
a specific class of nicotinamide adenine dinucleotide
(NAD+)-dependent deacetylase that targets several
protein substrates to carry out diverse biological functions, such
as oxidative stress regulation, metabolism, cell survival, division
and aging (6,7). SIRT3 is expressed as a full-length 44-kD
protein that is targeted to the mitochondria by its N-terminal
localization sequence, and it uniquely regulates apoptosis and cell
survival; it functions either as a tumor promoter or inhibitor
depending on the cell or tumor type and on the different types of
stress or cell apoptosis stimuli (8).
SIRT3 is expressed at low levels in human breast cancer and
lung cancer (9), acting as a
genomically expressed, mitochondrial-located tumor suppressor
(10,11). Similarly, aberrant low expression of
SIRT3 was demonstrated in patients with gastric cancer
(12). SIRT3 was also
indicated to be downregulated in prostate carcinoma, and
SIRT3 overexpression inhibited prostate cancer cell
proliferation and improved patient survival (13). The current study delivered the
SIRT3 gene with ultrasound-targeted microbubble destruction
(UTMD). UTMD has emerged as a novel gene therapy that appears to be
safe and repeatable, enables the plasmids in the bubbles to avoid
degradation in blood, and avoids safety issues of previous gene
delivery systems and focuses on gene delivery to target tissues
(14–16). UTMD has been documented to enhance
tissue or cell penetration and to serve as a supportive technique
for cancer therapies (17,18). The current study assessed the
biological mechanisms of the SIRT3 gene delivered by UTMD in
the malignant behavior of HOC cells to provide a theoretical
foundation for a novel target treatment for HOC.
Materials and methods
Cell culture
The HOC cell lines COC1, A2780 and SKOV3 and the
normal human ovarian epithelial cell line HOSEpiC were purchased
from National Collection of Authenticated Cell Cultures of The
Chinese Academy of Sciences and incubated in cell culture dishes at
a density of 1×105 cells/cm2. The cells were
cultured in RPMI-1640 medium (COC1 and SKOV3), DMEM (A2780) or
minimum essential medium (MEM; HOSEpiC) (all purchased from Gibco;
Thermo Fisher Scientific, Inc.) supplemented with 10% FBS (Hyclone;
Cytiva) at 37°C with 5% CO2 for 48 h. When confluence
reached 80–90%, cells were detached with 0.025% trypsin (Gibco;
Thermo Fisher Scientific, Inc.) and passaged.
SIRT3 recombinant vector
construction
The lentiviral vector pLVX–IRES-ZsGreen1, which
contains the green fluorescent protein (GFP) reporter gene, was
purchased from Clontech Laboratories, Inc. Human SIRT3 cDNA was
synthesized by Shanghai Sangon Biotech Co., Ltd. A SIRT3
target cDNA fragment was ligated into pLVX–IRES-ZsGreen1 vector to
generate pLVX-SIRT3-IRES-ZsGreen1 overexpression vector
(SIRT3 vector).
Microbubble (MB) preparation
Cationic lipid MBs were synthesized using
polyethylene glycol-40 stearate (1 mg/ml), 1-distearoyl
phosphatidylcholine (2 mg/ml), 1,2-dicarboxyl-3-trifluoromethyl
propane (0.4 mg/ml) and decafluorobutane gas (all purchased from
Avanti Polar Lipids, Inc.) by ultrasonic dispersion (220 V, 50 Hz,
100 W for 20 sec, at an interval of 10 sec; 10 cycles in total) in
aqueous phase. The MBs were observed under an optical microscope
(×400 magnification), and the particle size and electric potential
were measured using a Malvern Zetasizer Nano ZS90 (Malvern
Instruments, Inc.). The SIRT3 vector (4.0 µg) was incubated
in 50 µl MB suspension at 37°C for 30 min. The plasmids that did
not bind to the MBs were washed out with PBS. The
SIRT3-loaded MB was obtained and was resuspended in 500 µl
RPMI-1640 medium for subsequent experiments.
Cell transfection and grouping
Exponentially growing SKOV3 cells (1×105
cells/ml) at passage 3 were seeded into 6-well plates, and 50 µl
SIRT3-MB suspension was added to each well, followed by 1
MHz ultrasound treatment for 30 min at 2.5 W/cm2; these
cells were designated the SIRT3-MB group. The ultrasonic
therapeutic apparatus was provided by Institute of Ultrasound
Imaging of Chongqing Medical University (Chongqing, China)
(19). In addition, 4.0 µg
SIRT3 vector or empty vector was mixed with 2 µl
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) and further resuspended in 500 µl RPMI-1640
medium; 50 µl SIRT3 suspensions or empty vector suspensions
were loaded onto 6-well plates and named the SIRT3 group or
the negative control (NC) group, respectively. Moreover, SKOV3
cells mixed with 500 µl RPMI-1640 medium were used as the blank
group. At 24 h post transfection, GFP in cells was observed under
an Olympus FSX100 fluorescence microscope (×100 magnification;
Olympus Corporation), and the mRNA and protein expression levels of
SIRT3 in cells was measured using reverse
transcription-quantitative PCR (RT-qPCR) and western blot analysis,
respectively, to validate the transfection efficiency.
RT-qPCR
Total RNA from 5×107 HOC cells was
extracted using the TRIzol® (Invitrogen; Thermo Fisher
Scientific, Inc.) one-step method, and the RNA quality was
identified by formaldehyde denaturing gel electrophoresis. cDNA was
generated by RNA reverse transcriptase M-MLV (cat. no. RP1105,
Beijing Solarbio Science & Technology Co., Inc.); the reaction
conditions were as follows: 42°C for 60 min, and 95°C for 5 min.
SYBR Green (Takara Biotechnology Co., Ltd.) was used to conduct
qPCR, and PCR primers (Table I) were
designed and synthesized by Shanghai Sangon Biotech Co., Ltd., with
β-actin set as the internal reference. The PCR volume was 15 µl,
and the reaction conditions were as follows: Initial denaturation
at 96°C for 5 min; followed by 30 cycles of denaturation at 94°C
for 40 sec, annealing at 53°C for 45 sec and extension at 72°C for
1 min; and a final extension at 72°C for 10 min. The PCR product
was examined using agarose gel electrophoresis to test the length
and concentration of the amplified fragment (data not shown). Data
were analyzed using the 2−ΔΔCq method (20), where Cq is the quantitation cycle and
2−ΔΔCq refers to the ratio of the target gene expression
between the experimental group and the control group. The formula
is as follows: ΔΔCq=[Cq (target gene)-Cq (control
gene)]experiment group-[Cq (target gene)-Cq (control
gene)]control group.
 | Table I.Primer sequences used for reverse
transcription-quantitative PCR. |
Table I.
Primer sequences used for reverse
transcription-quantitative PCR.
| Gene | Primer sequence
(5′→3′) |
|---|
| SIRT | F:
CCGCTCGAGATGGCGTTCTGGGGTTGG |
| SIRT | R:
CGCGGATCCCTATTTGTCTGGTCCATCA |
| E-cadherin | F:
TCACATCCTACACTGCCCAG |
| E-cadherin | R:
AGTGTCCCTGTTCCAGTAGC |
| N-cadherin | F:
AGGGGACCTTTTCCTCAAGA |
| N-cadherin | R:
TCAAATGAAACCGGGCTATC |
| Vimentin | F:
GGACCAGCTAACCAACGACA |
| Vimentin | R:
AAGGTCAAGACGTGCCAGAG |
| HIF-1α | F:
TCCAGAAGGAGTTCTTATTCG |
| HIF-1α | R:
AAAATCTCATCCAAGAAGCC |
| β-actin | F:
ACAGTCAGCCGCATCTTCTT |
| β-actin | R:
GACAAGCTTCCCGTTCTCAG |
Western blot analysis
When the confluence of HOC cell lines reached about
70%, the cells were treated for 48 h with MB or recombinant
plasmid, and then protein was extracted using RIPA lysis buffer
(cat. no. P0013B; Beyotime Institute of Biotechnology), and protein
concentration was determined in accordance with the instructions of
the BCA kit (Qiagen GmbH). Extracted proteins (20–40 µg) were
separated by 12% SDS-PAGE at a voltage from 80 to 120 V, and
subsequently transferred onto PVDF membranes using a semidry method
(at 80 mV for 30–45 min). The membranes were blocked with 5% BSA
(cat. no. AR0185; Boster Biological Technology) for 1 h at room
temperature and subsequently incubated at 4°C overnight with
primary antibodies (all from Abcam) against the following antigens:
Cleaved caspase-3 (1:1,000; cat. no. ab2302), pro-caspase 3
(1:10,000; cat. no. ab32499), Bcl-2 (1:1,000; cat. no. ab32124),
cleaved caspase-9 (1:500; ab2324), pro-caspase 9 (1:1,000; cat. no.
ab138412), hypoxia inducible factor-1α (HIF-1α; 1:500; cat. no.
ab51608), Vimentin (1:1,000; cat. no. ab193555), E-cadherin (1:50;
cat. no. ab1416), N-cadherin (1:100; cat. no. ab18203) and β-actin
(1:5,000; cat. no. ab227387). Following three washes (5 min each)
with TBS + 0.05% Tween-20 (TBST), membranes were incubated with a
horseradish peroxidase (HRP)-labeled goat anti-rabbit IgG (H + L)
secondary antibody (1:1,000; cat. no. A0208; Beyotime Institute of
Biotechnology) at room temperature for 1 h and washed with TBST
three times (5 min each) before chemiluminescence development using
BeyoECL Plus (cat. no. P0018S; Beyotime Institute of
Biotechnology). A Bio-Rad Gel Doc EZ Imager (Bio-Rad Laboratories,
Inc.) was used to visualize the protein bands. The signal intensity
of target bands was analyzed using ImageJ software V1.8.0 [National
Institutes of Health (NIH)]. β-actin was used to normalized protein
expression levels.
Transwell invasion assay
The top chamber of each 24-well Transwell was
precoated with Matrigel (BD Biosciences) for 30 min, filled with 30
µl RPMI-1640 medium and placed in a CO2 incubator. The
SKOV3 cells were detached using trypsin, centrifuged at 800 × g for
3 min at room temperature, and then resuspended using serum-free
RPMI-1640 medium and dispersed into a cell suspension at a density
of 5×105 cells/ml. The lower chamber was filled with 500
µl 10% FBS-supplemented RPMI-1640 medium and loaded with 200 µl
cell suspension. The cells were cultured at 37°C with 5%
CO2 for 48 h. Subsequently, the medium in the chambers
was washed with PBS, and the cells were stained with 0.1% crystal
violet for 10 min at room temperature. Excess crystal violet was
washed away with running water, and cells on the upper membrane
were removed with a cotton swab, images of the cells on the lower
membrane were captured under an Olympus X71 inverted phase contrast
microscope (magnification, ×100; Olympus Corporation) for manual
cell number calculation.
Wound healing assay
Cell migration was examined using the scratch test.
HOC cells were seeded on 6-well plates at 5×105 cells
per well and cultured in complete medium with 10% FBS at 37°C with
5% CO2 for 24 h. When cells reached 90% confluency, a 1
ml pipette tip was used to produce a scratch on the cell monolayer
and the cells were washed with PBS and cultured with serum-free
medium at 37°C. Images of the wounds were captured under an Olympus
X71 inverted phase contrast microscope (magnification, ×100;
Olympus Corporation) at 0, 24 and 48 h. ImageJ software V1.8.0
(NIH) was used to analyze and calculate the scratch area. The ratio
of scratch area of differently treated cells to scratch area of the
same cells at 0 h represented the relative cell migration rate.
MTT assay
HOC cells from each group at logarithmic growth
phase were detached using trypsin and resuspended to
1×104 cells/ml single cell suspension in RPMI-1640
medium containing 10% FBS-. The suspension was mixed and plated on
96-well plates at 200 µl (2×103 cells) per well; wells
filled with culture medium and without cells were used as the blank
control. Five duplicated wells were set for each group. The cells
were cultured in a 37°C incubator with 5% CO2 for 0–3
days. MTT solution (20 µl of 5 mg/ml) was added at 0, 24, 48 and 72
h, and the culture was terminated 4 h later with the supernatant of
each well discarded. The well was filled with 200 µl dimethyl
sulfoxide to remove the purple formazan crystals, and the plate was
vortexed for 10 min to fully dissolve the purple formazan crystals
at room temperature. The optical density (OD) value of each well at
OD490 was detected using multifunctional enzyme label
analyzer (Molecular Devices, LLC).
5-Ethynyl-2′-deoxyuridine (EdU)
labeling assay
Exponentially growing SKOV3 cells at passage 3 were
collected, and the DNA replication ability of cells was detected
using a Cell-Light EdU Apollo567 In Vitro Kit (Guangzhou RiboBio
Co., Ltd.) following the manufacturer's instructions. Five views of
cells were randomly selected and observed under a fluorescence
microscope (magnification, ×100). The blue fluorescence (DAPI)
refers to all cells, and the red fluorescence indicates the
replicating cells stained by EdU. The EdU-positive cell rate in
five randomly selected fields was determined by ImageJ software
V1.8.0 (NIH).
Colony formation assay
SKOV3 cells in logarithmic growth phase were
collected, detached with 0.025% trypsin and seeded on 6-well plates
at a density of 1,000 cells/well for incubation at 37°C with 5%
CO2 for 14 days. Subsequently, the cells were fixed with
75% methanol for 30 min at room temperature, followed by staining
with 0.2% crystal violet at room temperature. Under an Olympus X71
inverted phase contrast microscope (magnification, ×100; Olympus
Corporation), 10 fields were randomly selected and the number of
clones with >50 cells in the field were counted; each experiment
was repeated three times.
Flow cytometry
Cell cycle and apoptosis were detected by cell cycle
and apoptosis detection kit (cat. no. C1052; Beyotime Institute of
Biotechnology). For the cell cycle assay, cells were detached at
37°C using trypsin into single cells, and the detachment was
terminated after adding the serum-contained medium; next, cells
were centrifuged at 800 × g at room temperature for 3 min, the
supernatant was discarded, and the pellet was resuspended and
washed with PBS to adjust the cell concentration to
1×106 cells/ml. The single cell suspension was
centrifuged at 1,000 × g for 5 min at room temperature and the
supernatant was discarded. Cells were mixed with 500 µl 70% cold
ethanol and fixed at 4°C overnight. After discarding the fixation
solution, the cells were further washed with 1 ml PBS and
centrifuged at 1,000 × g for 3 min at room temperature; the
supernatant was discarded. The cells were then treated with 100 µl
RNase A and incubated in a water bath at 37°C for 30 min.
Subsequently, the cells were mixed with 400 µl propidium iodide
(PI) at 4°C in the dark for 30 min for detection. The red
fluorescence at a wavelength of 480 nm was recorded using a flow
cytometer (Beckman Coulter, Inc.) and the data were analyzed using
CellQuest software v3.3 (BD Biosciences).
For the apoptosis assay, the cell suspension was
mixed with 2 µl Annexin V-FITC (20 µg/ml) and allowed to stand on
ice in the dark for 15 min. Next, the mixture was transferred into
the flow cytometry tube and mixed with 300 µl PBS, after which each
sample was further mixed with 1 µl PI (50 µg/ml) and detected
within 30 min using a flow cytometer (Beckman Coulter, Inc). Modfit
software v5.0 (Verity Software House, Inc.) was used for analysis.
The results were interpreted as follows: Annexin V-FITC was set as
the horizontal axis and PI was set as the vertical axis, with the
upper-left quadrant containing the mechanically injured cells, the
upper-right quadrant containing the late apoptotic cells or
necrotic cells, the lower-left quadrant containing the negative
normal cells and the lower-right quadrant containing the early
apoptotic cells. Total apoptotic rates were measured as the sum of
early- and late-stage apoptosis.
Xenograft tumors in nude mice
A total of 40 specific-pathogen-free grade BALB/c
nude mice [age, 4–6 weeks; weight, 20±2 g; purchased from Beijing
Vital River Laboratory Animal Technology Co., Ltd.; animal license.
no. SCXK (Beijing) 2015-0001] were numbered by weight and randomly
assigned into four groups (n=10 mice/group). The mice were
separately housed at a constant temperature (20±2°C) and humidity
(50-60%) with free access to food and water under a 12-h light-dark
cycle. The housing conditions met the requirements for
environmental facilities for medical laboratory animals. A total of
4×106 SKOV3 cells were suspended in 200 µl physiological
saline and subcutaneously injected into the nude mice through the
right axilla. At 7 days post-injection, the SIRT3 vector or NC
empty vector was diluted in 200 µl saline and then injected into
the mice through the caudal vein, and the groups of mice were
correspondingly named the SIRT3 group and the NC group. Similarly,
the SIRT3-MB vector was diluted in 200 µl saline and injected into
the mice and subjected to 10 min of ultrasound treatment at 10 MHz
at the cancer cell transplantation sites; this group was named the
SIRT3-MB group. A blank group was set as a control in which nude
mice were injected with an equal volume of saline. From the date of
transplantation, the behavior (excessive excitement, fighting,
abnormal calls or excessive fear), activity, response to external
stimulation (human touch, change of light), diet, defecation and
weight of nude mice were observed every day to monitor their health
and behavior. Tumor formation was also recorded every day. The
tumor volume of the mice was measured every 7 days and calculated
as follows: (length × width2)/2 for a total of 35 days
(21). After subcutaneous injection,
each mouse bore only one tumor. On day 35 following cell injection,
the tumors were 1–1.5 cm in length, accompanied by weight loss and
decreased response to external stimuli (Fig. S1). The mice were then euthanized by
intraperitoneal administration of overdose of pentobarbital (800
mg/kg) (22,23). After observation of pupil dilation and
complete cardiac arrest, the tumors were removed, weighed and
prepared for immunohistochemical assays.
Immunohistochemical assay
The extracted mouse tumor tissues of each group were
fixed using 4% paraformaldehyde at 4°C for 24 h, embedded in
paraffin, dewaxed and cut into five sections (4 µm each). The
sections were washed three times with PBS, and subsequently
incubated with 3 drops of 3% H2O2 at room
temperature for 15 min to eliminate the activity of endogenous
peroxidase. After three washes with PBS, the slices were incubated
with normal goat serum (cat. no. AR0009; Wuhan Boster Biological
Technology, Ltd.) blocking solution and allowed to stand at room
temperature for 15 min. Next, the slices were incubated at 4°C
overnight with the following primary antibodies obtained from Abcam
(50 µl each): Anti-Ki67 (1:500; cat. no. ab15580), anti-E-cadherin
(1:100; cat. no. ab1416) and anti-N-cadherin (1:100; cat. no.
ab18203). The slices were then washed with PBS three times,
followed by the incubation with HRP-AffiniPure Goat Anti-Rabbit IgG
(H + L) secondary antibodies (1:500; cat. no. 1:500; Wuhan Boster
Biological Technology, Ltd.) for 15 min at 37°C. Following three
PBS washes, the tissues were incubated with 40 µl HRP-labeled
streptomyces ovalbumin solution (2 µg/ml; cat. no. SY0746-CIK;
Beijing Biolab Technology Co., Ltd.) in PBS at 37°C for 15 min.
After three PBS washes, the slices were treated with
2,4-diaminobutyric acid (DAB) for color reaction for 3–5 min at
room temperature, after which they were washed with distilled water
and counterstained with hematoxylin for 30 sec at room temperature,
and finally dehydrated using an ascending series of ethanol and
sealed with neutral gum. Five non-overlapping fields were randomly
selected for each slice and observed under an LED5000 light
microscope (magnification, ×100; Leica Microsystems GmbH). The
number of positive cells was counted.
Dual luciferase reporter assay
HIF-1α is a possible target gene of SIRT3 (24). The mutant-type (MT) sequence and
wild-type (WT) sequence of the binding sites of HIF-1α and SIRT3
were designed. The MT sequence and WT sequence fragments were
cloned into the pmir-GLO vector (Promega Corporation). The pmir-GLO
vector containing the MT sequence was co-transfected with SIRT3
vector or empty vector NC into SKOV3 cells using the
Lipofectamine® 3000 (Invitrogen; Thermo Fisher
Scientific, Inc.) which were subsequently named the MT + SIRT3
group and the MT + NC group, respectively. Likewise, the pmir-GLO
vector containing the WT sequence was transfected in similar
combinations, and the groups were named the WT + SIRT3 group and
the WT + NC group. The fluorescence intensity of cells in each
group was measured with a Dual Luciferase Reporter Gene Detection
kit (Beijing Yuanpinghao Biotech Co., Ltd.) 48 h after
transfections using a GloMax 20/20 luminometer (Promega
Corporation). Renilla activity was used for
normalization.
Statistical analysis
The Statistical Package for the Social Sciences
version 21.0 (IBM Corp.) was used for data analysis. The normality
test was conducted using the Kolmogorov-Smirnov method. Measurement
data were in normal distribution and are expressed as mean ±
standard deviation. Differences among multiple groups were compared
using one-way followed by Tukey's post hoc test for pairwise
comparisons. The P-value was obtained from a two-tailed test and
P<0.05 was considered to indicate a statistically significant
difference.
Results
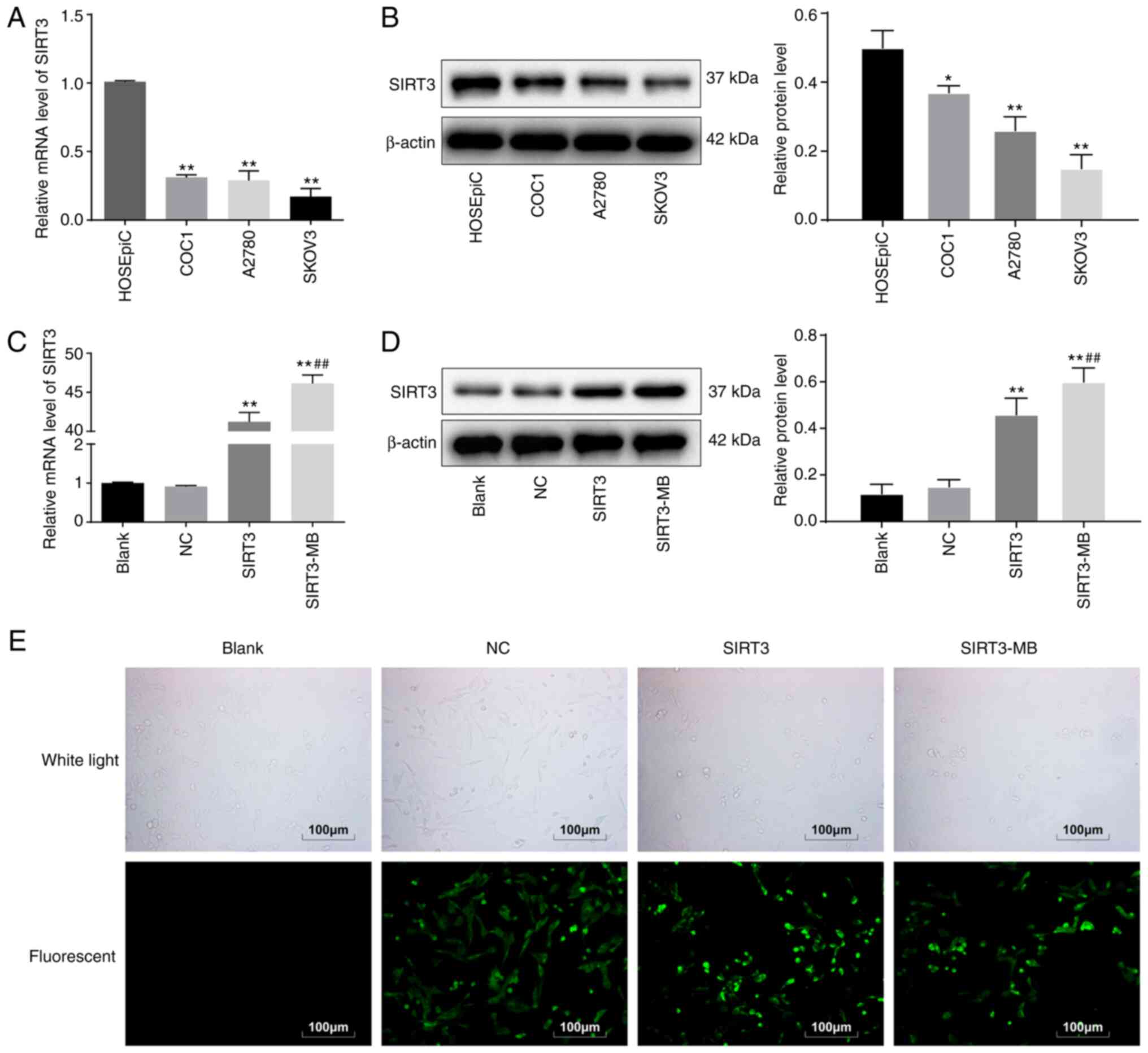
SIRT3 is expressed at low levels in
HOC cells
The expression of SIRT3 mRNA in the HOC cell lines
COC1, A2780 and SKOV3 and human ovarian epithelial cells HOSEpiC
was detected by RT-qPCR, the results of which indicated that the
SIRT3 mRNA levels in HOC cells were significantly lower
compared with that in HOSEpiC (P<0.01; Fig. 1A). SIRT3 protein expression in cells
exhibited a similar trend according to western blot analysis
(P<0.05; Fig. 1B). Among the cell
lines, SKOV3 cells had the lowest SIRT3 expression and were
selected for the subsequent experiments.
After transfecting the pLVX-SIRT3-IRES-ZsGreen1
vector into SKOV3 cells for 48 h, the mRNA expression of
SIRT3 was significantly increased compared with Blank
(P<0.01; Fig. 1C). Moreover,
western blot analysis demonstrated that SIRT3 protein expression
was also increased (P<0.01; Fig.
1D). Under a fluorescence microscope, the cells presented with
green fluorescence (Fig. 1E), and the
transfection efficiency exceeded 80%, indicating that a SKOV3 cell
line overexpressing SIRT3 was successfully constructed. When SKOV3
cells were treated with SIRT3-MB, SIRT3 mRNA (P<0.01; Fig. 1C) and protein (P<0.05; Fig. 1D) expression levels were further
increased compare with the SIRT3 group; the cells also exhibited
green fluorescence (Fig. 1E). It was
suggested that ultrasound microbubbles could further promote the
transfection efficiency of SIRT3 plasmid.
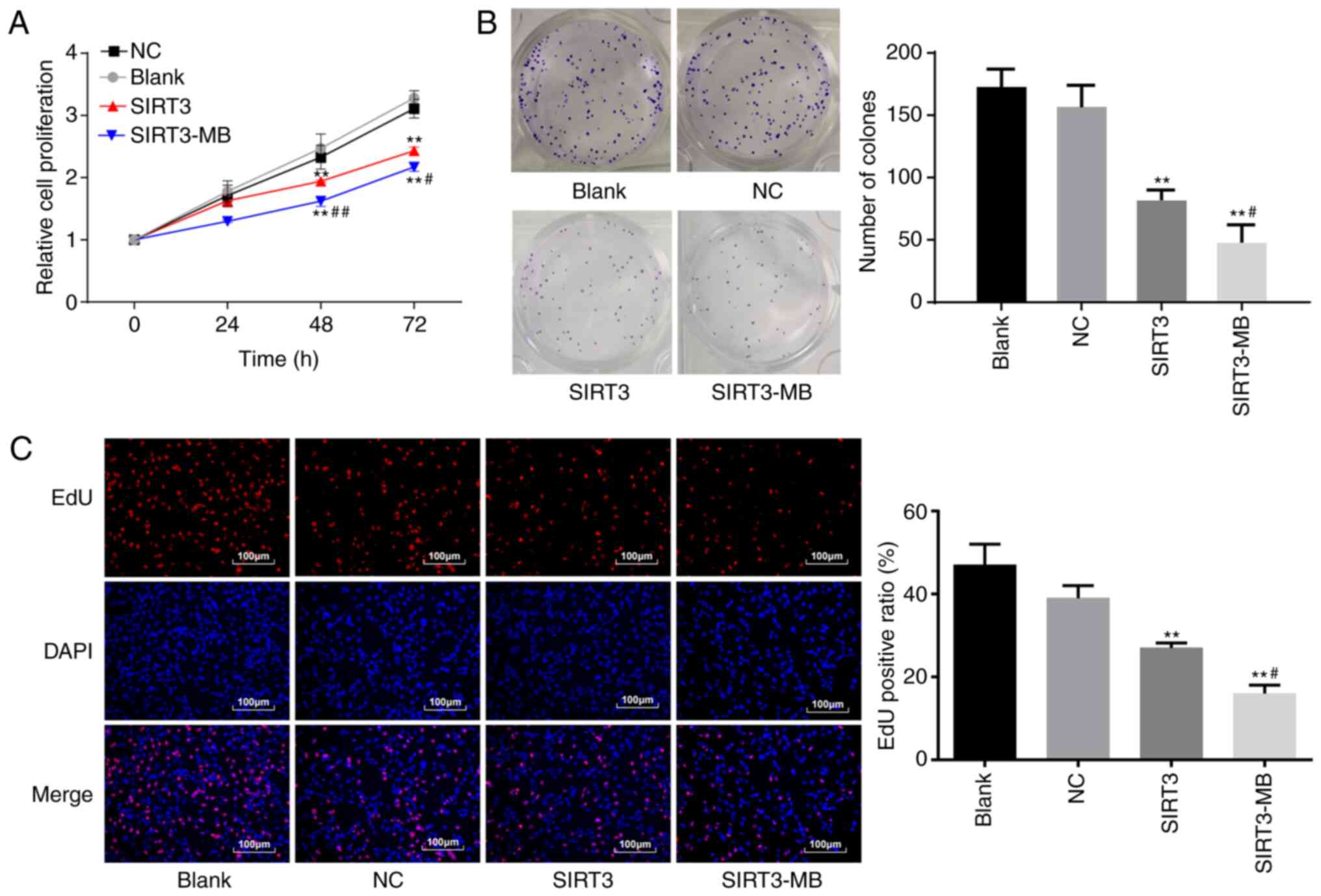
UTMD-mediated overexpression of SIRT3
inhibits the proliferation of SKOV3 cells in vitro
Cell proliferation was measured by an MTT assay. No
significant difference was observed between the NC group and the
blank group (P>0.05), whereas cell proliferation rates at 48
(P<0.01) and 72 h (P<0.01) were significantly lower in the
SIRT3 group compared with the blank and NC groups (Fig. 2A). Cells in the SIRT3-MB group after
UTMD treatment showed further reduced cell proliferation rates
compared with the SIRT3 group at 48 and 72 h (P<0.01 and
P<0.05, respectively).
The colony formation ability of SKOV3 cells was
measured (Fig. 2B). No significant
difference was observed in the number of clones between the blank
group and the NC group (P>0.05); however, compared with the
blank group, the number of clones was significantly reduced in the
SIRT3 group (P<0.01). After UTMD treatment, cells in the
SIRT3-MB group showed more significantly decreased cell clones
compared with the SIRT3 group (P<0.05; Fig. 2B). The results above indicated that
overexpression of SIRT3 could inhibit the proliferation of SKOV3
cells in vitro.
The EdU assay was used to measure the DNA
replication activity, the results of which demonstrated that the
DNA replication activity of cells in the blank and NC groups were
not significantly different in the rate of EdU-positive cells
(Fig. 2C). The DNA replication
activity in the cells overexpressing SIRT3 was significantly
decreased compared with the blank group (P<0.01). After UTMD
treatment, cells in the SIRT3-MB group showed a further decrease in
the number of EdU-positive cells compared with the SIRT3 group
(P<0.05; Fig. 2C).
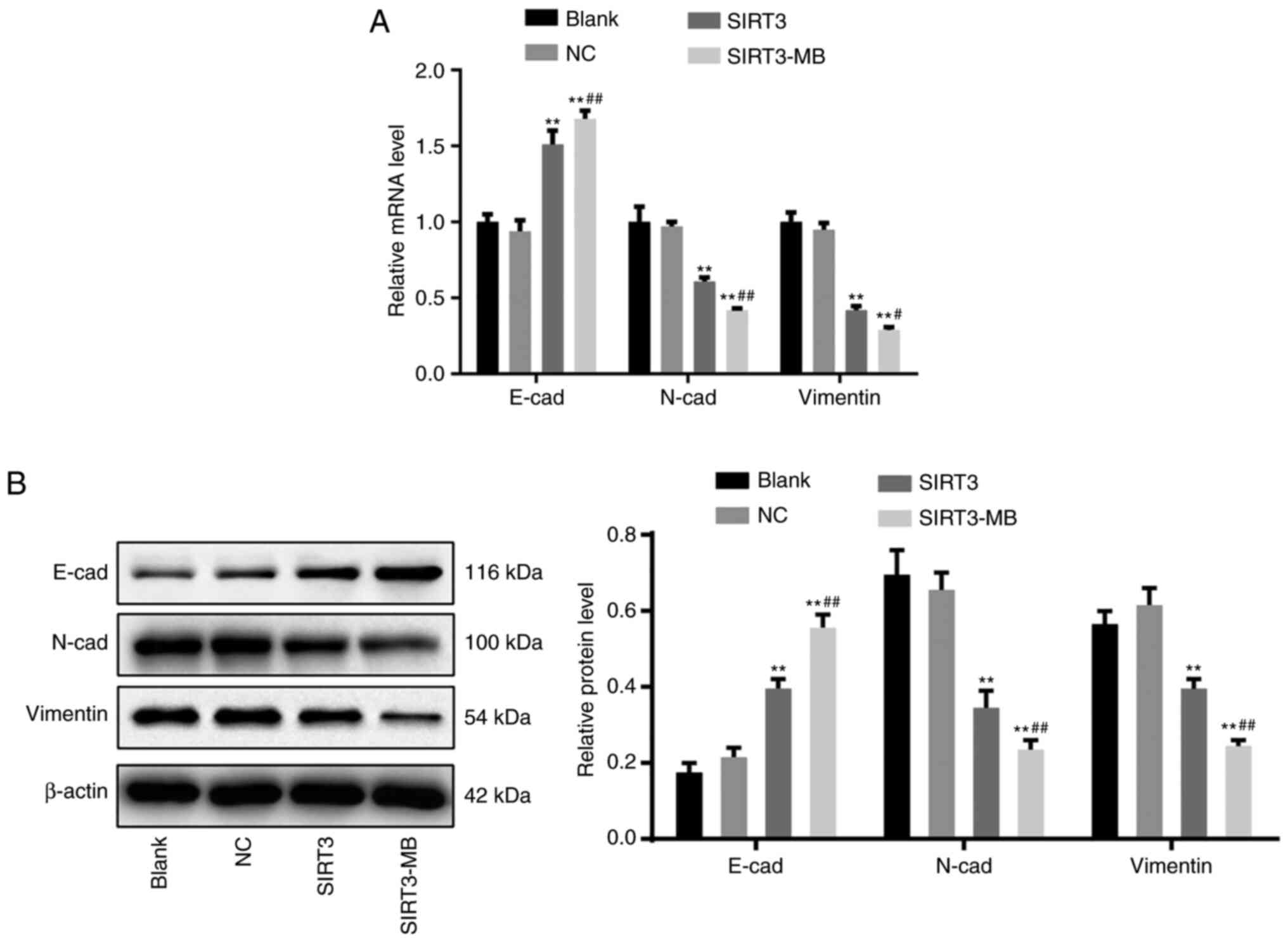
UTMD-mediated overexpression of SIRT3
inhibits the epithelial-mesenchymal transition (EMT) of SKOV3
cells
EMT is a phenotypic change closely related to tumor
metastasis. EMT is usually marked by decreased expression of
E-cadherin and increased expression of N-cadherin and Vimentin
(25). The results of RT-qPCR
revealed that the mRNA expression levels of E-cadherin in
the SIRT3 group were significantly increased compared with those in
the blank and NC groups (P<0.01), whereas the mRNA expression
levels of N-cadherin and Vimentin were significantly
decreased (P<0.01) (Fig. 3A).
Moreover, after UTMD treatment, cells in the SIRT3-MB group
demonstrated further elevated E-cadherin mRNA expression
(P<0.01), as well as further decreased N-cadherin
(P<0.01) and Vimentin (P<0.05) mRNA expression
compared with the SIRT3 group (Fig.
3A). Similar trends in protein expression levels were observed
by western blot analysis (Fig. 3B).
These results suggested that overexpression of SIRT3 may inhibit
EMT of SKOV3 cells.
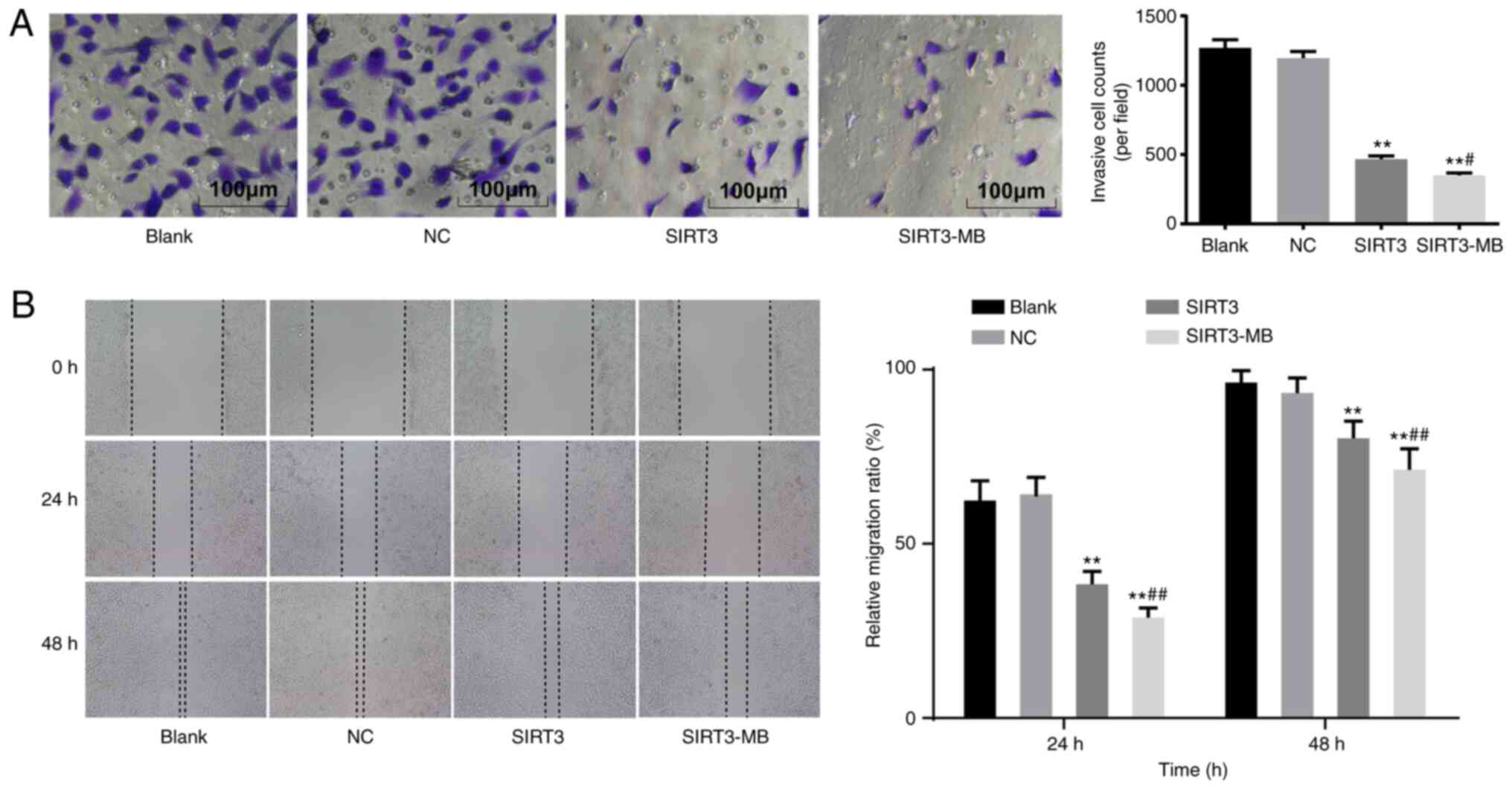
UTMD-mediated overexpression of SIRT3
suppresses the migration and invasion of SKOV3 cells
Malignant tumors are characterized by the ability to
invade and migrate (26). Therefore,
the invasive and migratory abilities of SKOV3 cells were determined
by in vitro Matrigel and wound healing experiments,
respectively. The results demonstrated that cell invasion
(P<0.01) and migration (P<0.01) were significantly decreased
in the SIRT3 group compared with the blank and NC groups (Fig. 4A and B). After UTMD treatment, SKOV3
cells in the SIRT3-MB group demonstrated further decreases in
invasion (P<0.05) and migration (P<0.01) compared with those
in the SIRT3 group (Fig. 4A and
B).
UTMD-mediated overexpression of SIRT3
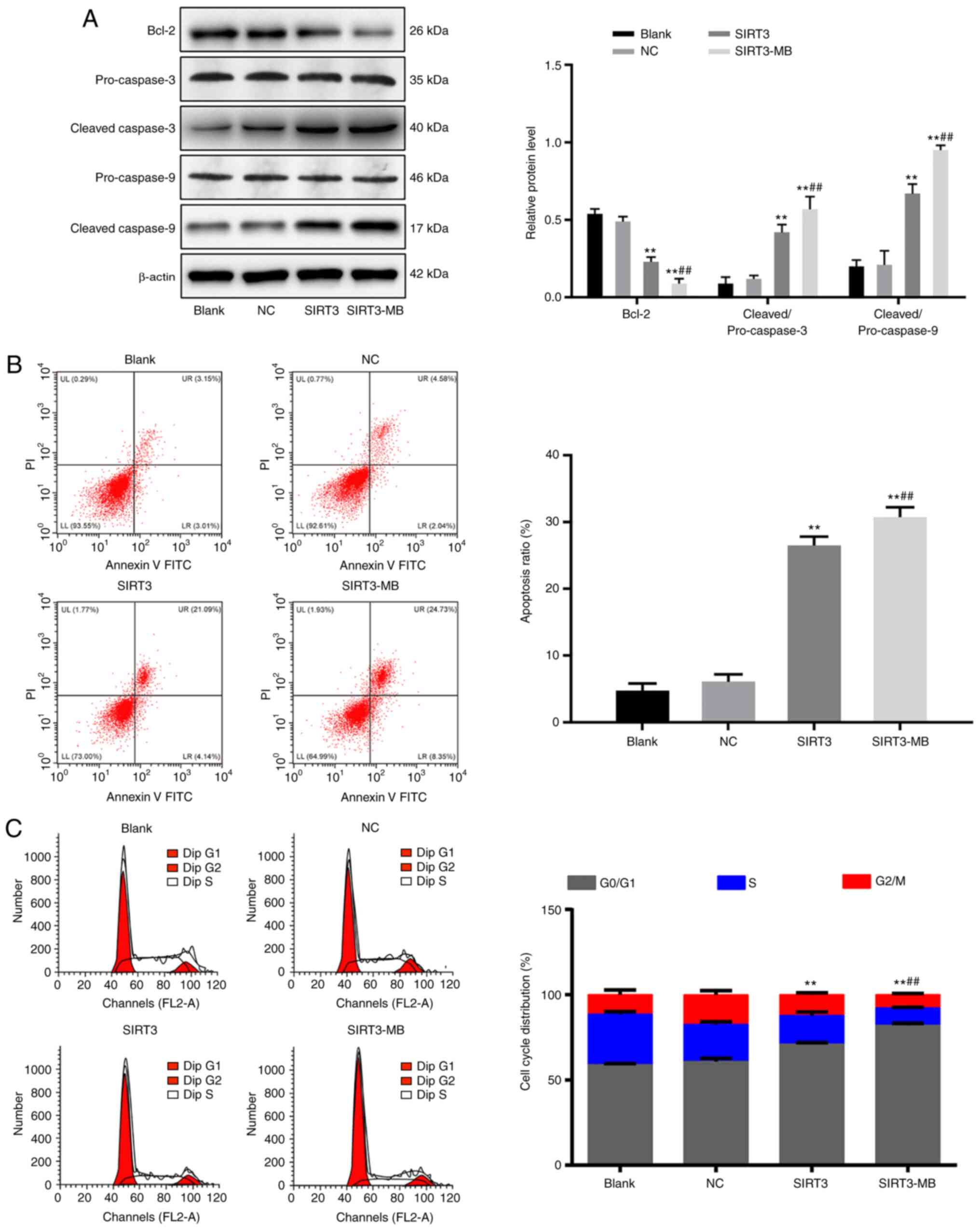
promotes apoptosis and cell cycle arrest in SKOV3 cells
The protein expression levels of cell
apoptosis-related proteins, including cleaved caspase-3,
pro-caspase-3, Bcl-2, cleaved caspase-9 and pro-caspase-9 were
measured by western blot analysis. No significant difference was
observed in the protein expression levels of the above factors
between the blank and NC groups (Fig.
5A). Compared with those in the blank group, the protein ratio
levels of cleaved/pro-caspase-3 and cleaved/pro-caspase-9 were
significantly increased in the SIRT3 group (P<0.01), whereas the
level of Bcl-2 was decreased (P<0.01). After UTMD treatment, the
protein ratio levels of cleaved/pro-caspase-3 and
cleaved/pro-caspase-9 were further increased, whereas the Bcl-2
protein level was further decreased in the SIRT3-MB group, compared
with the SIRT3 group (all P<0.01; Fig.
5A).
Flow cytometry was performed to detect cell
apoptosis (early + late) and cell cycle progression, and the
results showed that, compared with blank and NC groups, the number
of early apoptotic cells was increased (P<0.01; Fig. 5B), G2/M cells were decreased
(P<0.01), and G0/G1 cells were increased significantly in SIRT3
group (P<0.01) (Fig. 5C). Compared
with that in the SIRT3 group, cell apoptosis was significantly
increased in the SIRT3-MB group (P<0.01). Fewer cells were
detected in the G2/M phase (P<0.01), whereas significantly more
cells were found in G0/G1 phase (P<0.01).
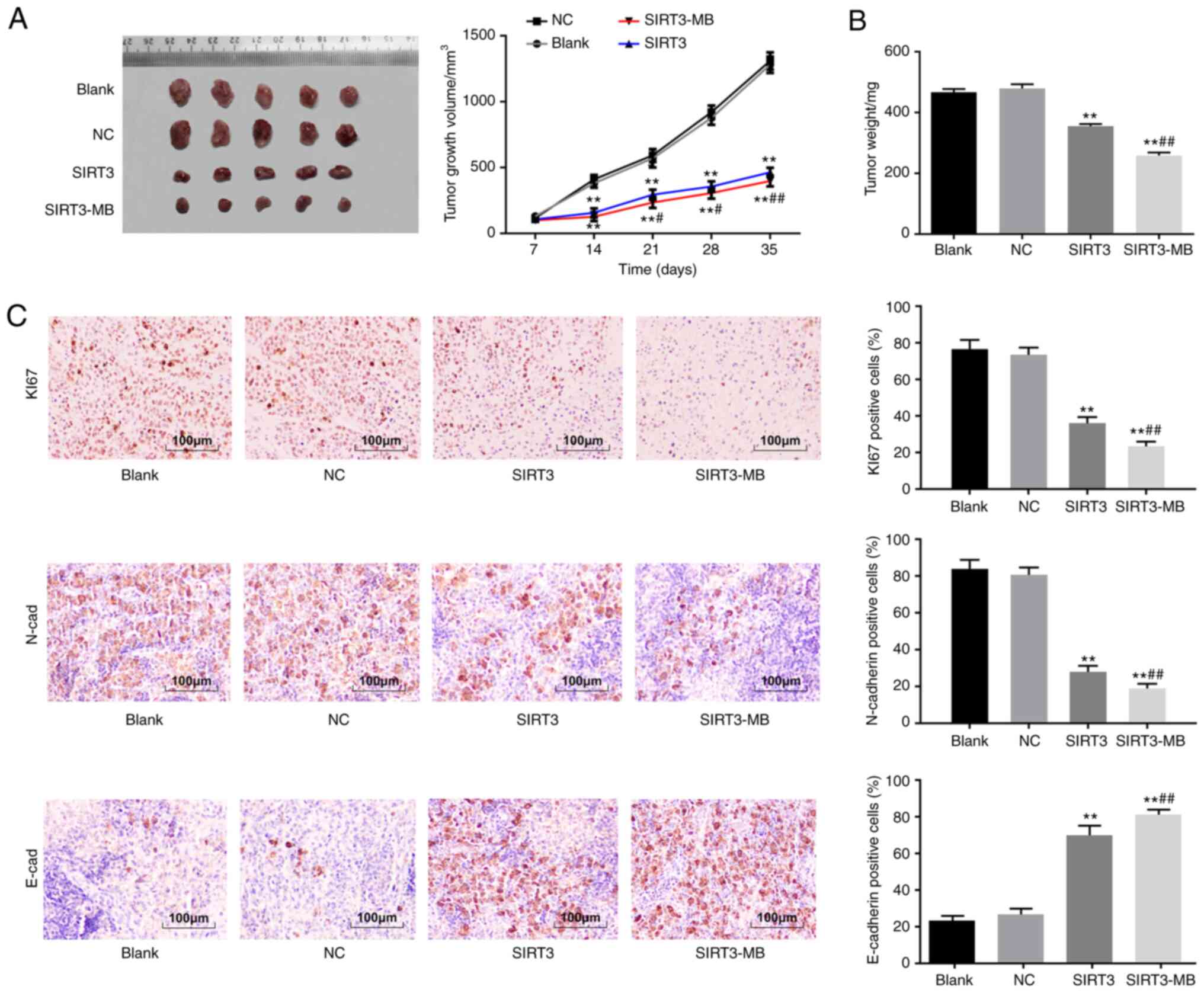
UTMD-mediated overexpression of SIRT3
inhibits tumorigenesis of SKOV3 cells in vivo
A nude mouse model with SKOV3 ×enograft tumors was
established, and the effect of SIRT3 on the in vivo growth
of these cells was evaluated by measuring tumor volume and weight.
The results revealed that the volume of xenograft tumors (after day
14) in the SIRT3 and SIRT3-MB groups was significantly smaller
compared with those in mice in the blank and NC groups; xenograft
tumor growth inhibition was more pronounced in the SIRT3-MB group
(all P<0.05; Fig. 6A). On day 35,
the largest tumor volume of nude mice was 1,430 mm3.
Tumor weight was inhibited in the SIRT3 group compared with the
blank and NC groups (P<0.01), and further inhibited in the
SIRT3-MB group compared with SIRT3 group (P<0.01) (Fig. 6B).
Ki67 is an antigen associated with cell
proliferation. The results of the immunohistochemistry demonstrated
that, compared with those in the blank and NC groups, the number of
Ki67- and N-cadherin-expressing cells were significantly decreased
in the SIRT3 group, whereas the number of E-cadherin-positive cells
was increased (P<0.01; Fig. 6C).
In the SIRT3-MB group, the number of Ki67- and N-cadherin-positive
cells in tumor tissues were further decreased, and the
positive-expression rate of E-cadherin was further increased
compared with that in the SIRT3 group (all P<0.01; Fig. 6C). These results indicated that
UTMD-mediated overexpression of SIRT3 may inhibit tumorigenesis of
SKOV3 cells in vivo.
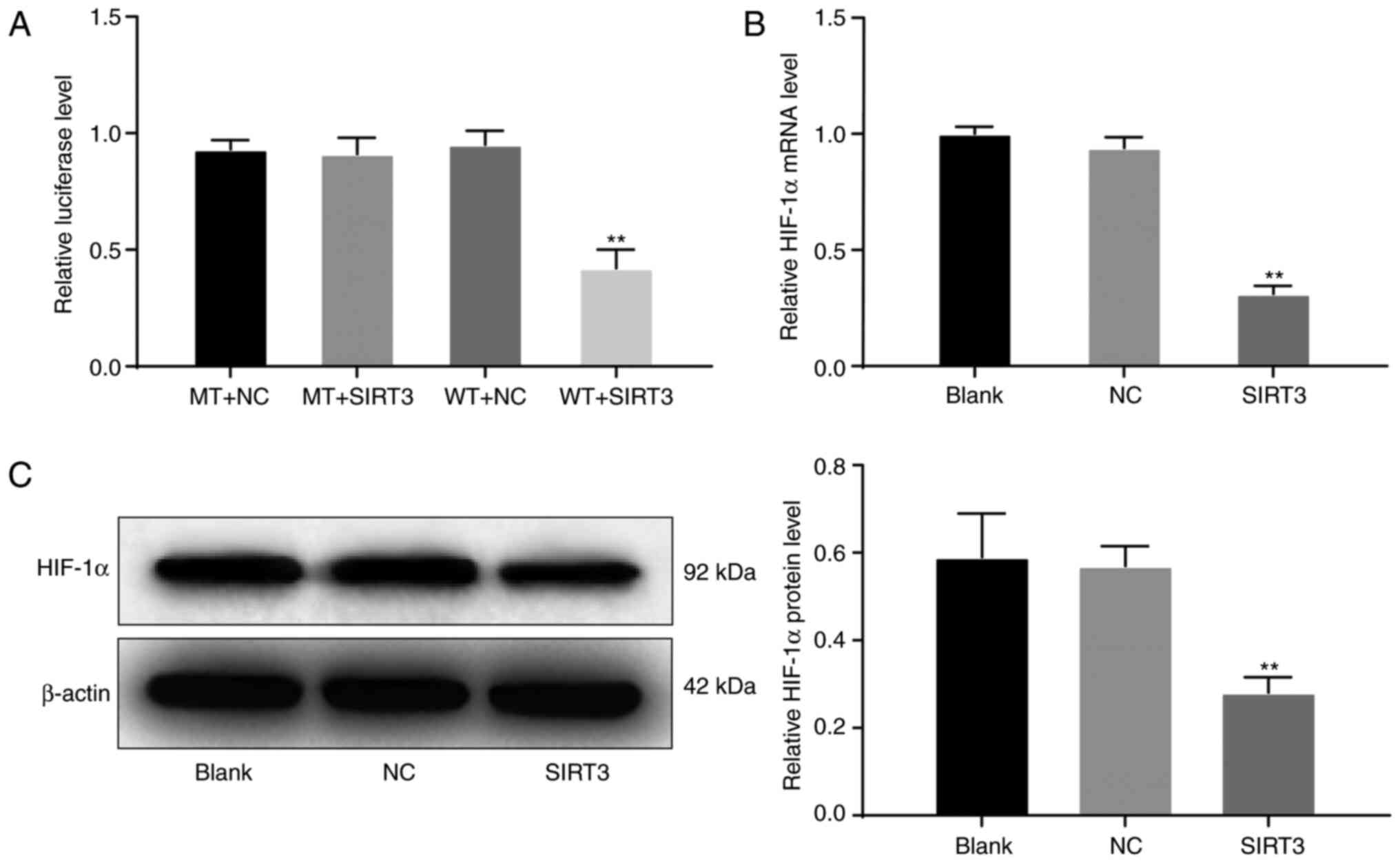
SIRT3 may inhibit the growth of HOC
SKOV3 cells by regulating HIF-1α
HIF-1α is a cytokine produced in response to tumor
hypoxia. Under hypoxic conditions, the HIF-1α signaling pathway is
activated, inducing a range of cellular activities, such as
metabolism, inflammatory response, angiogenesis, and tumor
formation. The HIF-1α signaling pathway was found in many tumors,
including HOC (27). Therefore,
combining these observations with those aforementioned, we
hypothesized that SIRT3 may inhibit the growth of HOC cells by
regulating the HIF-1α signaling pathway. The dual luciferase assay
results demonstrated that there was no significant difference in
luciferase activity among the MT + SIRT3, MT + NC and WT + NC
groups, whereas the luciferase activity in the WT + SIRT3 group was
significantly decreased (P<0.01; Fig.
7A). The results of RT-qPCR revealed that the mRNA expression
levels of HIF-1α in the SIRT3 group was significantly decreased
compared with that in the blank and NC groups (P<0.01; Fig. 7B). Similarly, western blot analysis
results revealed that, compared with the expression in the blank
and NC groups, the protein expression levels of HIF-1α in the SIRT3
group was significantly decreased (P<0.01; Fig. 7C). These results suggested that
SIRT3 may inhibit the growth of HOC cells by regulating the
HIF-1α signaling pathway.
Discussion
HOC remains one of the leading causes of
cancer-related mortalities among patients with gynecologic
malignancies (28). SIRT3 has roused
wide interest in cancer treatment as it could serve as either a
tumor suppressor or promoter in different cases (8). UTMD has been suggested as a promising
non-invasive tool for site-specific gene delivery allowing direct
transfection into cells (29,30). In the present study, the role of SIRT3
in the malignant behaviors of HOC cells was investigated, and it
was found that UTMD-mediated overexpression of SIRT3 could
reduce the malignant behaviors of SKOV3 cells, possibly by
downregulating HIF-1α.
In the present study, low expression of SIRT3
was demonstrated in SKOV3 cells. Following UTMD treatment,
SIRT3 expression was significantly elevated and led to
reduced cell proliferation, migration and invasion, as well as
decreased Ki67, Vimentin and N-cadherin expression and increased
E-cadherin expression, in addition to increased expression of
caspase-3 and caspase-9. However, decreased expression of Bcl-2 was
demonstrated. Ki67 is an important proliferation marker and is
activated during all phases in the cell cycle (31). E-cadherin is a cell junction protein
that is downregulated or silenced, whereas Vimentin expression is
usually increased during EMT progression (32). Bcl-2 is an anti-apoptotic protein
whose high expression has been suggested to be frequently presented
in human cancers and closely associated with resistance to
chemotherapies and cancer development (33); caspase-3 and caspase-9 are well-known
apoptosis inducers (34). Moreover,
the present study revealed that UTMD further prompted the
inhibitory effect of SIRT3 on SKOV3 cells and further suppressed
tumor growth in vivo. UTMD has emerged rapidly in recent
years as a novel technique for gene therapy that shows potential
for wide use in gene therapy and cell biology. For instance, UTMD
combined with liposomes has been suggested to promote short hairpin
(sh)RNA transfection efficiency into cells to inhibit metadherin
expression, thus leading to the downregulation of proliferation and
metastatic processes in MCF-7 breast cancer cells (35). Similarly, UTMD-mediated HIF-1 shRNA
transfection has served an enhancing role in tumor growth
inhibition (36). The results of the
present study demonstrated that UTMD upregulated SIRT3
expression and further suppressed the malignant behaviors of SKOV3
cells. SIRT3 has been demonstrated to serve as either a
tumor suppressor or tumor promoter in different malignancies
(37). For example, SIRT3 was found
to be expressed at low levels in human gastric cancer and to
inhibit cell proliferation (38). In
addition, SIRT3 has been reported to inhibit the development of
prostate cancer (13). Conversely,
its tumor promoting role has also been revealed in several
malignancies, including cervical cancer (39) and breast cancer cells (40). In the present study, SIRT3
inhibited SKOV3 HOC cell proliferation, invasion, migration, EMT
and resistance to apoptosis, with an inhibitory effect found on
tumor growth in nude mice. SIRT3 was previously reported to
be downregulated in HOC cell lines and inhibited in cell metastasis
and invasion (41). Similarly,
downregulation of SIRT3 has been found to promote OC
metastasis (42). Based on these
data, SIRT3 may serve as a potential therapeutic option for
HOC control and treatment.
These findings prompted the authors of the present
study to further identify the potential molecular mechanism
involved in HOC progression. Importantly, the dual luciferase
reporter gene assay results suggested that SIRT3 could
directly bind to HIF-1α. Similarly, RT-qPCR and western blot
analysis revealed that SIRT3 overexpression led to decreased
HIF-1α mRNA and protein expression in SKOV3 cells. HIF-1α can
initiate angiogenesis, and upregulation of the HIF-1α pathway has
been found in various cancers caused by different environmental
factors (43). HIF-1α is closely
associated with the malignancy of ovarian tumors, and the
downregulation of HIF-1α expression could result in a decreased
proliferation of ovarian cells (44).
Overexpressed SIRT3 has been demonstrated to reduce HIF-1α
protein stabilization in hypoxia and reverse the increases in the
transcriptional activity of HIF-1α to inhibit colon cancer
tumorigenesis (45). Hence, it could
be inferred that SIRT3 might inhibit the progression of HOC
through the downregulation of HIF-1α.
In summary, the present study demonstrated that
SIRT3 was expressed at low levels in HOC cells. Upregulation
of SIRT3 inhibited HOC cell proliferation, invasion, EMT and
in vivo tumor growth, and promoted cell apoptosis and cell
cycle arrest. UTMD further strengthened the inhibitory effect of
SIRT3 overexpression on HOC growth in vivo and in
vitro. Downregulation of HIF-1α might be involved in the above
events, as it was demonstrated that SIRT3 led to decreased
HIF-1α expression in SKOV3 cells. Future studies will explore more
detailed links between SIRT3 and HIF-1α expression and will
determine additional molecular mechanisms involved in the effect of
SIRT3 on HIF-1α. These findings may provide novel insights
into HOC control and treatment, and additional studies in this
field are required to develop additional therapeutic options for
HOC and other severe diseases.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
Not applicable.
Availability of data and materials
All data generated and analyzed during this study
are included in this published article.
Authors' contributions
LC and DZ designed the study. LC carried out the
analytical assays. WY performed the statistical analyses. LC
drafted the manuscript. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
The present study was performed with the approval of
the Ethics Committee of The Affiliated Hospital of Changchun
University of Traditional Chinese Medicine (Jilin, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Hua F, Li CH, Chen XG and Liu XP: Daidzein
exerts anticancer activity towards SKOV3 human ovarian cancer cells
by inducing apoptosis and cell cycle arrest and inhibiting the
Raf/MEK/ERK cascade. Int J Mol Med. 41:3485–3492. 2018.PubMed/NCBI
|
|
2
|
Bareiss PM, Paczulla A, Wang H, Schairer
R, Wiehr S, Kohlhofer U, Rothfuss OC, Fischer A, Perner S, Staebler
A, et al: SOX2 expression associates with stem cell state in human
ovarian carcinoma. Cancer Res. 73:5544–5555. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wolterink S, Moldenhauer G, Fogel M,
Kiefel H, Pfeifer M, Lüttgau S, Gouveia R, Costa J, Endell J,
Moebius U and Altevogt P: Therapeutic antibodies to human L1CAM:
Functional characterization and application in a mouse model for
ovarian carcinoma. Cancer Res. 70:2504–2515. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhang S, Xie B, Wang L, Yang H, Zhang H,
Chen Y, Wang F, Liu C and He H: Macrophage-mediated vascular
permeability via VLA4/VCAM1 pathway dictates ascites development in
ovarian cancer. J Clin Invest. 131:e1403152021. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Campbell S and Gentry-Maharaj A: The role
of transvaginal ultrasound in screening for ovarian cancer.
Climacteric. 21:221–226. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Varughese J, Cocco E, Bellone S, Bellone
M, Todeschini P, Carrara L, Schwartz PE, Rutherford TJ, Pecorelli S
and Santin AD: High-grade, chemotherapy-resistant primary ovarian
carcinoma cell lines overexpress human trophoblast cell-surface
marker (Trop-2) and are highly sensitive to immunotherapy with
hRS7, a humanized monoclonal anti-Trop-2 antibody. Gynecol Oncol.
122:171–177. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Alhazzazi TY, Kamarajan P, Joo N, Huang
JY, Verdin E, D'Silva NJ and Kapila YL: Sirtuin-3 (SIRT3), a novel
potential therapeutic target for oral cancer. Cancer.
117:1670–1678. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Taylor DM, Maxwell MM, Luthi-Carter R and
Kazantsev AG: Biological and potential therapeutic roles of sirtuin
deacetylases. Cell Mol Life Sci. 65:4000–4018. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Alhazzazi TY, Kamarajan P, Verdin E and
Kapila YL: SIRT3 and cancer: Tumor promoter or suppressor? Biochim
Biophys Acta. 1816:80–88. 2011.PubMed/NCBI
|
|
10
|
Desouki MM, Doubinskaia I, Gius D and
Abdulkadir SA: Decreased mitochondrial SIRT3 expression is a
potential molecular biomarker associated with poor outcome in
breast cancer. Hum Pathol. 45:1071–1077. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kim HS, Patel K, Muldoon-Jacobs K, Bisht
KS, Aykin-Burns N, Pennington JD, van der Meer R, Nguyen P, Savage
J, Owens KM, et al: SIRT3 is a mitochondria-localized tumor
suppressor required for maintenance of mitochondrial integrity and
metabolism during stress. Cancer Cell. 17:41–52. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yang B, Fu X, Shao L, Ding Y and Zeng D:
Aberrant expression of SIRT3 is conversely correlated with the
progression and prognosis of human gastric cancer. Biochem Biophys
Res Commun. 443:156–160. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Quan Y, Wang N, Chen Q, Xu J, Cheng W, Di
M, Xia W and Gao WQ: SIRT3 inhibits prostate cancer by
destabilizing oncoprotein c-MYC through regulation of the PI3K/Akt
pathway. Oncotarget. 6:26494–26507. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Carson AR, McTiernan CF, Lavery L, Hodnick
A, Grata M, Leng X, Wang J, Chen X, Modzelewski RA and Villanueva
FS: Gene therapy of carcinoma using ultrasound-targeted microbubble
destruction. Ultrasound Med Biol. 37:393–402. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chen S and Grayburn PA:
Ultrasound-targeted microbubble destruction for cardiac gene
delivery. Methods Mol Biol. 1521:205–218. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Villanueva FS: Ultrasound mediated
destruction of DNA-loaded microbubbles for enhancement of
cell-based therapies: New promise amidst a confluence of
uncertainties? JACC Cardiovasc Imaging. 2:880–882. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Gao F, Wu J, Niu S, Sun T, Li F, Bai Y,
Jin L, Lin L, Shi Q, Zhu LM and Du L: Biodegradable, pH-sensitive
hollow mesoporous organosilica nanoparticle (HMON) with controlled
release of pirfenidone and
ultrasound-target-microbubble-destruction (UTMD) for pancreatic
cancer treatment. Theranostics. 9:6002–6018. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Jing H, Cheng W, Li S, Wu B, Leng X, Xu S
and Tian J: Novel cell-penetrating peptide-loaded nanobubbles
synergized with ultrasound irradiation enhance EGFR siRNA delivery
for triple negative breast cancer therapy. Colloids Surf B
Biointerfaces. 146:387–395. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhong S, Shu S, Wang Z, Luo J, Zhong W,
Ran H, Zheng Y, Yin Y and Ling Z: Enhanced homing of mesenchymal
stem cells to the ischemic myocardium by ultrasound-targeted
microbubble destruction. Ultrasonics. 52:281–286. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Naito S, von Eschenbach AC, Giavazzi R and
Fidler IJ: Growth and metastasis of tumor cells isolated from a
human renal cell carcinoma implanted into different organs of nude
mice. Cancer Res. 46:4109–4115. 1986.PubMed/NCBI
|
|
22
|
Zatroch KK, Knight CG, Reimer JN and Pang
DS: Refinement of intraperitoneal injection of sodium pentobarbital
for euthanasia in laboratory rats (Rattus norvegicus). BMC Vet Res.
13:602017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kopaladze RA: Methods for the euthanasia
of experimental animals-the ethics, esthetics and personnel safety.
Usp Fiziol Nauk. 31:79–90. 2000.(In Russian). PubMed/NCBI
|
|
24
|
Yao W, Ji S, Qin Y, Yang J, Xu J, Zhang B,
Xu W, Liu J, Shi S, Liu L, et al: Profilin-1 suppresses
tumorigenicity in pancreatic cancer through regulation of the
SIRT3-HIF1α axis. Mol Cancer. 13:1872014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Paolillo M and Schinelli S: Extracellular
matrix alterations in metastatic processes. Int J Mol Sci.
20:49472019. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Brown GT and Murray GI: Current
mechanistic insights into the roles of matrix metalloproteinases in
tumour invasion and metastasis. J Pathol. 237:273–281. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Hu Y, Liu J and Huang H: Recent agents
targeting HIF-1α for cancer therapy. J Cell Biochem. 114:498–509.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hamanishi J, Mandai M, Ikeda T, Minami M,
Kawaguchi A, Murayama T, Kanai M, Mori Y, Matsumoto S, Chikuma S,
et al: Safety and antitumor activity of anti-PD-1 antibody,
nivolumab, in patients with platinum-resistant ovarian cancer. J
Clin Oncol. 33:4015–4022. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Chen ZY, Liang K, Sheng XJ, Si-Tu B, Sun
XF, Liu JQ, Qiu RX, Zhang H, Li YW, Zhou XX and Yu JX: Optimization
and apoptosis induction by RNAi with UTMD technology in
vitro. Oncol Lett. 3:1030–1036. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Su J, Wang J, Luo J and Li H:
Ultrasound-mediated destruction of vascular endothelial growth
factor (VEGF) targeted and paclitaxel loaded microbubbles for
inhibition of human breast cancer cell MCF-7 proliferation. Mol
Cell Probes. 46:1014152019. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Stromar IK and Jakic-Razumovic J: The
value of immunohistochemical determination of topoisomerase IIα and
Ki67 as markers of cell proliferation and malignant transformation
in colonic mucosa. Appl Immunohistochem Mol Morphol. 22:524–529.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhao J, Dong D and Sun L, Zhang G and Sun
L: Prognostic significance of the epithelial-to-mesenchymal
transition markers e-cadherin, vimentin and twist in bladder
cancer. Int Braz J Urol. 40:179–189. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Xiang XY, Kang JS, Yang XC, Su J, Wu Y,
Yan XY, Xue YN, Xu Y, Liu YH, Yu CY, et al: SIRT3 participates in
glucose metabolism interruption and apoptosis induced by BH3
mimetic S1 in ovarian cancer cells. Int J Oncol. 49:773–784. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Liu J, Yao Y, Ding H and Chen R:
Oxymatrine triggers apoptosis by regulating Bcl-2 family proteins
and activating caspase-3/caspase-9 pathway in human leukemia HL-60
cells. Tumour Biol. 35:5409–5415. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Xu J, Wang Y, Li Z, Wang Q, Zhou X and Wu
W: Ultrasound-targeted microbubble destruction (UTMD) combined with
liposome increases the effectiveness of suppressing proliferation,
migration, invasion, and epithelial-mesenchymal transition (EMT)
via targeting metadherin (MTDH) by ShRNA. Med Sci Monit.
25:2640–2648. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Liao Y, Luo H, He Z, Kuang Y, Chen P,
Zhang X, Chen J, Wen Q, Xie Y and Ding S: A combination of
UTMD-mediated HIF-1 α shRNA transfection and TAE in the treatment
of hepatic cancer. Biomed Res Int. 2019:19374602019. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Chen Y, Fu LL, Wen X, Wang XY, Liu J,
Cheng Y and Huang J: Sirtuin-3 (SIRT3), a therapeutic target with
oncogenic and tumor-suppressive function in cancer. Cell Death Dis.
5:e10472014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Wang L, Wang WY and Cao LP: SIRT3 inhibits
cell proliferation in human gastric cancer through down-regulation
of notch-1. Int J Clin Exp Med. 8:5263–5271. 2015.PubMed/NCBI
|
|
39
|
Xu LX, Hao LJ, Ma JQ, Liu JK and Hasim A:
SIRT3 promotes the invasion and metastasis of cervical cancer cells
by regulating fatty acid synthase. Mol Cell Biochem. 464:11–20.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Zhang L, Ren X, Cheng Y, Huber-Keener K,
Liu X, Zhang Y, Yuan YS, Yang JW, Liu CG and Yang JM:
Identification of sirtuin 3, a mitochondrial protein deacetylase,
as a new contributor to tamoxifen resistance in breast cancer
cells. Biochem Pharmacol. 86:726–733. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Dong XC, Jing LM, Wang WX and Gao YX:
Down-regulation of SIRT3 promotes ovarian carcinoma metastasis.
Biochem Biophys Res Commun. 475:245–250. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Wu Y, Gao WN, Xue YN, Zhang LC, Zhang JJ,
Lu SY, Yan XY, Yu HM, Su J and Sun LK: SIRT3 aggravates
metformin-induced energy stress and apoptosis in ovarian cancer
cells. Exp Cell Res. 367:137–149. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Boreddy SR, Sahu RP and Srivastava SK:
Benzyl isothiocyanate suppresses pancreatic tumor angiogenesis and
invasion by inhibiting HIF-α/VEGF/Rho-GTPases: Pivotal role of
STAT-3. PLoS One. 6:e257992011. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Fujita M, Yasuda M, Kitatani K, Miyazawa
M, Hirabayashi K, Takekoshi S, Iida T, Hirasawa T, Murakami M,
Mikami M, et al: An up-to-date anti-cancer treatment strategy
focusing on HIF-1alpha suppression: Its application for refractory
ovarian cancer. Acta Histochem Cytochem. 40:139–142. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Bell EL, Emerling BM, Ricoult SJ and
Guarente L: SirT3 suppresses hypoxia inducible factor 1α and tumor
growth by inhibiting mitochondrial ROS production. Oncogene.
30:2986–2996. 2011. View Article : Google Scholar : PubMed/NCBI
|