Introduction
Lung cancer is the leading cause of cancer-related
mortality worldwide. Non-small cell lung cancer (NSCLC) occurs in
~85% of patients with lung cancer (1). Recently, several driver gene
aberrations, including epidermal growth factor receptor mutations
and anaplastic lymphoma kinase gene rearrangements, have been
reported to play a crucial role in the occurrence of NSCLC, against
which corresponding targeted molecular therapeutics have been
developed. Targeted therapies against NSCLC harboring such
mutations have exhibited notable success (2); however, ~30% of patients with NSCLC
in Japan lacking driver oncogene aberrations do not obtain optimal
clinical benefits from these treatments (3). Platinum-based combination
chemotherapy, including cisplatin and pemetrexed, is still being
used as the standard therapy for patients with NSCLC lacking driver
mutations. However, the development of cisplatin resistance is a
major obstacle for cancer treatment. Accordingly, the development
of novel therapeutic strategies for NSCLC is of utmost importance
for overcoming cisplatin resistance.
Cisplatin resistance-related cellular mechanisms
have been revealed to be multifunctional, including decreased
cisplatin accumulation, increased intracellular detoxification, and
increased DNA repair ability (4,5).
Nucleotide excision repair (NER) is a principal and well-known
repair system for chemotherapy-induced DNA damage (6). The xeroderma pigmentosum
complementation group proteins (XPA, XPC, XPD, XPF, and XPG) are
involved in the NER pathway, including in the damage recognition,
unwinding, excision and refilling of DNA (7,8).
Previously, it has been reported that XPC protein contributes to
the sensitivity of colorectal cancer cells to cisplatin (9), and the overexpression of XPF and XPG
mRNA has been associated with the sensitivity of ovarian and colon
cancer cells to cisplatin (10).
Moreover, XPA, XPC and XPD proteins modulate the sensitivity of
gastric cancer cells to cisplatin (11). NER pathways that are involved in
the response or resistance to cisplatin have been reported in a
number of types of cancer, including testicular and ovarian cancer,
and NSCLC (7). However, the role
of DNA repair in cisplatin resistance is not yet fully understood
in cisplatin-resistant NSCLC, including multiple mechanisms of
cisplatin resistance.
Ecteinascidin (Et)743 (Trabectedin,
Yondelis®) has been approved for use in soft tissue
sarcoma and ovarian cancer. The unique mechanism of Et743 involves
double-strand breaks induced by the NER pathway following covalent
binding to the N2 of guanine located in the DNA minor groove
(12–14).
In the present study, the expression levels of NER
pathway-associated genes in cisplatin-resistant NSCLC cell lines
were investigated and the effects of Et analogs on
cisplatin-resistant NSCLC cell lines in which the NER pathway was
altered were evaluated.
Materials and methods
Chemical and reagents
Et770 and Renieramycin M (RmM) were isolated by the
authors from the Thai tunicate Ecteinascidia thurstoni
(15) and the Thai blue marine
sponge Xestospongia sp. (16), respectively. Renieramycin T (RmT)
was synthesized using RmM in the Department of Pharmaceutical
Chemistry, Meiji Pharmaceutical University (Tokyo, Japan). Their
chemical structures were determined as previously described
(17–19). For stock solutions, all chemicals
were dissolved in dimethyl sulfoxide at a concentration of 10 mM
and then further diluted to the working concentration prior to
use.
Cell lines
Three NSCLC cell lines, PC-7, PC-9 and PC-14, and
the cisplatin (CDDP)-resistant sublines (20), PC-7/CDDP, PC-9/CDDP, and PC-14/CDDP
were kindly provided by Professor Kazuto Nishi (Kinki University
School of Medicine, Osaka, Japan). The cells were maintained in
RPMI-1640 (Wako Pure Chemical Industries, Ltd.) supplemented with
10% fetal bovine serum (FBS) (Nichirei Biosciences Inc.), 100 U/ml
penicillin and 100 µg/ml streptomycin at 37°C with 5%
CO2 in a humidified incubator. These cell lines were
authenticated using STR DNA profiling analysis at the Japanese
Collection of Research Bioresources Cell Bank.
Cell proliferation assays
In order to evaluate the growth inhibitory effects
of Et770, RmT and RmM on cisplatin-resistant NSCLC cell lines, the
cells were seeded in 96-well microplates at a density of
2,000-3,500 cells/well in RPMI-1640 supplemented with 10% FBS, 100
units/ml penicillin and 100 µg/ml streptomycin. The cells were then
treated with various concentrations of the test samples for 96 h.
Cell viability was evaluated using a Cell Counting
Kit-8® (Dojindo Laboratories, Inc.) or
ATPlite® 1 step reagents (PerkinElmer, Inc.) according
to the manufacturer's instructions. The IC50 values were
determined using GraphPad Prism 7.04 software (GraphPad Software,
Inc.).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA from the cultured cells and
siRNA-transfected cells was isolated using the miRNeasy®
Mini kit (Qiagen GmbH). The purity and concentration of the total
RNA were assessed using a UV-visible spectrophotometer V730 (JASCO
Corporation). cDNA was synthesized from 1 µg total RNA using
ReverTra Ace® qPCR RT Master Mix (Toyobo Life Science)
at 37°C for 15 min, then 50°C for 5 min, followed by 80°C for 10
min. PCR was performed using converted cDNA and the following
reagents: TaqMan Gene Expression Assays® for human XPA
(assay ID Hs00166045_m1), XPD [excision repair
cross-complementation group (ERCC)2; assay ID Hs00361161_m1], XPF
(ERCC4; assay ID Hs00193342_m1), XPG (ERCC5; assay ID
Hs01557031_m1), excision repair cross-complementation group 1
(ERCC1; assay ID Hs01012156_m1), and GAPDH (assay ID Hs02758991_g1)
genes (Thermo Fisher Scientific Inc.), and the Luna Universal Probe
qPCR Master Mix (New England BioLabs, Inc.). XPA, XPD, XPF, XPG,
and ERCC1 mRNA expression levels were assessed by using RT-qPCR
with the Quant Studio® 5 Real-Time PCR System (Thermo
Fisher Scientific Inc.). The thermocycling conditions were as
follows: 95°C for 60 sec, followed by 40 cycles of 95°C for 15 sec
and 60°C for 60 sec. Relative gene expression levels were
determined using the comparative cycle threshold method
(2−ΔΔCq), using GAPDH as the reference gene. ΔCq was
calculated by subtracting the Cq of GAPDH from that the target gene
Cq (21).
SDS-PAGE and western blot
analysis
Soluble proteins were extracted from the PC-7, PC-9,
PC-14, PC-7/CDDP, PC-9/CDDP and PC-14/CDDP cells using RIPA buffer
(Nacalai Tesque Inc.), supplemented with a protease inhibitor
cocktail and a phosphatase inhibitor cocktail (Nacalai Tesque
Inc.). Sample protein concentration was determined using a BCA
protein assay kit (Thermo Fisher Scientific Inc.) with bovine serum
albumin as the standard. Briefly, 20 µg proteins/well were
separated using 5–20% SDS-polyacrylamide gels (ATTO Corporation)
and transferred to a nitrocellulose membrane (Wako Pure Chemical
Industries, Ltd.). After blocking with BlockAce (KAC Inc.) at room
temperature for 1 h, the membranes were incubated with the
following primary antibodies overnight at 4°C: XPA (dilution,
1:5,000; cat. no. sc-56813; Santa Cruz Biotechnology, Inc., XPD
(dilution, 1:3,000; cat. no. GTX108948) and XPF (dilution, 1:5,000;
cat. no. GTX129285) (both from GeneTex, Inc.), XPG (dilution,
1:1,000; cat. no. 11331-1-AP; Proteintech Group, Inc.), ERCC1
(dilution, 1:300; cat. no. A5291; ABclonal, Biotech Co., Ltd.) and
β-actin (clone C4; dilution, 1:5,000; cat. no. MAB1501; Merck
KGaA). All membranes were then incubated with HRP-labeled secondary
antibodies (dilution 1:10,000; anti-rabbit, cat no. PI-1000;
anti-mouse, PI-2000; Vector Laboratories, Inc.) at room temperature
for 1 h. After washing with 0.1% Tweew20 containing PBS, specific
signals were detected using an Immobilon Western Chemiluminescent
HRP substrate (Millipore Sigma). Each band intensity was calculated
using Image Lab. 5.2 software (Bio Rad Laboratories, Inc.).
siRNA transfection
RNAi knockdown was performed on the PC-14 and
PC-14/CDDP cellsy using Lipofectamine RNAiMAX (Thermo Fisher
Scientific Inc.) according to the manufacturer's instructions.
siRNAs against XPA (siRNA ID s532043 forward, GGGAGACGAUUGUUCAUCATT
and reverse, UGAUGAACAAUCGUCUCCCTT; s14926 forward,
GAAUUGCGGCGAGCAGUAATT and reverse, UUACUGCUCGCCGCAAUUCTT), XPD
(ERCC2; siRNA ID s4787 forward, GAUUCGUGAGAAUGACUUUTT and reverse,
AAAGUCAUUCUCACGAAUCTG; s230766 forward, GCAUAUCCGCUGGAGGUGATT and
reverse, UCACCUCCAGCGGAUAUGCTC), XPF (ERCC4; siRNA ID s535114
forward, GGAUAGCAAAGCUGAAGAATT and reverse, UUCUUCAGCUUUGCUAUCCTT;
s4799 forward, GGAUAUGCGUGAAUUUCGATT and reverse,
UCGAAAUUCACGCAUAUCCAC), XPG (ERCC5; siRNA ID s4803 forward,
GGAAUACCGUUUACUGCAATT and reverse, UUGCAGUAAACGGUAUUCCTT; s4802
forward, GCAUAACAAAUACCUUAGATT and reverse, UCUAAGGUAUUUGUUAUGCCT),
ERCC1 (siRNA ID s4785 forward, GCAAGGAAGAAAUUUGUGATT and reverse,
UCACAAAUUUCUUCCUUGCTG; s4784 forward GGAUCUCUGGAACAGCUCATT and
reverse, UGAGCUGUUCCAGAGAUCCAA), and non-target siRNA (Silencer
Select Negative Control #1) were obtained from Thermo Fisher
Scientific, Inc. A total of 20 pmol siRNA was diluted in a 150 µl
Opti-MEM medium in a six-well tissue culture plate. Thereafter, 3
µl Lipofectamine RNAiMAX were added to each well, containing the
diluted siRNA molecules. The solution was mixed gently followed by
incubation for 5 min at room temperature. Cells (4×105
per well) were seeded into a six-well plate containing siRNA and
liposome complexes and cultured at 37°C. Following overnight
culture, an equal volume of RPMI-1640 medium, supplemented with 10%
FBS, 100 U/ml penicillin and 100 µg/ml streptomycin was added. The
knockdown efficiency was determined using RT-qPCR and western blot
analysis at 72 and 120 h following RNAi knockdown.
Statistical analysis
Statistical analyses were performed using GraphPad
Prism 7.04 (GraphPad Software, Inc.) and R (The R Foundation). The
results are expressed as the mean ± SEM. The statistical
significance of the differences was examined using analysis of
variance (ANOVA) and Tukey's post hoc test. P<0.05 was
considered to indicate a statistically significant difference.
Pearson's correlation analysis was used for correlation
analysis.
Results
Expression of NER pathway-associated
genes is increased in cisplatin-resistant NSCLC cell lines
It has been reported that DNA damage induced by
cisplatin is generally repaired via the NER pathway (22). In the present study, the XPA, XPD,
XPF, XPG and ERCC1 gene expression levels in NSCLC cell lines,
PC-7, PC-9 and PC-14, and in the cisplatin-resistant sublines,
PC-7/CDDP, PC-9/CDDP and PC-14/CDDP, were investigated, in order to
elucidate the contribution of the NER pathway to cisplatin
resistance in NSCLC.
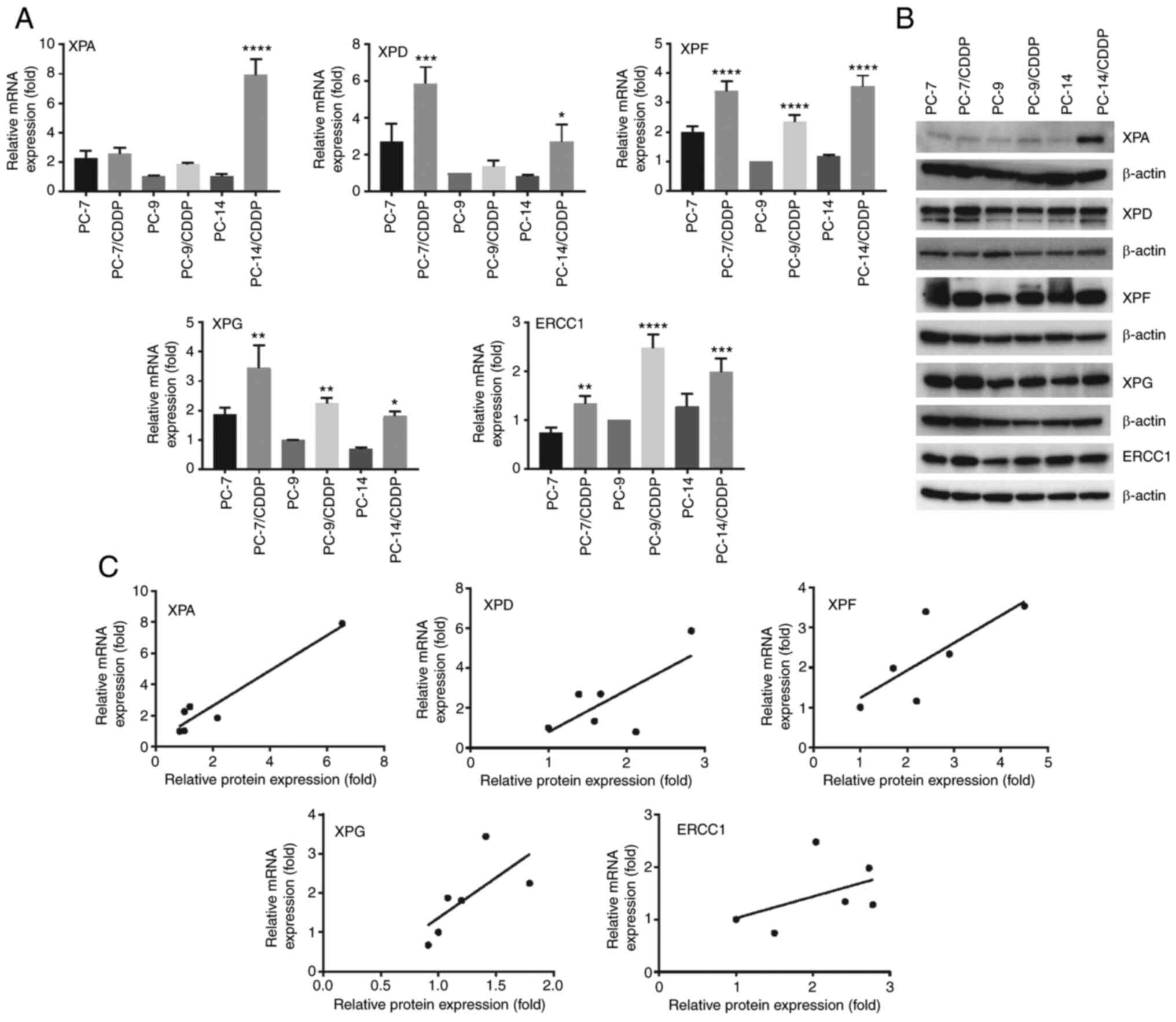
As depicted in Fig.
1A, the XPF expression levels were significantly higher in the
cisplatin-resistant cell lines than in each parental cell line
(P<0.0001). ERCC1 mRNA expression was also significantly
increased in the PC-7/CDDP (P=0.0015), PC-9/CDDP (P<0.0001) and
PC-14/CDDP (P=0.0002) cell lines in comparison with the
corresponding parental cell lines. Furthermore, the XPG expression
levels were also significantly increased in the PC-7/CDDP
(P=0.0012), PC-9/CDDP (P=0.0075) and PC-14/CDDP (P=0.0147) cell
lines compared with each respective parental cell line. On the
other hand, the XPD mRNA expression levels in the PC-9/CDDP cells
and the XPA mRNA expression levels in the PC-7/CDDP and PC-9/CDDP
cells did not differ significantly from those in the respective
parental cell lines. However, the XPD mRNA levels in the PC-7/CDDP
(P=0.0010) and PC-14/CDDP (P=0.0423) cells were significantly
increased in comparison with the respective parental cell lines.
The XPA mRNA expression levels in the PC-14/CDDP cells were
significantly increased as compared with the respective parental
cell line (P<0.0001). In addition, the expression levels of NER
pathway-associated proteins were also highly increased in
cisplatin-resistant sublines according to western blot analysis
(Figs. 1B and S1). In addition, correlations were
observed between the NER-related gene expression levels and NER
pathway-associated protein expression levels in these
cisplatin-resistant lung cancer cells (Fig. 1C).
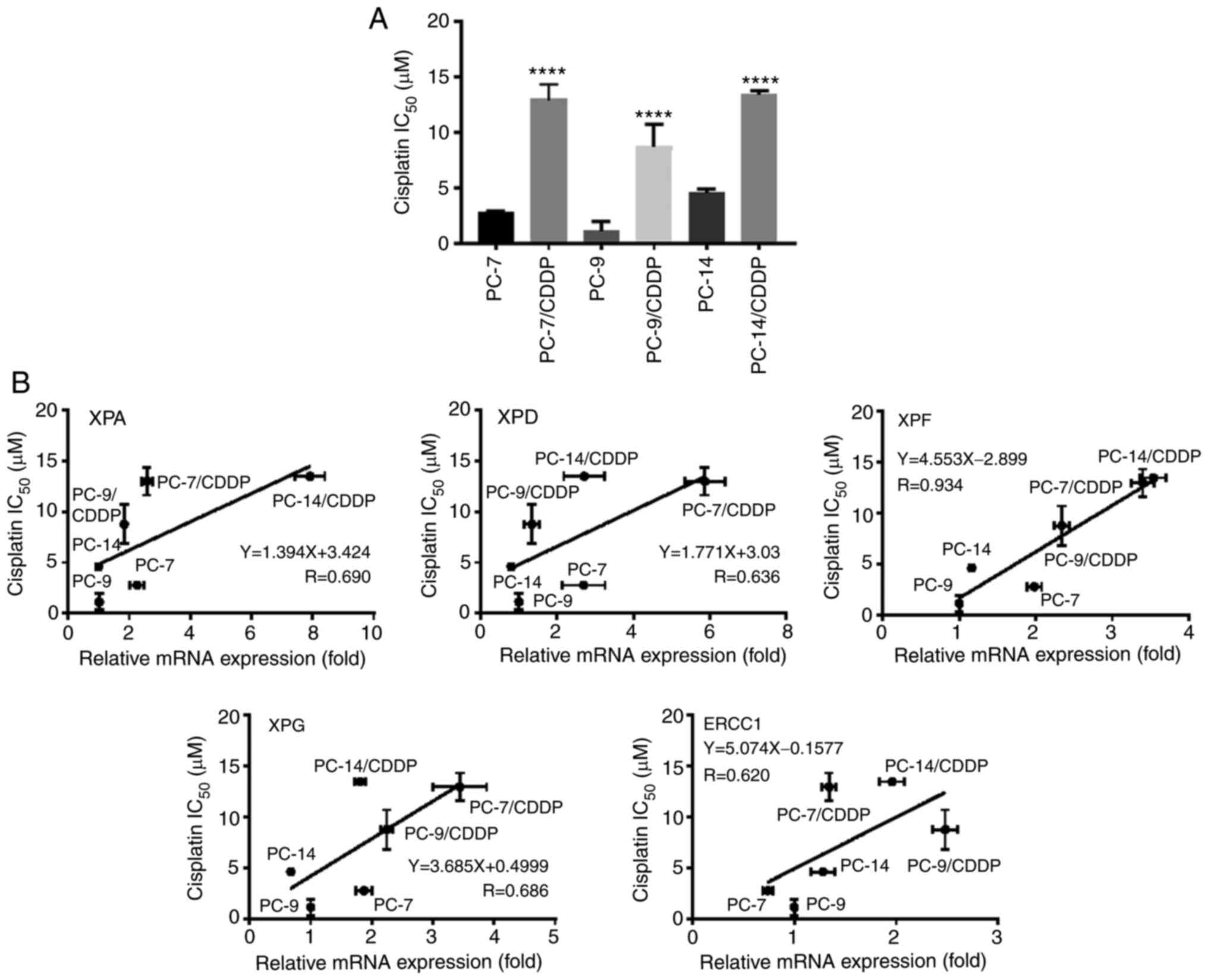
As presented in Fig.
2A, the cisplatin-resistant cell lines exhibited a 3- to 8-fold
resistance to cisplatin, in comparison with their respective
parental cell lines (P<0.0001). To explore the contribution of
the NER pathway to cisplatin resistance, Pearson's correlation
analysis was performed. As depicted in Fig. 2B, XPF mRNA expression level
strongly correlated with cisplatin sensitivity (R=0.934). The XPA
(R=0.690), XPD (R=0.636), XPG (R=0.686) and ERCC1(R=0.620)
expression levels also correlated with cisplatin sensitivity.
NER pathway-associated gene silencing
alters the sensitivity of cisplatin-resistant NSCLC cell lines to
cisplatin
To explore the contribution of the NER pathway to
the cisplatin resistance of NSCLC cell lines, cisplatin sensitivity
was analyzed following the knockdown of each molecule of the NER
pathway using siRNA. A shown in Fig.
1, the XPA and XPD expression levels were relatively decreased
in the PC-9/CDDP cell line and the XPA level was also relatively
decreased in the PC-7/CDDP cell line in comparison with the
PC-14/CDDP cells; thus, the PC-14/CDDP and PC-14 cell lines were
used for gene knockdown experiments. Following XPA, XPD, XPF, XPG
and ERCC1 mRNA silencing using siRNAs in the PC-14 and PC-14/CDDP
cell lines, the respective expression levels were significantly
decreased in comparison with siRNA negative control
(siNTC)-transfected cells (Fig.
3A). The knockdown efficiency was also investigated for XPA,
XPD, XPF, XPG and ERCC1 proteins using western blot analysis. The
protein expression levels were also suppressed following siRNA
transfection (Figs. 3B and
S2). Furthermore, cisplatin
sensitivity was evaluated following siRNA silencing. As presented
in Fig. 3C, the relative viability
of the PC-14/CDDP cells transfected with XPF siRNA 1 and 2, and
treated with 30 µM cisplatin was significantly decreased by 19.2
and 15.6%, respectively, compared with the siNTC-transfected cells
(48.2%; P<0.0001). XPD and XPG silencing in the PC-14/CDDP cells
also significantly decreased relative viability following treatment
with 30 µM cisplatin (P<0.0001). Moreover, the relative
viability of the XPD siRNA-transfected PC-14 cells treated with 10
mM cisplatin was also significantly decreased in comparison with
the siNTC-transfected cells (P<0.01).
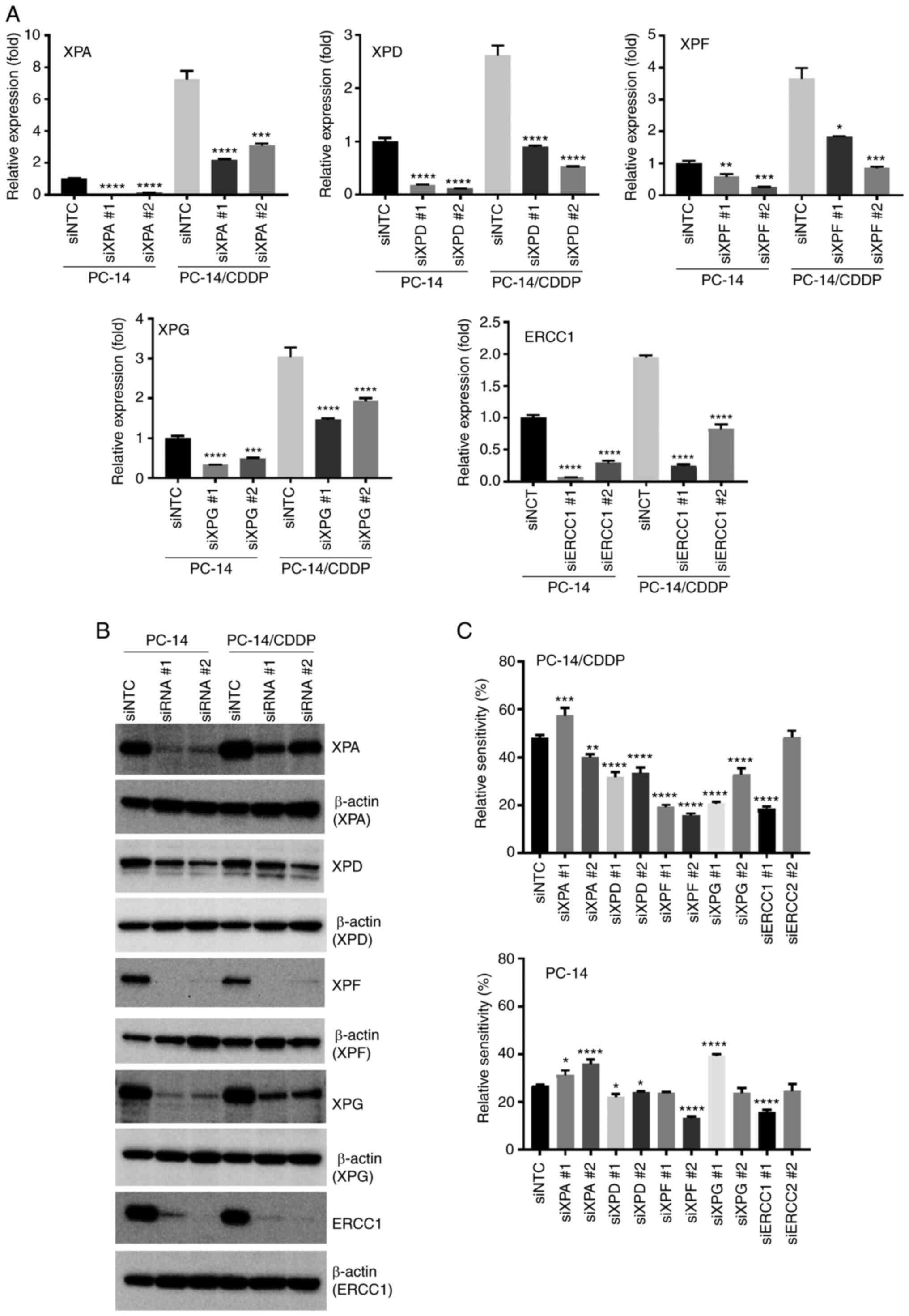 | Figure 3.Restoration of the sensitivity of
PC-14 and PC-14/CDDP cells to cisplatin by silencing a gene
associated with the NER pathway. (A) RT-qPCR analysis of NER
pathway-associated gene mRNA levels 72 h after siRNA transfection.
Data represent the mean + SEM. *P<0.05, **P<0.01,
***P<0.001 and ****P<0.0001, vs. siRNA negative control
(siNTC)-transfected cells. (B) Protein expression levels were
assessed using western blot analysis at 120 h following siRNA
transfection. (C) Relative viability of 30 µM cisplatin-exposed
PC-14/CDDP and 10 µM cisplatin-exposed PC-14 cells was assessed
using a Cell Counting Kit-8 after 96 h of incubation with
cisplatin. Data represent the mean + SEM. *P<0.05, **P<0.01,
***P<0.001 and ****P<0.0001 vs. siNTC-transfected cells.
CDDP, cisplatin; NER, nucleotide excision repair; ERCC1, ERCC1,
excision repair cross-complementation group 1; XP, xeroderma
pigmentosum group-complementing protein. |
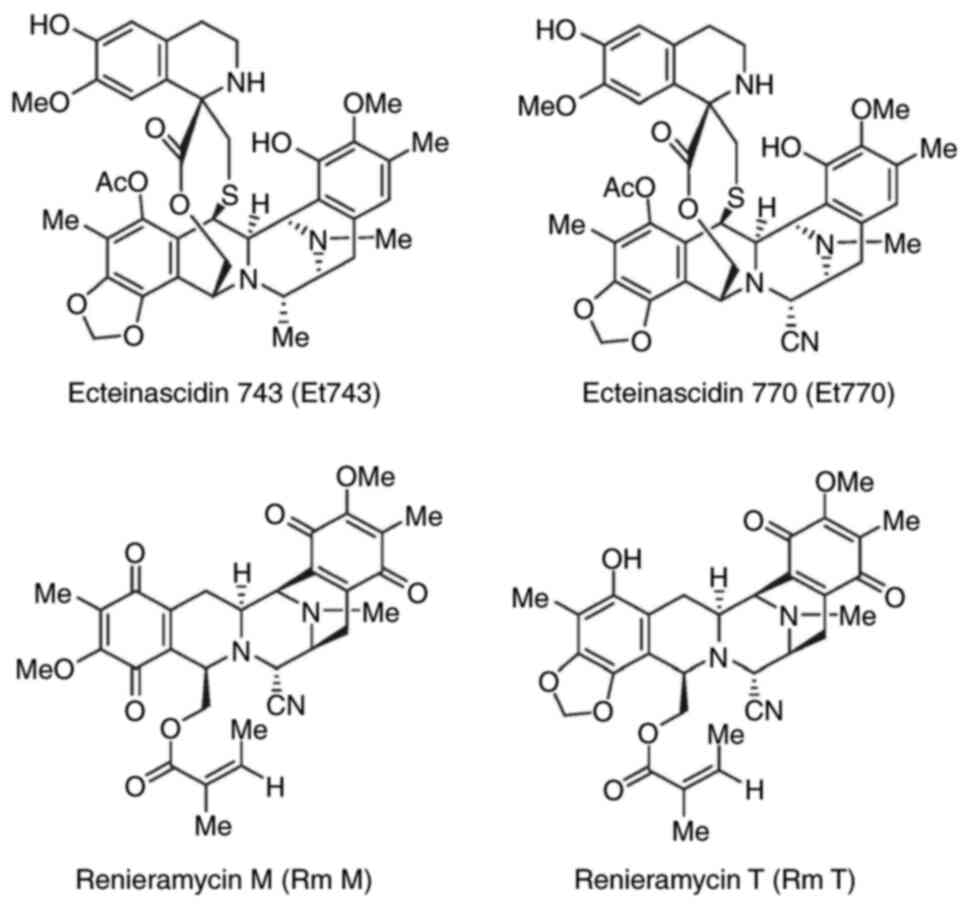
Et has the potential to overcome
cisplatin resistance in cisplatin-resistant NSCLC
It was demonstrated in the present study that the
NER pathway is a principal resistance factor in cisplatin-resistant
NSCLC cells. Et743 has been previously reported to be more active
in cells with an enhanced NER pathway (23). In a previously published study by
the authors, Et770 was purified as a stabilized derivative of
Et743, maintaining anticancer activities (24). RmM and RmT are structurally
analogous to Et. RmT refers to the hybrid compound of RmM and RmT
(Fig. 4). It has also been
previously reported by the authors that these compounds could
potentially be used as anticancer agents (25). In order to investigate the
potential of Et and its derivatives in overcoming cisplatin
resistance, the sensitivity of Et770, RmT and RmM in
cisplatin-resistant NSCLC cell lines with the enhanced expression
of NER pathway-related genes was evaluated. Et770, RmT and RmM
sensitivity were analyzed in the cisplatin-resistant NSCLC cell
line (PC-14/CDDP). As presented in Fig. 5A, Et770 treatment exerted growth
inhibitory effects on the PC-14/CDDP cells in comparison with the
PC-14 cells (P<0.01). However, similar effects were not observed
for the PC-7/CDDP and PC-9/CDDP cells with Et770 treatment.
Moreover, RmT and RmM treatment did not exert notable inhibitory
effects on the PC-14/CDDP cells in which NER pathway-related genes
were silenced.
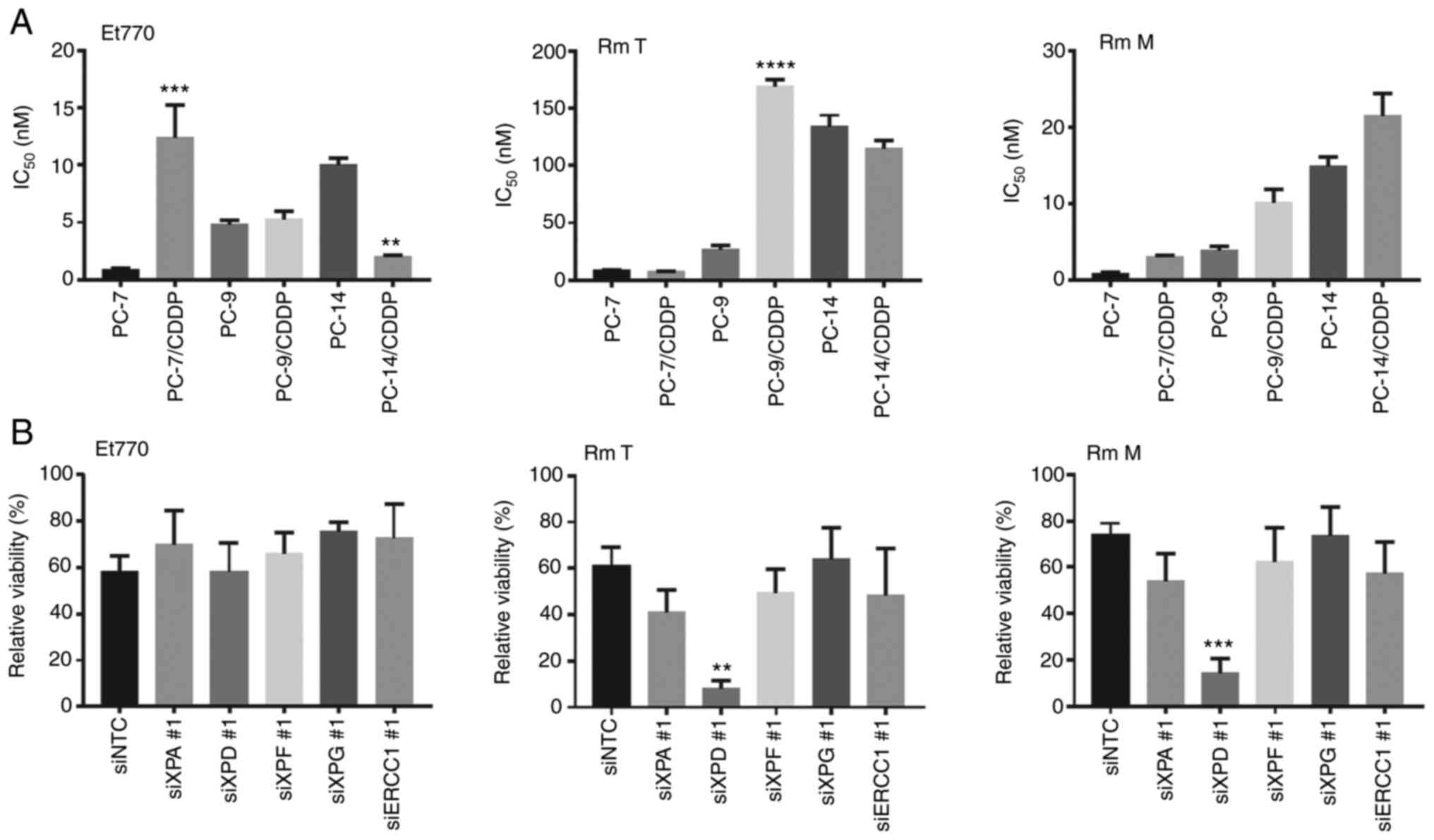 | Figure 5.Effect of nucleotide excision repair
enzyme activity for antiproliferation activity of ecteinascidin and
renieramycins. (A) Growth inhibitory effects of Et770 on PC-14 and
PC-14/CDDP cells. Cell viability was measured using a Cell Counting
Kit-8 after 96 h of incubation with 1.8 nM Et770. Data are
presented as the mean + SEM. **P<0.01, ***P<0.001 and
****P<0.0001, vs. parental cells. (B) The relative viability of
1.8 nM Et770-, 80nM RmT-, or 24 nM RmM-exposed PC-14/CDDP cells
were assessed using a Cell Counting Kit-8 after 96 h of incubation
of each compound. Data represent the mean + SEM. **P<0.01 and
***P<0.001, vs. siRNA negative control-transfected cells. Et,
Ecteinascidin; CDDP, cisplatin; RmM, Renieramycin M; RmT,
Renieramycin T; ERCC1, ERCC1, excision repair cross-complementation
group 1; XP, xeroderma pigmentosum group-complementing protein. |
Of note, the PC-14/CDDP cells exhibited
hypersensitivity to Et770, whereas the PC-14/CDDP cells exhibited a
slight resistance to RmM. Subsequently, in order to explore the
contribution of the NER pathway genes to Et anticancer activity in
cisplatin-resistant NSCLC, the sensitivity of the compounds was
analyzed following the silencing of NER pathway-associated genes in
PC-14/CDDP cells. As presented in Fig.
5B, although a significant difference was not observed, XPG
silencing slightly altered Et770 resistance as compared with the
siNTC-transfected cells (P=0.403). However, XPD silencing rather
resulted in an increased sensitivity to RmT (P<0.01) and RmM
(P<0.0001) in comparison with the siNTC-transfected cells.
Discussion
In the present study, it was observed that NER
pathway-associated gene expression levels were significantly
associated with cisplatin sensitivity in cisplatin-resistant and
parental NSCLC cell lines. The XPF mRNA expression level was the
most highly correlated factor with cisplatin sensitivity in these
cell lines (R=0.934). Furthermore, XPF or XPD knockdown markedly
restored cisplatin sensitivity in the cisplatin-resistant NSCLC
cell line PC-14/CDDP. Previously, it has been reported that
cisplatin resistance mechanisms in PC-14/CDDP are associated with
cisplatin accumulation, which is modulated by uptake mechanisms and
the upregulation of efflux transporters (26,27).
However, resistance to cisplatin is associated with several complex
mechanisms (4,28). In the present study, it was
revealed that NER pathway-associated molecules may also be related
to cisplatin resistance in the NSCLC cell lines, PC-7/CDDP,
PC-9/CDDP and PC-14/CDDP.
It is well-known that XPF-ERCC1 participates in
multiple DNA damage repair pathways, including NER and inter-strand
crosslink repair (6). XPF-ERCC1
also has alternative important functions, in addition to its
NER-related function, for the prevention of endogenous DNA damage
(29). XPF has been also reported
to be a predictor of platinum sensitivity and a promising biomarker
for PARP inhibitors (30). Thus,
various XPF features have been reported. Although NER pathways are
involved in cisplatin resistance, the contribution of XPF-ERCC1
remains unclear. The results of the present study also suggested
that XPF may be a principal factor in cisplatin-resistant
mechanisms.
In the present study, the XPA expression levels
significantly correlated with cisplatin sensitivity in all tested
cisplatin-resistant NSCLC cell lines (R=0.690). However, XPA
silencing slightly reduced sensitivity to cisplatin in the PC-14
cells, and consistent results were not obtained with the PC-14/CDDP
cells (Fig. 3C). The expression
level of XPA in the PC-14/CDDP cells was increased 8-fold in
comparison with the parental cell line, PC-14. Hence, when
suppressed using siRNA, XPA expression was decreased to ~25% of
that in siNTC-transfected cells; however, the levels were still
increased in comparison with those of the parental cell line. Due
to this increased residual activity, no clear results could be
obtained in cisplatin-resistant cell lines.
Cierna et al (31) reported that the XPA expression
levels are crucial for cisplatin resistance in germ cell tumors and
cell lines. A recent study suggested that the efficiency of XPA
activity may be impeded by enhancing DNA polymerase β activity
(32). The present study focused
on the XPA expression level in the context of NER system function;
thus, DNA polymerase expression levels and gene mutations were not
investigated. DNA polymerase may be involved in the mechanisms of
cisplatin resistance as a counterpart of XPA in cisplatin-resistant
NSCLC cells. Future studies are required to clarify the
contribution of the NER system, including the impact of DNA
polymerase on cisplatin resistance. Recently, Jian et al
(33) reported that XPD
overexpression significantly increased cisplatin sensitivity, and
revealed that XPD regulated the PI3K/AKT signaling pathway in
esophageal squamous cell carcinoma, this information also being
valuable for the further evaluation of cisplatin resistance.
However, Li et al (34)
reported that XPD and XPF polymorphisms may contribute to the risk
of NSCLC and the response to cisplatin-based chemotherapy in a
Chinese population. In the present study, XPD silencing enhanced
cisplatin sensitivity, and the XPF and XPD gene expression levels
correlated with cisplatin resistance in NSCLC cell lines.
Consequently, the results of the present study supported the
findings of the clinical study of Li et al (34), and suggested that these molecules
may be utilized as biomarkers for predicting cisplatin
sensitivity.
Et743 has been observed to be more active in cancer
cells with an enhanced NER pathway (23). The present study attempted to
clarify the Et770 anticancer activity in cisplatin-resistant cell
lines with in which NER pathway genes were enhanced (Fig. 5A). It was expected that Et770 would
overcome the effect of cisplatin resistance in all
cisplatin-resistant cell lines in which NER pathway genes were
silenced; however, Et770 only demonstrated hypersensitivity in a
PC-14/CDDP cell line in comparison with the parental cell line.
Following the silencing of NER pathway-associated genes in
PC-14/CDDP cells, Et770 sensitivity was slightly decreased in the
XPG-silenced cells (Fig. 5B). It
is likely that further studies on the effects of Et770 may
contribute to the elucidation of unknown cisplatin resistance
mechanisms. Due to the large structure of these compounds,
including Et770, it is difficult to clarify their biological
activity mechanism. Therefore, the effects on resistant cells
obtained in the present study may prove to be useful for future
therapeutic developmental research, since it is easy to assemble a
cell-based high-throughput screening system and due to its possible
contribution in finding new cisplatin resistance-overcoming
therapeutics.
In conclusion, in a previously published study by
the authors, it was clarified that the cisplatin resistance
mechanism of PC-14/CDDP could be dependent on the decrease of
cisplatin accumulation (4).
However, strong cell line resistance could not be fully explained
by cisplatin accumulation alterations. The present study focused on
DNA repair, also known to be involved in cisplatin resistance, and
NER-related molecules in these cell lines were analyzed. XPF was
predominantly expressed in cisplatin-resistant cell lines.
Furthermore, it was clarified that XPF may contribute to cisplatin
resistance in NSCLC cell line suppression experiments by using
siRNA and PC-14/CDDP cell lines.
Et may prove to be useful for overcoming cisplatin
resistance in NSCLC cells, via an enhanced NER pathway. However,
RmT and RmM did not demonstrate any activity in cisplatin-resistant
NSCLC cells with an enhanced NER pathway. Although cisplatin
resistance mechanisms are complex and the contribution ratio of
each resistance factor remains largely unknown, further evaluation
of resistant cell lines may contribute to the elucidation of the
cisplatin resistance mechanisms in future studies.
Supplementary Material
Supporting Data
Acknowledgements
The authors would like to thank Professor Kazuto
Nishio (Kinki University, School of Medicine, Osaka Japan) for the
provision of all cell lines.
Funding
The present study was supported by Grants-in-Aid for Scientific
Research KAKENHI from the Japan Society for the Promotion of
Science (grant nos. 15K08083 and 18K06561).
Availability of data and materials
The datasets used or analyzed in this study are
available from the corresponding author upon reasonable
request.
Authors' contributions
TS designed the study and wrote the manuscript. NSi
and RTN performed the chemical synthesis and biochemistry
experiments. AB and YE performed the biological experiments. NSa
and YO supervised the project and interpreted the data. All authors
confirm the authenticity of all the raw data and have read and
approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
CDDP
|
cisplatin
|
|
Et
|
ecteinascidin
|
|
ERCC1
|
excision repair cross-complementation
group 1
|
|
GAPDH
|
glyceraldehyde 3-phosphate
dehydrogenase
|
|
NER
|
nucleotide excision repair
|
|
RmM
|
renieramycin M
|
|
RmT
|
renieramycin T
|
|
XPF
|
xeroderma pigmentosum group F
|
References
|
1
|
Zappa C and Mousa SA: Non-small cell lung
cancer: Current treatment and future advances. Transl Lung Cancer
Res. 5:288–300. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Takano N, Ariyasu R, Koyama J, Sonoda T,
Saiki M, Kawashima Y, Oguri T, Hisakane K, Uchibori K, Nishikawa S,
et al: Improvement in the survival of patients with stage IV
non-small-cell lung cancer: Experience in a single institutional
1995–2017. Lung Cancer. 131:69–77. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Saito M, Shiraishi K, Kunitoh H,
Takenoshita S, Yokota J and Kohno T: Gene aberrations for precision
medicine against lung adenocarcinoma. Cancer Sci. 107:713–720.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Suzuki T, Nishio K and Tanabe S: The MRP
family and anticancer drug metabolism. Curr Drug Metab. 2:367–377.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Shen DW, Pouliot LM, Hall MD and Gottesman
MM: Cisplatin resistance: A cellular self-defense mechanism
resulting from multiple epigenetic and genetic changes. Pharmacol
Rev. 64:706–721. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Gilson P, Drouot G, Witz A, Merlin JL,
Becuwe P and Harlé A: Emerging roles of DDB2 in cancer. Int J Mol
Sci. 20:51682019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bowden NA: Nucleotide excision repair: Why
is it not used to predict response to platinum-based chemotherapy?
Cancer Lett. 346:163–171. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Spivak G: Nucleotide excision repair in
humans. DNA Repair (Amst). 36:13–18. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhang Y, Cao J, Meng Y, Qu C, Shen F and
Xu L: Overexpression of xeroderma pigmentosum group C decreases the
chemotherapeutic sensitivity of colorectal carcinoma cells to
cisplatin. Oncol Lett. 15:6336–6344. 2018.PubMed/NCBI
|
|
10
|
Stevens EV, Nishizuka S, Antony S, Reimers
M, Varma S, Young L, Munson PJ, Weinstein JN, Kohn EC and Pommier
Y: Predicting cisplatin and trabectedin drug sensitivity in ovarian
and colon cancers. Mol Cancer Ther. 7:10–18. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Pajuelo-Lozano N, Bargiela-Iparraguirre J,
Dominguez G, Quiroga AG, Perona R and Sanchez-Perez I: XPA, XPC,
and XPD modulate sensitivity in gastric cisplatin resistance cancer
cells. Front Pharmacol. 9:11972018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zewail-Foote M and Hurley LH:
Ecteinascidin 743: A minor groove alkylator that bends DNA toward
the major groove. J Med Chem. 42:2493–2497. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Takebayashi Y, Pourquier P, Zimonjic DB,
Nakayama K, Emmert S, Ueda T, Urasaki Y, Kanzaki A, Akiyama SI,
Popescu N, et al: Antiproliferative activity of ecteinascidin 743
is dependent upon transcription-coupled nucleotide-excision repair.
Nat Med. 7:961–966. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
14
|
Herrero AB, Martin-Castellanos C, Marco E,
Gago F and Moreno S: Cross-talk between nucleotide excision and
homologous recombination DNA repair pathways in the mechanism of
action of antitumor trabectedin. Cancer Res. 66:8155–8162. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Tsujimoto M, Lowtangkitcharoen W, Mori N,
Pangkruang W, Putongking P, Suwanborirux K and Saito N: Chemistry
of ecteinascidins. Part 4: Preparation of 2′-N-acyl ecteinascidin
770 derivatives with improved cytotoxicity profiles. Chem Pharm
Bull (Tokyo). 61:1052–1064. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Sirimangkalakitti N, Chamni S, Charupant
K, Chanvorachote P, Mori N, Saito N and Suwanborirux K: Chemistry
of renieramycins. 15. Synthesis of 22-O-ester derivatives of
jorunnamycin A and their cytotoxicity against non-small-cell lung
cancer cells. J Nat Prod. 79:2089–2093. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Suwanborirux K, Amnuoypol S, Plubrukarn A,
Pummangura S, Kubo A, Tanaka C and Saito N: Chemistry of
renieramycins. Part 3.(1) isolation and structure of stabilized
renieramycin type derivatives possessing antitumor activity from
Thai sponge Xestospongia species, pretreated with potassium
cyanide. J Nat Prod. 66:1441–1446. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Daikuhara N, Tada Y, Yamaki S, Charupant
K, Amnuoypol S, Suwanborirux K and Saito N: Chemistry of
renieramycins. Part 7: Renieramycins T and U, novel
renieramycin-ecteinascidin hybrid marine natural products from Thai
sponge Xestospongia sp. Tetrahedron Lett. 50:4276–4278.
2009. View Article : Google Scholar
|
|
19
|
Rinehart KL, Holt TG, Fregeau NL, Stroh
JG, Keifer PA, Sun F, Li LH and Martin DG: Ecteinascidins 729, 743,
745, 759A, 759B, and 770: Potent antitumor agents from the
Caribbean tunicate Ecteinascidia turbinata. J Org Chem.
55:4512–4515. 1990. View Article : Google Scholar
|
|
20
|
Morikage T, Ohmori T, Nishio K, Fujiwara
Y, Takeda Y and Saijo N: Modulation of cisplatin sensitivity and
accumulation by amphotericin B in cisplatin-resistant human lung
cancer cell lines. Cancer Res. 53:3302–3307. 1993.PubMed/NCBI
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chen SH and Chang JY: New insights into
mechanisms of cisplatin resistance: From tumor cell to
microenvironment. Int J Mol Sci. 20:41362019. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Le VH, Inai M, Williams RM and Kan T:
Ecteinascidins. A review of the chemistry, biology and clinical
utility of potent tetrahydroisoquinoline antitumor antibiotics. Nat
Prod Rep. 32:328–347. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Suwanborirux K, Charupant K, Amnuoypol S,
Pummangura S, Kubo A and Saito N: Ecteinascidins 770 and 786 from
the Thai tunicate Ecteinascidia thurstoni. J Nat Prod.
65:935–937. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yokoya M, Toyoshima R, Suzuki T, Le VH,
Williams RM and Saito N: Stereoselective total synthesis of
(−)-renieramycin T. J Org Chem. 81:4039–4047. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ohmori T, Morikage T, Sugimoto Y, Fujiwara
Y, Kasahara K, Nishio K, Ohta S, Sasaki Y, Takahashi T and Saijo N:
The mechanism of the difference in cellular uptake of platinum
derivatives in non-small cell lung cancer cell line (PC-14) and its
cisplatin-resistant subline (PC-14/CDDP). Jpn J Cancer Res.
84:83–92. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Suzuki T, Nishio K, Sasaki H, Kurokawa H,
Saito-Ohara F, Ikeuchi T, Tanabe S, Terada M and Saijo N: cDNA
cloning of a short type of multidrug resistance protein homologue,
SMRP, from a human lung cancer cell line. Biochem Biophys Res
Commun. 238:790–794. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Nishio K, Nakamura T, Koh Y, Suzuki T,
Fukumoto H and Saijo N: Drug resistance in lung cancer. Curr Opin
Oncol. 11:109–115. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Jones M, Beuron F, Borg A, Nans A, Earl
CP, Briggs DC, Snijders AP, Bowles M, Morris EP, Linch M and
McDonald NQ: Cryo-EM structures of the XPF-ERCC1 endonuclease
reveal how DNA-junction engagement disrupts an auto-inhibited
conformation. Nat Commun. 11:11202020. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Mesquita KA, Alabdullah M, Griffin M, Toss
MS, Fatah TMAA, Alblihy A, Moseley P, Chan SYT, Rakha EA and
Madhusudan S: ERCC1-XPF deficiency is a predictor of olaparib
induced synthetic lethality and platinum sensitivity in epithelial
ovarian cancers. Gynecol Oncol. 153:416–424. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Cierna Z, Miskovska V, Roska J,
Jurkovicova D, Pulzova LB, Sestakova Z, Hurbanova L, Machalekova K,
Chovanec M, Rejlekova K, et al: Increased levels of XPA might be
the basis of cisplatin resistance in germ cell tumours. BMC Cancer.
20:172020. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wang M, Li E, Lin L, Kumar AK, Pan F, He
L, Zhang J, Hu Z and Guo Z: Enhanced activity of variant DNA
polymerase β (D160G) contributes to cisplatin therapy by impeding
the efficiency of NER. Mol Cancer Res. 17:2077–2088. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Jian J, Li S, Liu LZ, Zhen L, Yao L, Gan
LH, Huang YQ and Fang N: XPD inhibits cell growth and invasion and
enhances chemosensitivity in esophageal squamous cell carcinoma by
regulating the PI3K/AKT signaling pathway. Int J Mol Med.
46:201–210. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Li M, Chen R, Ji B, Fan C, Wang G, Yue C
and Li G: Contribution of XPD and XPF polymorphisms to
susceptibility of non-small cell lung cancer in high-altitude
areas. Public Health Genomics. 24:189–198. 2021. View Article : Google Scholar : PubMed/NCBI
|