Introduction
Endometrial carcinoma (EC) is the sixth leading
cause of cancer-related deaths among women, resulting in an
estimated 11,350 deaths annually (1). Despite effective control of the
overall incidence of cancer, the number of newly diagnosed ECs has
increased steadily over the years (2). Although a number of patients are
diagnosed with early-stage disease, 15–20% of patients with EC
experience recurrence after primary surgery for metastatic disease.
Thus, more effective biomarkers and targets for EC diagnosis or
treatment strategies are urgently needed (3–5).
Post-translational modifications (PTMs) have
critical roles in cancer progression, among which ubiquitin and
ubiquitin-like (UBL) protein modifications are the most abundant
PTMs that determine protein fate (6,7).
Recently, the UBL system has received increasing attention, but
breakthroughs are still lacking (8–10). A
panel of UBL proteins has been identified, including small UBL
modifiers (11), neural precursor
cell expressed, developmentally downregulated 8 (12), autophagy-related protein (ATG)8 and
ATG12, and interferon-stimulated gene 15 (ISG15) (13,14).
ISG15 is a 15 kDa protein that reportedly regulates antiviral
defense and provides immunomodulation (15,16).
Aberrant ISG15 expression has been reported in various types of
cancer, including nasopharyngeal carcinoma, hepatocellular
carcinoma and breast cancer (17–19).
Activation of the type I interferon (IFN) signaling
pathway is a key component of anticancer innate immunity (20). The transcription of ISG15 is
induced by type I IFN signals and IFN-induced enhancer elements
mediate the effect of IFN-α on ISG15 transcription (21). ISG15 expression is related to the
induction of IFN and the activation of immune cells, both of which
are important mediators of tumor immunity (22). Previous studies showed that
melanoma cells secrete free ISG15 to regulate the phenotype of
tumor-infiltrating dendritic cells (DCs) (23,24).
High ISG15 levels have also been detected in the culture medium of
melanoma, along with the strong positive expression of ISG15 in the
cytoplasm of melanoma specimens (25). Thus, ISG15 plays a tumor-promoting
role in tumor immunogenicity. When ISG15 is secreted, free ISG15
can act as a cytokine to regulate the immune response (26). Moreover, free ISG15 can activate
natural killer cells, enhance lymphokine-activated killer-like
activity, stimulate IFN-γ production, induce DC maturation and
recruit neutrophils (27).
The expression pattern, biological functions and
underlying mechanisms of ISG15 in EC remain unclear. In this study,
the expression levels of ISG15 in EC and the relationship of ISG15
with clinicopathological features and patient survival were
investigated. Furthermore, the potential molecular mechanism of
ISG15 in promoting tumorigenesis was also explored.
Materials and methods
Samples
All ISG15 expression data in clinical samples,
including both RNA and protein levels, were derived from the
bioinformatics analysis. Cell-related functional and mechanistic
experiments, including polymerase chain reaction (PCR), western
blotting and flow cytometry, were performed at the Fourth Hospital
of Hebei Medical University (Shijiazhuang, China). This study was
approved by the ethics committee of the Fourth Hospital of Hebei
Medical University (approval no. 2020KY065). All patients who
contributed to the research provided their written consent.
Gene expression analysis in tumor
samples from the public database
Differential RNA expression data between tumor and
normal tissues collected from The Cancer Genome Atlas (TCGA)
database were downloaded (28) and
analyzed using the online tools MEXPRESS (https://mexpress.be/) and Gene Expression Profiling
Interactive Analysis (GEPIA; http://gepia.cancer-pku.cn/) (29). Transcription factor analysis
against ISG15 was performed online using the Cistrome website
(http://cistrome.org/db); the purpose of this
database is to use ChIP sequence data to analyze the lacation of
binding sites of transcription throughout the genome in vivo
(30). The proteomic data for EC
were obtained from the online Uniprot website (http://www.uniprot.org/). Protein IHC analysis, mRNA
(FPKM) and corresponding clinical information data were downloaded
from The Human Protein Atlas (https://www.proteinatlas.org/).
Immunohistochemistry (IHC)
Tissue array with 11 normal endometrial and 34 EC
tissues were provided by National Human Genetic Resources Sharing
Service Platform (China; HUteA045PG01). Written informed consent
was obtained from the patients undergoing surgery. The clinical
characteristics of the patients are described in Table SI. The tissue was fixed with 4%
paraformaldehyde at room temperature for 72 h, then embedded in
paraffin and cut into 4-µm serial sections for immunohistochemical
study. For IHC analysis, the tissue sections were deparaffinized
with xylene at room temperature and rehydrated using a gradient
ethanol solution. Endogenous catalase activity was blocked using 3%
hydrogen peroxide for 10 min in the dark at room temperature.
Tissue sections underwent antigen retrieval in 0.01 M sodium
citrate buffer for 5 min with a pressure cooker and were cooled at
room temperature. Primary antibodies against ISG15 (catalog no.
ab285367; Abcam) were diluted in phosphate-buffered saline (PBS) at
1:200 and incubated with the slices overnight at 4°C. The reactions
were visualized according to the manufacturer's instructions using
the Enhanced Polymer DAB detection kit (OriGene Technologies, Inc.)
and an optical microscope.
Tumor IMmune Estimation Resource
(TIMER) analysis
TIMER2.0 (http://timer.cistrome.org/) provides a more robust
estimation of immune infiltration levels for TCGA or user-provided
tumor profiles using six state-of-the-art algorithms (31). TIMER was used in this study to
evaluate the correlation between the expression of ISG15 and the
level of immune infiltration. The somatic copy number alteration
(SCNA) module in the TIMER database can relate the rate of tumor
infiltration of a gene to various changes in somatic copy number.
SCNA incorporates high amplification (2), arm-level gain (1), normal diploid (0), arm-level deletion
(−1) and deep deletion (−2). For each SCNA group, the degree of
infiltration was compared to the average using a two-sided Wilcoxon
test.
Cell culture
Human EC HEC-1A and Ishikawa cells were obtained
from Peking Union Medical College (Beijing, China). Both cell lines
were cultured in DMEM (Gibco; Thermo Fisher Scientific, Inc.)
containing 100 U/ml penicillin and 100 U/ml streptomycin (Beijing
Solarbio Science & Technology Co., Ltd.) and supplemented with
10% fetal bovine serum (Biological Industries). All cell lines were
incubated at 37°C in a 5% CO2 atmosphere.
RNA isolation and reverse
transcription-quantitative PCR (RT-qPCR) analysis
Total RNA was extracted from EC cell lines using
TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.). cDNA
was synthesized using random primers and M-MLV reverse
transcriptase (Promega Corporation) according to the manufacturer's
instructions. qPCR was performed using SYBR Premix Ex Taq kit
(Takara Bio, Inc.) according to the manufacturer's instructions.
The reaction conditions: Pre-denaturation at 95°C for 2 min;
followed by 40 cycles at 95°C for 15s and 60°C for 1 min; and
melting curve analysis at 95°C for 1 min, 55°C for 30 sec and 95°C
for 30 sec. Data were analyzed using the 2−∆∆cq method
(32). The primers used were as
reported in the literature (28):
ISG15 forward, 5′-GCGCAGATCACCCAGAAGAT-3′ and reverse,
5′-GTTCGTCGCATTTGTCCACC-3′; and GAPDH forward,
5′-CAAATTCCATGGCACCGTCA-3′ and reverse,
5′-GACTCCACGACGTACTCAGC-3′.
Western blotting
Total protein was extracted from EC cells using RIPA
lysis buffer and a protease inhibitor cocktail (Applygen
Technologies, Inc.). Protein concentrations were determined using a
BCA Kit (Thermo Fisher Scientific, Inc.). A total of 50 µg
protein/lane was electrophoresed via 10% SDS-PAGE gels and then
transferred onto polyvinylidene difluoride (PVDF) membranes.
Blocking was performed using 5% skimmed milk for 1 h at room
temperature. The following primary antibodies were used: Anti-ISG15
(catalog no. ab285367; Abcam), anti-tubulin (catalog no. 5335; Cell
Signaling Technology, Inc.), anti-β-actin (catalog no. 3700; Cell
Signaling Technology, Inc.) and anti-phosphorylated
(p)-retinoblastoma-associated protein (RB1; Ser807/811) (catalog
no. 8516; Cell Signaling Technology, Inc.); the antibodies and TBST
(20% Tween) were diluted at a ratio of 1:1,000 and incubated
overnight at 4°C. Horseradish peroxidase (HRP)-conjugated secondary
antibodies (catalog no. 9003-99-0) were purchased from Applygen
Technologies, Inc., diluted in 5% skimmed milk and incubated at
room temperature for 1 h. After incubation with chemiluminescence
solution (catalog no. WBKLS0050; Merck KGaA), the signals of the
protein bands were acquired using an ImageQuant 800 system
(Cytiva).
Small interfering (si)RNA transfection
assays
siRNA sequences against human ISG15 mRNA were
purchased from Shanghai GenePharma Co., Ltd., and the detailed
sequences were downloaded from the published literature (33): si-negative control (NC),
5′-AATTCTCCGAACGTGTCACGT-3′; siRNA-1, 5′-TCCTGGTGAGGAATAACAA-3′;
and siRNA-2, 5′-GGTGGACAAATGCGACGAA-3′. The transfection assay was
performed using Lipofectamine® 3000 reagent (Invitrogen;
Thermo Fisher Scientific, Inc.) according to the manufacturer's
instructions. A total of 2 µg siRNA was used to transfect the
HEC-1A and Ishikawa cells when the cell density reached 80%. At 48
h after transfection, the cells were harvested for RT-qPCR assays
and further functional experiments.
Cell proliferation assay
The proliferation rate of cells was determined using
the xCELLigence Real-Time Cell Analyzer (RTCA)-MP system (Agilent
Technologies, Inc.) according to the manufacturer's instructions.
The cell culture well was pre-balanced by adding 50 µl medium;
HEC-1A and Ishikawa cells were inoculated into the cell plate, and
3,000 cells in 100 µl medium were then added into each well, and
the cell index was measured every h.
Colony formation assay
HEC-1A and Ishikawa cells transfected with
ISG15-specific siRNA or control siRNA (si-NC) were inoculated into
six-well plates at a density of 1,500 cells per well and cultured
for 2 weeks until visible clones appeared. After removing the
supernatant, the cells were fixed with 4% paraformaldehyde for 20
min at room temperature and stained with 0.1% crystal violet at
room temperature. After washing with PBS, images were collected and
the results were recorded. Finally, the number of colonies (>50
cells) was counted using Adobe Photoshop 2021 (Adobe Systems, Inc.)
Data were normalized to each control group and to 100%.
Cell cycle assay
HEC-1A cells and Ishikawa cells in logarithmic
growth phase were resuscitated with complete culture medium and
made into suspension with a density of 5×104 per ml. The
suspension was inoculated into a petri dish with a diameter of 6 cm
according to 3 ml per plate and cultured for 24 h. The cells were
divided into three groups: i) si-NC; ii) si-1; and iii) si-2. Three
parallel dishes were set up in each group. The cells were collected
after 48 h of culture, fixed overnight at 4°C with 70% ethanol,
then washed twice with pre-cooled PBS (pH, 7.4), re-suspended with
RNaseA reagent (Beijing Solarbio Science & Technology Co.,
Ltd.) (100 µl), and incubated in a water bath at 37°C for 30 min.
PI staining solution [500 µl; Hangzhou Multi Sciences (Lianke)
Biotech Co., Ltd.] was added, mixed, incubated for 30 min at 4°C in
the dark, and detected by flow cytometry (FACSCalibur; BD
Biosciences), using FlowJoV10 software (BD Biosciences) for data
analysis.
Gene Set Enrichment Analysis
(GSEA)
GSEA was used to determine the effects of ISG15
genes on endometrial cancer-related pathways, with phenotypes
marked as positive and negative for ISG15 expression. The number of
random combinations of gene sets per analysis was set at 1,000,
P<0.05 was used to indicate statistical significance and gene
sets with a false-discovery rate of <0.25 were considered
enriched.
Statistical analysis
All the experiments were repeated three times. All
data were analyzed with GraphPad Prism 6 for Windows (GraphPad
Software, Inc.) and SPSS for Windows, version 16.0 (SPSS, Inc.).
Data are shown as the mean ± SD or mean ± SEM. One-way ANOVA and
Bonferroni's correction were performed for multiple comparisons
between groups, and Pearson's correlation coefficient was used to
determine the correlations between methylation scores and ISG15
mRNA levels, and various cell markers. Spearmen's correlation
analysis was also applied. Survival analysis was performed using
the Kaplan-Meier method to determine whether the ISG15 gene was
associated with survival in patients with endometrial cancer, and
P-values were calculated by the log-rank test. P<0.05 was
considered to indicate a statistically significant difference.
Results
ISG15 is highly expressed and may be a
marker of EC tumor progression
The mRNA expression data of ISG15 in normal and
tumor tissues for different cancers were obtained from the GEPIA
database. As shown in Fig. 1A,
ISG15 levels were significantly elevated in cancers, including EC,
esophageal squamous cell carcinoma (ESCC), gastric cancer, liver
cancer and breast cancer. When all cancer types were ranked
according to the mRNA levels of ISG15, most gynecological
malignancies, including uterine corpus EC, cervical squamous cell
carcinoma and endocervical adenocarcinoma, ovarian cancer and
uterine carcinosarcoma, showed elevated ISG15 levels. For EC, the
tumor tissues had >10-fold higher ISG15 expression than the
corresponding normal tissues (Fig.
1B).
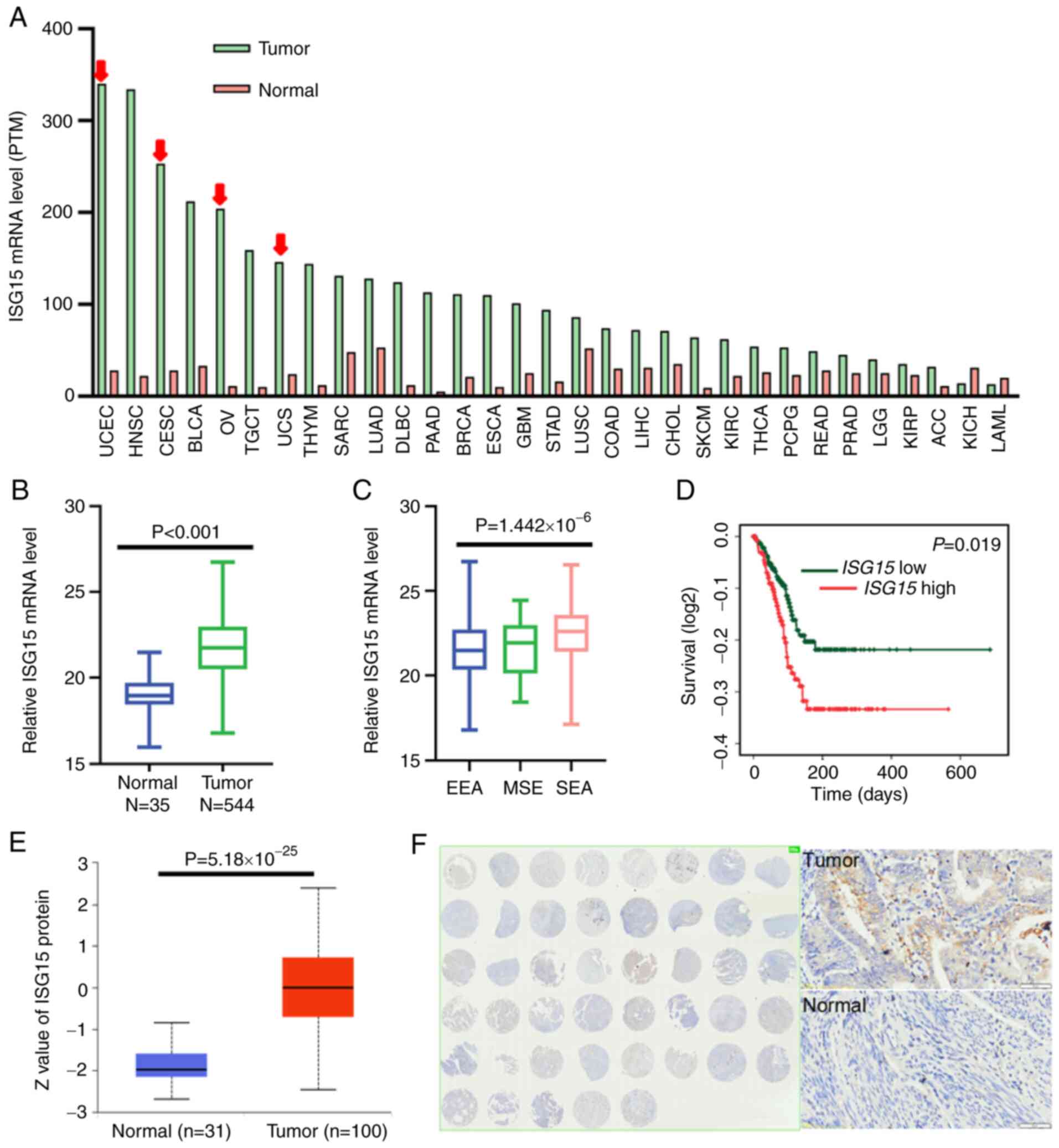 | Figure 1.ISG15 is a marker of tumor
progression and prognosis in EC. (A) The RNA expression of ISG15
was most significantly increased in gynecologic tumors, with
expression levels decreasing from left to right. Gynecological
tumors are indicated by the red arrow. (B) mRNA expression levels
of ISG15 in tumor vs. normal tissue in cases of EC in TCGA
database. Green and blue represent tumor and normal tissue,
respectively. (C) mRNA expression levels of ISG15 in different
subtypes of EC in cases of EC in TCGA database. (D) Overall
survival of patients in the ISG15 high and low expression groups in
TCGA EC samples. The mRNA (FPKM) and corresponding clinical
information data were downloaded from The Human Protein Atlas and
analyzed using SPSS software. Kaplan-Meier survival curve was
constructed, which was analyzed using a log-rank test. (E) Protein
expression levels of ISG15 in EC based on quantified ISG15 protein
data from the UALCAN public database. (F) Representative
immunohistochemistry results of ISG15 in EC, showing the results
from an EC tissue assay. ISG15, interferon-stimulated gene 15; EC,
endometrial carcinoma; TCGA, The Cancer Genome Atlas; FPKM,
Fragments Per Kilobase of transcript per Million; UCEC, uterine
corpus endometrial carcinoma; CESC, cervical squamous cell
carcinoma and endocervical adenocarcinoma; OV, ovarian cancer; UCS,
uterine carcinosarcoma; EEA, endometrioid endometrial
adenocarcinoma; MSE, mixed serous and endometrioid; SEA, serous
endometrial adenocarcinoma. |
Next, the association between high ISG15 expression
and ES subtypes was investigated. ISG15 expression in EC was not
significantly associated with age or tumor recurrence, but did
differ significantly among molecular subtypes (Fig. S1A). Serous endometrial
adenocarcinoma cancer subtypes had the highest level of ISG15
expression, followed by the mixed-type, while adenocarcinoma-type
endometrial cancer tissues showed the lowest ISG15 mRNA level
(Fig. 1C). Different pathological
subtypes of EC have different risk factors and degrees of
malignancy (34). As the
expression of ISG15 was significantly associated with pathological
subtypes, the association between ISG15 expression and EC prognosis
was further investigated. It was found that patients with EC with
high ISG15 mRNA expression showed significantly shorter overall
survival (OS; Fig. 1D).
Next, the protein levels of ISG15 in ECs were
evaluated. First, analysis of the proteomic data (Uniprot;
http://www.uniprot.org/) in 100 EC samples and 31
normal samples indicated that the protein levels of ISG15 were also
significantly elevated in EC samples (Fig. 1E). Furthermore, IHC of a tissue
array of EC and normal tissues showed that 55.9% of EC tissues (19
of 34) expressed notable levels of ISG protein, while ISG15
expression could not be detected in all 11 normal endometrial
tissues (Fig. 1F).
Taken together, the results of the mRNA and protein
expression analyses showed that ISG15 upregulation was most
significant in EC, representing a potential tumor marker of EC.
Moreover, ISG15 may be a marker of pathological progression of EC
with a significant prognostic value.
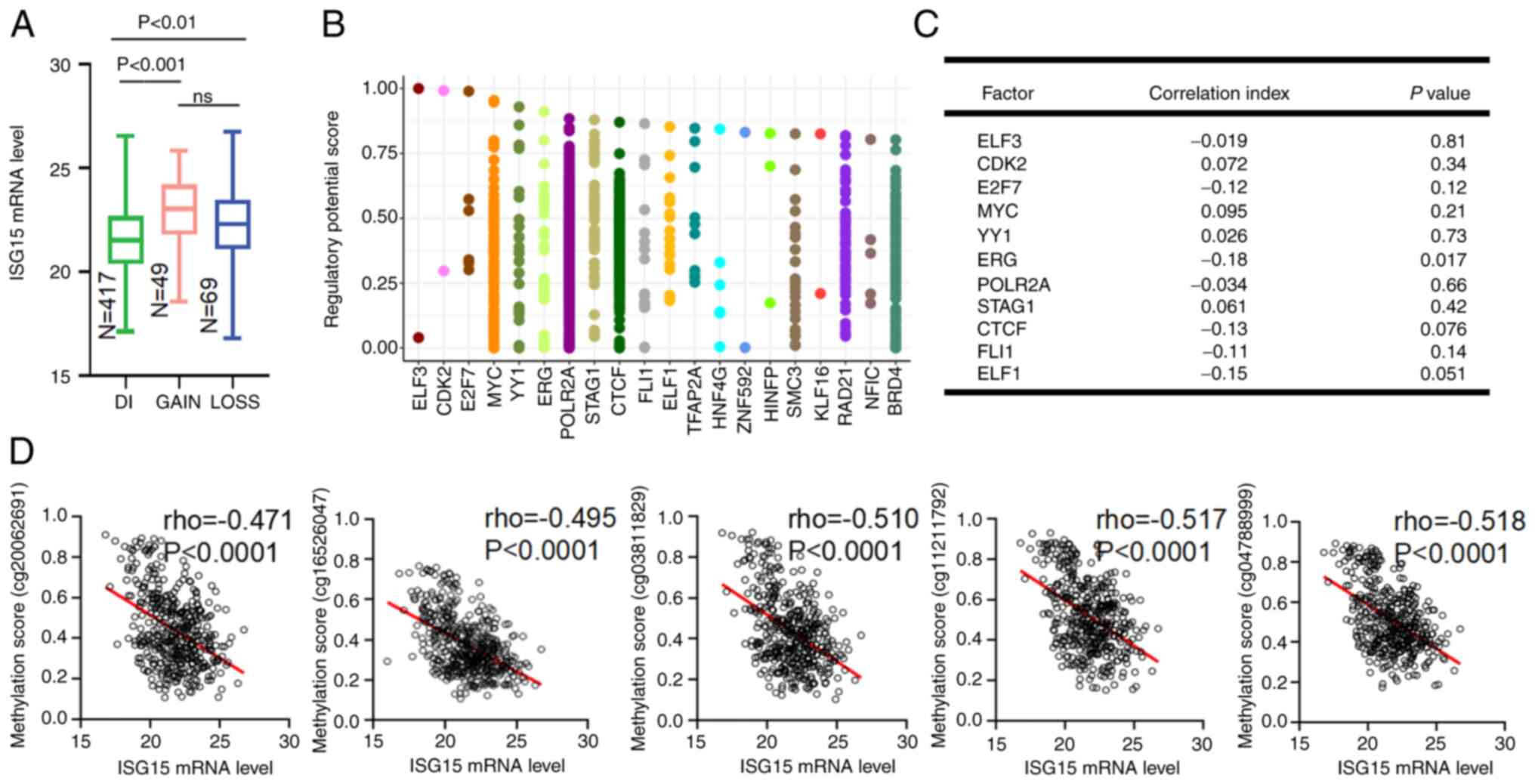
Abnormal DNA methylation leads to
ISG15 upregulation in EC
Next, the molecular mechanisms leading to the
upregulation of ISG15 were explored. Surprisingly, increased ISG15
gene copy number was detected in <10% of the samples and there
was no significant association between the increased ISG15 copy
number and mRNA levels (Fig. 2A).
Therefore, ISG15 upregulation in EC was not caused by gene
amplification.
Then, it was considered whether dysregulation of
ISG15-specific transcription factors was involved in ISG15
upregulation. The Cistrome database was used, which allows the
analysis of in vivo genome-wide location of transcription
factor binding sites to screen for transcription factors that may
bind to the ISG15 promoter. It was found that a series of
well-known tumor-associated transcription factors could bind to
this promoter in chromatin immunoprecipitation experiments,
including MYC proto-oncogene protein (MYC), E74-like factor 3 and
cyclin-dependent kinase 2 (CDK2) (Fig.
2B). However, none of the top 10 transcription factors with the
highest binding score to the ISG15 promoter were significantly
correlated with ISG15 expression (Fig.
2C). Abnormal DNA methylation plays a regulatory role in the
expression of oncogenes in human cancers (35). Notably, the methylation of the CpG
sites located in the promoter region of ISG15 also showed no
correlation with ISG15 mRNA expression in the present study
(Fig. S1B). However, the
methylation states of five CpG sites located in the gene body of
ISG15 were significantly negatively correlated with ISG15 mRNA
levels (R=−0.471 for Chr11014012; R=−0.510 for Chr11014069;
R=−0.517 for Chr11014254; R=−0.518 for Chr11014471; and R=−0.495
for Chr11014514; Fig. 2D).
Therefore, it was concluded that the upregulation of ISG15 in EC
was caused by the dysregulation of DNA methylation in the ISG15
gene body.
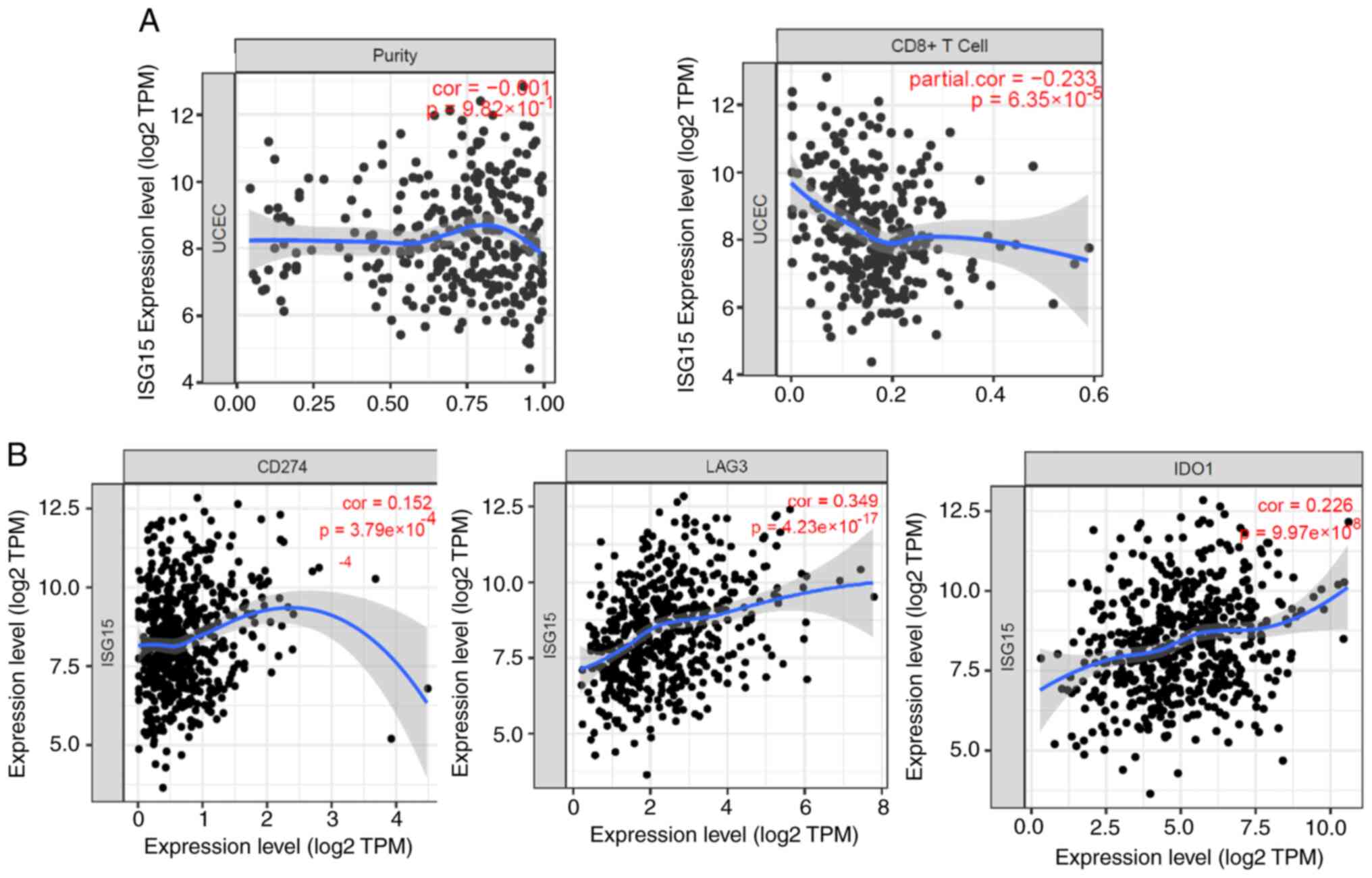
ISG15 inhibits T cell infiltration and
promotes immune escape
ISG15 is an IFN-stimulating gene. IFN can
significantly induce the transcriptional expression of this gene in
an inflammatory environment (20).
Therefore, in the present study it was considered whether the high
ISG15 expression in EC was related to the tumor immune
microenvironment. Effective infiltration of CD8+ T cells
into tumors is the basis for antitumor immunity. Inhibited
infiltration of CD8+ T cells is also a fundamental
mechanism for tumor immune escape (36). Correlation analysis showed that the
ISG15 expression level was significantly negatively correlated with
the number of CD8+ T cells in EC samples; in other
words, patients with EC with high ISG15 levels tended to have fewer
CD8+ T cells (Fig. 3A),
suggesting that it is easier to form an immune microenvironment for
‘cold’ tumors (without infiltrating lymphocytes). Among T cells
that infiltrate the tumor, tumor cells also express T cell
inhibitory factors to send an inhibition signal to reduce the
killing activity of the T cells (37). In the current study, analysis of
the effects of ISG15 on the expression of T cell inhibitory factors
showed that ISG15 was highly positively correlated with the
expression of most T cell inhibitory molecules, such as programmed
death-ligand 1 (PD-L1), indoleamine 2′3-dioxygenase 1 (IDO1) and
lymphocyte-activation gene 3 (Fig.
3B). Therefore, the tumor-killing activity of CD8+ T
cells was inhibited in ECs with high ISG15 expression.
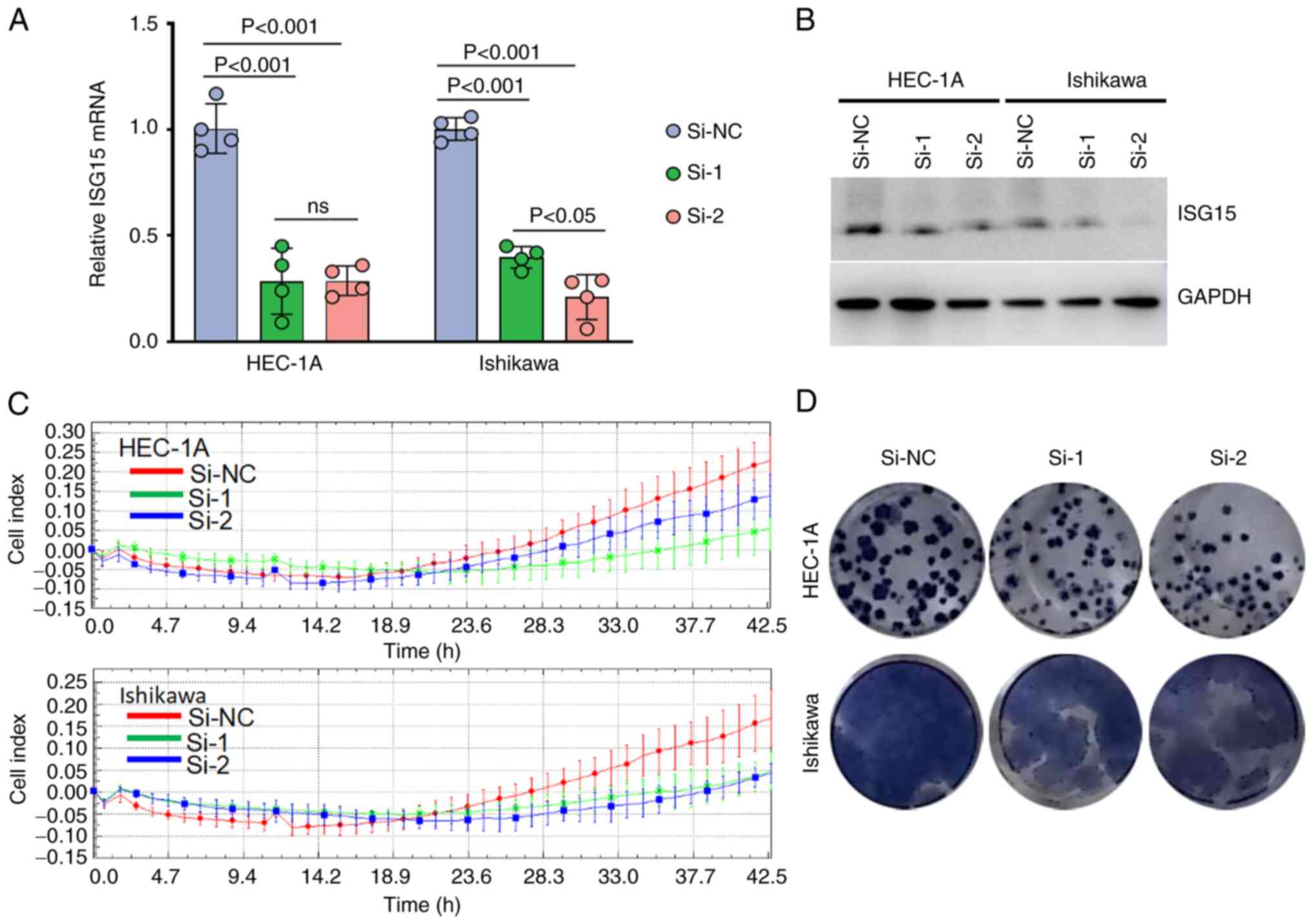
ISG15 promotes EC cell proliferation
and colony formation
Next, the possible function of ISG15 in EC cells was
investigated. First, the successful ISG15 knockdown in the two cell
lines using ISG15-specific siRNAs was confirmed (Fig. 4A and B). Cell proliferation was
measured using the RTCA-MP system. According to the growth curve
drawn by the dynamic cell index, the inhibition of ISG15 markedly
slowed EC cell proliferation (Fig.
4C), suggesting that ISG15 had a growth-promoting function in
EC. The anti-anoikis capacity is vital for tumor formation
(38). Therefore, the function of
ISG15 in cell colony formation was examined. Transfection with
ISG15 siRNA resulted in a notable decrease in colony number
(Fig. 4D). Taken together, the
direct association between ISG15 expression and cell proliferation
or colony formation ability indicated the important role of ISG15
in EC tumor cell growth.
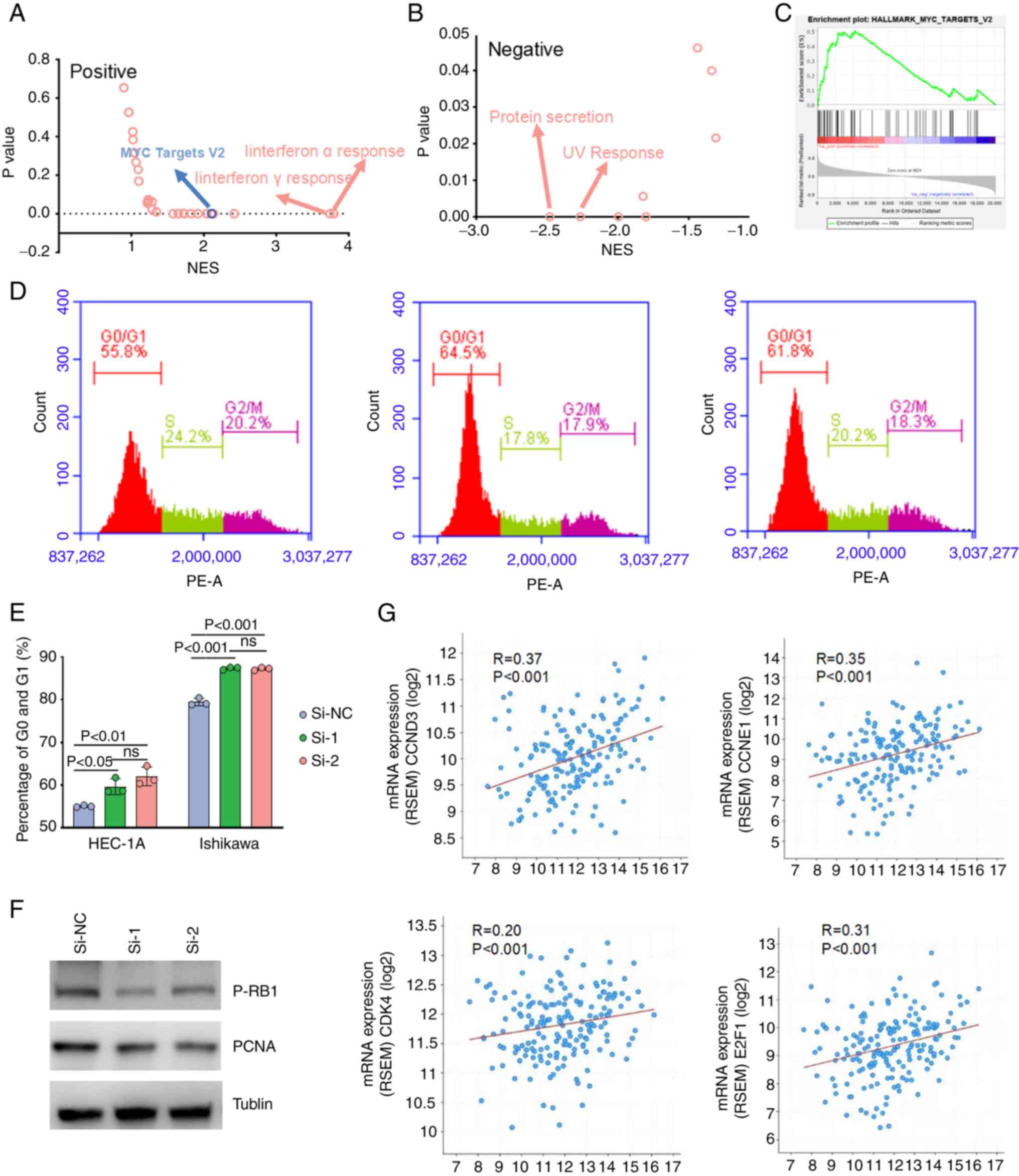
ISG15 contributes to EC development
via the MYC pathway and cell cycle regulation
The molecular mechanism of ISG15 in promoting EC
progression was then explored using Gene Set Enrichment Analysis
(GSEA). A total of 14 and nine pathways were positively and
negatively correlated with ISG15 expression, respectively (Tables SII and SIII). In addition to the well-studied
ISG15-associated pathway in immune responses, such as IFN-α
response and IFN-γ response signaling pathways (39) (Fig. 5A
and B), the ‘hallmark_MYC_targets_V2’ pathway was also
evaluated as MYC target activation is involved in numerous
processes necessary for cancer development, especially cell
proliferation (40). It was found
that MYC expression was significantly associated with ISG15
expression (Fig. 5C). Thus, ISG15
may function via the MYC pathway. Additionally, the activation of
MYC targets is generally considered a marker of cell cycle
activation (40). Therefore, the
role of ISG15 in the cell cycle was investigated. The results
showed that ISG15 knockdown increased the number of cells in the
G1/G0 phase of both EC cell lines (Fig. 5D and E), which may be responsible
for the inhibition of EC cell proliferation with a low expression
of ISG15. To further confirm the function of ISG15 in the cell
cycle G1/S transition, the expression of p-RB1, a standard protein
marker representing the activation of the cell cycle G1/S
transition (41), was examined. As
shown in Fig. 5F, ISG15 knockdown
reduced the expression of p-RB1 and proliferating cell nuclear
antigen (PCNA) in both EC cell lines. According to TCGA tumor mRNA
expression database, ISG15 was significantly positively correlated
with G1/S-specific cyclin-D3 (CCND3), G1/S-specific cyclin-E1
(CCNE1), CDK4 and transcription factor E2F1 (E2F1) (R=0.37 for
CCND3; R=0.35, P<0.001 for CCNE1; R=0.20 for CDK4; and R=0.31
for E2F1; Fig. 5G). Taken
together, these results suggested that ISG15 promoted the G1/S cell
cycle transition in EC cells via the MYC pathway.
Discussion
Despite advances in diagnostic and clinical
management, the prognosis of EC remains poor (35,42).
The molecular mechanisms underlying EC progression remain unclear.
Thus, there is a need to further explore the molecular mechanisms
underlying the occurrence and development of EC and identify new
and meaningful prognostic markers. ISG15, a UBL protein involved in
protein modification, reportedly plays an essential role in tumor
development. Some investigations have revealed a strong association
between aberrant ISG15 expression and the incidence of some human
malignancies, including esophageal squamous cancer and liver cancer
(43,44). However, the relative expression
status and function of ISG15 in ECs remain to be elucidated.
In the present study, the upregulation of ISG15 in
EC was demonstrated by re-analyzing the public transcriptome
sequencing, proteomics and IHC data. In addition, the Kaplan-Meier
survival curves showed the association of high ISG15 levels with an
unfavorable prognosis. These results suggested that ISG15 may play
an important role in the development of EC. It is worth noting that
the expression of ISG15 in tumors was specific to gynecologic
tumors. Among the five types of tumors with the highest ISG15
expression levels, three were gynecologic tumors (EC, cervical
cancer and ovarian cancer), with EC ranking first. The occurrence
and progression of gynecologic tumors are closely related to
abnormal hormone levels (45),
suggesting a regulatory relationship between ISG15 and hormones,
which requires clarification in future studies.
The results of the clinical correlation analysis in
the current study showed that elevated ISG15 expression was highly
correlated with the pathological stage (Fig. S1C), indicating that ISG15 may be a
promising marker of EC progression. Furthermore, it was found that
ISG15 knockdown significantly inhibited the malignant phenotypes of
EC cells, including proliferation and colony formation, suggesting
the central role of ISG15 in the malignant progression of EC
cells.
Furthermore, as ISG15 is an IFN-induced gene, the
relationship between ISG15 and immunity status in EC was analyzed.
It was found that ISG15 was negatively correlated with
CD8+ T cell infiltration and positively correlated with
CD274, ATG3 and IDO1. PD-L1, also called B7-H1 or CD274, is a
ligand of PD-1. This axis affects immune checkpoints to mediate T
cell depletion, characterized by a loss of cytokines, impaired
proliferation, lack of cytotoxic activity, and, ultimately,
inhibition of an effective immune response (46). In cancer, PD-L1 upregulates the
immune defense of similarly hijacked hosts to promote T cell
depletion, resulting in immune resistance (47–49).
ATG3 is a key gene involved in autophagy and its homologous genes
are common in eukaryotes. Lawson et al (50) screened the whole-genome Clustered
regularly interspaced short palindromic repeats of Renca cells and
restored the established suppressors. Cytotoxic T lymphocyte (CTL)
escape mutations in numerous types of cancers were identified in
this screen, including genes annotated in the autophagy pathway
(ATG3, ATG5, ATG7, ATG10, ATG12 and ATG14), confirming the
importance of IFN-γ response to the innate CTL escape phenotype
(50). The study showed that
disturbances in autophagy and peroxisome pathways are some of the
most sensitive mutations in wild-type Renca cells when IFN-γ (such
as interferon-induced transmembrane protein 2) is used alone. These
results highlight the profound effects of the autophagy pathway in
regulating the inherent CTL escape of cancer cells (50). The inhibition of autophagy also
affects some aspects of the immune system, such as the memory
formation of virus-specific CD8+ T cells and the
activation of CD4+ T cells by DCs, which may inhibit the
death of immunogenic cells induced by chemotherapy (51). IDO1 is a cytoplasmic monomer
enzyme-containing heme, the most crucial inducer of which is the
cytokine IFN-γ. IDO1 is considered an effector molecule that can
mediate a survival strategy based on tryptophan deprivation and
catalyze the initial rate-limiting step of the degradation of the
essential amino acid tryptophan in the canine pathway, through
which IDO1 plays an essential role in maintaining maternal T cell
tolerance (52). Thus, IDO1 is now
considered an immunomodulator in autoimmune diseases, chronic
inflammation and tumor immunity (53). Taken together with the present
results, it can be speculated that high ISG15 expression negatively
regulates antitumor immunity and is positively associated with
tumor immune escape.
ISG15 functions not only as a modifier of target
proteins, but also as a free protein that promotes cancer
progression (54,55). ISG15 can conjugate with Ki-Ras to
reverse the malignant phenotypes of breast cancer (56). Falvey et al (57) reported that the depletion of ISG15
expression promoted autophagy. Thus, ISG15 is also a novel
inhibitor of autophagy, potentially influencing
5-Fluorouracil-mediated chemosensitivity in esophageal cancer cells
(43). High ISG15 mRNA expression
has been observed in ESCC tissues and may serve as a novel
prognostic biomarker for ESCC among alcohol drinkers (43). However, the specific mechanism of
ISG15 in EC has not yet been fully elucidated. In the present
study, the potential molecular mechanism by which ISG15 promotes
tumorigenesis was explored. The combination of data from
bioinformatics analysis, flow cytometry and immunoblotting analyses
indicated that ISG15 regulated the G1/S transition and cell
proliferation via the MYC pathway. However, the function of ISG15
as a free protein or protein-modifying factor in EC requires
further investigation.
DNA methylation is indicative of gene expression,
with gene body methylation being a more useful indicator than
promoter methylation (58). The
results of the current study showed that ISG15 upregulation in EC
was caused by a dysregulation of DNA methylation in the ISG15 gene
body, suggesting that ISG15 deregulation in EC is related to DNA
methylation and that regulation of methylation may be a strategy to
decrease ISG15 expression.
Our research work has certain reference significance
for understanding the pathogenesis of EC, including cell
proliferation immune escape. The PTM of proteins has gradually
become a research hotspot in the development of tumor-targeted
therapeutic drugs (59).
Considering the significant clinical value of ISG15 in EC, we
speculate that ISG15 may also be a promising therapeutic target for
EC in the future. However, the current study has certain
limitations. For example, the investigation of the methylation
modification of the ISG15 promoter region was limited at the
phenotypic level in its infancy, and the experiments investigating
the immune escape of ISG15 lacked further cytology and animal
experiments. Thus, we will perform animal experiments to further
verify the immunotherapy value of ISG15, and explore the detailed
mechanism of ISG15 in upstream and downstream pathways in different
types of EC cells.
In conclusion, the results of this study
demonstrated that dysregulation of the methylation of ISG15 led to
its high expression and was related to poor clinical outcomes and
pathological stage of EC. The knockdown of ISG15 expression
attenuated the malignant phenotypes of EC cell lines. Moreover, it
was also found that the MYC pathway was part of the potential
mechanism through which ISG15 promoted EC tumorigenesis.
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
This study was supported by the Natural Science Foundation of
Hebei Province (grant nos. H2019206697, H2020206131 and
H2020206549).
Availability of data and materials
This datasets used and/or analyzed during the
current study are available from the corresponding author on
reasonable request.
Authors' contributions
LZ and XZ conceived and designed the experiments. XZ
and JW completed the experiments and explained the experimental
data. YW and MZ collected specimens for the experiments and
participated in the drafting of the manuscript. WZ and HZ analyzed
the database information and substantially revised the content of
the manuscript. LZ and XZ integrated all experimental data for data
analysis and statistical analysis and calibrated the publication of
the final version. LZ and XZ confirm the authenticity of all the
raw data. All the authors have read and approved the final
manuscript.
Ethics approval and consent to
participate
This study was approved by the ethics committee of
the Fourth Hospital of Hebei Medical University (Shijiazhuang,
China). All patients who contributed to the research provided their
written consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bell DW and Ellenson LH: Molecular
genetics of endometrial carcinoma. Annu Rev Pathol. 14:339–367.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Shiozawa T and Konishi I: Early
endometrial carcinoma: Clinicopathology, hormonal aspects,
molecular genetics, diagnosis, and treatment. Int J Clin Oncol.
11:13–21. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Buhtoiarova TN, Brenner CA and Singh M:
Endometrial carcinoma: Role of current and emerging biomarkers in
resolving persistent clinical dilemmas. Am J Clin Pathol. 145:8–21.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
MacKintosh ML and Crosbie EJ: Prevention
strategies in endometrial carcinoma. Curr Oncol Rep. 20:1012018.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lu H, Ju DD, Yang GD, Zhu LY, Yang XM, Li
J, Song WW, Wang JH, Zhang CC, Zhang ZG and Zhang R: Targeting
cancer stem cell signature gene SMOC-2 Overcomes chemoresistance
and inhibits cell proliferation of endometrial carcinoma.
EBioMedicine. 40:276–289. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Vu LD, Gevaert K and De Smet I: Protein
language: Post-translational modifications talking to each other.
Trends Plant Sci. 23:1068–1080. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Loboda AP, Soond SM, Piacentini M and
Barlev NA: Lysine-specific post-translational modifications of
proteins in the life cycle of viruses. Cell Cycle. 18:1995–2005.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Akimov V, Barrio-Hernandez I, Hansen SVF,
Hallenborg P, Pedersen AK, Bekker-Jensen DB, Puglia M, Christensen
SDK, Vanselow JT, Nielsen MM, et al: UbiSite approach for
comprehensive mapping of lysine and N-terminal ubiquitination
sites. Nat Struct Mol Biol. 25:631–640. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Shin D, Mukherjee R, Liu Y, Gonzalez A,
Bonn F, Liu Y, Rogov VV, Heinz M, Stolz A, Hummer G, et al:
Regulation of phosphoribosyl-linked serine ubiquitination by
deubiquitinases DupA and DupB. Mol Cell. 77:164–179.e6. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ribet D and Cossart P: Ubiquitin, SUMO,
and NEDD8: Key targets of bacterial pathogens. Trends Cell Biol.
28:926–940. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Cossec JC, Theurillat I, Chica C, Aguín
SB, Gaume X, Andrieux A, Iturbide A, Jouvion G, Li H, Bossis G, et
al: SUMO safeguards somatic and pluripotent cell identities by
enforcing distinct chromatin states. Cell Stem Cell. 23:742–757.e8.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Baek K, Krist DT, Prabu JR, Hill S, Klügel
M, Neumaier LM, von Gronau S, Kleiger G and Schulman BA: NEDD8
nucleates a multivalent cullin-RING-UBE2D ubiquitin ligation
assembly. Nature. 578:461–466. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Fischer S, Rijal R, Frommolt P, Wagle P,
Konertz R, Faix J, Meßling S and Eichinger L: Functional
characterization of ubiquitin-like core autophagy protein ATG12 in
dictyostelium discoideum. Cells. 8:722019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Marshall RS, Hua Z, Mali S, McLoughlin F
and Vierstra RD: ATG8-binding UIM proteins define a new class of
autophagy adaptors and receptors. Cell. 177:766–781.e24. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Gómez CE, Perdiguero B, Falqui M, Marín
MQ, Bécares M, Sorzano COS, García-Arriaza J, Esteban M and Guerra
S: Enhancement of HIV-1 env-specific CD8 T cell responses using
interferon-stimulated gene 15 as an immune adjuvant. J Virol.
95:e01155–e01120. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Sandy Z, da Costa IC and Schmidt CK: More
than meets the ISG15: Emerging roles in the DNA damage response and
beyond. Biomolecules. 10:15572020. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chen RH, Du Y, Han P, Wang HB, Liang FY,
Feng GK, Zhou AJ, Cai MY, Zhong Q, Zeng MS and Huang XM: ISG15
predicts poor prognosis and promotes cancer stem cell phenotype in
nasopharyngeal carcinoma. Oncotarget. 7:16910–16922. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Desai SD, Haas AL, Wood LM, Tsai YC,
Pestka S, Rubin EH, Saleem A, Nur-E-Kamal A and Liu LF: Elevated
expression of ISG15 in tumor cells interferes with the
ubiquitin/26S proteasome pathway. Cancer Res. 66:921–928. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Tecalco-Cruz AC, Cortes-Gonzalez CC,
Cruz-Ramos E, Jarquín JOR, Romero-Mandujano AK and Sosa-Garrocho M:
Interplay between interferon-stimulated gene 15/ISGylation and
interferon gamma signaling in breast cancer cells. Cell Signal.
54:91–101. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Parker BS, Rautela J and Hertzog PJ:
Antitumour actions of interferons: Implications for cancer therapy.
Nat Rev Cancer. 16:131–144. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Reich N, Evans B, Levy D, Fahey D, Knight
E Jr and Darnell JE Jr: Interferon-induced transcription of a gene
encoding a 15-kDa protein depends on an upstream enhancer element.
Proc Natl Acad Sci USA. 84:6394–6398. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Liang KC, Suzuki Y, Kumagai Y and Nakai K:
Analysis of changes in transcription start site distribution by a
classification approach. Gene. 537:29–40. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Padovan E, Terracciano L, Certa U, Jacobs
B, Reschner A, Bolli M, Spagnoli GC, Borden EC and Heberer M:
Interferon stimulated gene 15 constitutively produced by melanoma
cells induces e-cadherin expression on human dendritic cells.
Cancer Res. 62:3453–3458. 2002.PubMed/NCBI
|
|
24
|
Remmel E, Terracciano L, Noppen C, Zajac
P, Heberer M, Spagnoli GC and Padovan E: Modulation of dendritic
cell phenotype and mobility by tumor cells in vitro. Hum Immunol.
62:39–49. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lamberti MJ, Mentucci FM, Roselli E, Araya
P, Rivarola VA, Vittar NBR and Maccioni M: Photodynamic modulation
of type 1 interferon pathway on melanoma cells promotes dendritic
cell activation. Front Immunol. 10:26142019. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Liu G, Lee JH, Parker ZM, Acharya D,
Chiang JJ, van Gent M, Riedl W, Davis-Gardner ME, Wies E, Chiang C
and Gack MU: ISG15-dependent activation of the sensor MDA5 is
antagonized by the SARS-CoV-2 papain-like protease to evade host
innate immunity. Nat Microbiol. 6:467–478. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
D'Cunha J, Knight E Jr, Haas AL, Truitt RL
and Borden EC: Immunoregulatory properties of ISG15, an
interferon-induced cytokine. Proc Natl Acad Sci USA. 93:211–215.
1996. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Tang Z, Li C, Kang B, Gao G, Li C and
Zhang Z: GEPIA: A web server for cancer and normal gene expression
profiling and interactive analyses. Nucleic Acids Res. 45:W98–W102.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Koch A, De Meyer T, Jeschke J and Van
Criekinge W: MEXPRESS: Visualizing expression, DNA methylation and
clinical TCGA data. BMC Genomics. 16:6362015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zheng R, Wan C, Mei S, Qin Q, Wu Q, Sun H,
Chen CH, Brown M, Zhang X, Meyer CA and Liu XS: Cistrome data
browser: Expanded datasets and new tools for gene regulatory
analysis. Nucleic Acids Res. 47:D729–D735. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Li T, Fu J, Zeng Z, Cohen D, Li J, Chen Q,
Li B and Liu XS: TIMER2.0 for analysis of tumor-infiltrating immune
cells. Nucleic Acids Res. 48:W509–W514. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yuan H, Zhou W, Yang Y, Xue L, Liu L and
Song Y: ISG15 promotes esophageal squamous cell carcinoma
tumorigenesis via c-MET/Fyn/beta-catenin signaling pathway. Exp
Cell Res. 367:47–55. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zang Y, Dong M, Zhang K, Tian W, Wang Y
and Xue F: Bioinformatics analysis of key differentially expressed
genes in well and poorly differentiated endometrial carcinoma. Mol
Med Rep. 18:467–476. 2018.PubMed/NCBI
|
|
35
|
Zhao X, Wei X, Zhao L, Shi L, Cheng J,
Kang S, Zhang H, Zhang J, Li L, Zhang H and Zhao W: The rs6983267
SNP and long non-coding RNA CARLo-5 are associated with endometrial
carcinoma. Environ Mol Mutagen. 57:508–515. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wang G, Yu Y, Li ZM, Zhu ZM, Wang ZJ and
Tao MF: Triterpenoids of Rhus Chinensis supressed colorectal cancer
progress by enhancing antitumor immunity and CD8 + T cells tumor
infiltration. Nutr Cancer. 6:1–15. 2021. View Article : Google Scholar
|
|
37
|
Lentz RW, Colton MD, Mitra SS and
Messersmith WA: Innate immune checkpoint inhibitors: The next
breakthrough in medical oncology? Mol Cancer Ther. 20:961–974.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Su H, Si XY, Tang WR and Luo Y: The
regulation of anoikis in tumor invasion and metastasis. Yi Chuan.
35:10–16. 2013.(In Chinese). View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhao Z, Collins MN, Hsiang TY and Krug RM:
Interferon-induced ISG15 pathway: An ongoing virus-host battle.
Trends Microbiol. 21:181–186. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Bretones G, Delgado MD and León J: Myc and
cell cycle control. Biochim Biophys Acta. 1849:506–516. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Ihle MA, Huss S, Jeske W, Hartmann W,
Merkelbach-Bruse S, Schildhaus HU, Büttner R, Sihto H, Hall KS,
Eriksson M, et al: Expression of cell cycle regulators and
frequency of TP53 mutations in high risk gastrointestinal stromal
tumors prior to adjuvant imatinib treatment. PLoS One.
13:e01930482018. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Tran AQ and Gehrig P: Recent advances in
endometrial cancer. F1000Res. 6:812017. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Tao J, Ping H, Wen J, Hu Y, Yang H and Xie
X: Prognostic value of ISG15 mRNA level in drinkers with esophageal
squamous cell cancers. Int J Clin Exp Pathol. 8:10975–10984.
2015.PubMed/NCBI
|
|
44
|
Li C, Wang J, Zhang H, Zhu M, Chen F, Hu
Y, Liu H and Zhu H: Interferon-stimulated gene 15 (ISG15) is a
trigger for tumorigenesis and metastasis of hepatocellular
carcinoma. Oncotarget. 5:8429–8441. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Edey KA, Rundle S and Hickey M: Hormone
replacement therapy for women previously treated for endometrial
cancer. Cochrane Database Syst Rev. May 15–2018.(Epub ahead of
print). View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Zhao Y, Shao Q and Peng G: Exhaustion and
senescence: Two crucial dysfunctional states of T cells in the
tumor microenvironment. Cell Mol Immunol. 17:27–35. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
He X and Xu C: PD-1: A driver or passenger
of T cell exhaustion? Mol Cell. 77:930–931. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Pardoll DM: The blockade of immune
checkpoints in cancer immunotherapy. Nat Rev Cancer. 12:252–264.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Seidel JA, Otsuka A and Kabashima K:
Anti-PD-1 and Anti-CTLA-4 therapies in cancer: Mechanisms of
action, efficacy, and limitations. Front Oncol. 8:862018.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Lawson KA, Sousa CM, Zhang X, Kim E,
Akthar R, Caumanns JJ, Yao Y, Mikolajewicz N, Ross C, Brown KR, et
al: Functional genomic landscape of cancer-intrinsic evasion of
killing by T cells. Nature. 586:120–126. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Yamamoto K, Venida A, Yano J, Biancur DE,
Kakiuchi M, Gupta S, Sohn ASW, Mukhopadhyay S, Lin EY, Parker SJ,
et al: Autophagy promotes immune evasion of pancreatic cancer by
degrading MHC-I. Nature. 581:100–105. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Lewis-Ballester A, Pham KN, Batabyal D,
Karkashon S, Bonanno JB, Poulos TL and Yeh SR: Structural insights
into substrate and inhibitor binding sites in human indoleamine
2,3-dioxygenase 1. Nat Commun. 8:16932017. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Pallotta MT, Rossini S, Suvieri C, Coletti
A, Orabona C, Macchiarulo A, Volpi C and Grohmann U: Indoleamine
2,3-dioxygenase 1 (IDO1): An up-to-date overview of an eclectic
immunoregulatory enzyme. FEBS J. Jun 19–2021.(Epub ahead of print).
doi: 10.1111/febs.16086, 2021. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Zhang Q, Wang J, Qiao H, Huyan L, Liu B,
Li C, Jiang J, Zhao F, Wang H and Yan J: ISG15 is downregulated by
KLF12 and implicated in maintenance of cancer stem cell-like
features in cisplatin-resistant ovarian cancer. J Cell Mol Med.
25:4395–4407. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Andersen JB, Aaboe M, Borden EC, Goloubeva
OG, Hassel BA and Orntoft TF: Stage-associated overexpression of
the ubiquitin-like protein, ISG15, in bladder cancer. Br J Cancer.
94:1465–1471. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Burks J, Reed RE and Desai SD: ISGylation
governs the oncogenic function of Ki-Ras in breast cancer.
Oncogene. 33:794–803. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Falvey CM, O'Donovan TR, El-Mashed S,
Nyhan MJ, O'Reilly S and McKenna SL: UBE2L6/UBCH8 and ISG15
attenuate autophagy in esophageal cancer cells. Oncotarget.
8:23479–23491. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Mehdi A and Rabbani SA: Role of
methylation in pro- and anti-cancer immunity. Cancers (Basel).
13:5452021. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Zhang H, He J, Hu G, Zhu F, Jiang H, Gao
J, Zhou H, Lin H, Wang Y, Chen K, et al: Dynamics of
post-translational modification inspires drug design in the kinase
family. J Med Chem. 64:15111–15125. 2021. View Article : Google Scholar : PubMed/NCBI
|