Digestive system cancers are the leading cause of
cancer-related death worldwide and represent a global health
challenge, with cancer cells participating in the occurrence and
progression of cancer by altering cellular molecular and cellular
biological processes (1). A variety
of treatments for gastric cancer (GC), including chemotherapy,
radiotherapy, surgery, immunotherapy and targeted therapy, have
been clinically proven to be effective against GC, but poorly
differentiated gastric adenocarcinoma and gastric adenocarcinoma
without immunotherapy active marker subtypes still require
effective treatment (2). Systemic
therapies for hepatocellular carcinoma (HCC) are currently in
clinical use, while checkpoint inhibitors and tyrosine kinase
inhibitors or anti-VEGF therapies are still being tested (3). Screening for pancreatic cancer (PC) is
difficult; as a result, numerous patients are diagnosed with
advanced or metastatic cancer. Surgically assisted chemotherapy and
chemotherapy may be used for early-stage patients, while multidrug
radiotherapy may be used for advanced and metastatic patients to
improve patient survival (4,5).
Colorectal cancer (CRC) is the fourth most deadly cancer in the
world and is closely related to poor lifestyle habits. The most
current treatment regimens may only improve patient survival due to
patients not exhibiting clinical symptoms until the later stages
(6). Therefore, the early screening
and diagnosis of CRC urgently require breakthroughs. Intrahepatic
cholangiocarcinoma (ICC) is the second most common primary liver
cancer and its high lethality and increasing incidence year by year
are of increasing concern (7).
Gallbladder cancer (GBC) is a common malignancy in the biliary
system and current treatment modalities include surgical resection,
chemotherapy and radiotherapy. Immunotherapy, targeted therapy and
nanoparticle therapy are advancing in clinical trials (8). Esophageal squamous cell carcinoma
(ESCC) is the most common form of esophageal cancer and despite
innovations and changes in clinical treatment, its five-year
survival rate remains low (9).
Ubiquitination is a post-translational modification
that is involved in the degradation of proteins and regulates a
variety of cellular physiological processes. Ubiquitination has a
specific role in neurodegenerative diseases, including Parkinson's
disease, Alzheimer's disease and Huntington's disease. These
neurodegenerative disorders are immunoreactive for anti-ubiquitin
antibodies (10). In addition,
ubiquitination is involved in regulating the tumor necrosis factor
signaling pathway, IL-1β and Toll-like receptor signaling (11). Ubiquitination also has a surprising
relationship with cellular autophagy. Ubiquitination degrades the
associated proteasome and regulates the expression of
autophagy-related factors (12).
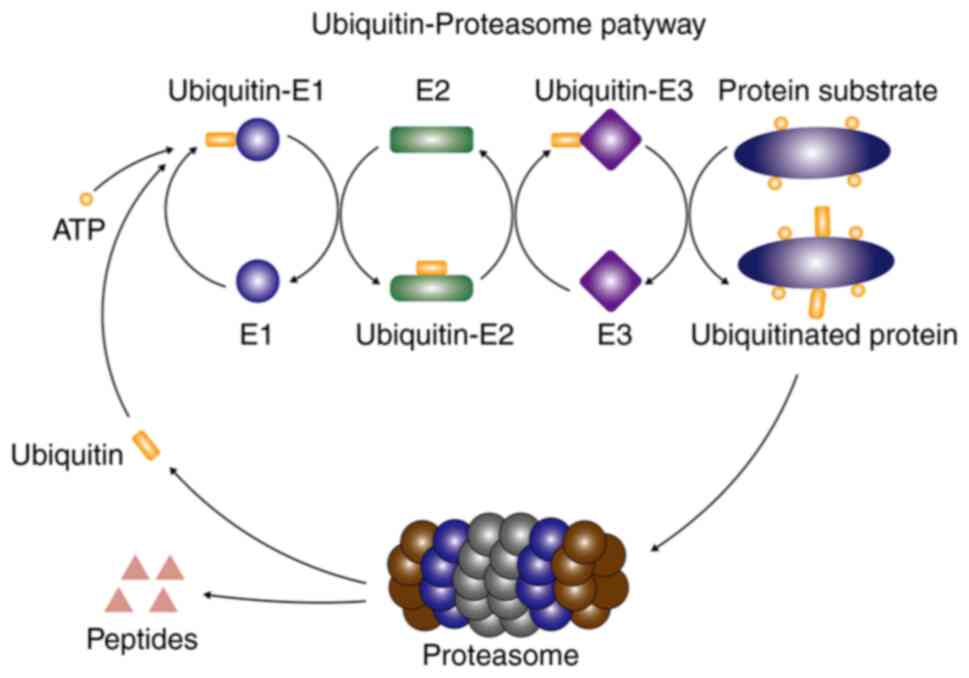
The ubiquitin-proteasome system is the main pathway
for protein degradation. The ubiquitin-proteasome system is a
combination of ubiquitin, ubiquitin-activating enzyme (E1),
ubiquitin-conjugating enzyme (E2) and ubiquitin-ligase enzyme (E3)
to complete the degradation of target proteins (13). In the ubiquitin-proteasome pathway,
E1 transfers the activated ubiquitin molecules to the Cys residues
of E2. Subsequently, an E2 may mark different types of E3 to
catalyze specific substrate ubiquitination. In this process, E3 is
responsible for the transfer of ubiquitin molecules from E2 to the
substrate protein (14–16). The ubiquitin-proteasome pathway is
presented in Fig. 1. The
ubiquitin-proteasome system has a role in a variety of tumors and
multiple small molecule inhibitors targeting the
ubiquitin-proteasome pathway are already in clinical trials
(17). Prior to this, the function
of E2s in different cancers has been previously reviewed in
corresponding articles (18). In
addition, the regulation of autophagic degradation pathways by E2s
and their possible use as potential therapeutic targets in cancers
have also been summarized (12,19).
However, E2s also have multiple roles in digestive system cancers.
The present article provides a detailed review of the expression
and roles of E2s in digestive system cancers, which may provide a
reference for E2s to become molecular therapeutic targets in
digestive system cancers.
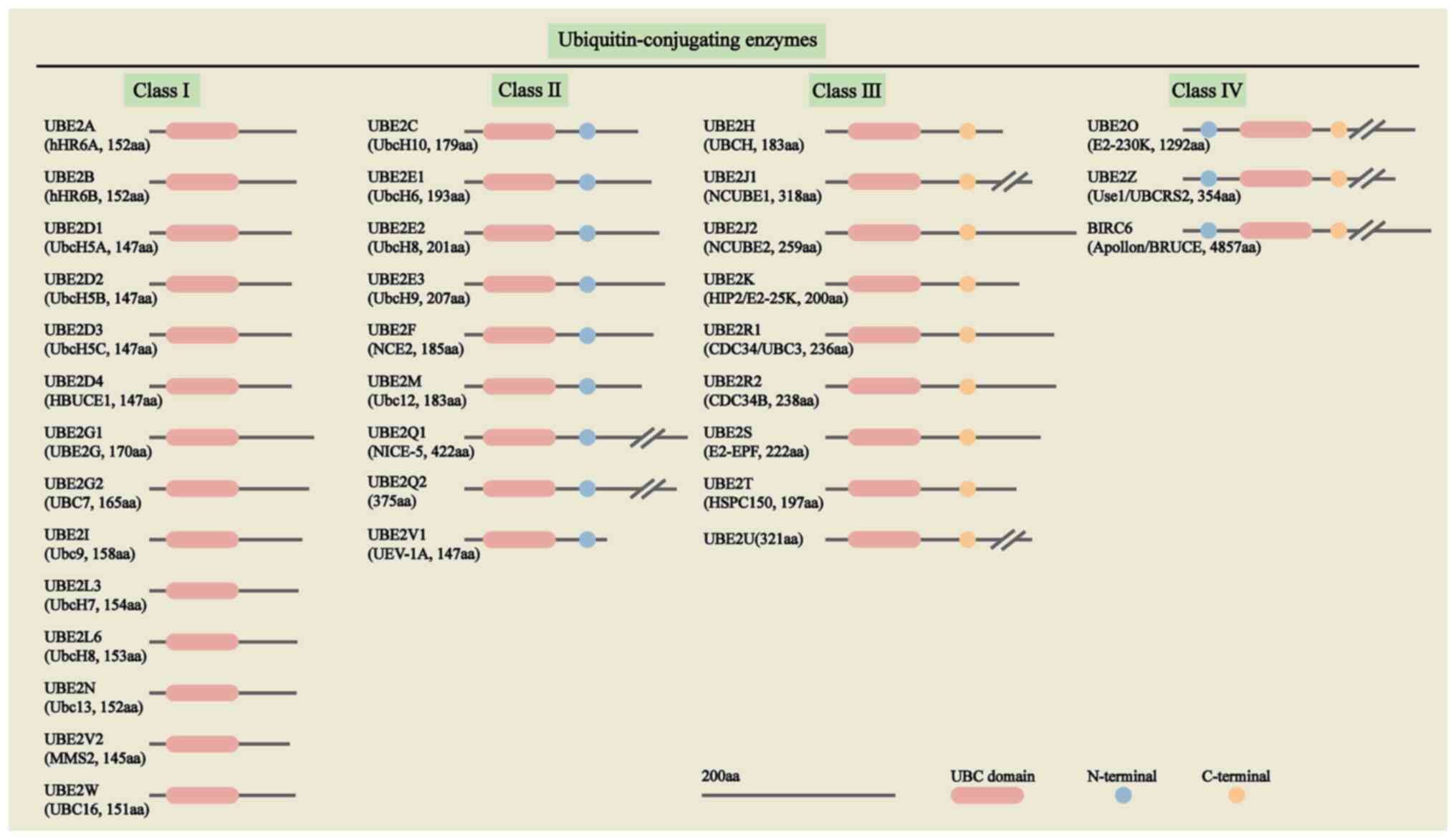
E2s belong to a multi-gene family with different
numbers of members in different species. There are nearly 40 E2s in
humans, which are divided into four categories according to the
highly conserved ubiquitin-binding catalytic (UBC) folding
structural domain and the presence or absence of N-terminal and
C-terminal, of which the class I E2s consists of the catalytic
folding structural domain only. Class II E2s consist of the
catalytic folding domain and the N terminal. Class III E2s consist
of the catalytic folding domain and the C-terminal. Class IV E2s
consist of the catalytic folding structural domain, N-terminal and
C-terminal together (18,20), as presented in Fig. 2.
E2s are involved in a variety of physiological
functions in cells. E2s and the ubiquitin ligase Parkin protein
mediate the mitochondrial autophagic pathway in cells. Furthermore,
E2s containing Baculovirus IAP repeats may inhibit the
expression of autophagic fluxes (12). The ratio of charged to uncharged E2s
and the concentration of free ubiquitin determine whether the OTU
deubiquitinase, ubiquitin aldehyde binding 1-E2 complex can
function as a deubiquitinating enzymes or polyubiquitination
inhibitor (21). Of note, E2s may
themselves be ubiquitinated and their ubiquitination levels may be
regulated by their protein levels (22). E2s also have a non-negligible role
in digestive system cancers, and importantly, molecular inhibitors
targeting E2s are currently under investigation, while molecular
inhibitors applied to immune strategy therapy of cancer are still
waiting to be developed, and E2s, as important platforms in the
ubiquitin system, have an incalculable role as targets for clinical
treatment (19,23).
Initial studies showed that intron 6 and exon 7 of
ubiquitin-conjugating enzyme D1 (UBE2D1) are highly homologous to
the sequence of SFT, a membrane protein for iron transport.
Compared to normal control tissue, the liver of patients with
hereditary hemochromatosis expressed higher levels of UBE2D1
(24). UBE2D1 is highly expressed
in GC tissues and UBE2D1 is involved in gastric carcinogenesis; as
a target of microRNA (miR)-144-3p, it is negatively correlated with
miR-144-3p and its expression is closely related to the resistance
of patients with GC to cisplatin therapy (25). Xie et al (26) used lentivirus to transfect GC cells
and found that silencing UBE2D1 downregulated the expression of
epithelial-mesenchymal transition (EMT) proteins MMP2 and MMP9,
thereby inhibiting the migratory ability of GC cells. In
vivo experiments also demonstrated that the knockdown of UBE2D1
inhibited the metastasis of xenograft tumors in mice. In addition,
UBE2D1 regulates the TGF-β/SMAD4 signaling pathway through the
ubiquitination of SMAD4, which modulates the development of GC
(26).
UBE2A is the first class of E2s and previous studies
have indicated its involvement in the DNA damage repair pathway
(27). UBE2A was not only highly
expressed in HCC tissues but also elevated in 6 HCC cell lines, and
its expression was mainly located in the cytoplasmic sites of HCC
cells. UBE2A was significantly associated with tumor stage,
vascular invasion and patient survival in HCC (28). However, the potential mechanism of
UBE2A affecting HCC requires to be further studied. UBE2D1 belongs
to the UBE2D family of E2s. It has been experimentally proven to
ubiquitinate and degrade p53 in vivo and in vitro
(29,30). UBE2D1 is highly expressed in HCC
tissues and HCC cells, which may reduce the survival time of
patients with HCC. High expression of UBE2D1 promotes the
proliferation of HCC cells and also exacerbates the progression of
HCC through ubiquitination and degradation of p53 (31). UBE2I is essential for positive
selection and the late maturation process of thymocytes. In
addition, UBE2I-mediated sumoylation has a beneficial effect on
cardiac function in transgenic mice after cardiac damage (32,33).
UBE2I is upregulated in numerous cancers, including breast cancer,
as well as glioma (34,35). Fang et al (36) examined the expression levels of UBC9
(UBE2I) in HCC tissues and HCC cells and determined that UBC9
expression was increased in HCC tissues and also in HCC cells. In
addition, UBC9 exhibited higher sensitivity to adriamycin in HCC.
Of note, UBC9 is able to bind to HDAC4 and be acetylated. The
expression level of acetylated UBC9 is negatively correlated with
HDAC4 and targeting this pathway increases radiation-induced
mortality of HCC cells (37). Cell
functional experiments demonstrated that UBE2I promoted the
proliferation, migration and invasion of HCC cells, which is
regulated by the upstream gene miR-195-3p. UBE2I may regulate the
expression of autophagy-related proteins, including microtubule
associated protein 1 light chain 3 α/β, Beclin-1 and
autophagy-related 16 like 1. Of note, high UBE2I expression reduces
overall survival (OS) and relapse-free survival in patients as
analyzed by the The Cancer Genome Atlas (TCGA) database. Therefore,
UBE2I may promote the occurrence and malignant progression of HCC
through the autophagy pathway (38,39).
However, the specific autophagy pathway has not been studied and
confirmed. UBE2L3 regulates the proportion of cells in S-phase and
regulates the proliferation of HeLa cells. UBE2L3 may ubiquitously
degrade p53, thereby reducing the p53 damage response to DNA
(40,41). Furthermore, UBE2L3 regulates the
proliferation of HCC cells by regulating the activation of p65
through an ubiquitin-mediated proteasome degradation pathway. At
the same time, in situ tumorigenesis was performed in mice
and it was found that tumor growth was inhibited by knocking down
UBE2L3. Of note, analysis of liver cancer and normal tissue samples
revealed that high UBE2L3 expression shortened the OS time and
disease-free survival (DFS) time of patients (42). UBE2N is a cancer-associated E2,
which is known to promote the growth of melanoma (43). UBE2N mRNA expression levels are
upregulated in HCC tissues and HCC lines. MiR-147b regulates UBE2N
to promote the proliferation of HCC cells. Whether it may be used
as a therapeutic target for liver cancer still requires to be
proven (44).
UBE2N was highly expressed in pancreatic ductal
carcinoma and elevated in 4 types of PC cells, including SW1990.
Tripartite motif containing 11 (TRIM11) is an important protein
that may regulate ferritin phagocytosis and sensitivity to
gemcitabine in pancreatic ductal carcinoma. Of note, UBE2N has a
similar role to TRIM11 in regulating the autophagic pathway. It was
demonstrated that UBE2N interacts with TRIM11, thereby promoting
TRIM11-mediated ferritin phagocytosis and resistance to gemcitabine
in PC (45).
It has been demonstrated that UBE2B ubiquitinates
β-catenin in MCF10A breast cancer cells and that ubiquitination of
β-catenin is reduced by inhibition of UBE2B expression (46). Of note, UBE2B is highly expressed in
CRC cells with ionizing radiation resistance. The use of the UBE2B
inhibitor significantly reduced the resistance of CRC cells to
ionizing radiation. In addition, the expression of UBE2B in
patients with CRC was closely associated with certain clinical
indicators, such as vascular invasion and tumor regression grade
(47).
UBE2C expression was upregulated in pancreatic
ductal carcinoma tissue, as well as in pancreatic ductal carcinoma
cells, and the OS time was shorter in patients with high levels of
UBE2C expression. It promotes the progression of pancreatic ductal
carcinoma by regulating the cell cycle, cell proliferation and EMT.
In addition, knockdown of UBE2C inhibited tumor growth in nude
mice, which may suggest UBE2C as a new target for PC therapy
(63). Recent studies have
indicated that UBE2C is a potential therapeutic target for PC. The
small molecule inhibitor DHPO, a sesquiterpene lactone compound,
has been reported to inhibit the proliferation and migration of PC
cells in vitro and inhibit the growth and metastasis of
pancreatic tumors in vivo. DHPO inhibits the malignant
progression of PC. DHPO provides a new candidate for the precise
treatment of PC (64).
Relative to GES1 in gastric epithelial cells,
Huntingtin-interacting protein 2 (HIP2) was highly expressed at the
mRNA and protein level in AGS and other six GC cell lines.
Furthermore, the expression of HIP2 was also higher in GC tissues
than in paracancerous tissues. In addition, the OS rate was higher
in the HIP2 low-expression patient group. The results of in
vivo experiments indicated that high expression of HIP2
promoted the proliferation, migration and invasion of GC (85). UBE2T has a proliferative effect in
cells, as it may monoubiquitinate Fanconi anemia (FA)
complementation group D2 and participate in the regulation of the
FA pathway (86). UBE2T was highly
expressed in GC tissues, as well as in the 9 GC cells tested, and
associated with the OS of patients with GC. It promotes the
proliferation of GC cells in vitro and participates in the
regulation of the cell cycle, as well as the process of EMT of GC
cells (87). In addition, the
knockdown of UBE2T inhibits the growth of xenograft tumors in nude
mice. UBE2T may activate the Wnt/β-catenin pathway, inhibit c-Myc
expression and promote GC progression (88). Of note, Yu et al (89) found that UBE2T regulates the
ubiquitination of receptor for activated C kinase 1 as an upstream
gene independently of ubiquitin ligase, thereby activating the
Wnt/β-catenin pathway to promote the progression of GC. By
contrast, the UBE2T inhibitor M435-1279 blocks the Wnt/β-catenin
pathway to inhibit the progression of GC (89). In conclusion, Wnt/β-catenin and
PI3K/Akt signaling pathways are involved in the progression of G;
these E2s and their related miRNAs have important roles in the
carcinogenesis and progression of GC.
UBE2S has an essential role in mitosis. Studies have
indicated that it works with the ubiquitin ligase APC/C to promote
the ubiquitination of substrates (90). Both UBE2S and UBE2T belong to the
third class of E2s, both of which have a larger role in HCC. By
bioinformatics analysis, Zhang and Yang (91) found that UBE2S, UBE2T and UBE2C are
associated with HCC progression. UBE2C is highly expressed in
patients with TP53-mutated HCC, as are UBE2S and UBE2T. The results
of this analysis of clinical patient data were confirmed by other
experiments (91). Expression of
UBE2S is associated with survival, metastasis and recurrence in
patients with HCC and analysis of TCGA data indicated that the OS
and DFS time were shorter in patients with high UBE2S expression.
By contrast, in vivo experiments have indicated that UBE2S
regulates the p53 signaling pathway through ubiquitination to
affect cell cycle, proliferation and migration (92,93).
Furthermore, UBE2S may also be transcriptionally activated by
forkhead box M1, facilitate ubiquitination of phosphatase and
tensin homolog and phosphorylates AKT, thereby enhancing the
resistance of HCC cells to 5-fluorouracil and oxaliplatin (94). The effect of UBE2S on other
chemotherapeutic drugs remains elusive. UBE2T is widely regulated
in HCC and its expression is significantly associated with the
grade of HCC, vascular invasion and poor clinical prognosis of
patients with HCC. UBE2T may regulate the cell cycle and apoptosis,
which may also be regulated by miRNA to affect the malignant
phenotype of HCC cells. For instance, regulation by miR-212-5p
affects cell proliferation, migration and invasion. MiR-543 may
also regulate UBE2T ubiquitination and degrades p53 protein.
MiR-1305 is also able to target it to inhibit the Akt signaling
pathway and inhibit the self-renewal and tumorigenesis of liver
cancer stem cells (95–100). UBE2T is the sumoylation target of
SUMO-specific peptidase 1, which mediates its carcinogenic effects
(101). Furthermore, patients with
HCC with high UBE2T expression had shorter OS and DFS. UBE2T
interacts with ubiquitin ligase ring finger protein 8 to
monoubiquitin H2AX, activating phosphorylation of checkpoint kinase
1 and enhancing radioresistance in HCC (102).
The expression of UBE2S and UBE2T was observed to be
elevated in pancreatic ductal carcinoma tissues and cells, and they
promoted cell proliferation and migration. In addition, high UBE2S
expression was associated with reduced OS time. UBE2S may promote
EMT in PC cells. Furthermore, UBE2S is negatively correlated with
von Hippel-Lindau tumor suppressor expression, activates
hypoxia-inducible factor 1α, regulates the levels of phosphorylated
STAT3 and promotes the malignant progression of PC under hypoxic
conditions. UBE2T regulates protein expression during EMT in PC
cells (103,104). The roles of E2s in PC with high
mortality remain largely elusive.
UBE2S regulates the proliferation and migration of
CRC cells. The Wnt/β-catenin signaling pathway is involved in
several key cellular processes. Furthermore, studies suggest that
UBE2S and β-catenin may have a common interaction molecule, SOX2.
Experimental results demonstrated that UBE2S ubiquitinated
β-catenin at the position of the K19 residue, thus stabilizing its
protein expression. Therefore, UBE2S may be an emerging target for
CRC treatment (105). Luo and Zhou
(106) and Wu et al
(107) demonstrated that UBE2T
expression was elevated in CRC tissues and samples and that
elevated UBE2T was associated with reduced OS time in clinical
patients. It promotes the proliferation, migration and invasion of
CRC cells and inhibits apoptosis. In vivo experiments
suggested that knockdown of UBE2T inhibits the growth of
transplanted tumors (108). These
studies suggest that UBE2S and UBE2T have carcinogenic effects in
CRC and they may be used as a screening index for early-onset
CRC.
UBE2T is considered to be a biomarker associated
with the pathological features, clinical staging and survival
prognosis of GBC (109). UBE2T has
a pro-cancer function in a variety of cancers. Yu et al
(110) investigated the expression
levels of UBE2T in normal intrahepatic ductal tissue and ICC tissue
and assessed their clinical prognosis. The results of the survival
analysis indicated that high UBE2T expression was an independent
risk factor for time to tumor recurrence and OS (110). Similarly, Liu et al
(111) demonstrated that the
expression of UBE2T was higher in cholangiocarcinoma tissue samples
than in paracancerous tissues. In vitro experiments
suggested that the expression of UBE2T was higher in HuCCT1, QBC939
and RBE, 3 cholangiocarcinoma cell lines, than in the normal human
intrahepatic bile duct epithelial cell line HIBEpiC. Furthermore,
in vitro experiments suggested that UBE2T regulates the
proliferation, cell cycle and EMT process of cholangiocarcinoma. Of
note, the mTOR inhibitor rapamycin suppressed the malignant
behavior of UBE2T, promoting cholangiocarcinoma by inhibiting mTOR
(111).
Class IV E2s in HCC. The expression of UBE2O is
higher in HCC than in paracancerous tissues. The follow-up results
are consistent with the TCGA data suggesting longer OS in patients
with HCC with low levels of UBE2O. Its influence on the
proliferation, migration and invasion of HCC cells is not
negligible. In addition, UBE2O negatively regulates protein kinase
adenosine monophosphate-activated catalytic subunit α2 and
modulates the mTOR signaling pathway to promote malignant
progression of HCC (113). Shi
et al (114) analyzed data
from patients with HCC and found that high UBE2Z expression was
associated with decreased OS and DFS time. It was negatively
correlated with the expression of MMP2 and MMP9 proteins, thus
regulating the proliferation and migration of HCC cells. In
addition, UBE2Z may regulate the ERK and STAT3 signaling pathways
to promote HCC progression (114).
The existing evidence suggests that E2s have a carcinogenic role in
HCC and promote its malignant progression.
In summary, class II and class III E2 have been more
studied in digestive system tumors than class I and IV E2,
especially in regulating digestive system tumors signaling
pathways. Among them, the inhibitors of UBE2C and UBE2T have been
studied experimentally. Class I E2s and class IV E2s are less
maturely studied in digestive system tumors and additional
experimental results are required as proof. However, there is a
non-negligible role of E2s in digestive system tumors. The present
study describes a number of E2s in tumors with poor clinical
prognoses. The upregulation of E2 expression and its involvement in
the mechanistic and physiological processes of digestive system
cancers is summarized in Table
I.
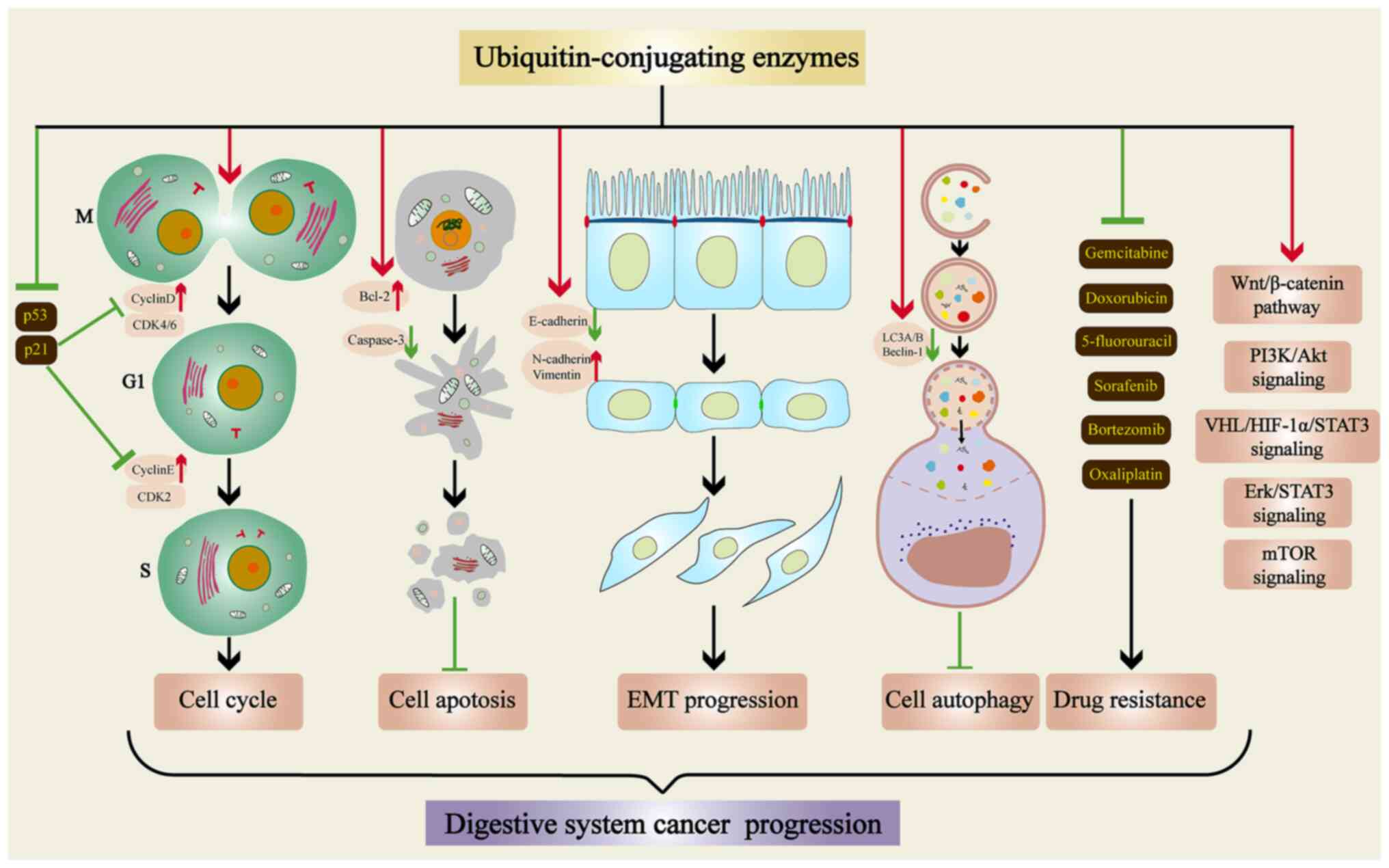
The roles of E2s in various physiological processes
of tumor cells are presented in Fig.
3. E2s may increase Cyclin D and Cyclin E expression and
inhibit p21, as well as p53 expression. Thus, they regulate the
cell cycle, promote the proliferation of tumor cells and regulate
the development of digestive system cancers. E2s inhibit tumor cell
death and promote tumor cell multiplication by regulating apoptosis
and autophagy protein expression. In the EMT pathway, E2s inhibit
E-cadherin and upregulate N-cadherin, as well as Vimentin
expression, increasing the ability of tumor cells to migrate. As a
result of E2 upregulation, tumor cells may resist sorafenib,
bortezomib and other drugs, increasing their viability. E2s are
also involved in the signaling pathways that promote tumor
progression in digestive system cancers. In conclusion, E2s are
involved in proliferation, migration, invasion, apoptosis, cycling,
autophagy, drug resistance and cell signaling pathways of digestive
system cancer cells.
Therefore, E2s may potentially be used as part of a
molecular therapeutics strategy to treat digestive system cancers.
Targeting E2s to inhibit their expression, thereby attenuating
their involvement in multiple physiological processes in tumor
cells, may be achieved to suppress the malignant progression of
digestive system cancers. There is certainly more work to be done
to explore the feasibility of E2s as potential targets, as well as
their sensitivity. E2s require to be more effective, sensitive and
safe as therapeutic targets for digestive system cancers.
Not applicable.
This study was supported by grants from the National Natural
Science Foundation of China (grant no. 81874049); the
Co-construction of Provincial and Department Project of the Health
Commission of Zhejiang Province (WKJ-ZJ-2204); the Co-construction
of Provincial and Department Project of the Health Commission of
Zhejiang Province (WKJ-ZJ-2205); the Natural Science Foundation of
Zhejiang Province (LQ22H160062); the Medical and Health Science
Technology Project of Zhejiang Province (2022KY516).
Not applicable.
XL and XH: Conception of the review and writing of
the manuscript. QL and WF: Charting. WS and QM: Literature search.
DH and QX: Review of manuscript. All authors have read and agreed
to the final manuscript. Data authentication is not applicable.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
|
1
|
Grady WM, Yu M and Markowitz SD:
Epigenetic alterations in the gastrointestinal tract: Current and
emerging use for biomarkers of cancer. Gastroenterology.
160:690–709. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Joshi SS and Badgwell BD: Current
treatment and recent progress in gastric cancer. CA Cancer J Clin.
71:264–279. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Llovet JM, Kelley RK, Villanueva A, Singal
AG, Pikarsky E, Roayaie S, Lencioni R, Koike K, Zucman-Rossi J and
Finn RS: Hepatocellular carcinoma. Nat Rev Dis Primers. 7:62021.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Park W, Chawla A and O'Reilly EM:
Pancreatic cancer: A review. JAMA. 326:851–862. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
US Preventive Services Task Force, . Owens
DK, Davidson KW, Krist AH, Barry MJ, Cabana M, Caughey AB, Curry
SJ, Doubeni CA, Epling JW Jr, et al: Screening for pancreatic
cancer: US Preventive Services Task Force Reaffirmation
Recommendation statement. JAMA. 322:438–444. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Dekker E, Tanis PJ, Vleugels JLA, Kasi PM
and Wallace MB: Colorectal cancer. Lancet. 394:1467–1480. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sirica AE, Strazzabosco M and Cadamuro M:
Intrahepatic cholangiocarcinoma: Morpho-molecular pathology, tumor
reactive microenvironment, and malignant progression. Adv Cancer
Res. 149:321–387. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Song X, Hu Y, Li Y, Shao R, Liu F and Liu
Y: Overview of current targeted therapy in gallbladder cancer.
Signal Transduct Target Ther. 5:2302020. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Reichenbach ZW, Murray MG, Saxena R,
Farkas D, Karassik EG, Klochkova A, Patel K, Tice C, Hall TM, Gang
J, et al: Clinical and translational advances in esophageal
squamous cell carcinoma. Adv Cancer Res. 144:95–135. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Popovic D, Vucic D and Dikic I:
Ubiquitination in disease pathogenesis and treatment. Nat Med.
20:1242–1253. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Cockram PE, Kist M, Prakash S, Chen SH,
Wertz IE and Vucic D: Ubiquitination in the regulation of
inflammatory cell death and cancer. Cell Death Differ. 28:591–605.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ikeda F: Ubiquitin conjugating enzymes in
the regulation of the autophagy-dependent degradation pathway.
Matrix Biol. 100–101. 23–29. 2021.PubMed/NCBI
|
|
13
|
Hershko A and Ciechanover A: The ubiquitin
system. Annu Rev Biochem. 67:425–479. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Liu W, Tang X, Qi X, Fu X, Ghimire S, Ma
R, Li S, Zhang N and Si H: The ubiquitin conjugating enzyme: An
important ubiquitin transfer platform in ubiquitin-proteasome
system. Int J Mol Sci. 21:28942020. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ye Y and Rape M: Building ubiquitin
chains: E2 enzymes at work. Nat Rev Mol Cell Biol. 10:755–764.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hofmann RM and Pickart CM: In vitro
assembly and recognition of Lys-63 polyubiquitin chains. J Biol
Chem. 276:27936–27943. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Park J, Cho J and Song EJ:
Ubiquitin-proteasome system (UPS) as a target for anticancer
treatment. Arch Pharm Res. 43:1144–1161. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Bui QT, Hong JH, Kwak M, Lee JY and Lee
PC: Ubiquitin-Conjugating Enzymes in Cancer. Cells. 10:13832021.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Du X, Song H, Shen N, Hua R and Yang G:
The molecular basis of ubiquitin-conjugating enzymes (E2s) as a
potential target for cancer therapy. Int J Mol Sci. 22:34402021.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
van Wijk SJ and Timmers HT: The family of
ubiquitin-conjugating enzymes (E2s): Deciding between life and
death of proteins. FASEB J. 24:981–993. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wiener R, DiBello AT, Lombardi PM, Guzzo
CM, Zhang X, Matunis MJ and Wolberger C: E2 ubiquitin-conjugating
enzymes regulate the deubiquitinating activity of OTUB1. Nat Struct
Mol Biol. 20:1033–1039. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Stewart MD, Ritterhoff T, Klevit RE and
Brzovic PS: E2 enzymes: More than just middle men. Cell Res.
26:423–440. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hosseini SM, Okoye I, Chaleshtari MG,
Hazhirkarzar B, Mohamadnejad J, Azizi G, Hojjat-Farsangi M,
Mohammadi H, Shotorbani SS and Jadidi-Niaragh F: E2
ubiquitin-conjugating enzymes in cancer: Implications for
immunotherapeutic interventions. Clin Chim Acta. 498:126–134. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Gehrke SG, Riedel HD, Herrmann T,
Hadaschik B, Bents K, Veltkamp C and Stremmel W: UbcH5A, a member
of human E2 ubiquitin-conjugating enzymes, is closely related to
SFT, a stimulator of iron transport, and is up-regulated in
hereditary hemochromatosis. Blood. 101:3288–3293. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Li Y, Wang L, Xu X, Sun H and Wu L: LncRNA
HLA Complex Group 11 knockdown alleviates cisplatin resistance in
gastric cancer by targeting the miR-144-3p/UBE2D1 Axis. Cancer
Manag Res. 13:7543–7557. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Xie H, He Y, Wu Y and Lu Q: Silencing of
UBE2D1 inhibited cell migration in gastric cancer, decreasing
ubiquitination of SMAD4. Infect Agent Cancer. 16:632021. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Bergink S and Jentsch S: Principles of
ubiquitin and SUMO modifications in DNA repair. Nature.
458:461–467. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Shen JD, Fu SZ, Ju LL, Wang YF, Dai F, Liu
ZX, Ji HZ, Shao JG and Bian ZL: High expression of
ubiquitin-conjugating enzyme E2A predicts poor prognosis in
hepatocellular carcinoma. Oncol Lett. 15:7362–7368. 2018.PubMed/NCBI
|
|
29
|
Tokumoto M, Fujiwara Y, Shimada A,
Hasegawa T, Seko Y, Nagase H and Satoh M: Cadmium toxicity is
caused by accumulation of p53 through the down-regulation of Ube2d
family genes in vitro and in vivo. J Toxicol Sci. 36:191–200. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Saville MK, Sparks A, Xirodimas DP,
Wardrop J, Stevenson LF, Bourdon JC, Woods YL and Lane DP:
Regulation of p53 by the ubiquitin-conjugating enzymes UbcH5B/C in
vivo. J Biol Chem. 279:42169–42181. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhou C, Bi F, Yuan J, Yang F and Sun S:
Gain of UBE2D1 facilitates hepatocellular carcinoma progression and
is associated with DNA damage caused by continuous IL-6. J Exp Clin
Cancer Res. 37:2902018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wang A, Ding X, Demarque M, Liu X, Pan D,
Xin H, Zhong B, Wang X, Dejean A, Jin W and Dong C: Ubc9 is
required for positive selection and late-stage maturation of
thymocytes. J Immunol. 198:3461–3470. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Gupta MK, McLendon PM, Gulick J, James J,
Khalili K and Robbins J: UBC9-mediated sumoylation favorably
impacts cardiac function in compromised hearts. Circ Res.
118:1894–1905. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhao Z, Tan X, Zhao A, Zhu L, Yin B, Yuan
J, Qiang B and Peng X: microRNA-214-mediated UBC9 expression in
glioma. BMB Rep. 45:641–646. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhu S, Sachdeva M, Wu F, Lu Z and Mo YY:
Ubc9 promotes breast cell invasion and metastasis in a
sumoylation-independent manner. Oncogene. 29:1763–1772. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Fang S, Qiu J, Wu Z, Bai T and Guo W:
Down-regulation of UBC9 increases the sensitivity of hepatocellular
carcinoma to doxorubicin. Oncotarget. 8:49783–49795. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Tsai CL, Liu WL, Hsu FM, Yang PS, Yen RF,
Tzen KY, Cheng AL, Chen PJ and Cheng JC: Targeting histone
deacetylase 4/ubiquitin-conjugating enzyme 9 impairs DNA repair for
radiosensitization of hepatocellular carcinoma cells in mice.
Hepatology. 67:586–599. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Yang H, Gao S, Chen J and Lou W: UBE2I
promotes metastasis and correlates with poor prognosis in
hepatocellular carcinoma. Cancer Cell Int. 20:2342020. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Wang XK, Liao XW, Zhou X, Han CY, Chen ZJ,
Yang CK, Huang JL, Wang JY, Liu JQ, Huang HS, et al: Oncogene UBE2I
enhances cellular invasion, migration and proliferation abilities
via autophagy-related pathway resulting in poor prognosis in
hepatocellular carcinoma. Am J Cancer Res. 10:4178–4197.
2020.PubMed/NCBI
|
|
40
|
Reyes-Hernandez OD, Mejia-Garcia A,
Sanchez-Ocampo EM, Cabañas-Cortés MA, Ramírez P, Chávez-González L,
Gonzalez FJ and Elizondo G: Ube2l3 gene expression is modulated by
activation of the aryl hydrocarbon receptor: Implications for p53
ubiquitination. Biochem Pharmacol. 80:932–940. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Whitcomb EA, Dudek EJ, Liu Q and Taylor A:
Novel control of S phase of the cell cycle by ubiquitin-conjugating
enzyme H7. Mol Biol Cell. 20:1–9. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Tao NN, Zhang ZZ, Ren JH, Zhang J, Zhou
YJ, Wai Wong VK, Kwan Law BY, Cheng ST, Zhou HZ, Chen WX, et al:
Overexpression of ubiquitin-conjugating enzyme E2 L3 in
hepatocellular carcinoma potentiates apoptosis evasion by
inhibiting the GSK3β/p65 pathway. Cancer Lett. 481:1–14. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Dikshit A, Jin YJ, Degan S, Hwang J,
Foster MW, Li CY and Zhang JY: UBE2N promotes melanoma growth via
MEK/FRA1/SOX10 signaling. Cancer Res. 78:6462–6472. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Zhang E, Liu Q, Wang Y, Wang H, He L, Jin
X and Li N: MicroRNA miR-147b promotes tumor growth via targeting
UBE2N in hepatocellular carcinoma. Oncotarget. 8:114072–114080.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Shang M, Weng L, Xu G, Wu S, Liu B, Yin X,
Mao A, Zou X and Wang Z: TRIM11 suppresses ferritinophagy and
gemcitabine sensitivity through UBE2N/TAX1BP1 signaling in
pancreatic ductal adenocarcinoma. J Cell Physiol. 236:6868–6883.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Shekhar MP, Gerard B, Pauley RJ, Williams
BO and Tait L: Rad6B is a positive regulator of beta-catenin
stabilization. Cancer Res. 68:1741–1750. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Huang WL, Luo CW, Chou CL, Yang CC, Chen
TJ, Li CF and Pan MR: High expression of UBE2B as a Poor prognosis
factor in patients with rectal cancer following chemoradiotherapy.
Anticancer Res. 40:6305–6317. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Okamoto Y, Ozaki T, Miyazaki K, Aoyama M,
Miyazaki M and Nakagawara A: UbcH10 Is the Cancer-related E2
Ubiquitin-conjugating Enzyme. Cancer Res. 63:4167–4173.
2003.PubMed/NCBI
|
|
49
|
Zhang H, Zhao G, Ke B, Ma G, Liu GL, Liang
H, Liu LR and Hao XS: Overexpression of UBE2C correlates with poor
prognosis in gastric cancer patients. Eur Rev Med Pharmacol Sci.
22:1665–1671. 2018.PubMed/NCBI
|
|
50
|
Wang Y, Huang F, Liu M and Zhao Q: UBE2C
mRNA expression controlled by miR-300 and HuR determines its
oncogenic role in gastric cancer. Biochem Biophys Res Commun.
534:597–603. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Zhang Y, Han T, Wei G and Wang Y:
Inhibition of microRNA-17/20a suppresses cell proliferation in
gastric cancer by modulating UBE2C expression. Oncol Rep.
33:2529–2536. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Wang R, Song Y, Liu X, Wang Q, Wang Y, Li
L, Kang C and Zhang Q: UBE2C induces EMT through Wnt/β-catenin and
PI3K/Akt signaling pathways by regulating phosphorylation levels of
Aurora-A. Int J Oncol. 50:1116–1126. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Yang M, Qu Y, Shi R, Wu X, Su C, Hu Z,
Chang Q, Liu S, Pan G, Lei M, et al: Ubiquitin-conjugating enzyme
UbcH10 promotes gastric cancer growth and is a potential biomarker
for gastric cancer. Oncol Rep. 36:779–786. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Ieta K, Ojima E, Tanaka F, Nakamura Y,
Haraguchi N, Mimori K, Inoue H, Kuwano H and Mori M: Identification
of overexpressed genes in hepatocellular carcinoma, with special
reference to ubiquitin-conjugating enzyme E2C gene expression. Int
J Cancer. 121:33–38. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Zhu M, Wu M, Bian S, Song Q, Xiao M, Huang
H, You L, Zhang J, Zhang J, Cheng C, et al: DNA primase subunit 1
deteriorated progression of hepatocellular carcinoma by activating
AKT/mTOR signaling and UBE2C-mediated P53 ubiquitination. Cell
Biosci. 11:422021. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Wei Z, Liu Y, Qiao S, Li X, Li Q, Zhao J,
Hu J, Wei Z, Shan A, Sun X and Xu B: Identification of the
potential therapeutic target gene UBE2C in human hepatocellular
carcinoma: An investigation based on GEO and TCGA databases. Oncol
Lett. 17:5409–5418. 2019.PubMed/NCBI
|
|
57
|
Xiong Y, Lu J, Fang Q, Lu Y, Xie C, Wu H
and Yin Z: UBE2C functions as a potential oncogene by enhancing
cell proliferation, migration, invasion, and drug resistance in
hepatocellular carcinoma cells. Biosci Rep. 39:BSR201823842019.
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Zhou W, Xu J, Tan M, Li H, Li H, Wei W and
Sun Y: UBE2M is a stress-inducible dual E2 for neddylation and
ubiquitylation that promotes targeted degradation of UBE2F. Mol
Cell. 70:1008–1024. e62018. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Zhang GC, Yu XN, Sun JL, Xiong J, Yang YJ,
Jiang XM and Zhu JM: UBE2M promotes cell proliferation via the
β-catenin/cyclin D1 signaling in hepatocellular carcinoma. Aging
(Albany NY). 12:2373–2392. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Zhang B, Deng C, Wang L, Zhou F, Zhang S,
Kang W, Zhan P, Chen J, Shen S, Guo H, et al: Upregulation of
UBE2Q1 via gene copy number gain in hepatocellular carcinoma
promotes cancer progression through β-catenin-EGFR-PI3K-Akt-mTOR
signaling pathway. Mol Carcinog. 57:201–215. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Chang R, Wei L, Lu Y, Cui X, Lu C, Liu L,
Jiang D, Xiong Y, Wang G, Wan C and Qian H: Upregulated expression
of ubiquitin-conjugating enzyme E2Q1 (UBE2Q1) is associated with
enhanced cell proliferation and poor prognosis in human
hapatocellular carcinoma. J Mol Histol. 46:45–56. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Hu N, Fan XP, Fan YC, Chen LY, Qiao CY,
Han LY and Wang K: Hypomethylated ubiquitin-conjugating enzyme2 Q1
(UBE2Q1) gene promoter in the serum is a promising biomarker for
hepatitis B Virus-associated hepatocellular carcinoma. Tohoku J Exp
Med. 242:93–100. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Wang X, Yin L, Yang L, Zheng Y, Liu S,
Yang J, Cui H and Wang H: Silencing ubiquitin-conjugating enzyme 2C
inhibits proliferation and epithelial-mesenchymal transition in
pancreatic ductal adenocarcinoma. FEBS J. 286:4889–4909. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Qi S, Guan X, Zhang J, Yu D, Yu X, Li Q,
Yin W, Cheng XD, Zhang W and Qin JJ: Targeting E2
ubiquitin-conjugating enzyme UbcH5c by small molecule inhibitor
suppresses pancreatic cancer growth and metastasis. Mol Cancer.
21:702022. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Chen S, Chen Y, Hu C, Jing H, Cao Y and
Liu X: Association of clinicopathological features with UbcH10
expression in colorectal cancer. J Cancer Res Clin Oncol.
136:419–426. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Fujita T, Ikeda H, Taira N, Hatoh S, Naito
M and Doihara H: Overexpression of UbcH10 alternates the cell cycle
profile and accelerate the tumor proliferation in colon cancer. BMC
Cancer. 9:872009. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Cacciola NA, Calabrese C, Malapelle U,
Pellino G, De Stefano A, Sepe R, Sgariglia R, Quintavalle C,
Federico A, Bianco A, et al: UbcH10 expression can predict
prognosis and sensitivity to the antineoplastic treatment for
colorectal cancer patients. Mol Carcinog. 55:793–807. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Li SZ, Song Y, Zhang HH, Jin BX, Liu Y,
Liu WB, Zhang XD and Du RL: UbcH10 overexpression increases
carcinogenesis and blocks ALLN susceptibility in colorectal cancer.
Sci Rep. 4:69102014. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Chen SM, Jiang CY, Wu JY, Liu B, Chen YJ,
Hu CJ and Liu XX: RNA interference-mediated silencing of UBCH10
gene inhibits colorectal cancer cell growth in vitro and in vivo.
Clin Exp Pharmacol Physiol. 37:525–529. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Zhang Y, Tian S, Li X, Ji Y, Wang Z and
Liu C: UBE2C promotes rectal carcinoma via miR-381. Cancer Biol
Ther. 19:230–238. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Bavi P, Uddin S, Ahmed M, Jehan Z, Bu R,
Abubaker J, Sultana M, Al-Sanea N, Abduljabbar A, Ashari LH, et al:
Bortezomib stabilizes mitotic cyclins and prevents cell cycle
progression via inhibition of UBE2C in colorectal carcinoma. Am J
Pathol. 178:2109–2120. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Shafiee SM, Seghatoleslam A, Nikseresht M,
Hosseini SV, Alizadeh-Naeeni M, Safaei A and Owji AA: UBE2Q1
expression in human colorectal tumors and cell lines. Mol Biol Rep.
40:7045–7051. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Fahmidehkar MA, Shafiee SM, Eftekhar E,
Mahbudi L and Seghatoleslam A: Induction of cell proliferation,
clonogenicity and cell accumulation in S phase as a consequence of
human UBE2Q1 overexpression. Oncol Lett. 12:2169–2174. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Shafiee S, Seghatoleslam A, Nikseresht M,
Hosseini SV, Alizadeh-Naeeni M, Safaei A and Owji AA: Expression
status of UBE2Q2 in colorectal primary tumors and cell lines. Iran
J Med Sci. 39 (Suppl 2):S196–S202. 2014.PubMed/NCBI
|
|
75
|
Mokarram P, Shakiba-Jam F, Kavousipour S,
Sarabi MM and Seghatoleslam A: Promoter methylation status of two
novel human genes, UBE2Q1 and UBE2Q2, in colorectal cancer: A new
finding in Iranian patients. Asian Pac J Cancer Prev. 16:8247–8252.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Shen T, Cai LD, Liu YH, Li S, Gan WJ, Li
XM, Wang JR, Guo PD, Zhou Q, Lu XX, et al: Ube2v1-mediated
ubiquitination and degradation of Sirt1 promotes metastasis of
colorectal cancer by epigenetically suppressing autophagy. J
Hematol Oncol. 11:952018. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Washiro M, Ohtsuka M, Kimura F, Shimizu H,
Yoshidome H, Sugimoto T, Seki N and Miyazaki M: Upregulation of
topoisomerase IIalpha expression in advanced gallbladder carcinoma:
A potential chemotherapeutic target. J Cancer Res Clin Oncol.
134:793–801. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Zhao B, Gao C, Shi D, Mao J, Zhao J, Guo
L, Guo J and Jiao Z: Knockdown of Nedd8conjugating enzyme UBE2M
suppresses the proliferation and induces the apoptosis of
intrahepatic cholangiocarcinoma cells. Oncol Rep. 42:2670–2679.
2019.PubMed/NCBI
|
|
79
|
Wang X, Li G, Luo Q and Gan C:
Identification of crucial genes associated with esophageal squamous
cell carcinoma by gene expression profile analysis. Oncol Lett.
15:8983–8990. 2018.PubMed/NCBI
|
|
80
|
Dastsooz H, Cereda M, Donna D and Oliviero
S: A comprehensive bioinformatics analysis of UBE2C in cancers. Int
J Mol Sci. 20:22282019. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Li J, Xie Y, Wang X, Jiang C, Yuan X,
Zhang A, Yang L, Liu C, Zou H, Li F and Hu J: Identification of hub
genes associated with esophageal cancer progression using
bioinformatics analysis. Oncol Lett. 20:2142020. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Matsumoto A, Ishibashi Y, Urashima M,
Omura N, Nakada K, Nishikawa K, Shida A, Takada K, Kashiwagi H and
Yanaga K: High UBCH10 protein expression as a marker of poor
prognosis in esophageal squamous cell carcinoma. Anticancer Res.
34:955–961. 2014.PubMed/NCBI
|
|
83
|
Li R, Pang XF, Huang ZG, Yang LH, Peng ZG,
Ma J and He RQ: Overexpression of UBE2C in esophageal squamous cell
carcinoma tissues and molecular analysis. BMC Cancer. 21:9962021.
View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Li L, Li X, Wang W, Gao T and Shi Z: UBE2C
is involved in the functions of ECRG4 on esophageal squamous cell
carcinoma. Biomed Pharmacother. 98:201–206. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Wu J, Tian B, Yang J, Huo H, Song Z, Yu J
and Gu Y: Reduction of Hip2 suppresses gastric cancer cell
proliferation, migration, invasion and tumorigenesis. Transl Cancer
Res. 9:774–785. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Machida YJ, Machida Y, Chen Y, Gurtan AM,
Kupfer GM, D'Andrea AD and Dutta A: UBE2T is the E2 in the Fanconi
anemia pathway and undergoes negative autoregulation. Mol Cell.
23:589–596. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Yu H, Xiang P, Pan Q, Huang Y, Xie N and
Zhu W: Ubiquitin-Conjugating Enzyme E2T is an independent
prognostic factor and promotes gastric cancer progression. Tumour
Biol. 37:11723–11732. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Luo C, Yao Y, Yu Z, Zhou H, Guo L, Zhang
J, Cao H, Zhang G, Li Y and Jiao Z: UBE2T knockdown inhibits
gastric cancer progression. Oncotarget. 8:32639–32654. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Yu Z, Jiang X, Qin L, Deng H, Wang J, Ren
W, Li H, Zhao L, Liu H, Yan H, et al: A novel UBE2T inhibitor
suppresses Wnt/β-catenin signaling hyperactivation and gastric
cancer progression by blocking RACK1 ubiquitination. Oncogene.
40:1027–1042. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Garnett MJ, Mansfeld J, Godwin C,
Matsusaka T, Wu J, Russell P, Pines J and Venkitaraman AR: UBE2S
elongates ubiquitin chains on APC/C substrates to promote mitotic
exit. Nat Cell Biol. 11:1363–1369. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Zhang CY and Yang M: Functions of three
ubiquitin-conjugating enzyme 2 genes in hepatocellular carcinoma
diagnosis and prognosis. World J Hepatol. 14:956–971. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Pan YH, Yang M, Liu LP, Wu DC, Li MY and
Su SG: UBE2S enhances the ubiquitination of p53 and exerts
oncogenic activities in hepatocellular carcinoma. Biochem Biophys
Res Commun. 503:895–902. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Ma Y, Li K, Li S, Liang B, Liu Q and Mo Z:
Prognostic value of ubiquitin-conjugating enzyme E2 S
overexpression in hepatocellular carcinoma. Int J Biol Macromol.
119:225–231. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Gui L, Zhang S, Xu Y, Zhang H, Zhu Y and
Kong L: UBE2S promotes cell chemoresistance through PTEN-AKT
signaling in hepatocellular carcinoma. Cell Death Discov.
7:3572021. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Zhao X, Weng W, Jin M, Li S, Chen Q, Li B,
Zhou Z, Lan C and Yang Y: Identification of biomarkers based on
bioinformatics analysis: The expression of ubiquitin-conjugating
enzyme E2T (UBE2T) in the carcinogenesis and progression of
hepatocellular carcinoma. Med Sci Monit. 27:e9290232021.PubMed/NCBI
|
|
96
|
Ren X, Li A, Ying E, Fang J, Li M and Yu
J: Upregulation of ubiquitin-conjugating enzyme E2T (UBE2T)
predicts poor prognosis and promotes hepatocellular carcinoma
progression. Bioengineered. 12:1530–1542. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Wei X, You X, Zhang J and Zhou C:
MicroRNA-1305 inhibits the stemness of LCSCs and tumorigenesis by
repressing the UBE2T-Dependent akt-signaling pathway. Mol Ther
Nucleic Acids. 16:721–732. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Liu LL, Zhu JM, Yu XN, Zhu HR, Shi X,
Bilegsaikhan E, Guo HY, Wu J and Shen XZ: UBE2T promotes
proliferation via G2/M checkpoint in hepatocellular carcinoma.
Cancer Manag Res. 11:8359–8370. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Guo J, Wang M, Wang J and Wu C:
Ubiquitin-conjugating enzyme E2T knockdown suppresses
hepatocellular tumorigenesis via inducing cell cycle arrest and
apoptosis. World J Gastroenterol. 25:6386–6403. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Liu LP, Yang M, Peng QZ, Li MY, Zhang YS,
Guo YH, Chen Y and Bao SY: UBE2T promotes hepatocellular carcinoma
cell growth via ubiquitination of p53. Biochem Biophys Res Commun.
493:20–27. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Tao Y, Li R, Shen C, Li J, Zhang Q, Ma Z,
Wang F and Wang Z: SENP1 is a crucial promotor for hepatocellular
carcinoma through deSUMOylation of UBE2T. Aging (Albany NY).
12:1563–1576. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Sun J, Zhu Z, Li W, Shen M, Cao C, Sun Q,
Guo Z, Liu L and Wu D: UBE2T-regulated H2AX monoubiquitination
induces hepatocellular carcinoma radioresistance by facilitating
CHK1 activation. J Exp Clin Cancer Res. 39:2222020. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Zheng YW, Gao PF, Ma MZ, Chen Y and Li CY:
Role of ubiquitin-conjugating enzyme E2T in the carcinogenesis and
progression of pancreatic cancer. Oncol Lett. 20:1462–1468. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Wang L, Liang Y, Li P, Liang Q, Sun H, Xu
D and Hu W: Oncogenic Activities Of UBE2S Mediated By VHL/HIF-1α
/STAT3 Signal Via the ubiquitin-proteasome system In PDAC. Onco
Targets Ther. 12:9767–9781. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Li Z, Wang Y, Li Y, Yin W, Mo L, Qian X,
Zhang Y, Wang G, Bu F, Zhang Z, et al: Ube2s stabilizes
beta-Catenin through K11-linked polyubiquitination to promote
mesendoderm specification and colorectal cancer development. Cell
Death Dis. 9:4562018. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Luo M and Zhou Y: Comprehensive analysis
of differentially expressed genes reveals the promotive effects of
UBE2T on colorectal cancer cell proliferation. Oncol Lett.
22:7142021. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Wu X, Liu G, Liu R, He J, Wang G, Zhang H,
Liu T, Bai J, Cheng N and Qiu J: Expression of
ubiquitin-conjugating enzyme E2T in colorectal cancers and clinical
implications. Oncol Lett. 20:2752020. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Wu M, Li X, Huang W, Chen Y, Wang B and
Liu X: Ubiquitin-conjugating enzyme E2T(UBE2T) promotes colorectal
cancer progression by facilitating ubiquitination and degradation
of p53. Clin Res Hepatol Gastroenterol. 45:1014932021. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Zhu X, Li T, Niu X, Chen L and Ge C:
Identification of UBE2T as an independent prognostic biomarker for
gallbladder cancer. Oncol Lett. 20:442020. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Yu H, Wang H, Dong W, Cao ZY, Li R, Yang
C, Cong WM, Dong H and Jin GZ: The diagnostic and prognostic value
of UBE2T in intrahepatic cholangiocarcinoma. PeerJ. 8:e84542020.
View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Liu F, Zhu C, Gao P, Zheng S and Li C:
Ubiquitin-conjugating enzyme E2T regulates cell proliferation and
migration in cholangiocarcinoma. Anticancer Drugs. 31:836–846.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Wang X, Liu Y, Leng X, Cao K, Sun W, Zhu J
and Ma J: UBE2T contributes to the prognosis of esophageal squamous
cell carcinoma. Pathol Oncol Res. 27:6325312021. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Shi Z, Liu R, Lu Q, Zeng Z, Liu Y, Zhao J,
Liu X, Li L, Huang H, Yao Y, et al: UBE2O promotes hepatocellular
carcinoma cell proliferation and invasion by regulating the
AMPKalpha2/mTOR pathway. Int J Med Sci. 18:3749–3758. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Shi X, Wang B, Chen X, Zheng Y, Ding Y and
Wang C: Upregulation of ubiquitin-conjugating enzyme E2Z is
associated with human hepatocellular carcinoma. Biochem Biophys Res
Commun. 523:25–32. 2020. View Article : Google Scholar : PubMed/NCBI
|