Introduction
The family of adult-type diffuse glioma constitutes
the most common tumors of the central nervous system (1). The World Health Organization (WHO)
classifies these tumors based on the presence of certain biomarkers
and histological characteristics (1). Within this family, glioblastoma
multiforme (GBM) is a highly aggressive grade 4 tumor with elevated
mitotic activity, vascularization and necrosis (1). The molecular characteristics that
define this disease are: Wild-type isocitrate dehydrogenase (IDH)
enzymes, mutation in the telomerase reverse transcriptase
(Tert) promoter, the combination of gain of chromosome 7 and
loss of chromosome 10 and epidermal growth factor receptor
amplification (2). GBM is
considered one of the most lethal tumors in humans. Patients with
recurrent GBM have only a 1 year life expectancy after diagnosis
with a median survival time of 3–9 months (3). Despite therapeutic efforts,
chemotherapy and surgical advances, there is currently no cure for
GBM (4).
The main challenges of GBM are its intracranial
location combined with fast growth and infiltrative nature that
lead to difficult complete surgical resection; another key issue is
the development of therapy resistance caused by high genetic
instability (5). This instability
generates cell populations resistant to conventional therapies that
non-specifically target dividing cells. Consequently, novel
therapeutic strategies based on targeted therapies are required to
improve patient outcomes (5).
PIN1 is linked to tumor development and progression
and is commonly overexpressed or overactivated in numerous types of
cancer, including GBM (6,7). Atabay et al (8) reported that GBM U251 cells with PIN1
knockdown, have revealed that the downregulation of this protein
induces apoptosis and also decreases cell proliferation and
migration. Therefore, PIN1 is a potential molecular target for GBM
treatment (8).
PIN1 is an 18 kDa protein that contains two
functional domains: N-terminal WW domain capable of binding
specifically to phosphorylated Serine/Threonine-Proline domains and
a C-terminal peptidyl-prolyl isomerase domain, which catalyzes
cis/trans proline isomerization. It is through this isomerization
mechanism that PIN1 is able to generate modifications in a protein
after being phosphorylated. Such modifications cause a
conformational change that affects the target protein by promoting
its activation or inhibition, modifying its folding, altering
intracellular location, increasing its stability or promoting its
degradation and altering which proteins it interacts with (9,10). By
these mechanisms, PIN1 can regulate cellular processes and
signaling pathways, including Wnt/β-catenin, c-MYC and NF-κB
(11–14). PIN1 inhibition has been reported to
decrease activation of NF-κB pathway and its effectors in a GBM
cell model (11). NF-κB is a
transcription factor that serves a key role in regulating
expression of many genes, including inducible and constitutive IL-8
(15). Furthermore, it has been
reported that treatment of U251 GBM cells with IL-8 increases the
invasive potential of this cell line (16). In line with this, downregulation of
NF-κB activity decreases GBM cell invasion, partly mediated by a
decrease in IL-8 transcription (17). PIN1 has been reported to regulate
cellular processes such as proliferation, invasion/metastasis,
angiogenesis, cell death resistance, immune system evasion, cell
cycle progression, metabolism and replicative immortality (7).
One of the main characteristics of tumor cells is
replicative immortality (18). In a
non-pathological context during normal cell division, replicative
machinery duplicates each chromosome incompletely, resulting in
telomere shortening. Telomeres are nucleoprotein structures found
at the ends of each chromosome with the function of maintaining
genomic stability. Telomeres constitute repeated DNA sequences
joined to a protein complex called sheltering composed of telomeric
repeat binding factor (TRF) 1 and 2, protector of telomeres 1,
proteins that associate with these telomeric DNA binding actors as
TRF1-interacting protein 2, Repressor/activator protein 1 and
telomere protection protein 1 (18). Telomere sequence shortens with each
cell replication event. This process is essential for tissue
degeneration and aging in somatic cells, and its correct
functioning prevents malignant cell transformation (19). The major mechanism by which tumor
cells achieve unlimited replication is through telomerase enzyme
expression (20). Telomerase is a
ribonucleoprotein and; its main components are the catalytic
subunit human TERT (htert) and human telomerase RNA
(hTR) which acts as a primer for the addition of telomeric
sequences at the DNA 3′ end (20).
PIN1 modulates telomere maintenance via TRF1, one of
the main components of sheltering, by regulating the half-life of
this protein. Accordingly, the inhibition of PIN1 prevents the
degradation of TRF1, resulting in its accumulation in telomeres
(21). This blocks access to
telomerase, causing progressive telomere shortening (21). One of the genes regulated by NF-κB
pathway, and therefore by PIN1, is htert, which serves a key
role in replicative immortality caused by telomere elongation in
tumor cells (20). In addition, in
a leukemia cell model, PIN1 regulates expression of htert by
activating NF-κB signaling (22).
PIN1 inhibition may be involved in telomere maintenance in GBM not
only by TRF1 modulation but also by altering htert
expression and telomerase activity.
Several reports describe the role of PIN1 in cancer
development, both for its role in regulating signaling pathways and
its ability to modulate telomeric stability (7,8,15,21).
Although these processes have been identified in GBM, there is
little information on their association with PIN1 in this disease
(23,24). Therefore, the present study aimed to
determine the role of PIN1 in telomere maintenance and GBM tumor
progression, considering this protein and its molecular mechanism
may be an attractive target for the development of novel
therapies.
Materials and methods
Cell lines and culture
Human GBM cell line LN-229 was purchased from the
American Type Culture Collection (cat. no. CRL-2611). Cells were
cultured and maintained at 37°C as a monolayer with DMEM (Thermo
Fisher Scientific, Inc.) supplemented with 10% fetal bovine serum
(FBS, Sigma Aldrich; Merck KGaA) previously inactivated by heat, 2
mM glutamine and 80 mg/ml gentamicin in a 5% CO2
atmosphere. Cell cultures were routinely subcultured twice a week
by trypsinization according to the manufacturer's instructions.
For the relative determination of telomeric length,
senescence and apoptosis, cells at ≥9 passages were used. For in
vivo tumor progression assay, cells at <6 passages were
used.
CRISPR/Cas9-PIN1 plasmid cloning
Specific oligonucleotides for Pin1 gene as guide
RNAs (sgRNA) were in house designed to target exon 1 using CRISPRon
software v1.0 (25), corresponding
to ww domain of PIN1 protein. Sequences were as follows: Forward,
5′-CACCGCATCACTAACGCCAGCCAG-3′ and reverse,
5′-AAACCTGGCTGGCGTTAGTGATGC-3′. The commercial plasmid
Thy-1-CRISPR-gRNA#2-(pSpCas9(BB)-2A-Puro (PX459) V2.0) (cat. no.
#124284; Addgene, Inc.) was used as the backbone for the
CRISPR-PIN1 plasmid construction, according to the manufacturer's
protocol. Briefly, plasmid digestion was performed using
BbsI enzyme. The ligation product was transfected into TOP10
electrocompetent bacteria, and transformed bacteria were grown on a
plate with Luria-Bertani medium with ampicillin as selection
pressure at 10 µg/ml for selection and maintenance. Plasmid
amplification was performed from a positive clone. Correct ligation
was confirmed by observing the digestion pattern of double plasmid
digestion (BbsI and AgeI).
LN PIN1 KO cell line generation
Following CRISPR-PIN1 plasmid construction,
transfection was performed using TransIntro™ reagent (TransGen
Biotech Co., Ltd.) according to the manufacturer's instructions
with 1 µg of DNA and 5×105 LN-229 seeded in a 6-well
plate at 37°C for 6 h.
The selection process was performed 48 h after
transfection. For selecting cells containing the plasmid, 2 µg/ml
puromycin (Thermo Fisher Scientific, Inc.) was added to DMEM with
10% FBS for 5 days. After that, the surviving cells were cultured
without puromycin (DMEM 10% FBS) for the development of subsequent
experimentation. Surviving cells were cultured without puromycin
(DMEM 10% FBS) for the development of subsequent
experimentation.
LN PIN1 KO validation by flow
cytometry
LN-229 and LN PIN1 KO cells were harvested by
trypsinization, collected by centrifugation (5 min at 250 × g) and
resuspended in PBS. Cells were fixed with 4% paraformaldehyde in
PBS and 1% FBS (Thermo Fisher Scientific, Inc.) for 15 min at 4°C.
Then, cells were permeabilized using a 0.1% PBS-Tween solution with
1% FBS for 30 min at room temperature. Cells were collected by
centrifugation (5 min at 250 × g at room temperature) and
resuspended in 50 µl PBS with anti-PIN1 antibody (cat. no.
sc-46660; Santa Cruz Biotechnology, Inc.; 1:50) and incubated for
30 min at room temperature. After washing with PBS, cells were
incubated for 30 min at 4°C in dark with the secondary FITC
anti-mouse IgG2a (cat. no. 562028; BD Pharmingen; BD Biosciences,
1:100) and resuspended in 200 µl PBS. Data were collected and
analyzed with BD FACSCalibur cytometer and FlowJo7.6.2 software (BD
Biosciences).
Western blotting
For protein extraction, 1×106 LN-229 and
LN PIN1 KO were lysed using RIPA buffer with protease inhibitor
(Sigma Aldrich; Merck KGaA). The isolated protein was quantified by
BCA assay. A total of 20 µg/lane total proteins were analyzed by
western blotting to determine protein levels of PIN1 for LN PIN1 KO
validation, and Cyclin D1 for cell cycle analysis. A 10%
polyacrylamide gel was used and proteins were transferred to a PDVF
membrane, which was blocked using a suspension of milk powder in
0.1% TBS-T at room temperature for 1 h. Then, the membrane was
incubated overnight at 4°C with primary anti-PIN1 (cat. no.
sc-46660; Santa Cruz Biotechnology, Inc.; 1:500), anti-Cyclin D1
(kindly provided by Dra. MF Rubio; cat. no. sc-718; Santa Cruz
Biotechnology, Inc.; 1:1,500) and anti-β-tubulin as loading control
(cat. no. 22833; Thermo Fisher Scientific, Inc.; 1:5,000).
Following three 0.1% TBS-T washes, bands were visualized using a
horseradish peroxidase-conjugated secondary anti-mouse antibody
(cat. no. 1662408EDU; Bio-Rad Laboratories, Inc.: 1:10,000) with a
bioluminescence kit (Productos Bio-Lógicos) in a C-digit blot
scanner with Image Studio software v5.2.5 (LI-COR Biosciences).
Determination of active levels of
NF-κB
To evaluate the status of NF-κB in LN PIN1 KO cells,
the active levels of this protein were determined using a
commercial plasmid, pHAGE NFKB-TA-LUC-UBC-tdTomato-W (Addgene,
Inc.; cat. no. 49335). 1×105 LN-229 and LN PIN1 KO cells
were transfected in 24-well plates with this plasmid using
TransIntro™ transfection reagent (TransGene), according to the
manufacturer's instructions with 500 ng of DNA at 37°C for 6 h. At
72 h post-transfection, induction of the NF-κB pathway was
performed, at 37°C by adding 10 ng/ml LPS (Sigma Aldrich; MercK
KGaA). After 6 h, luciferin (Sigma Aldrich; MercK KGaA) was added
to the culture medium at 0.1 mM for 10 min at room temperature;
luminescence associated with active levels of NF-κB was measured
using Cytation 5 plate reader with Gen5 software v3.11 (Biotek
Instruments Inc.).
RNA extraction and copy (c)DNA
synthesis
Total RNA was isolated from LN-299 and LN PIN1 KO
cells using BioZol (Productos Bio-Lógicos), according to the
manufacturer's protocol. The extracted RNA concentration and purity
were determined with NanoDrop 1000 (Thermo Fisher Scientific, Inc.)
spectrophotometer by calculating the ratio of optical density at
wavelengths of 260/280 and 260/230 nm. cDNA was synthesized from 1
µg total RNA in 20 µl reaction volume using oligodT18 (Productos
Bio-Lógicos) and Superscript III (Thermo Fisher Scientific, Inc.)
according to the manufacturer's instructions.
Reverse transcription-quantitative
(RT-q)PCR of IL-8, htert and cyclin D1
IL-8, htert and cyclin D1 specific
primers were designed using Primer Express® Software
Version 3.0 (Thermo Fisher Scientific, Inc.). The assay was
performed on StepOne™ System using SYBR-Green detection reagent
(both Thermo Fisher Scientific, Inc.). β-actin was used as a
loading control. Analysis of relative gene expression data was
performed by the ΔΔCq method (26).
Sequences of primers were as follows: htert forward,
5′-CTACTCCTCAGGCGACAAGG-3′ and reverse, 5′-TGGAACCCAGAAAGATGGTC-3′;
β-actin forward, 5′-GGACTTCGAGCAAGAGATGG-3′ and reverse,
5′-AGGAAGGAAGGCTGGAAGAG-3′; il-8 forward,
5′-TAAAAAGCCACCGGAGCACT-3′ and reverse, 5′-ATCAGGAAGGCTGCCAAGAG-3′
and cyclin D1 forward, 5′-TGGTGAACAAGCTCAAGTGG-3′ reverse,
5′-CTGGCATTTTGGAGAGGAAG-3′. Thermocycling conditions were as
follows: 10 min at 95°C for initial denaturation, followed by 40
cycles of PCR (95°C for 15 sec and 60°C for 1 min).
Telomerase activity determination
Telomerase activity was determined by real-time
quantitative telomerase repeat amplification protocol (RQ-TRAP)
assay using SYBRGreen (StepOne™ System; Thermo Fisher Scientific,
Inc.). LN-229 and LN PIN1 KO cells (2×106) were first
washed with PBS and centrifuged at 450 × g for 8 min at room
temperature in a 1.5 ml Eppendorf tube. The pellets were
resuspended in 200 μl of CHAPS {[3-(3-Cholamidopropyl)
dimethylammonio-1-propanesulfonate]} lysis buffer at 0.5% p/v with
RNaseOUT (Thermo Fisher Scientific, Inc.) and protease inhibitor
(Sigma Aldrich; MercK KGaA). Protein concentration was measured by
Micro BCA Protein Assay kit (Thermo Fisher Scientific, Inc.)
according to the manufacturer's instructions and stored at −20°C
until use.
Briefly, telomerase activity was measured in a final
volume of 10 µl, using 2 µl lysate as a template, Power SYBR Green
Master Mix 1X (Thermo Fisher Scientific), 250 nM alternative
complementary (ACX) primer (5′-GCGCGGCTTACCCTTACCCTTACCCTAACC-3′)
and 800 nM telomerase substrate (TS) primer
(5′-AATCCGTCGAGCAGAGTT-3′). First, 20 min incubation at 25°C was
performed for telomerase mediated extension of the TS primer.
Samples were subjected to an initial denaturation at 90°C for 10
min, followed by 40 cycles of PCR (95°C for 15 sec and 60°C for 10
sec). Finally, the reaction ended with melt curve analysis with a
linear increase in temperature of 0.2°C/sec from 55 to 95°C.
StepOne Software v2.3 (Thermo Fisher Scientific, Inc.) was used to
analyze results (27).
Relative telomere length
determination
Telomere length was evaluated according to the
protocol for telomere measurements by qPCR described by Cawthon
(28). Pure gDNA kit (Productos
Bio-Lógicos) was used to extract high molecular weight DNA from
LN-229 and LN PIN1 KO cells, according to the manufacturer's
protocol. Extracted DNA was quantified at 230, 260 and 280 nm
absorbance using NanoDrop 1000 (Thermo Fisher Scientific, Inc.)
spectrophotometer. Specific primers for the repetitive telomere
sequence were used to quantify telomere length. To determine the
genome copies on the samples specific primers for the single copy
gene ribosomal protein lateral stalk subunit P0 (rplp0) were
used. Primer sequences and concentrations were as follows: Telomere
length (500 nM) forward,
5′-CGGTTTGTTTGGGTTTGGGTTTGGGTTTGGGTTTGGGTT-3′ and reverse,
5′-GGCTTGCCTTACCCTTACCCTTACCCTTACCCTTACCCT-3′ and single copy gene
rplp0 (250 nM) forward, 5′-CAGCAAGTGGGAAGGTGTAATCC-3′ and
reverse, 5′-CCCATTCTATCATCAACGGGTACAA-3′. Thermocycling conditions
for the telomere amplification were 90°C for 10 min followed by 40
two-step PCR cycles of 95°C for 15 sec and 60°C for 10 sec. Results
were analyzed using StepOne software v2.3 (Thermo Fisher
Scientific, Inc.).
Cell cycle progression
Cell cycle distribution of LN-229 and LN PIN1 KO
cells was evaluated by flow cytometry with propidium bromide. Both
cell lines were seeded (3×106) in p100 plates in
complete DMEM (Thermo Fisher Scientific, Inc.) with 10% FBS (Sigma
Aldrich; Merck KGaA). Once 50–60% confluence was reached, the
medium was replaced with fresh serum-free DMEM for 48 h at 37°C to
synchronize the cells. Following starvation, 10% FBS was added to
only one plate for both lines; after 20 h at 37°C, the cells were
trypsinized, resuspended in 0.1% FBS-PBS, fixed in cold 70% v/v
methanol for 30 min at −20°C, treated with 1 µg/ml RNase A (Sigma
Aldrich; MercK KGaA) and stained with propidium iodide (100 µg/ml)
for 30 min at 37°C protected from light. A total of
1×104 events was recorded on a FACSCalibur (BD
Biosciences) flow cytometer. Analysis was performed with
FlowJo7.6.2 software (BD Biosciences).
Doubling time determination
A total of 1.5×104 LN-229 and LN PIN1 KO
cells was seeded in 24-well plates. Cells were cultured at 37°C for
24, 48, 72 or 96 h. Then, cells were stained and fixed with crystal
violet-methanol colorimetry method. Finally, cells were resuspended
in ethanol:acetic solution (3:1) and measured at 570 nm.
Duplication time was calculated using exponential growth phase
values by comparing non-linear fit curves using GraphPad Prism 6
software (GraphPad Software, Inc.).
Cell migration
LN-229 and LN PIN KO cells were cultured in DMEM
with 10% FBS to 60–70% confluence and then washed with PBS. Fresh
serum-free DMEM was added; 24 h post-starvation, cells were
resuspended using Dissociation Buffer (Gibco; Thermo Fisher
Scientific, Inc.) and were manually counted and incubated for 30
min at 37°C for recovery. For migration assay, Transwell insert
plates with 8.0 µm membrane (Guangzhou Jet Bio-Filtration Co.,
Ltd.) were used. A total of 500 µl DMEM with 10% FBS was added to
the lower chamber and 300 µl serum-free medium was added to the
upper chamber containing 1.5×105 cells. The plate was
incubated at 37°C. After 20 h, cells were fixed for 10 min with
methanol solution and stained for 15 min with crystal violet
solution, both at room temperature. Migration was quantified by
direct manually counting under an inverted light microscope (Leica
Microsystems, Inc.) with magnification of ×100.
Evaluation of senescence
induction
Senescence-associated β-galactosidase (SA-β-gal)
activity was measured in LN-229 and LN PIN1 KO cells. Fixation and
staining were performed using the Senescence β-galactosidase
Staining Kit (Cell Signaling Technology, Inc.) according to the
manufacturer's instructions. SA-β-gal-positive cells were manually
counted in four randomly selected fields/well under an inverted
light microscope (Leica Microsystems) at a magnification of
100×.
Evaluation of apoptosis induction
Apoptosis in protein lysates of LN-229 and LN PIN1
KO cells was determined through Caspase 3 activity measurement
using CaspACE™ Assay System, according to the manufacturer's
instructions (Promega Corporation).
Animals
A total of 20 8-week-old (weight, 20 g) inbred
athymic and immunocompetent BALB/c AnN N:NIH(S)-nu female mice were
purchased from National University of La Plata (Buenos Aires,
Argentina) and, after randomization to LN-229 (n=10) and LN PIN1 KO
(n=10), were housed (5 mice/cage). Mice were housed at constant
temperature (24°C) and relative humidity (40%), under a 12/12-h
light/dark schedule. Food and water were supplied ad
libitum. The general health of the animals was monitored daily.
The present protocol was approved by the Institutional Commission
for the Care and Use of Laboratory Animals (approval no. CICUAL UNQ
011-15).
Tumor progression
LN-229 and LN PIN1 KO cells were collected by
trypsinization once they reached 80% confluence in monolayer
culture. Cells were manually quantified by hemocytometer and
resuspended in RPMI medium (Thermo Fisher Scientific, Inc.). After
30 min at 37°C, the cell suspension was mixed with Vitrogel
(TheWell Bioscience, Inc.) in a 2:3 ratio. A 250 µl mixture
containing 5×106 cells was injected into nude mice
subcutaneously. The following day, in order to establish the
starting tumor volume, the remaining volume of injection with
Vitrogel was determined. During a 69-day follow-up period, the size
of tumors was measured twice/week. The shortest and longest
diameters of tumors were measured using a caliper and tumor volume
was calculated as follows: Tumor volume (mm3)=(longest
diameter) × (shortest diameter)2 ×0.5 (29). The tumor progression was assessed
twice with 5 animals/experimental group.
Mice were anaesthetized with 5% isoflurane and
sacrificed by cervical dislocation when tumor volume reached 1,000
mm3 If the indicated volume was not reached after 69
days, the mice were sacrificed as humane endpoint. The animals were
necropsied in order to excise tumors, which were fixed overnight in
4% Paraformaldehyde at room temperature and embedded in paraffin at
50°C for 8 h. Tumors were stored at room temperature for subsequent
histological analysis.
Histological analysis
The fixed tumors were cut into 5-µm-thick sections,
which were deparaffinized. The sections were stained at room
temperature with hematoxylin (Biopack) for 5 min, soaked in tap
water for 5 min, and counterstained for 3 min with eosin (Biopack).
Color brightfield images of hematoxylin and eosin (H&E)-stained
tumor sections were acquired using Cytation 5 Cell Imaging
Multi-Mode Reader (BioTek Instruments Inc.) at 2.5× magnification.
Images were collected using a 4×5 grid and stitching was performed
with the Image Montage function of the Gen5 Image software (BioTek
Instruments Inc.) setting a tile overlap of 10%.
Necrotic area
The necrotic area in tissue stitched images was
measured with ImageJ 1.5p Software (30). Tumor necrosis was identified as
sections with increased eosinophilia; quantification of both
necrotic and viable tissue was performed using the ‘Color
Threshold’ tool, setting the units in µm using the scale bar in
each image as reference.
Mitosis
H&E-stained tissue sections were analyzed for
mitotic count calculation. Mitotic figures in all basic stages of
mitosis were manually counted using a high-power field
(magnification, ×400) and 10 images/experimental group.
Statistical analysis
All data are presented as the mean ± SEM (n=3).
Comparisons between >2 was performed using one-way ANOVA
followed by post hoc Tukey's multiple comparisons test. For two
groups, differences were determined by unpaired Student's t or
Mann-Whitney test as appropriate. The analysis was performed using
GraphPad Prism 6 (GraphPad Software, Inc.; Dotmatics). P<0.05
was considered to indicate a statistically significant
difference.
Results
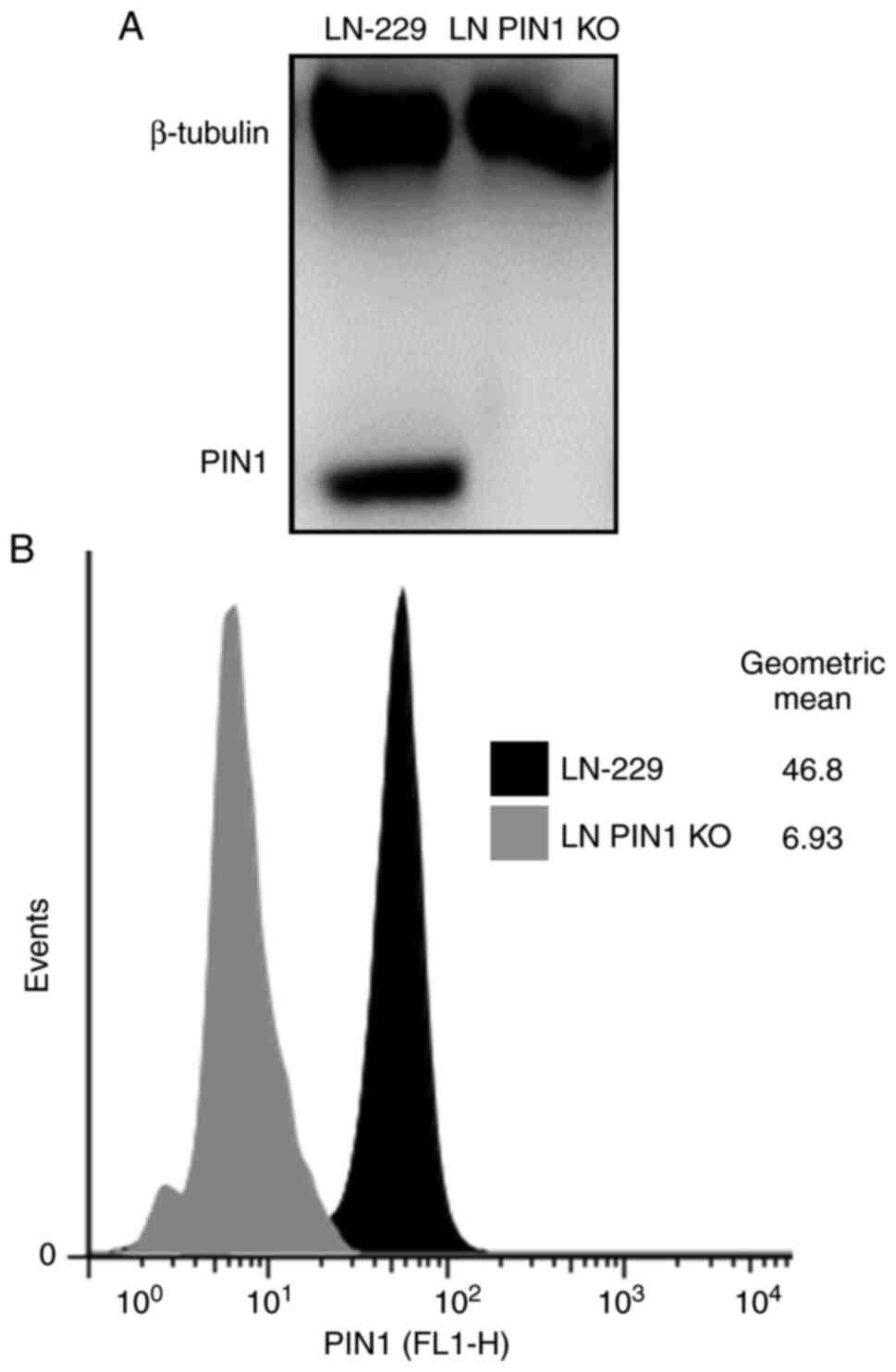
Validation of PIN1 KO
To assess PIN1 KO by CRISPRCas9, PIN1 levels were
determined in control and transfected LN-229 cells using western
blotting and flow cytometry. In western blotting and flow cytometry
analysis PIN 1 was not detected on LN PIN1 KO cell line (Fig. 1A and B, respectively). Thus,
successful induction of LN PIN1 KO was confirmed.
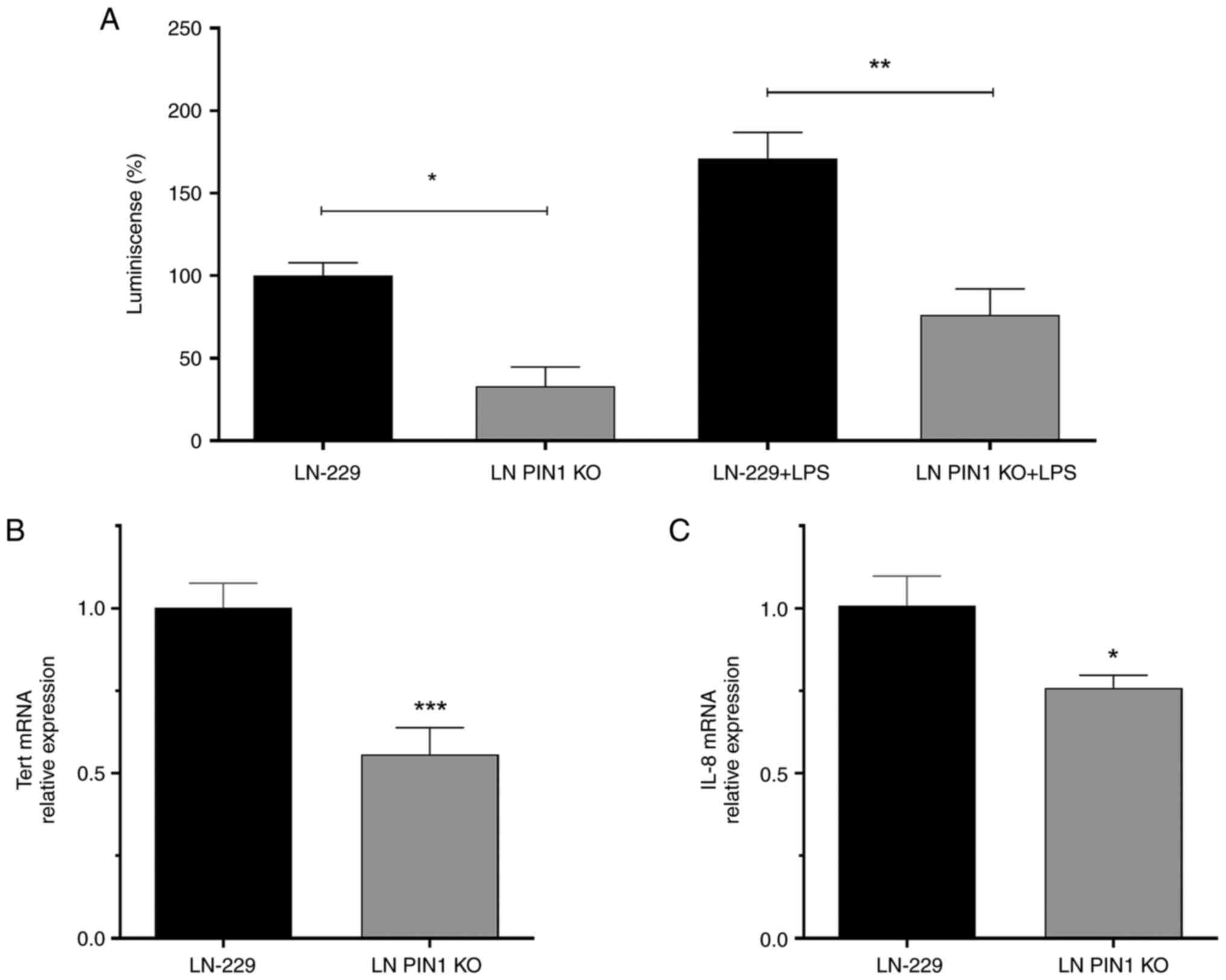
PIN1 KO inhibits NF-κB pathway
signaling
As previously described, NF-κB pathway signaling
promotes proliferation and invasion in GBM (15). A GBM model showed that decreased
PIN1 levels inhibits the NF-κB pathway (15); therefore, the present study
evaluated the status of this signaling pathway in LN PIN1 KO cells.
A commercial plasmid that contains a promoter sequence recognized
by active NF-κB associated with a reporter gene was used. In LN
PIN1 KO cells, active levels of NF-κB decreased significantly in
comparison with LN-229 cells (Fig.
2A).
PIN1 KO reduces both htert and IL-8
gene expression
As PIN1 has been reported to regulate htert
and IL-8 gene expression by NF-κB activation (15,22),
the relative expression of both genes was quantified by the ΔΔCq
method using β-actin as endogenous loading control. The
relative gene expression of htert and IL-8 was
significantly decreased by 50% in LN PIN1 KO cells compared with
LN-229 cells (Fig. 2B and C).
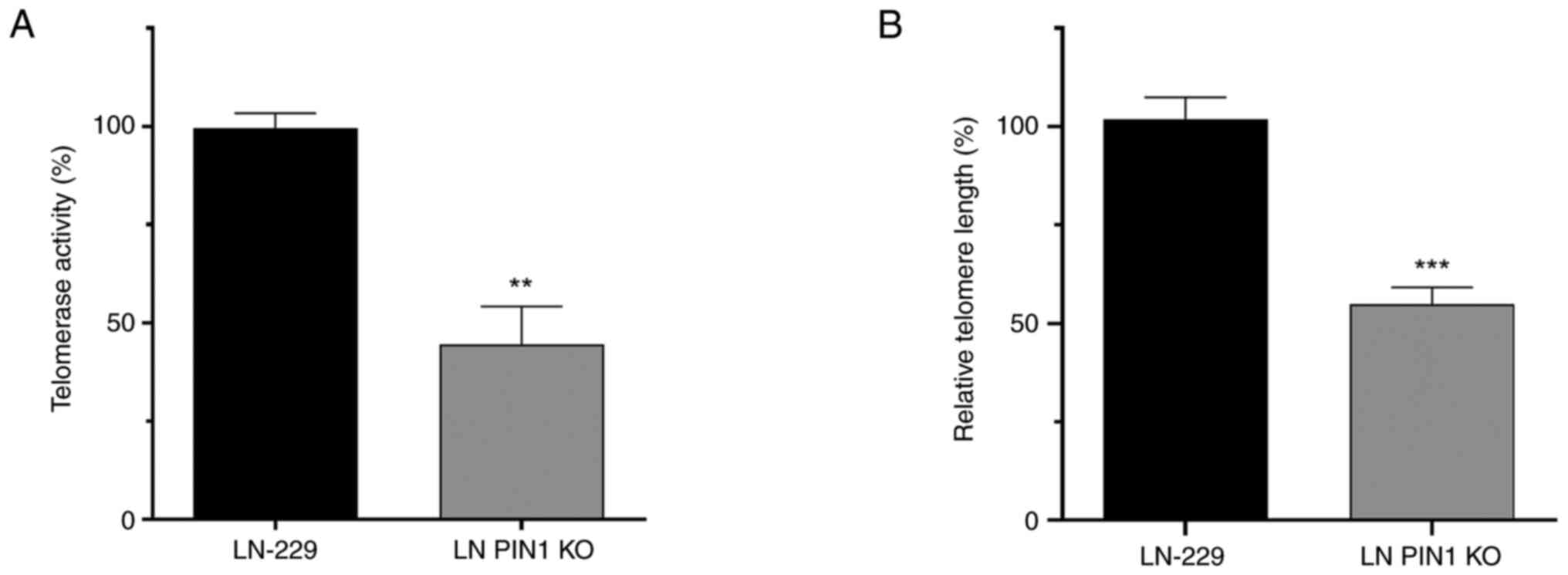
LN PIN1 KO cells exhibit decreased
telomerase activity
Due to changes observed in htert
transcription levels, it was hypothesized that LN PIN1 KO cells
would exhibit lower telomerase activity. Using RQ-TRAP assay, a
significant drop to 44.5% telomerase activity was confirmed in LN
PIN1 KO cells compared with its parental cell line LN-229 (Fig. 3A).
PIN1 KO induces telomere
shortening
Relative telomeric length in both cell lines was
determined by qPCR with specific primers for the telomeric sequence
and rplp0 gene for normalization. LN PIN1 KO cells underwent
telomeric shortening, showing a significant decrease of 47.04% in
length of their telomeres after at least 9 passages (Fig. 3B).
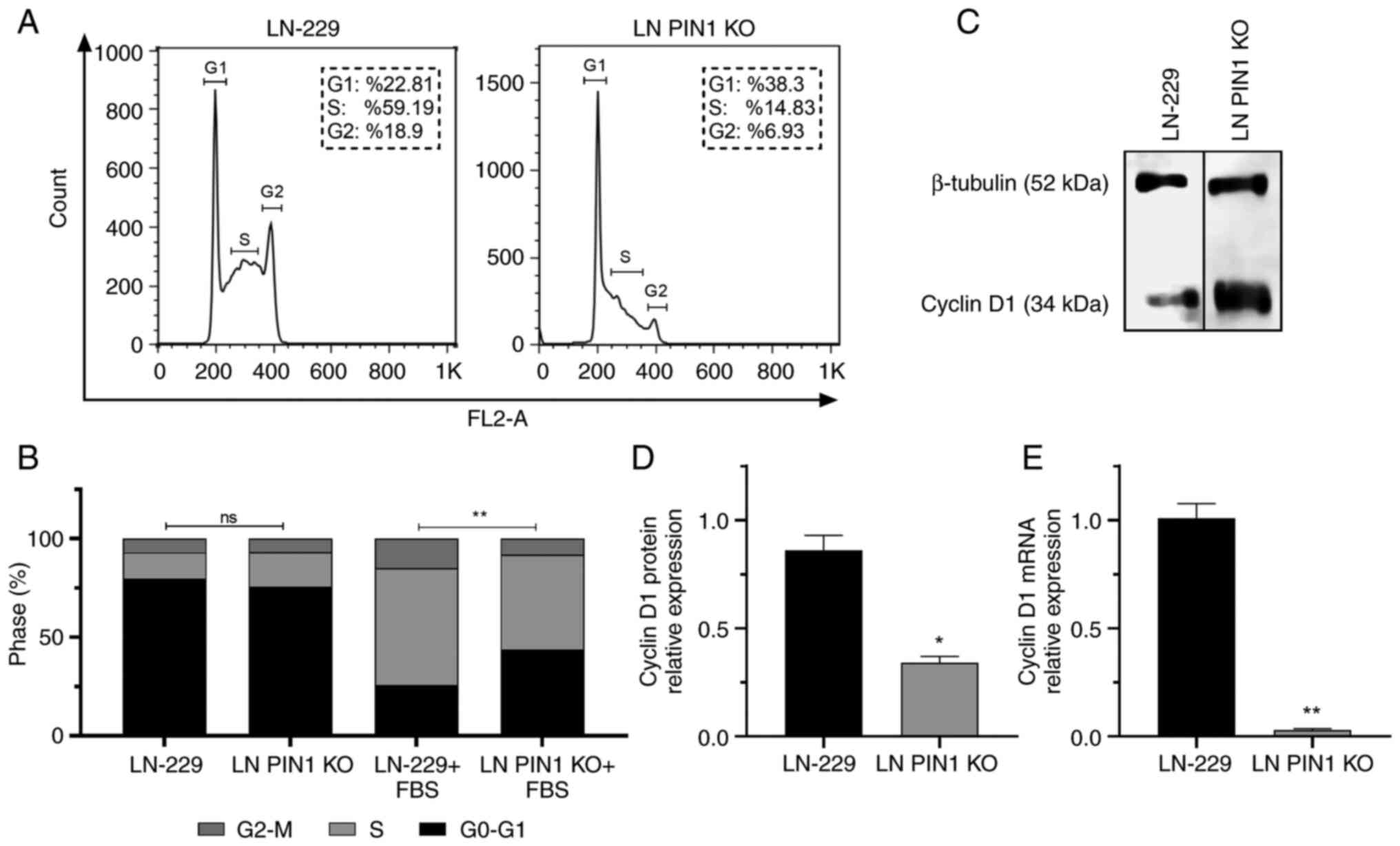
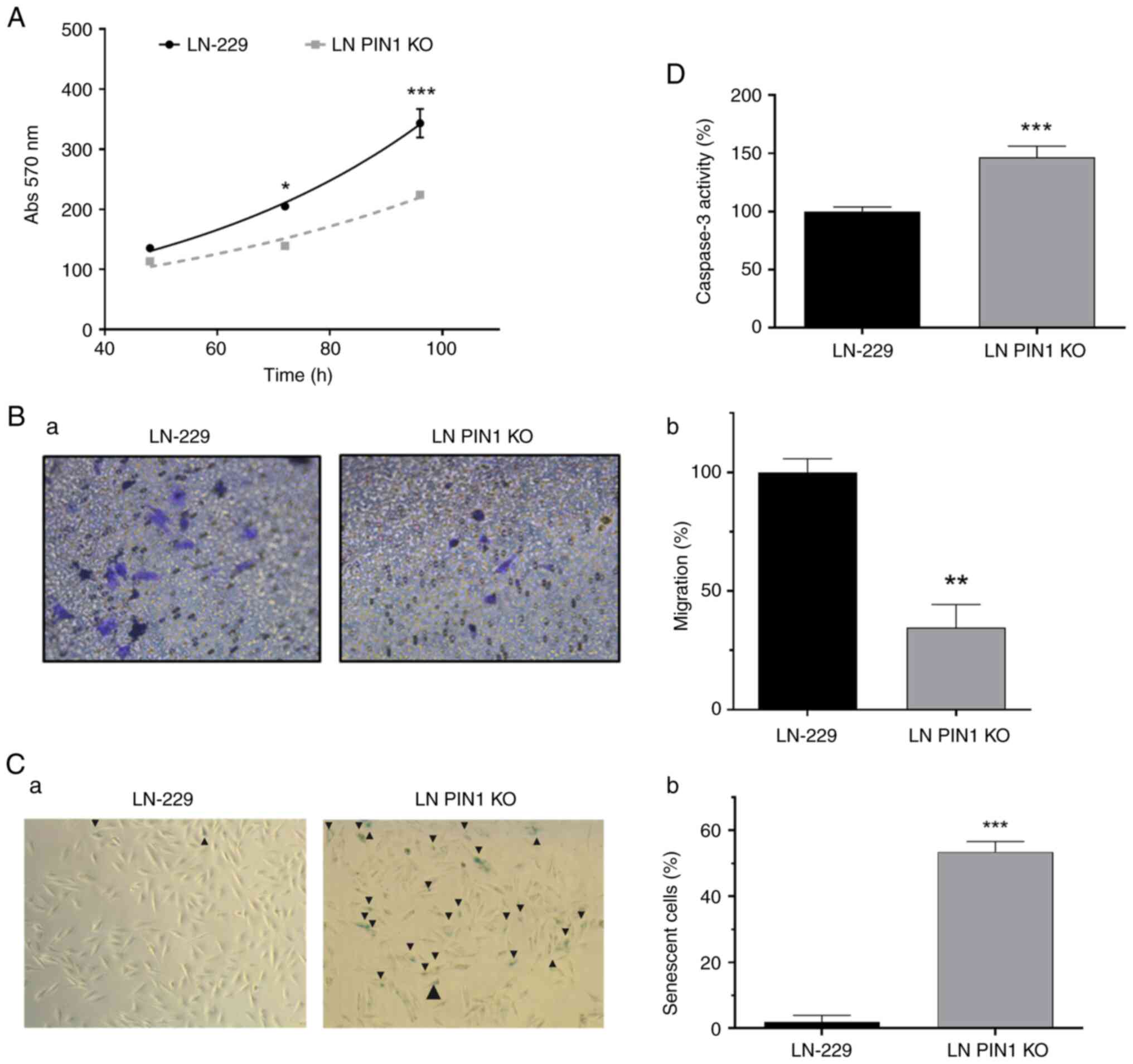
Proliferation decreases in LN PIN1 KO
cells
The present study evaluated the impact of PIN1 KO on
cell cycle progression by flow cytometry. The percentage of cells
in each phase of the cell cycle was determined. A significant
difference was observed in response to FBS stimulus; LN PIN1 KO
cells showed a lower response to FBS stimulus than the parental
cell line LN-229 (Fig. 4B).
The percentage of cells in G0-G1 phase increased by
67.9% in LN PIN1 KO cells compared with LN-229. Furthermore, the
percentage of LN PIN1 KO cells in S and G2-M phases decreased by
75.00 and 63.34%, respectively, compared with LN-229 (Fig. 4A and B). Additionally, PIN1 induces
expression of Cyclin D1, a key cell cycle regulator (31,32).
Therefore, levels of cyclin D1 were evaluated. A significant
decrease in Cyclin D1 mRNA and protein levels were determined for
LN PIN1 KO compared with LN-229 cells (Fig. 4C and D).
LN PIN1 KO increases cell doubling
time
As PIN1 is involved in cell cycle regulation
(31,32), the effect of PIN1 KO on cell
doubling time was determined (Fig.
5A). For LN PIN1 KO, a higher mean doubling time of 47.13 h was
obtained, in comparison with LN-229 doubling time which was 34.6
h.
PIN1 KO reduces cell migration
Given the key role of PIN1 importance in cell
migration (17), the LN PIN1 KO
cell migratory capacity was evaluated using Transwell assays. The
migratory capacity of the PIN1 KO cells decreased to 28.04%
compared with LN22-9 cells (Fig. 5Ba
and b).
PIN1 KO induces senescence and
apoptosis
The role of PIN1 in senescence and apoptosis was
evaluated. The percentage of senescent cells was calculated using
the Senescence β-Gal Staining kit. The mean stained LN-229
cells/field was 1.97%; LN PIN1 KO exhibited ~53.46% β-gal-positive
cells (Fig. 5C).
To evaluate whether PIN1 KO triggers apoptosis,
caspase 3 activity was measured by CaspACE Assay System apoptosis
kit. LN PIN1 KO cells showed a significant 46.4% increase in
Caspase 3 activity compared with LN-229 (Fig. 5D).
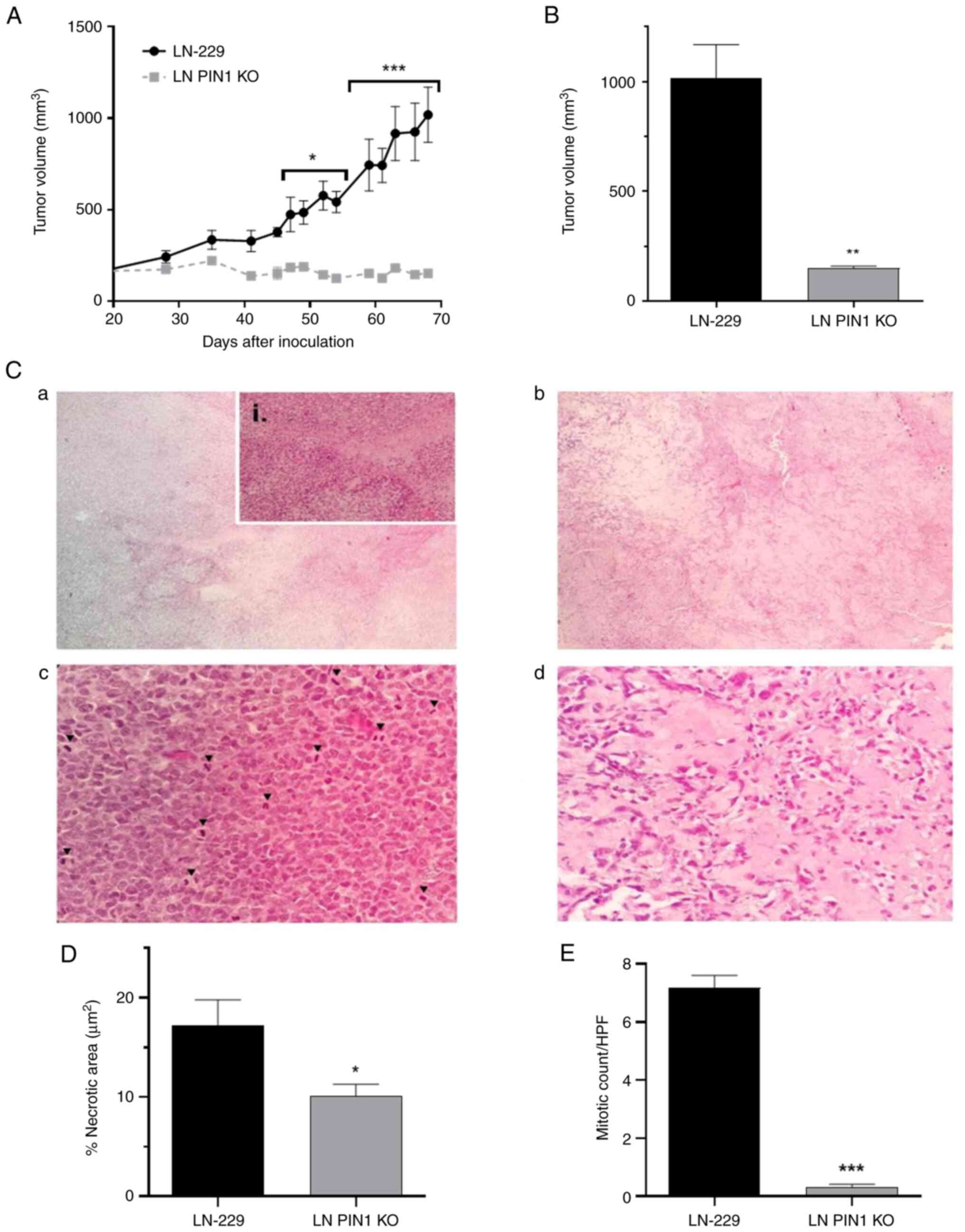
PIN1 KO decreases tumorigenic
potential in vivo
Finally, a nude mouse xenograft model was
established to validate the functional effect of PIN1 KO. Tumors in
control mice injected with the LN-229 cells grew significantly at
day 52 (data not shown); however, the LN PIN1 KO group showed no
notable tumor growth throughout the trial. In addition, the
difference in tumor volume of both groups was significant starting
at day 47 (Fig. 6A). Since Vitrogel
is not completely reabsorbed, a threshold of 400 mm3 was
used to determine tumor incidence (data not shown). The LN-229
group generated tumors between days 41 and 52, while 100% of the
group inoculated with the LN PIN1 KO remained tumor free to the
final of the protocol on day 69. Finally, on day 69 tumor volumes
were calculated. Tumors in the LN-229 group presented a mean volume
of 1,018 mm3; mean volume for the group inoculated with
LN PIN1 KO was 152.4 mm3, (Figs. 6B and S1).
LN PIN KO inhibits tumor
development
H&E staining was used to examine histological
features of tumors generated by both cell lines. While LN-229
tumors showed marked hypercellularity, large necrotic foci area and
high mitotic cell count (Fig. 6Ca and
b), tumors corresponding to LN PIN KO cells showed lower cell
density and a decrease in mitotic cell count (Fig. 6Cc and d). LN-229 cell tumors
exhibited a mean necrotic area of 17.22%; necrotic area in LN PIN1
KO cell tumors was 10.12% (Fig.
6D). The mean mitotic cells/field for LN PIN1 KO was 0.32; for
LN-229, the mean was significantly higher at 7.18 (Fig. 6D). Overall LN PIN1 KO tumors
presented a less proliferative phenotype than tumors obtained from
the parental cell line LN-229.
Discussion
PIN1 serves a role in multiple types of cancer due
to its ability to interact with several signaling pathways and
regulate a wide range of cellular processes linked to tumor
development and progression (7,33,34).
PIN1 is highly expressed in most types of cancer and increased PIN1
expression is usually associated with poorer cancer prognosis
(12). Although the full role of
PIN1 activity in cancer remains to be defined, several molecular
mechanisms associated with PIN1 activity, including regulation of
different transcription factors responsive to growth-inducing
signals, have been described (35–37).
These underlying mechanisms result in enhanced proliferative
signaling, resistance to cancer cell death, replicative immortality
and invasion and metastasis (35).
PIN1 has also been associated with glioma genesis and progression
since it is involved in pro-survival mechanisms, increased invasion
and angiogenic potential. In addition, PIN1 has been reported to
provide metabolic advantages to glioma cells (36,38,39).
PIN1 is expressed in glioma stem-like cells (GSCs) and its
silencing or inhibition abrogates GSC viability and mitigates
GSC-driven tumor progression (40).
Therefore, PIN1 has emerged as an attractive molecular target for
development of novel treatments in GBM. Thus, the aim of the
present work was to determine the role of PIN1 in a GBM model to
postulate it as a novel target for treatment of this disease.
PIN1-targeted small molecule compounds have already
been described (35). Juglone is a
pharmacological inhibitor of PIN1 but its potential application for
cancer treatment is limited due to specificity issues (41,42).
Additionally, other inhibitory molecules have been described, such
as PiB (43) and KPT-6566 (44), that show a promising clinical
application. United States Food and Drug Administration-approved
all-trans retinoic acid (ATRA) acts as a PIN1 inhibitor in acute
promyelocytic leukemia and breast cancer (45). Further, combination therapy with
ATRA and arsenic trioxide (ATO) has shown a cooperative effect
affecting primarily PIN1 activity (46). These pharmacological inhibitors
demonstrate that PIN1 is a feasible target for cancer therapy;
validation studies are key to establish which types of cancer may
benefit from this therapeutic approach.
KO model generation for specific genes is a widely
employed tool in basic research to validate a gene of interest as a
molecular target (47,48). CRISPR/Cas9 facilitates both
identification and validation of new molecular targets for specific
drug development and provides a rapid method to generate KO cell
and animal models (49).
The present study used CRISPR/Cas9 to construct a LN
PIN1 KO cell model and validate PIN1 as a molecular target. PIN1
protein was not detected in PIN1 KO cell lysate by western blot
analysis, indicating successful model construction by CRISPR/Cas9.
This result was verified by evaluation of PIN1 expression by flow
cytometry, where no PIN1 signal was observed in LN PIN1 KO
cells.
To the best of our knowledge, the present study is
the first to use a PIN1 KO cell model in GBM to investigate the
roles of PIN1 in this disease. By using this model, PIN1
involvement in telomere maintenance and oncogenic behavior in GBM
were studied by comparing KO cells with the LN-229 cell line. The
present work comprises a single cell line of GBM. Future studies
should use a larger cell line panel and patient-derived cells to
confirm PIN1 role in GBM. Nevertheless, the results of the present
study contribute to evidence of the role of PIN1 in GBM.
PIN1 inhibition decreases activation of NF-κB
pathway signaling and its effectors in a GBM cell model (15). One of the genes regulated by this
pathway is htert, which serves a key role in replicative
immortality by promoting telomere elongation in tumor cells
(50). PIN1 enhances htert
expression by activating NF-κB in GBM, as seen in another disease
model (22). PIN1 inhibition may be
involved in telomere maintenance in GBM, not only by TRF1
modulation but also by altering htert expression and telomerase
activity. The results obtained support this hypothesis: The present
study evaluated the active levels of NF-κB in LN PIN1 KO cells and
observed that PIN1 KO promoted inhibition of this pathway, as seen
in another GBM cell model (15).
Here, htert transcription also decreased significantly in LN
PIN1 KO cells.
Here, decreased htert expression was observed
to trigger a decrease in activity of the holoenzyme telomerase,
which generates progressive telomere shortening in the PIN1 KO cell
model. Therefore, it was hypothesized that this telomeric
imbalance, generated by PIN1 deletion, contributes to the entry of
LN PIN1 KO cells into senescence and cell apoptosis. However,
telomere shortening may not be the only process mediated by PIN1
that generates senescence and apoptosis considering that this
protein also regulates cell cycle progression and survival signals
(34).
Here, LN PIN1 KO cells presented a longer doubling
time, with an increase of almost 40% compared with LN-229 cells.
This was consistent with cell cycle analysis: LN PIN1 KO cells
showed cell cycle arrest in G0-G1 phase. This was due at least in
part to decreased Cyclin D1 levels in LN PIN1 KO cells (as
indicated by western blot and RT-qPCR analysis).
GBM presents an invasive phenotype that is one of
the characteristics of this type of tumor that hinder its treatment
at the clinical level (32). PIN1
has also been reported to promote migration in different tumor
types, including GBM (33). It has
been reported that the activation of NF-κB promotes migration in
GBM via the expression of IL-8 (15,51).
The present model exhibited a decrease in the active levels of
NF-κB; therefore levels of IL-8 transcription were
evaluated, as well as the migratory capacity of LN PIN1 KO cells.
LN PIN1 KO cells exhibited a decrease in IL-8 transcription
and migratory capacity compared with LN-229 cells.
Here, PIN1 deletion also appears to suppress the
ability to form tumors in vivo in a murine xenogeneic model.
Compared with LN-229 cells, xenografts generated from LN PIN1 KO
cells grew at a markedly slower rate and some tumors did not
progress from their initial volume. These results were consistent
with histological analysis of the tumors, where LN-229 cells showed
a high percentage of necrotic area accompanied by high cellularity
and mitotic body count. All these features are typical histological
characteristics of developed GBM tumors (2). By contrast, these aspects were not
observed in LN PIN1 KO group, suggesting a higher number of
senescent and/or apoptotic cells. Similar results have been
reported in other tumors, where depletion or inhibition of PIN1
leads to modulation of cellular processes, such as cell migration,
cell cycle progression and in vivo tumor growth (2,13,52).
Taken together, the results of the present study
show the contribution of PIN1 in telomeric regulation and other key
cellular processes for development and tumor progression in GBM.
Although the ability of PIN1 to regulate telomeres via TRF1 has
been reported (21), to the best of
our knowledge, the present results support a novel mechanism that
involves regulation of htert expression and lower telomerase
activity.
In summary, loss of PIN1 in LN-229 cells leads to
decreased malignant behavior and tumorigenicity, both in
vitro and in vivo, supporting a key role of PIN1 in the
present GBM model. Furthermore, the present study demonstrated that
the presence of PIN1 affects telomeric dynamics by downregulating
htert expression and telomerase activity in GBM. The present
results provide a basis for design and development of new therapies
for GBM based on PIN1 as a novel target for treatment of this
disease.
Supplementary Material
Supporting Data
Acknowledgements
The authors would like to thank Dr Fernanda Rubio
(Molecular Biology and Apoptosis Laboratory, Medical Investigation
Institute, UBA-CONICET, Buenos Aires, Argentina) for providing
cyclin D1 antibody and Ms Isabel Cardoso (Molecular Oncology Unit,
Center of Molecular and Translational Oncology, National University
of Quilmes. Buenos Aires, Argentina) for technical support in qPCR
and western blotting assays.
Funding
The present study was supported by Quilmes National University
(grant no. PUNQ EXPTE 1297/19), the National Agency for the
Promotion of Science and Technology (grant no. PICT 2018 2372),
Cancer National Institute (grant no. EXPTE 827-1533/18; Argentina)
and Commission of Scientific Investigation from Buenos Aires (grant
no. PDCT2022/GomezDE).
Availability of data and materials
The datasets used and analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
JM, GAC, DEG and DLMG designed the study. JM, RGA
and LB performed in vitro experiments. JM and NTS performed
in vivo experiments. JM, GAC, LB and DLMG wrote the
manuscript. DEG and DLMG confirm the authenticity of all the raw
data. All authors have read and approved the final manuscript.
Ethics approval and consent to
participate
Animal experimentation protocol was approved by the
Institutional Commission for the Care and Use of Laboratory Animals
(approval no. CICUAL UNQ 011-15).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Huttner A: Overview of primary brain
tumors: Pathologic classification, epidemiology, molecular biology,
and prognostic markers. Hematol Oncol Clin North Am. 26:715–732.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Louis DN, Perry A, Wesseling P, Brat DJ,
Cree IA, Figarella-Branger D, Hawkins C, Ng HK, Pfister SM,
Reifenberger G, et al: The 2021 WHO classification of tumors of the
central nervous system: A summary. Neuro Oncol. 23:1231–1251. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Vredenburgh JJ, Desjardins A, Herndon JE
II, Dowell JM, Reardon DA, Quinn JA, Rich JN, Sathornsumetee S,
Gururangan S, Wagner M, et al: Phase II trial of bevacizumab and
irinotecan in recurrent malignant glioma. Clin Cancer Res.
13:1253–1259. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Anjum K, Shagufta BI, Abbas SQ, Patel S,
Khan I, Shah SAA, Akhter N and Hassan SSU: Current status and
future therapeutic perspectives of glioblastoma multiforme (GBM)
therapy: A review. Biomed Pharmacother. 92:681–689. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zong H, Parada LF and Baker SJ: Cell of
origin for malignant gliomas and its implication in therapeutic
development. Cold Spring Harb Perspect Biol. 7:a0206102015.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bao L, Kimzey A, Sauter G, Sowadski JM, Lu
KP and Wang DG: Prevalent overexpression of prolyl isomerase Pin1
in human cancers. Am J Pathol. 164:1727–1737. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Chen Y, Wu YR, Yang HY, Li XZ, Jie MM, Hu
CJ, Wu YY, Yang SM and Yang YB: Prolyl isomerase Pin1: A promoter
of cancer and a target for therapy. Cell Death Dis. 9:8832018.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Atabay KD, Yildiz MT, Avsar T, Karabay A
and Kiliç T: Knockdown of Pin1 leads to reduced angiogenic
potential and tumorigenicity in glioblastoma cells. Oncol Lett.
10:2385–2389. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lu KP, Finn G, Lee TH and Nicholson LK:
Prolyl cis-trans isomerization as a molecular timer. Nat Chem Biol.
3:619–629. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Takahashi K, Uchida C, Shin RW, Shimazaki
K and Uchida T: Prolyl isomerase, Pin1: New findings of
post-translational modifications and physiological substrates in
cancer, asthma and Alzheimer's disease. Cell Mol Life Sci.
65:359–375. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Reichert M, Steinbach JP, Supra P and
Weller M: Modulation of growth and radiochemosensitivity of human
malignant glioma cells by acidosis. Cancer. 95:1113–1119. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhu Z, Zhang H, Lang F, Liu G, Gao D, Li B
and Liu Y: Pin1 promotes prostate cancer cell proliferation and
migration through activation of Wnt/β-catenin signaling. Clin
Transl Oncol. 18:792–797. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang Z, Yu W, Zheng M, Liao X, Wang J,
Yang D, Lu W, Wang L, Zhang S, Liu H, et al: Pin1 inhibition
potently suppresses gastric cancer growth and blocks PI3K/AKT and
Wnt/β-catenin oncogenic pathways. Mol Carcinog. 58:1450–1464. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Farrell AS, Pelz C, Wang X, Daniel CJ,
Wang Z, Su Y, Janghorban M, Zhang X, Morgan C, Impey S and Sears
RC: Pin1 regulates the dynamics of c-Myc DNA binding to facilitate
target gene regulation and oncogenesis. Mol Cell Biol.
33:2930–2949. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Atkinson GP, Nozell SE, Harrison DK,
Stonecypher MS, Chen D and Benveniste EN: The prolyl isomerase Pin1
regulates the NF-kappaB signaling pathway and interleukin-8
expression in glioblastoma. Oncogene. 28:3735–3745. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Raychaudhuri B, Han Y, Lu T and Vogelbaum
MA: Aberrant constitutive activation of nuclear factor kappaB in
glioblastoma multiforme drives invasive phenotype. J Neurooncol.
85:39–47. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wakabayashi K, Kambe F, Cao X, Murakami R,
Mitsuyama H, Nagaya T, Saito K, Yoshida J and Seo H: Inhibitory
effects of cyclosporin A on calcium mobilization-dependent
interleukin-8 expression and invasive potential of human
glioblastoma U251MG cells. Oncogene. 23:6924–6932. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Turner KJ, Vasu V and Griffin DK: Telomere
biology and human phenotype. Cells. 8:732019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Armando RG, Mengual Gomez DL, Maggio J,
Sanmartin MC and Gomez DE: Telomeropathies: Etiology, diagnosis,
treatment and follow-up. Ethical and legal considerations. Clin
Genet. 96:3–16. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Akincilar SC, Unal B and Tergaonkar V:
Reactivation of telomerase in cancer. Cell Mol Life Sci.
73:1659–1670. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lee TH, Tun-Kyi A, Shi R, Lim J, Soohoo C,
Finn G, Balastik M, Pastorino L, Wulf G, Zhou XZ and Lu KP:
Essential role of Pin1 in the regulation of TRF1 stability and
telomere maintenance. Nat Cell Biol. 11:97–105. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ghaffari SH, Momeny M, Bashash D, Mirzaei
R, Ghavamzadeh A and Alimoghaddam K: Cytotoxic effect of arsenic
trioxide on acute promyelocytic leukemia cells through suppression
of NFkβ-dependent induction of hTERT due to down-regulation of Pin1
transcription. Hematology. 17:198–206. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Naderlinger E and Holzmann K: Epigenetic
regulation of telomere maintenance for therapeutic interventions in
gliomas. Genes (Basel). 8:1452017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Nasser MM and Mehdipour P: Exploration of
involved key genes and signaling diversity in brain tumors. Cell
Mol Neurobiol. 38:393–419. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Xiang X, Corsi GI, Anthon C, Qu K, Pan X,
Liang X, Han P, Dong Z, Liu L, Zhong J, et al: Enhancing
CRISPR-Cas9 gRNA efficiency prediction by data integration and deep
learning. Nat Commun. 12:32382021. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Armando RG, Gomez DM and Gomez DE: AZT
exerts its antitumoral effect by telomeric and non-telomeric
effects in a mammary adenocarcinoma model. Oncol Rep. 36:2731–2736.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Cawthon RM: Telomere measurement by
quantitative PCR. Nucleic Acids Res. 30:e472002. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Tomayko MM and Reynolds CP: Determination
of subcutaneous tumor size in athymic (nude) mice. Cancer Chemother
Pharmacol. 24:148–154. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Schneider CA, Rasband WS and Eliceiri KW:
NIH image to ImageJ: 25 Years of image analysis. Nat Methods.
9:671–675. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Nakashima M, Meirmanov S, Naruke Y, Kondo
H, Saenko V, Rogounovitch T, Shimizu-Yoshida Y, Takamura N, Namba
H, Ito M, et al: Cyclin D1 overexpression in thyroid tumours from a
radio-contaminated area and its correlation with Pin1 and aberrant
beta-catenin expression. J Pathol. 202:446–455. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wulf GM, Ryo A, Wulf GG, Lee SW, Niu T,
Petkova V and Lu KP: Pin1 is overexpressed in breast cancer and
cooperates with Ras signaling in increasing the transcriptional
activity of c-Jun towards cyclin D1. EMBO J. 20:3459–3472. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wang J, Liu K, Wang XF and Sun DJ: Juglone
reduces growth and migration of U251 glioblastoma cells and
disrupts angiogenesis. Oncol Rep. 38:1959–1966. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhou XZ and Lu KP: The isomerase PIN1
controls numerous cancer-driving pathways and is a unique drug
target. Nat Rev Cancer. 16:463–478. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Chuang HH, Zhen YY, Tsai YC, Chuang CH,
Huang MS, Hsiao M and Yang CJ: Targeting Pin1 for modulation of
cell motility and cancer therapy. Biomedicines. 9:3592021.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Pu W, Zheng Y and Peng Y: Prolyl isomerase
Pin1 in human cancer: Function, mechanism, and significance. Front
Cell Dev Biol. 8:1682020. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Cheng CW and Tse E: PIN1 in cell cycle
control and cancer. Front Pharmacol. 9:13672018. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Sang Y, Li Y, Zhang Y, Alvarez AA, Yu B,
Zhang W, Hu B, Cheng SY and Feng H: CDK5-dependent phosphorylation
and nuclear translocation of TRIM59 promotes macroH2A1
ubiquitination and tumorigenicity. Nat Commun. 10:40132019.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Yang W and Lu Z: Nuclear PKM2 regulates
the Warburg effect. Cell Cycle. 12:3154–3158. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Zhang A, Tao W, Zhai K, Fang X, Huang Z,
Yu JS, Sloan AE, Rich JN, Zhou W and Bao S: Protein sumoylation
with SUMO1 promoted by Pin1 in glioma stem cells augments
glioblastoma malignancy. Neuro Oncol. 22:1809–1821. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Mesalam AA, El-Sheikh M, Joo MD, Khalil
AAK, Mesalam A, Ahn MJ and Kong IK: Induction of oxidative stress
and mitochondrial dysfunction by juglone affects the development of
bovine oocytes. Int J Mol Sci. 22:1682020. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Paulsen MT and Ljungman M: The natural
toxin juglone causes degradation of p53 and induces rapid H2AX
phosphorylation and cell death in human fibroblasts. Toxicol Appl
Pharmacol. 209:1–9. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Uchida T, Takamiya M, Takahashi M,
Miyashita H, Ikeda H, Terada T, Matsuo Y, Shirouzu M, Yokoyama S,
Fujimori F and Hunter T: Pin1 and Par14 peptidyl prolyl isomerase
inhibitors block cell proliferation. Chem Biol. 10:15–24. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Campaner E, Rustighi A, Zannini A,
Cristiani A, Piazza S, Ciani Y, Kalid O, Golan G, Baloglu E,
Shacham S, et al: A covalent PIN1 inhibitor selectively targets
cancer cells by a dual mechanism of action. Nat Commun.
8:157722017. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Wei S, Kozono S, Kats L, Nechama M, Li W,
Guarnerio J, Luo M, You MH, Yao Y, Kondo A, et al: Active Pin1 is a
key target of all-trans retinoic acid in acute promyelocytic
leukemia and breast cancer. Nat Med. 21:457–466. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Kozono S, Lin YM, Seo HS, Pinch B, Lian X,
Qiu C, Herbert MK, Chen CH, Tan L, Gao ZJ, et al: Arsenic targets
Pin1 and cooperates with retinoic acid to inhibit cancer-driving
pathways and tumor-initiating cells. Nat Commun. 9:30692018.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Chen S, Sun H, Miao K and Deng CX:
CRISPR-Cas9: From genome editing to cancer research. Int J Biol
Sci. 12:1427–1436. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Sánchez-Rivera FJ and Jacks T:
Applications of the CRISPR-Cas9 system in cancer biology. Nat Rev
Cancer. 15:387–395. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Lu Q, Livi GP, Modha S, Yusa K, Macarrón R
and Dow DJ: Applications of CRISPR genome editing technology in
drug target identification and validation. Expert Opin Drug Discov.
12:541–552. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Shalem-Cohavi N, Beery E, Nordenberg J,
Rozovski U, Raanani P, Lahav M and Uziel O: The effects of
proteasome inhibitors on telomerase activity and regulation in
multiple myeloma cells. Int J Mol Sci. 20:25092019. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Nagai S, Washiyama K, Kurimoto M, Takaku
A, Endo S and Kumanishi T: Aberrant nuclear factor-kappaB activity
and its participation in the growth of human malignant astrocytoma.
J Neurosurg. 96:909–917. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Lian X, Lin YM, Kozono S, Herbert MK, Li
X, Yuan X, Guo J, Guo Y, Tang M, Lin J, et al: Pin1 inhibition
exerts potent activity against acute myeloid leukemia through
blocking multiple cancer-driving pathways. J Hematol Oncol.
11:732018. View Article : Google Scholar : PubMed/NCBI
|