Introduction
Dickkopf-1 (DKK1) is a negative regulator of the
Wnt/β-catenin signaling pathway, which has a significant role in a
variety of cellular processes, including differentiation,
proliferation, cell motility and apoptosis (1). Increased DKK1 expression levels have
been identified in patients with Wilms’ tumors, hepatoblastoma,
multiple myeloma and breast cancer, suggesting a potential role for
DKK1 in carcinogenesis (2,3). Studies have reported that the
expression and roles of DKK1 differ between types of cancer, and
that increased expression of DKK1 is common amongst numerous
malignant tumors, including breast and lung cancer, as well as
esophageal carcinomas (4,5), supporting the hypothesis for a
potential oncogenic function of DKK1 (6).
Inhibition of the Wnt pathway by secreted DKK1 has
been shown to initiate carciogenesis in vertebrate embryos
(7), and the overexpression of
DKK1 has been described in multiple myeloma, hepatoblastoma and
Wilms’ tumor, as well as prostate, kidney, breast, lung (3) and esophageal cancers (4). The classification of types of lung
cancer is based on multiple clinicopathological features (8). However, such clinical information may
be incomplete or misleading in the determination of patient
prognosis (9).
Increasing evidence indicates that non-small cell
lung cancer (NSCLC) is one of the most common malignancies in China
(10–12). Studies regarding patients with
NSCLC have reported the potential predictive implication of
biological and molecular parameters, including Kirsten rat sarcoma
viral oncogene homolog mutations (13), c-erbB2 overexpression (14) and p53 mutations (15). Despite significant advances in
cancer treatment, the enduring survival of NSCLC cells has remained
elusive. To the best of our knowledge, few studies have been
published investigating the consequences of DKK1 expression in
NSCLC. The present study therefore aimed to reveal the expression
pattern of DKK1 and its role as a carcinogenic factor in human
NSCLC cell lines and tissues.
Materials and methods
Patient samples
NSCLC tissue samples were collected from 123
patients from The First Affiliated Hospital of Nanchang University
(Nanchang, China) in 2010 and 2013, and formalin fixed
(Sigma-Aldrich, St. Louis, MO, USA) for immunohistochemical
staining. Age-matched normal tissue samples from 18 patients with
primary NSCLC were used as a control. Tissue samples were stored
and ground in liquid nitrogen to isolate total RNA and protein.
Written informed consent was provided by all patients who
participated in the study. The study was approved by the ethics
committee of The First Affiliated Hospital of Nanchang University
(Nanchang, China) and protocols were performed according to their
ethical guidelines.
Cell culture
The lung squamous cell line YTMLC-9, carcinoma cell
lines A549, SPC-A-1, LTEP-a-2, GLC82, A2 and PC-9 as well as
large-cell lung carcinoma cell lines NCI-H460, 95C and 95D were
purchased from the American Type Culture Collection (Manassas, VA,
USA). Cells were cultured in RPMI-1640 medium (Invitrogen Life
Technologies, Carlsbad, CA, USA), supplemented with 10% fetal
bovine serum (Gibco-BRL, Invitrogen Life Technologies), 100 IU/ml
penicillin and 100 mg/ml streptomycin (Life Technologies, Carlsbad,
CA, USA), maintained at 37°C in 5% CO2 atmospheric air.
The cells were cultured to 80% confluence prior to transfection
with recombined eukaryotic or empty vectors using
Lipofectamine® 2000 (Invitrogen Life Technologies)
according to the manufacturer’s instructions. For the transfection
of cells, the pZERO-mcs mammalian expression vector was used
(Invitrogen Life Technologies). This vector contains a hybrid
EF1a/HTLV promoter, allowing efficient transcription of the
recombinant DKK1-cDNA, and the puromycin resistance gene for easy
selection of the transfected cells. The full-length sequence of
DKK1 cDNA was isolated from the recombinant TA plasmid used for
subcloning (Invitrogen Life Technologies) with the following
primers: Forward, 5′-CAAGGGGATCCCCCTGCAGTCAGGACTCTGGGAC-3′ and
reverse, 5-GTGTTCTGCTAGCTAGGTATTATTAATTTATTGGAAAC-3 (including
BamHI and NheI sites). The amplified DKK1 full-length
cDNA was subsequently subcloned into the pZERO-mcs expression
vector. The PCR product was gel-purified and digested with
BamHI and NheI, then ligated into the plasmid
pZERO-mcs, resulting in the recombinant plasmid pZERO-mcs-Dkk1.
Transfection of the cells with either the DKK1-cDNA or an empty
vector was performed using Lipofectamine 2000 (Invitrogen Life
Technologies), according to the manufacturer’s instructions.
Western blot analysis
The protein expression levels of DKK1 were evaluated
by western blot analysis. Proteins extracted from NSCLC specimens
and cell lines were used for this analysis. Tissue samples and
cells were lysed in radioimmunoprecipitation buffer (Cell Signaling
Technology, Danvers, MA, USA) at 4°C for 30 min. The lysate was
incubated on ice for 20 min, followed by sonication for 30 sec,
after which the lysate was incubated on ice for a further 15 min.
The lysate was then centrifuged at 10,000 x g for 10 min in a
microcentrifuge tube, and the supernatant was maintained at −80°C.
The protein concentration was quantified using the Bradford method
(16). The soluble proteins (10–20
µg) were separated by 10% SDS-PAGE and blotted onto polyvinylidene
fluoride membranes (Sigma-Aldrich). Subsequently, the membranes
were blocked with 5% nonfat dry milk in 1X Tris-buffered saline and
Tween 20 buffer (TBST) at room temperature for 1 h, prior to
incubation with rabbit anti-human DKK1 polyclonal antibody
(1:1,000; cat. no. LS-A2867; LifeSpan Biosciences, Inc., Seattle,
WA, USA) and rabbit polyclonal anti-β-actin (1:1,000; cat. no.
A2066; Sigma-Aldrich) at 25°C for 90 min. Following rinsing with
TBST containing 3% bovine serum albmin (BSA; Bio-Rad Laboratories,
Inc., Hercules, CA, USA), the membranes were incubated with
secondary antibody (Santa Cruz Biotechnology, Dallas, TX, USA) for
1 h at room temperature and the immunostained bands were
subsequently visualized using enhanced chemiluminescence (Pierce,
Thermo Fisher Scientific, Rockford, IL, USA). The intensity of each
band was normalized to β-actin, the internal control, and the
relative intensities were analyzed with ImageJ 1.0 software
(National Institutes of Health, Bethesda, MD, USA).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was extracted from the NSCLC cells using
Quick-RNA™ MicroPrep solution (Zymo Research Corp., Orange, CA,
USA) according to the manufacturer’s instructions. Subsequently,
the purified total RNA was reverse transcribed with the iScript™
Reverse Transcription supermix for PCR (Bio-Rad Laboratories, Inc.)
according to the manufacturer’s instructions. The reverse
transcribed RNA was subjected to PCR using the SsoFast™
EvaGreen® supermix (Bio-Rad Laboratories, Inc.). Primer
sequences were designed using the OligoPerfect™ Designer 1.0
software (Invitrogen Life Technologies), according to the
manufacturer’s instructions for optimal primer design, and were
synthesized commercially (Generay Biotech Co., Ltd., Shanghai,
China). The primer sequences used in the present study are listed
in Table I. The PCR cycling
conditions were as follows: Initial densturation at 35°C for 30
sec, followed by 30 cycles at 95°C for 30 sec, 55°C for 60 sec and
68°C for 2 min, and final extension at 68°C for 10 min. Each
reaction was performed in triplicate and three independent
experiments were conducted. A standard curve was constructed using
serial dilutions of a reference sample and was included in each run
to correct potential variations in amplification efficiency. The
relative copy numbers were obtained from the standard curve and
normalized to the values for β-actin. The fold-change in expression
was calculated using the 2−∆∆CT method.
 | Table IPrimer sequences used for polymerase
chain reaction analysis. |
Table I
Primer sequences used for polymerase
chain reaction analysis.
| Gene name | Primer sequence
(5′-3′) | Product size
(bp) |
|---|
| DKK1 |
CAACGCTATCAAGAACCTGC
GATCTTGGACCAGAAGTGTC | 168 |
| β-actin |
CCAACCGCGAGAAGATGA
CCAGAGGCGTACAGGGATAG | 200 |
| Cyclin D1 |
ACGAAGGTCTGCGCGTGTT
CCGCTGGCCATGAACTACCT | 323 |
| Bcl-2 |
TCCGCATCAGGAAGGCTAGA
AGGACCAGGCCTCCAAGCT | 113 |
| BAX |
GGGTGGTTGGGTGAGACTC
AGACACGTAAGGAAAACGCATTA | 199 |
| Akt-1 |
GCACAAACGAGGGGAGTACAT
CCTCACGTTGGTCCACATC | 113 |
| MMP2 |
GGCCCTGTCACTCCTGAGAT
GGCATCCAGGTTATCGGGGA | 474 |
| VEGFC |
AGGAGGGCAGAATCATCACG
TATGTGCTGGCCTFGGTGAG | 405 |
Histopathological analysis
Histopathological analyses were performed using
immunohistochemical and immunofluorescent staining methods. The
NSCLC samples collected from patients had previously been embedded
in paraffin blocks. The paraffin-embedded sections were
deparaffinized with xylene (Sigma-Aldrich) and rehydrated in graded
ethanol solutions. Endogenous peroxidase activity was blocked by
incubation with 3% H2O2 (Sigma-Aldrich) for
15 min at room temperature. Sections were subsequently heated with
0.01 M citrate (pH 6.0; Sigma-Aldrich) at 95°C for 15 min in a
microwave (220 watts) for antigen retrieval. Following incubation
with rabbit polyclonal anti-DKK1 antibody (Abcam Inc., Cambridge,
MA, USA) for 2 h at room temperature the sections were incubated
with horseradish peroxidase-labeled anti-rabbit immunoglobulin G
secondary antibody. The primary antibody was omitted in the
negative controls. The intensity of DKK1 staining was evaluated in
tumor cells in the cytoplasm, thereby determining the
immunoreactivity of DKK1 in the tumor cells.
TE13 cells were cultured on glass coverslips, fixed
with 4% paraformaldehyde and permeabilized with 0.2% Triton X-100
in phosphate-buffered saline (PBS) for 10 min at room temperature.
Cells were subsequently blocked with 3% BSAfor 30 min at room
temperature, and incubated with primary antibodies diluted in PBS
supplemented with 3% BSA for 60 min at room temperature. Following
washing with PBS, the cells were stained with fluorescein
isothiocyanate-conjugated secondary antibody (Santa Cruz
Biotechnology, Inc.) for 30 min at 37°C. Finally, the coverslips
were washed with PBS and the nuclei were stained with DAPI
(Sigma-Aldrich) or rhodamine B (Sigma-Aldrich) and visualized with
a confocal laser scanning microscope (Leica SP5; Leica
Microsystems, Wetzlar, Germany). The images were analyzed using
Leica LAS AF software (Leica Microsystems).
Statistical analysis
Statistical analyses were performed using SPSS
version 14.0 (SPSS Inc., Chicago, IL, USA) software. Values are
presented as the mean ± standard deviation. Student’s t-test,
one-way analysis of variance and χ2 test were performed.
P<0.05 was considered to indicate a statistically significant
difference. All experiments were performed at least in
triplicate.
Results
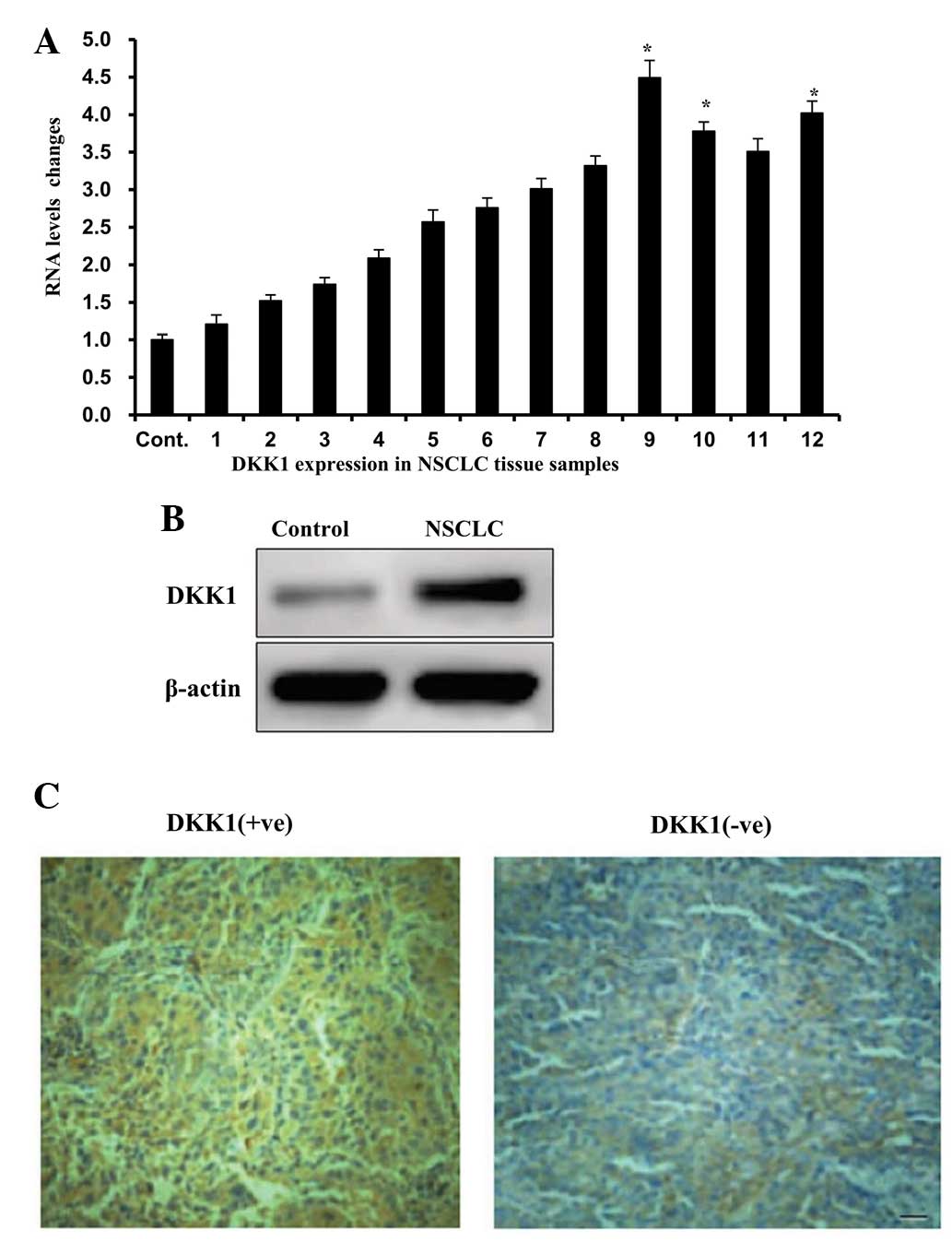
DKK1 is expressed in NSCLC tissues
DKK1 expression levels in the NSCLC samples
collected from 123 patients were evaluated and compared with those
of age-matched normal tissue from patients with low-grade NSCLC.
Evaluation included quantification of DKK1 gene and protein
expression levels and immunohistochemical analyses. For the
analysis of protein and gene expression, 25 samples of cancerous
tissue and 15 samples of age-matched normal tissue were evaluated
(17). The mRNA expression levels
of DKK1 in cancer tissues were found to be two- to six-fold greater
than those in normal tissues (Fig.
1A). Similarly, DKK1 protein expression levels were found to be
significantly greater in cancer tissue samples than those of the
normal tissue samples (Fig. 1B).
These results suggested a potential role for DKK1 in cancer
progression. Clinical samples were obtained from 98 patients were
subjected to immunohistochemical analysis. Of the 98 samples
analyzed, 62 (63.3%) were positively stained for DKK1 and 36 (37%)
demonstrated negative staining (Fig.
1C). Furthermore, no immunohistopathological staining for DKK1
was observed in normal tissues. These results indicated that
increased expression of DKK1 is common in human NSCLC, which may
potentially be associated with the metastasis of NSCLC.
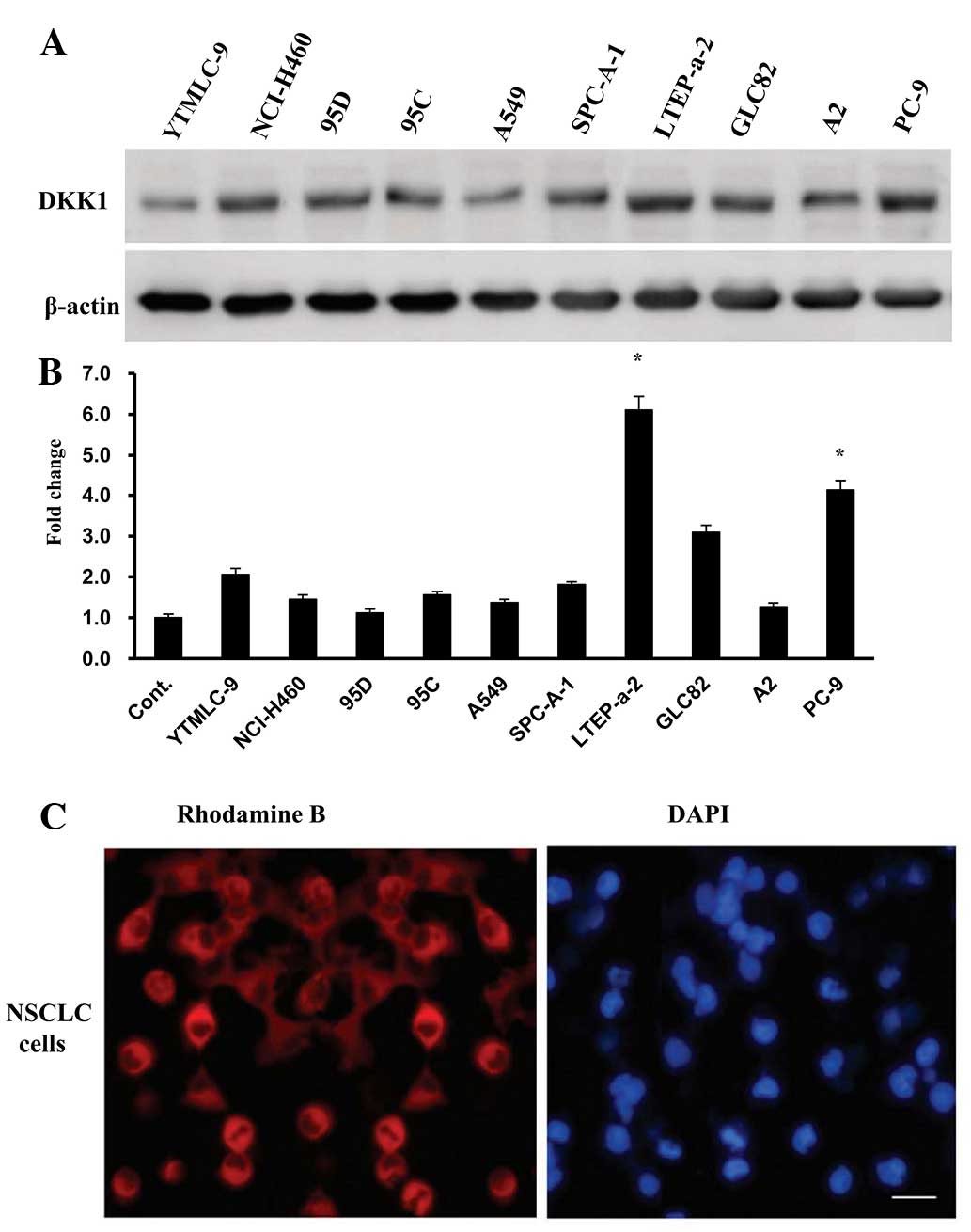
DKK1 is differentially expressed amongst
various NSCLC cell lines
DKK1 expression was evaluated in ten human NSCLC
cell lines. Western blot analysis revealed that DKK1 protein was
detected in all ten cell lines, but that the expression levels
varied amongst the cell lines. A significantly increased level of
DKK1 protein was observed in the LTEP-a-2, GLC-82 and PC-9 cell
lines. The lowest levels of DKK1 protein were observed in the A2
and 95C cell lines (Fig. 2A). The
mRNA expression levels of DKK1 were quantified using RT-qPCR, and
the results were concurrent with those of the protein expression
levels of corresponding cell lines (Fig. 2B). The LTEP-a-2 cell line
demonstrated significantly (six-fold) increased mRNA expression
levels. However, the minimum mRNA expression levels of DKK1 were
observed in 95D cells, compared with those of the other cell lines.
Immunofluorescent staining with rhodamine B revealed the
sub-cellular localization of DKK1 protein, which was determined by
the detection of rhodamine B-stained, fine-grained particles in the
cytoplasm (Fig. 2C). The
corresponding cell nuclei were stained with DAPI.
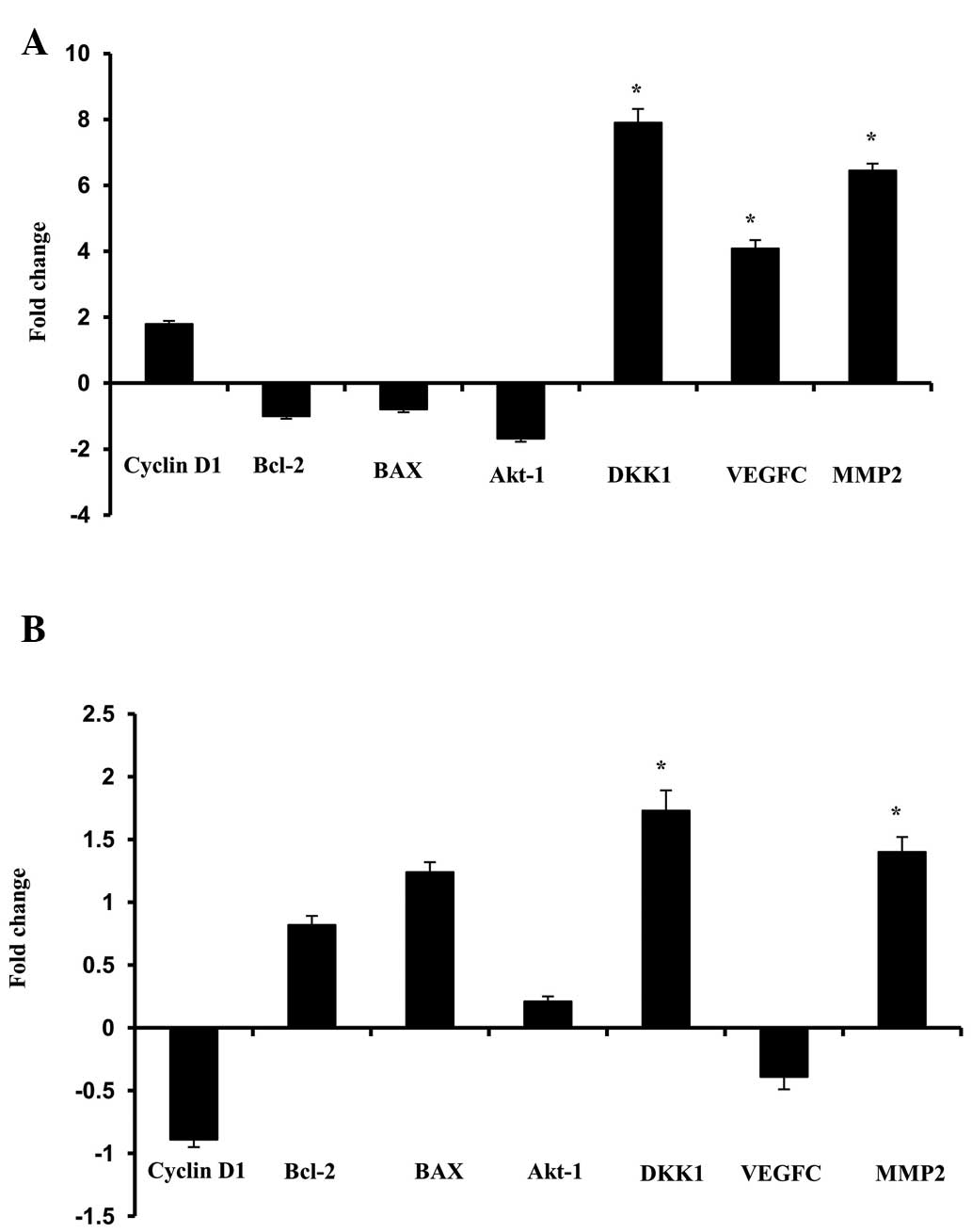
Regulatory effect of DKK1 in NSCLC cell
lines
The involvement of DKK1 in the cell cycle (cyclin
D1), apoptosis (Bcl-2, BAX), signaling pathways (Akt-1), as well as
invasion and metastasis (MMP2, VEGC) in selected cell lines with
high (LTEP-a-2) and low (95D) mRNA expression of DKK1, were
evaluated. A relative differential gene expression profile with
respect to the expression of DKK1 was constructed for the two cell
lines (Fig. 3). The gene
expression profile in LYEP-a-2 cells revealed increased mRNA levels
of cyclin D1, MMP2 and VEGC. However, the levels of Bcl-2, BAX and
Akt-1 were found to be significantly decreased (Fig. 3A). By contrast, the gene expression
profile in 95D cells revealed a contrary expression with respect to
the low levels of DKK1 expression. The mRNA expression levels of
cyclin D1, MMP2 and VEGC were decreased concurrently with the
decreased levels of DKK1. However, the levels of Bcl-2, BAX and
Akt1 were found to be increased (Fig.
3B). These results demonstrated a regulatory effect of the DKK1
gene in the suppression of cell cycle, signaling pathways and
apoptosis in NSCLC cells. In addition, enhanced DKK1 gene
expression facilitates the cell cycle, invasion and metastasis
(6).
Discussion
The secreted protein DKK1, an antagonist of the
Wnt/β-catenin signaling pathway, has been implicated in tumor
progression (4) and found to be
expressed in multiple types of human cancer (4). The role of DKK1 in tumor progression
may differ depending on the cell type (5). DKK1 is upregulated in certain types
of human cancer, including NSCLC, hepatocellular carcinoma and
pancreatic cancer (5,18).
Emerging studies have focused on elucidating the
significance of DKK1 in mediating tumor progression; however, the
biological effects of DKK1 in NSCLC cells have remained elusive.
The present study aimed to identify the differential expression of
DKK1 in NSCCLC cells, including those obtained from human cancerous
tissue specimens and in certain NSCLC cell lines. The present study
also aimed to confirm the involvement of DKK1 in the regulation of
tumor progression via the modulation of key genes involved in the
cell cycle, apoptosis, cell invasion and metastasis. DKK1
expression was therefore evaluated in human NSCLC tissue specimens
and ten NSCLC cell lines. DKK1 protein was expressed in almost all
the NSCLC cancer cells evaluated; however, the expression levels
varied between tissues and cell lines. DKK1 protein and gene were
expressed in analogous patterns, with respect to the type of tissue
or cell line. Immunohistochemical analysis of clinical samples from
98 NSCLC patients revealed DKK1-positive staining in 63.3% of the
samples and the remaining 37% with negative staining. Furthermore,
no staining for DKK1 was detected in non-cancerous or normal cells.
These results implicated DKK1 in the progession of NSCLC, which was
consistent with the results of previous studies (4,5,19).
Studies previously hypothesized that DKK1 was
involved in the downstream targeting of β-catenin/T-cell factor and
participated in a negative feedback loop within the Wnt signaling
pathway of colon cancer cells (1,20).
In addition, studies revealed that the overexpression of DKK1 was
associated with poor patient prognosis (5,21,22),
whereas no clear evidence regarding the function of DKK1 in NSCLC
was identified.
The present study aimed to elucidate the molecular
mechanisms underlying the effects of DKK1 in tumor progression. Two
NSCLC cell lines, which demonstrated significantly increased
(LYEP-a-2 cells) and decreased (95D cells) levels of DKK1 mRNA
expression were selected. The relative expression levels of key
proteins involved in the cell cycle (cyclin D1), apoptosis (Bcl-2,
BAX), signaling pathway (Akt-1), invasion and metastasis (MMP2,
VEGC) were examined with respect to DKK1 expression levels. Of
note, a proportional increase in genes associated with the cell
cycle, as well as invasion and metastasis, were detected
concurrently with high levels of DKK1 gene expression in LYEP-a-2
cells. Furthermore, the high levels of DKK1 in LYEP-a-2 cells were
associated with decreased expression levels of genes involved in
apoptosis and signaling pathways. These results were supported by
those obtained from the analysis of gene expression in 95D cells,
where low DKK1 expression levels were associated with inverse
alterations in gene expression. Low expression levels of DKK1
induced increases in the mRNA expression levels of apoptotic genes
Bcl-2 and BAX, as well as signaling pathway-associated Akt-1. The
results therefore indicated that increased DKK1 gene expression may
enhance the cell cycle, as well as invasion and metastasis of NSCLC
cells. However, further studies are required to elucidate the
specific mechanisms underlying this biological effect.
Conversely, previous studies have demonstrated that
DKK1 suppressed cell growth and migration (6,23),
suggesting that DKK1 may have diverse biological roles in distinct
types of cancer cell. To date, few studies have been published
regarding the role of DKK1 in NSCLC, and these have mainly focused
on elucidating its diagnostic or prognostic value (24–26).
Further studies are required to elucidate the mechanisms underlying
the differential and relative expression of DKK1 in NSCLC.
In conclusion, the present study confirmed the
involvement of DKK1 in NSCLC progression. The results revealed
differential expression of DKK1 in NSCLC cells, which may provide a
potential therapeutic target for cancer prevention, particularly in
NSCLC.
References
|
1
|
Niida A, Hiroko T, Kasai M, et al: DKK1, a
negative regulator of Wnt signaling, is a target of the
beta-catenin/TCF pathway. Oncogene. 23:8520–8526. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Gavriatopoulou M, Dimopoulos MA,
Christoulas D, et al: Dickkopf-1: a suitable target for the
management of myeloma bone disease. Expert Opin Ther Targets.
13:839–848. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Forget MA, Turcotte S, Beauseigle D, et
al: The Wnt pathway regulator DKK1 is preferentially expressed in
hormone-resistant breast tumours and in some common cancer types.
Br J Cancer. 96:646–653. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Darlavoix T, Seelentag W, Yan P, Bachmann
A and Bosman FT: Altered expression of CD44 and DKK1 in the
progression of Barrett’s esophagus to esophageal adenocarcinoma.
Virchows Arch. 454:629–637. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Makino T, Yamasaki M, Takemasa I, et al:
Dickkopf-1 expression as a marker for predicting clinical outcome
in esophageal squamous cell carcinoma. Ann Surg Oncol.
16:2058–2064. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wirths O, Waha A, Weggen S, Schirmacher P,
Kühne T, et al: Overexpression of human Dickkopf-1, an antagonist
of wingless/WNT signaling, in human hepatoblastomas and Wilms’
tumors. Lab Invest. 83:429–434. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Marvin MJ, Di Rocco G, Gardiner A, et al:
Inhibition of Wnt activity induces heart formation from posterior
mesoderm. Genes Dev. 15:316–327. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Mountain CF: Revisions in the
international system for staging lung cancer. Chest. 111:1710–1717.
1997. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Martini N, Bains MS, Burt ME, et al:
Incidence of local recurrence and second primary tumors in resected
stage I lung cancer. J Thorac Cardiovasc Surg. 109:120–129. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chen WQ, Zheng RS and Zeng HM: Bayesian
age-period-cohort prediction of lung cancer incidence in China.
Thoracic Cancer. 2:149–155. 2011. View Article : Google Scholar
|
|
11
|
Chen W, Zheng R, Zhang S, Zou X, Zhao P,
et al: Lung cancer incidence and mortality in China, 2009. Thoracic
Cancer. 4:102–108. 2013. View Article : Google Scholar
|
|
12
|
Zeng H, Zheng R, Zhang S, He J and Chen W:
Lung cancer incidence and mortality in China, 2008. Thoracic
Cancer. 4:53–58. 2013. View Article : Google Scholar
|
|
13
|
Fukuyama Y, Mitsudomi T, Sugio K, et al:
K-ras and p53 mutations are an independent unfavourable prognostic
indicator in patients with non-small-cell lung cancer. Br J Cancer.
75:1125–1130. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Osaki T, Mitsudomi T, Oyama T, Nakanishi R
and Yasumoto K: Serum level and tissue expression of c-erbB-2
protein in lung adenocarcinoma. Chest. 108:157–162. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Oyama T, Osaki T, Mitsudomi T, et al: p53
alteration, proliferating cell nuclear antigen and nucleolar
organizer regions in thymic epithelial tumors. Int J Mol Med.
1:823–826. 1998.PubMed/NCBI
|
|
16
|
Bradford MM: A rapid and sensitive method
for the quantitation of microgram quantities of protein utilizing
the principle of protein-dye binding. Anal Biochem. 72:248–254.
1976. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Tammemägi MC, Katki HA, Hocking WG, et al:
Selection criteria for lung-cancer screening. New Engl J Med.
368:728–736. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Takahashi N, Fukushima T, Yorita K, Tanaka
H, Chijiiwa K, et al: Dickkopf-1 is overexpressed in human
pancreatic ductal adenocarcinoma cells and is involved in invasive
growth. Int J Cancer. 126:1611–1620. 2010.
|
|
19
|
Yamabuki T, Takano A, Hayama S, Ishikawa
N, Kato T, et al: Dikkopf-1 as a novel serologic and prognostic
biomarker for lung and esophageal carcinomas. Cancer Res.
67:2517–2525. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
González-Sancho JM, Aguilera O, García JM,
et al: The Wnt antagonist DICKKOPF-1 gene is a downstream target of
beta-catenin/TCF and is downregulated in human colon cancer.
Oncogene. 24:1098–1103. 2005. View Article : Google Scholar
|
|
21
|
Yu B, Yang X, Xu Y, Yao G, Shu H, et al:
Elevated expression of DKK1 is associated with cytoplasmic/nuclear
beta-catenin accumulation and poor prognosis in hepatocellular
carcinomas. J Hepatol. 50:948–957. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Sheng SL, Huang G, Yu B and Qin WX:
Clinical significance and prognostic value of serum Dickkopf-1
concentrations in patients with lung cancer. Clin Chem.
55:1656–1664. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Qin X, Zhang H, Zhou X, Wang C, Zhang X,
et al: Proliferation and migration mediated by
Dkk-1/Wnt/beta-catenin cascade in a model of hepatocellular
carcinoma cells. Transl Res. 150:281–294. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yao X, Jiang H, Zhang C, Wang H, Yang L,
et al: Dickkopf-1 autoantibody is a novel serological biomarker for
non-small cell lung cancer. Biomarkers. 15:128–134. 2010.
View Article : Google Scholar
|
|
25
|
Osada H, Tomida S, Yatabe Y, Tatematsu Y,
Takeuchi T, et al: Roles of achaetescute homologue 1 in DKK1 and
E-cadherin repression and neuroendocrine differentiation in lung
cancer. Cancer Res. 68:1647–1655. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Licchesi JD, Westra WH, Hooker CM, Machida
EO, Baylin SB, et al: Epigenetic alteration of Wnt pathway
antagonists in progressive glandular neoplasia of the lung.
Carcinogenesis. 29:895–904. 2008. View Article : Google Scholar : PubMed/NCBI
|