Introduction
Renal cell carcinoma (RCC) is the most frequent
neoplasm that occurs in the adult kidney (1), and the 5 year survival rates are 98%
for stage I disease and 50% for stage III disease (2). These survival rates strengthen the
significance of early identification and timely treatment of RCC.
The mortality rate has not been substantially improved in the past
two decades, although certain progress has been achieved in early
recognition of RCC (3,4). RCC is characterized by aberrantly
enhanced proliferation of the cancer cells, and as a result, RCC is
relatively resistant to the conventional treatment of the disease,
including radiotherapy and chemotherapy (5). Although options are available in the
management of RCC, the clinical outcome is usually limited due to
its fast growth (4). Therefore, an
improved understanding of the underlying molecular mechanism and
identification of novel and pivotal diagnostic and therapeutic
molecules are warranted.
A growing body of evidence demonstrated that micro
(mi)RNAs, a class of non-protein-coding small RNAs, are associated
with the progression and proliferation of cancer. miRNAs
post-transcriptionally modulate gene expression via binding to the
3′-untranslated region (UTR) of target mRNAs, resulting in
suppression of translation and/or degradation of mRNA (6). miRNAs may be differentially expressed
between normal and tumor tissue, and miRNA expression profiles may
be specific in certain cancer types, which may be responsible for
the development of malignancy, including RCC (7). Discovery of differentially expressed
miRNA and the identification its downstream signaling pathway may
provide the possibility to develop novel therapeutic methods for
the treatment of RCC.
It has been previously reported that miRNAs are
responsible for the abnormally enhanced proliferation of RCC cells,
and miR-141, miR-187, miR-206, miR-218, miR-335 and miR-204 have
been identified to be differentially expressed in RCC based on
miRNA microarray analysis (8). The
present study performed confirmatory reverse
transcription-quantitative polymerase chain reaction (RT-qPCR) to
identify the expression level of each of those miRNAs in RCC tumor
tissue sample compared with the adjacent non-cancerous control
tissue samples. The results revealed that miR-218 was significantly
downregulated in RCC tissue. The present study also identified
B-cell lymphoma (BCL)9 as a target of miR-218 by computational
analysis, which was confirmed by luciferase assay.
Materials and methods
Patients and sample collection
The present study was approved by the Ethics
Committee of Yushan Central Hospital of Linshu County, (Yushan,
China). Written informed consent was obtained from all patients. A
total of 25 pairs of tumor tissue samples and adjacent
non-cancerous tissue samples, at least 2 cm away from the tumor
tissue, were collected from patients who were diagnosed with RCC
and received nephron-sparing surgery or radical nephrectomy for RCC
at the Department of Surgery, Yushan Central Hospital of Linshu
County. Patients treated with chemotherapy or radiotherapy prior to
surgery were excluded from the present study. Soon after removal,
the tissue samples were frozen in liquid nitrogen for future
use.
RT-qPCR
The mirVana™ miRNA isolation kit (Ambion, USA) was
used to extract the total RNA from the tissues or the cells,
according to the manufacturer's protocol. A NanoDrop
spectrophotometer (ND-1000; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) was used to identify the quantity and purity of
the RNA. The RNA samples were maintained at −80°C for future use.
The synthesis of cDNA from the total RNA was performed by using
miRNA-specific RT primers from the TaqMan MicroRNA assay (Applied
Biosystems, Foster City, CA, USA) at 65°C for 5 min, 25°C for 10
min, 42°C for 1 h and 75°C for 10 min. Subsequently, using the
TaqMan MicroRNA assay, qPCR was performed on the ABI PRISM 7900HT
system (Applied Biosystems). The conditions were as follows: 95°C
for 5 min followed by 40 cycles of 95°C for 10 sec and 60°C for 60
sec. Each experiment was repeated at least three times. The primer
sequences were as follows: Forward, 5′-TCAGTTTGCTGTTCTGGGTG-3′ and
reverse, 5′-CGGTTGGCTGGAAAGGAG-3′ for GAPDH, and forward,
5′-GAGCACTGCAGTGTCGTGAC-3′ and reverse, 5′-TCCCCTTGCCCTTGGCACCA-3′
for the miRNA-specific primers. SDS software (version 2.4; Applied
Biosystems; Thermo Fisher Scientific, Inc.) was used to perform
quantitative analysis according to the manufacturer's protocol. The
software dissociation curve analysis feature was used to verify and
determine the specificity of the PCR products. The
2−ΔΔCq method was used to analyze the relative
quantification, as described previously (9).
Oligonucleotides and transfection
The miR-218 mimics, miR-218 inhibitor and
oligonucleotide negative control (NC) miRNA were obtained from
GenePharma (Shanghai, China). The transfection of oligonucleotides
were performed using Lipofectamine 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.), according to the manufacturer's protocol. The
A498 cell line was purchased from American Type Culture Collection
(Manassas, VA, USA), and the cells were incubated in each well of a
96-well plate overnight.
Colorimetric
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT)
assay
To investigate cell proliferation, MTT (Promega
Corporation, Madison, WI, USA) was pipetted into all wells at
different time points. The cells were cultured at 37.8°C for 3 h.
Subsequently, the cells were centrifuged at 13,000 x g for 15 min
at 4°C to remove cell debris and insoluble material. A total of 100
µl dimethyl sulfoxide was used to dissolve the formazan
precipitates. A spectrophotometer (Dinex Technologies, Chantilly,
VA, USA) was used to measure the absorbance at a wavelength of 550
nm.
Western blotting
Following washing with cold phosphate-buffered
saline, the cells were lysed in radioimmunoprecipitation buffer
(BioTeke Corporation, Beijing, China) comprising 5 mM EDTA, 150 mM
NaCl, 50 mM Tris Cl (pH 7.4), 0.1% sodium dodecyl sulfate (SDS)/1%
aprotinin, 1% sodium deoxycholate, 1% Nonidet P-40, 0.1 mM
Na3VO4 and 50 mM NaF, supplemented with a
Protease Inhibitor Cocktail (Promega Corporation) on ice for 30
min. Following protein extraction, the samples were centrifuged at
13,000 x g for 15 min at 4°C. A bicinchoninic acid (BCA) assay kit
(BioTeke Corporation) was used to determine the concentration of
the protein, according to the manufacturer's protocol. The proteins
were heated with loading buffer (BioTeke Corporation) at 95.8°C for
5 min, and were separated by using 10% SDS-polyacrylamide gel
electrophoresis. The proteins were transferred onto a
polyvinylidene difluoride membrane (Bio-Rad Laboratories, Hercules,
CA, USA). The membrane was blocked in blocking buffer (Merck
Millipore, Darmstadt, Germany) with 5% skim milk for 60 min at room
temperature and subsequently incubated overnight at 4°C with mouse
monoclonal antibodies against β-actin (1:2,000; Santa Cruz
Biotechnology, Inc., Santa Cruz, CA, USA; cat. no. sc-47778) and
BCL9 (1:1,000; Santa Cruz Biotechnology, Inc.; cat. no. sc-81199).
Following primary antibody incubation, the membranes were incubated
with goat anti-mouse horseradish peroxidase-conjugated secondary
antibody (1:3,000; Santa Cruz Biotechnology, Inc.; cat. no.
sc-2005) for 1 h at room temperature. The percentage of the
expression of target proteins was normalized against β-actin using
ImageJ software (imagej.nih.gov/ij).
Luciferase assay
A498 cells were seeded into a 12-well plate at a
density of ~0.8×105 cells for 24 h. The cells were
subsequently transfected with 100 nmol/l miR-218 mimics using
Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) and
50 ng target sequence (wild type or mutant BCL9 3′-UTR) expression
clone (pGL3-BCL9 3′UTR). After incubation for 36 h, a luciferase
assay system (Promega Corporation) was used to determine the
activities of firefly luciferase, according to the manufacturer's
protocol. An LD400 luminometer (Beckman Coulter, Brea, CA, USA) was
used to collect data.
Apoptosis assay
Transfection was performed when cell confluency
reached 80% in the 48-well plates. The cells were serum starved and
10 µM camptothecin (Sigma-Aldrich) in serum-free media was
used to induce apoptosis as previously described (10). Propidium iodide (Invitrogen; Thermo
Fisher Scientific, Inc.) and Annexin V-fluorescein isothiocyanate
(Invitrogen; Thermo Fisher Scientific, Inc.) were used to stain the
cells, and FACScan flow cytometer (BD Biosciences, Franklin Lakes,
NJ, USA) was used to analyze the apoptosis of A498 cells, and
CellQuest Software (BD Biosciences) was used to analyze the data
according to the manufacturer's protocol. Each test was repeated
three times.
Statistical analysis
All statistical analyses were performed using SPSS
21.0 (IBM SPSS, Chicago, IL, USA). The data are expressed as the
mean ± standard deviation. The comparison between two groups was
assessed using an independent t-test, and the comparison among
multiple groups (>2 groups) were performed by using one-way
analysis of variance. P<0.05 was considered to indicate a
statistically significant difference.
Results
It has been previously reported that miRNAs are
responsible for the abnormally enhanced proliferation of RCC cells
(8). The present study was focused
on the differential expression of miRNAs, as well as its role in
tumorigenesis of RCC. A total of 25 pairs of tumor tissue samples
and adjacent non-cancerous controls were obtained from patients
with confirmed diagnosis of RCC. RT-qPCR was performed for the
tissue samples to determine and compare the expression of miRNAs
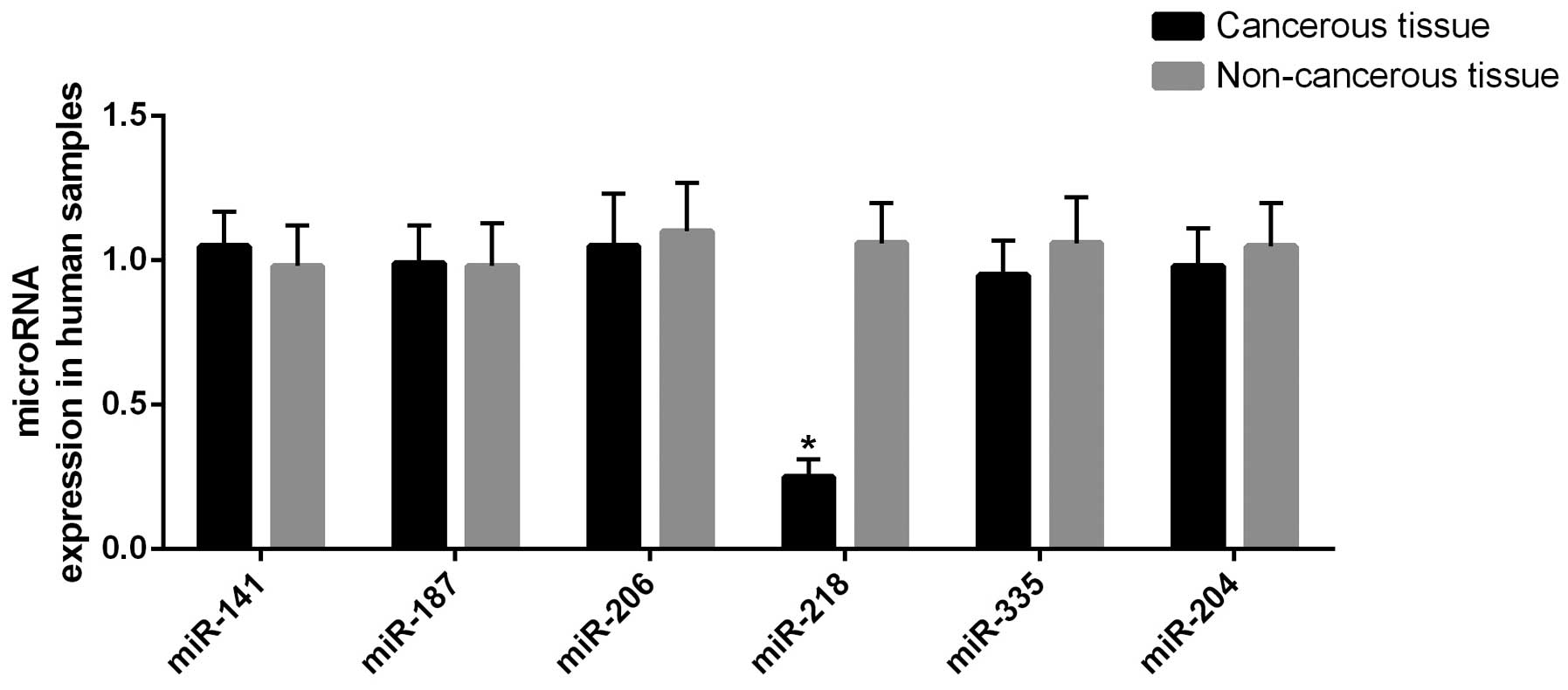
(miR-141, miR-187, miR-206, miR-218, miR-335 and miR-204) (8). miR-218 was revealed to be
significantly (P<0.01) downregulated in the tumor tissue
compared with the controls, as shown in Fig. 1.
The present study used online publicly available
algorithms (mirdb.org) to predict targets of
miR-218. The results revealed the putative binding site of miR-218
in the 3′UTR of BCL9 (Fig. 2). To
confirm this, the present study constructed pGL3-BCL9-3′UTR,
containing the miR-218 binding site or the mutant binding site
(Fig. 2), and co-transfected the
RCC cells with the construct and miR-218 mimics or scramble
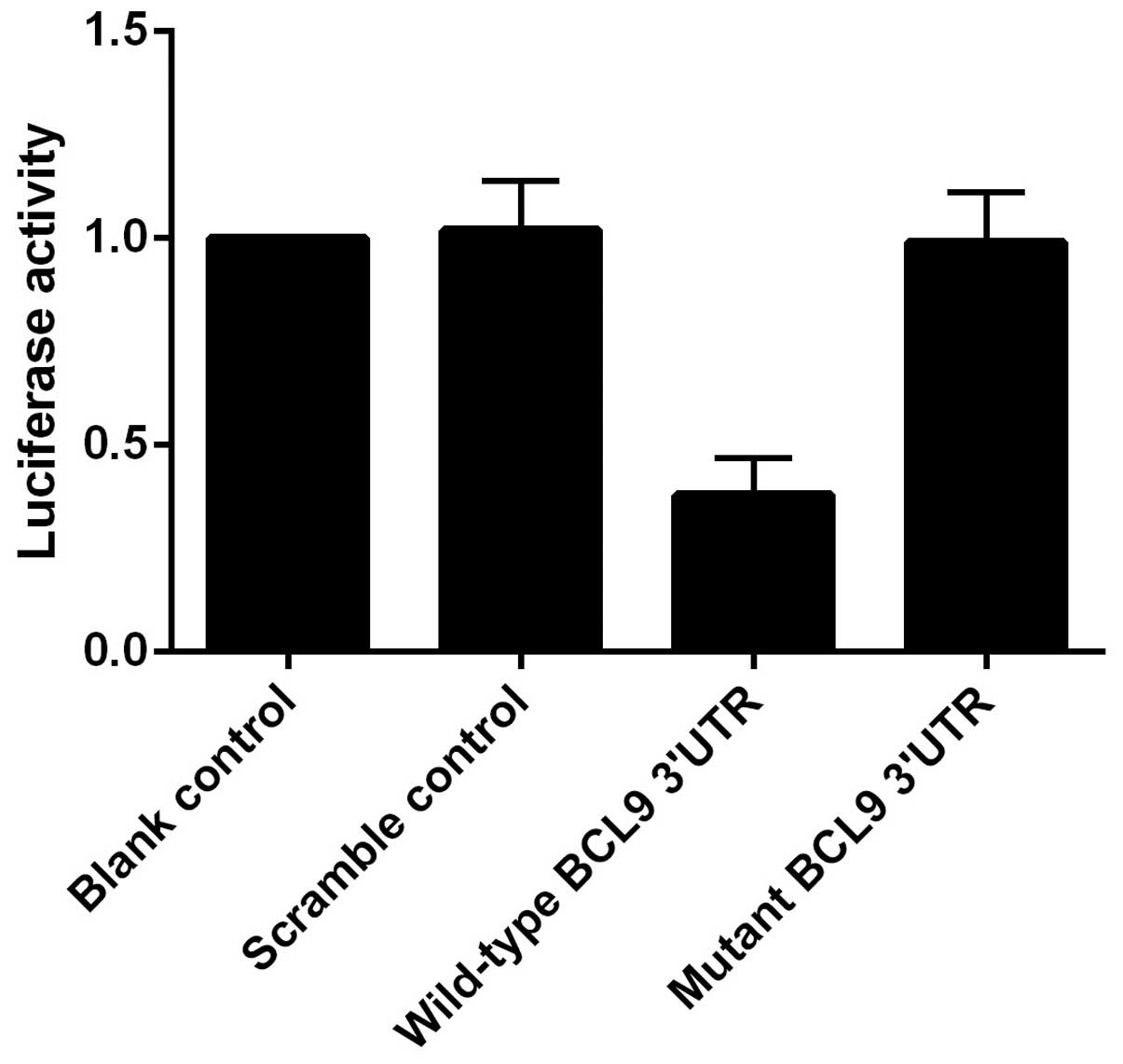
controls. As showed in Fig. 3, the
luciferase activity from pGL3-BCL9-3′UTR was significantly
inhibited by miR-218, but was not affected by the scramble
controls. These findings indicated that miR-218 inhibited the
expression of BCL9 by binding to complementary sequences in the
3′UTR of BCL9.
Given the negative regulatory association between
miR-218 and BCL9 in RCC cells, the present study further examined
the expression of BCL9 in the tumor samples and the controls,
finding that the mRNA expression of BCL9 was substantially
upregulated in the tumor tissue samples compared with the adjacent
non-cancerous controls (Fig. 4A).
In addition, western blot analysis was performed to determine the
protein expression levels of BCL9, and ImageJ software was used to
evaluate the relative density of the target bands normalized
against β-actin. As shown in Fig.
4B, the protein expression of BCL9 was significantly higher in
the cancerous tissue compared with the non-cancerous controls. To
further study whether miR-218 affects the expression of BCL9,
miR-218 mimics or its inhibitors were transfected into RCC cells
and were harvested 48 h later. RT-qPCR and western blotting
revealed that both the mRNA and protein expression levels of BCL9
were significantly downregulated by the miR-218 mimics (Fig. 5A). However, the inhibition of
miR-218 upregulated BCL9 levels (Fig.
5B).
Next, the present study explored the potential
impact of miR-218 on the proliferation of RCC cells. The cells were
transfected with the miR-218 mimics, inhibitors or the scramble
controls, and were analyzed 48 h later. An MTT proliferation assay
revealed that cell proliferation was suppressed in miR-218
mimic-transfected RCC cells compared with the control cells;
however, cell proliferation was significantly promoted in the RCC
cells transfected with miR-218 inhibitors compared with the
controls (Fig. 6A). Subsequently,
apoptosis analysis revealed that miR-218 overexpression caused more
cell death in RCC cells compared with the NC cells, while
introduction of miR-218 inhibitors significantly suppressed the
apoptosis in RCC cells compared with the control cells (Fig. 6B).
Discussion
Dysregulation of numerous miRNAs in RCC were
observed in several cancer types, demonstrating that miRNAs have a
regulatory role in ordinary cancer pathways. miRNAs can act on
genes that are associated with cancer pathogenesis and their
actions as tumor inhibitor genes or oncogenes has been demonstrated
previously (5). An improved
knowledge of the effects of miRNAs in these cellular processes may
assist with the development of novel therapies for the treatment of
cancer. In the present study, 25 pairs of tumor tissue samples and
adjacent non-cancerous controls were obtained from patients with
confirmed diagnosis of RCC. RT-qPCR was performed to determine and
compare the expression levels of miR-141, miR-187, miR-206,
miR-218, miR-335 and miR-204 (8).
miR-218 was significantly downregulated in the tumor tissue samples
compared with the controls, as shown in Fig. 1 (P<0.01).
The role of miR-218 has been well studied, and the
data from previous functional studies of miR-218 in a variety of
cancer types revealed the inhibition of cancer cell proliferation
and invasion by miR-218. For instance, underexpression of miR-218
was revealed to be correlated with lymph node metastasis, advanced
clinical stage and unfavorable prognosis (11). In non-small cell lung carcinoma,
the expression of miR-218 may act as an independent signal for
relapse-free and overall survival (12). Numerous previous studies have
successfully identified direct targets of miR-218, including
asIKK-B, ECOP, PXN, LASP1, Survivin, RICTOR and ROBO1 (13–17).
These findings indicated that the expression of miR-218 may be an
effective indicator for tumor diagnosis and prognosis, and miR-218
acts as a tumor inhibitor in certain other cancer types. In the
present study, online publicly available algorithms (mirdb.org) were used to predict miR-218 targets. A
putative binding site of miR-218 was identified in the 3′UTR of
BCL9. To confirm this regulatory association, the present study
constructed pGL3-BCL9-3′UTR, containing the miR-218 binding site or
the mutant one, and co-transfected the RCC cells with the construct
and miR-218 mimics or scramble controls. It was revealed that the
luciferase activity from pGL3-BCL9-3′UTR was significantly
inhibited by miR-218, but was not affected by the scramble
controls. These findings indicated that miR-218 inhibits the
expression of BCL9 by binding to complementary sequences in the
3′UTR of BCL9.
BCL9 is a crucial co-mediator of the transcriptional
activity of Wnt/β-catenin, and previous studies have demonstrated
its function as novel therapeutic target of malignancies (18,19).
Previous studies have demonstrated that BCL9 functions as oncogene,
which can promote tumor progression (epithelial-mesenchymal
transition, invasion, migration and metastasis) and cell
proliferation by regulating the expression of cyclin D1. By
contrast, knocking down BCL9 improves survival outcomes in
xenograft mouse models of multiple myeloma and colon cancer
(18). Notably, knocking out BCL9
in germ cells increased apoptotic cells by 10-fold (18). BCL9 has also been previously
reported to be regulated by miRNAs. For instance, miR-30 was
revealed to be involved in the control of BCL9 in multiple myeloma
(MM), and a reverse correlation exists between miR-30s and the mRNA
expression levels of BCL9 (20).
Elevated expression of miR-30s in MM cell lines results in a
decrease in proliferation, migration, survival, invasion and colony
formation (21). In the present
study, it was revealed that the protein expression of BCL9 was
significantly higher in the cancerous tissue compared with the
non-cancerous controls, and both the mRNA and protein expression
levels of BCL9 were significantly downregulated by miR-218 mimics.
However, inhibition of miR-218 upregulated BCL9 levels.
Furthermore, the present study assessed the
potential impact of miR-218 in RCC cell proliferation. The cells
were transfected with miR-218 mimics, inhibitors or the scramble
controls, and were analyzed 48 h later. An MTT proliferation assay
demonstrated that cell proliferation was suppressed in miR-218
mimic-transfected RCC cells compared with the control cells;
however, cell proliferation was significantly promoted in the RCC
cells transfected with miR-218 inhibitors compared with the
controls. Subsequent apoptosis analysis revealed that miR-218
overexpression caused increased death to RCC cells compared with NC
cells, while introduction of miR-218 inhibitors significantly
suppressed the apoptosis in RCC cells compared with the
control.
In conclusion, significant downregulation of miR-218
was observed in RCC cells. The expression of miR-218 was decreased
in clinical specimens of RCC, and had an inhibitory role in the
control of proliferation of RCC. The present results revealed that
oncogenic BCL9 may be upregulated as a result of the downregulation
of tumor suppressive miR-218 in the progression of human RCC. This
molecular network is likely important in oncogenesis of RCC and may
act as a novel therapeutic option for patients with RCC.
References
|
1
|
Weng L, Wu X, Gao H, Mu B, Li X, Wang JH,
Guo C, Jin JM, Chen Z, Covarrubias M, et al: MicroRNA profiling of
clear cell renal cell carcinoma by whole-genome small RNA deep
sequencing of paired frozen and formalin-fixed, paraffin-embedded
tissue specimens. J Pathol. 222:41–51. 2010.PubMed/NCBI
|
|
2
|
Devita VT Jr, Hellman S and Rosenberg SA:
Cancer principles and practice of oncology. 8th edition. Lippincott
Williams & Wilkins; Philadelphia, PA: 2008
|
|
3
|
Fridman E, Dotan Z, Barshack I, David MB,
Dov A, Tabak S, Zion O, Benjamin S, Benjamin H, Kuker H, et al:
Accurate molecular classification of renal tumors using microRNA
expression. J Mol Diagn. 12:687–696. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Aben KK, Luth TK, Janssen-Heijnen ML,
Mulders PF, Kiemeney LA and van Spronsen DJ: No improvement in
renal cell carcinoma survival: A population-based study in the
Netherlands. Eur J Cancer. 44:1701–1709. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Capitanio U, Cloutier V, Zini L, Isbarn H,
Jeldres C, Shariat SF, Perrotte P, Antebi E, Patard JJ, Montorsi F
and Karakiewicz PI: A critical assessment of the prognostic value
of clear cell, papillary and chromophobe histological subtypes in
renal cell carcinoma: A population-based study. BJU Int.
103:1496–1500. 2009. View Article : Google Scholar
|
|
6
|
Slaby O, Jancovicova J, Lakomy R, Svoboda
M, Poprach A, Fabian P, Kren L, Michalek J and Vyzula R: Expression
of miRNA-106b in conventional renal cell carcinoma is a potential
marker for prediction of early metastasis after nephrectomy. J Exp
Clin Cancer Res. 29:902010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Volinia S, Calin GA, Liu CG, Ambs S,
Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, et
al: A microRNA expression signature of human solid tumors defines
cancer gene targets. Proc Natl Acad Sci USA. 103:2257–2261. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hidaka H, Seki N, Yoshino H, Yamasaki T,
Yamada Y, Nohata N, Fuse M, Nakagawa M and Enokida H: Tumor
suppressive microRNA-1285 regulates novel molecular targets:
Aberrant expression and functional significance in renal cell
carcinoma. Oncotarget. 3:44–57. 2012.PubMed/NCBI
|
|
9
|
Vermes I, Haanen C, Steffens-Nakken H and
Reutelingsperger C: A novel assay for apoptosis. Flow cytometric
detection of phosphatidylserine expression on early apoptotic cells
using fluorescein labelled Annexin V. J Immunol Methods. 184:39–51.
PubMed/NCBI
|
|
10
|
Annaratone L, Volante M, Asioli S, Rangel
N and Bussolati G: Characterization of neuroendocrine tumors of the
pancreas by real-time quantitative polymerase chain reaction. A
methodological approach. Endocr Pathol. 24:83–91. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Tie J, Pan Y, Zhao L, Wu K, Liu J, Sun S,
Guo X, Wang B, Gang Y, Zhang Y, et al: MiR-218 inhibits invasion
and metastasis of gastric cancer by targeting the Robo1 receptor.
PLoS Genet. 6:e10008792010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Smilenov LB, Mikhailov A, Pelham RJ,
Marcantonio EE and Gundersen GG: Focal adhesion motility revealed
in stationary fibroblasts. Science. 286:1172–1174. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Alajez NM, Lenarduzzi M, Ito E, Hui AB,
Shi W, Bruce J, Yue S, Huang SH, Xu W, Waldron J, et al: MiR-218
suppresses nasopharyngeal cancer progression through downregulation
of survivin and the SLIT2-ROBO1 pathway. Cancer Res. 71:2381–2391.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Chiyomaru T, Enokida H, Tatarano S,
Kawahara K, Uchida Y, Nishiyama K, Fujimura L, Kikkawa N, Seki N
and Nakagawa M: miR-145 and miR-133a function as tumour suppressors
and directly regulate FSCN1 expression in bladder cancer. Br J
Cancer. 102:883–891. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Song L, Huang Q, Chen K, Liu L, Lin C, Dai
T, Yu C, Wu Z and Li J: miR-218 inhibits the invasive ability of
glioma cells by direct downregulation of IKK-β. Biochem Biophys Res
Commun. 402:135–140. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wu DW, Cheng YW, Wang J, Chen CY and Lee
H: Paxillin predicts survival and relapse in non-small cell lung
cancer by microRNA-218 targeting. Cancer Res. 70:10392–10401. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Gao C, Zhang Z, Liu W, Xiao S, Gu W and Lu
H: Reduced microRNA-218 expression is associated with high nuclear
factor kappa B activation in gastric cancer. Cancer. 116:41–49.
2010.
|
|
18
|
Mani M, Carrasco DE, Zhang Y, Takada K,
Gatt ME, Dutta-Simmons J, Ikeda H, Diaz-Griffero F, Pena-Cruz V,
Bertagnolli M, et al: BCL9 promotes tumor progression by conferring
enhanced proliferative, metastatic and angiogenic properties to
cancer cells. Cancer Res. 69:7577–7586. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Takada K, Zhu D, Bird GH, Sukhdeo K, Zhao
JJ, Mani M, Lemieux M, Carrasco DE, Ryan J, Horst D, et al:
Targeted disruption of the BCL9/β-catenin complex inhibits
oncogenic Wnt signaling. Sci Transl Med. 4:148ra1172012. View Article : Google Scholar
|
|
20
|
Nantel F, Monaco L, Foulkes NS, Masquilier
D, LeMeur M, Henriksén K, Dierich A, Parvinen M and Sassone-Corsi
P: Spermiogenesis deficiency and germ-cell apoptosis in CREM-mutant
mice. Nature. 380:159–162. 1996. View
Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhao JJ, Lin J, Zhu D, Wang X, Brooks D,
Chen M, Chu ZB, Takada K, Ciccarelli B, Admin S, et al: miR-30-5p
functions as a tumor suppressor and novel therapeutic tool by
targeting the oncogenic Wnt/β-catenin/BCL9 pathway. Cancer Res.
74:1801–1813. 2014. View Article : Google Scholar : PubMed/NCBI
|