Introduction
Primary lung cancer is the most common type of
malignancy after non-melanocytic skin cancer, and the leading cause
of human cancer-related fatality worldwide (1). While it has been the most important
cause of cancer-related mortality in men since the 1960s, it and
breast cancer have been identified as the most important causes of
mortality in women (2). Lung
cancer is increasing in prevalence and mortality rates worldwide
(3). In developed countries, the
latter has begun to decline in men, reflecting a decrease in
smoking, and is reaching a plateau for women in the majority of
European countries and in the United States, where lung cancer
mortality rates in women are approaching those in men (4). Lung cancer fatalities in women were
expected to increase (+7%) in the EU in 2012 (5). Non-small cell lung cancer (NSCLC)
accounts for 80–85% of all lung cancer cases (6). The management of NSCLC requires a
multidisciplinary approach. Patients generally require a
combination of surgery, radiotherapy and/or chemotherapy, depending
on the stage, resectability and overall performance status
(7). Chemotherapy is now
recognized as an important component in the treatment of all stages
of the disease, including in patients with completely resected,
early stage disease, who benefit with improved survival rates when
adjuvant platinum-based chemotherapy is administered (8). However, the majority of patients are
not diagnosed until the disease has spread beyond the primary tumor
site. Moreover, the occurrence rate is increasing annually
(9). Therefore, there is a high
unmet need for innovative approaches to prevent the occurrence and
migration of NSCLC.
CD44 is a 90-kDa transmembrane glycoprotein that is
widely distributed on the surface of T cells, granulocytes,
monocytes, fibroblasts, keratinocytes and epithelial cells, and is
involved in various cell adhesion events, including lymphocyte
migration, hematopoiesis and tumor metastasis (10). It is expressed on cell surfaces in
several isoforms, which are generated by alternative splicing of
mRNA (11). Expression of CD44 and
its variants has been shown to be associated with tumor progression
in various types of human malignancy (12). CD44 is a broadly distributed cell
surface protein hypothesized to mediate cell attachment to
extracellular matrix components or specific cell surface ligands
(13). The CD44 family of surface
receptors regulates adhesion, movement and activation of normal and
neoplastic cells (14). A previous
study demonstrated that the expression of the CD44 variant exon 6
is associated with lymph node metastasis in NSCLC (15). Furthermore, standard and variant
CD44 isoforms are commonly expressed in lung cancer of the
non-small cell type but not of the small cell type (16). In addition, CD44 stimulation is
known to downregulate Fas expression and Fas-mediated apoptosis of
lung cancer cells (17). CD44
standard and CD44 variant 6 levels in patients with NSCLC were not
significantly different from those in patients with benign lung
disease (18,19).
To the best of our knowledge, no studies have been
conducted regarding the significance of CD44 expression with
outcome and migration of NSCLC. In order to explore the
significance of CD44 expression in human NSCLC, the expression
pattern of CD44 was investigated using reverse
transcription-quantitative polymerase chain reaction (RT-qPCR) and
DNA sequencing. Followed by Gene Ontology (GO) term and Kyoto
Encyclopedia of Genes and Genomes (KEGG) pathway enrichment
analysis.
Materials and methods
Patients and samples
NSCLC tissue wax block samples of 36 cases were
obtained from the Department of Thoracic Surgery in the First
Hospital of Jilin University (Changchun, China) between January
2002 and July 2003. The samples were obtained from 28 men and 8
women, with a mean age of 63.5 years (range, 43–73 years). Normal
lung tissues (5 cm away from the cancer tissue) of 10 cases were
taken as negative controls. The present study was approved by the
First Hospital of Jilin University and written informed consent was
obtained from the patients.
RT-qPCR
RNA was extracted from tumor and normal tissues
using the Agilent Technologies Total RNA Isolation Mini kit
(Agilent Technologies, Palo Alto, CA, USA) according to the
manufacturer's protocol. Spectrophotometric methods were used to
assess the quality and quantity of the RNA samples. Reverse
transcription was performed using 1 μg of total RNA and the
Advantage RT-for-PCR kit (Takara Bio, Inc., Otsu, Japan) according
to the manufacturer's instructions. Conventional PCR primers were
designed to allow amplification of regions that have no overlap
with other known genes and span at least one intron. The following
primers were synthesized by BioAsia (Hyderabad, India): Forward:
5′-ACAACTGGTGATGGAGACTCATCC-3′ and reverse:
5′-GATTCCAGAGTGGCTTATCATCTTGG-3′ for CD44; and forward
5′-TGGAATCCTGTGGCATCCATGAAAC-3′ and reverse
5′-TAAAACGCAGCTCAGTAACAGTCCG-3′ for glyceraldehyde 2-phosphate
dehydrogenase (GAPDH). Conventional PCR was performed using cDNA
from tissues together with the Real-time qPCR Master mix-SYBR
Advantage qPCR Premix (Takara Bio, Inc.) using respective primers
in a PTC-100 thermocycler (Bio-Rad Laboratories, Inc., Hercules,
CA, USA). The reaction conditions were as follows: 95°C for 5 min,
94°C for 1 min, 55°C for 45 sec, 72°C for 30 sec and a final
extension phase at 72°C for 7 min for 40 cycles. The PCR products
were separated on a 2% agarose gel and stained with 5 μl
ethidium bromide prior to examination under UV light and images
were captured (GIS-700D digital imaging analysis system; Shanghai
Tianneng Electronics Co., Ltd., Shanghai, China). Fluorescence data
was collected at the extension step using a UV-254 ultraviolet
transmission and reflection analyzer (Beijing Dingguo Changsheng
Biotechnology, Co., Ltd., Beijing, China). The relative expression
of the target gene was determined using the 2−ΔΔCq
method (20). Optical density was
analyzed by Imagemaster VDS (Amersham Biosciences, Freiburg,
Germany) image analysis software. The CD44 mRNA expression level
was expressed as the CD44/β-actin optical density ratio.
CD44:β-actin >0.5 was considered to indicate positive
expression.
DNA sequencing
The DNA sequence represents a single format onto
which a broad range of biological phenomena can be projected for
high-throughput data collection (21). Tumor and normal tissues were
digested with appropriate volume of phosphate-buffered saline (PBS)
and proteinase K. Then DNA was extracted by saturated phenol and
the mixed liquor of phenol, chloroform and isoamylol. The extracted
DNA was precipitated with absolute ethyl alcohol, washed with 70%
ethanol and dissolved in cell lysis buffer after drying. Primers
were designed and synthesized by the method reported by Guldberg
et al (22) and the
sequences were as follows: 5′-ACAACTGGTGATGGAGACTCATCC-3′ and
5′-GATTCCAGAGTGGCTTATCATCTTGG-3′. PCR was conducted using an ABI
GeneAmp 2700 Mastercycler Personal (Applied Biosystems, Stockholm,
Sweden). The total volume of the PCR reaction system was 25
μl, containing 1 μl template DNA, 0.25 mmol/l dNTP,
0.25 μl Taq DNA polymerase, 0.8 μmol/l primer and 2.5
mmol/l Mg2+. The reaction conditions were as follows:
Initial denaturation was performed at 94°C for 5 min, 94°C for 30
sec, 52°C for 30 sec, 72°C for 60 sec for 40 cycles, and a final
extension phase at 72°C for 10 min. The PCR products of 10
μl were selected for electrophoresis on a 2% agarose gel.
Electrophoresis products were recycled and purified. DNA sequencing
was performed by the 310 style DNA sequencer (Applied Biosystems,
Foster City, CA, USA). The results were analyzed with DNASIS MAX V
3.0 software (MiraiBio; Hitachi Corporation, Tokyo, Japan).
Functional enrichment analysis and
pathway enrichment analysis
In order to facilitate the functional analysis of
the large lists of genes in the results, the Biological Networks
Gene Ontology tool (BiNGO, plugin of Cytoscape) for GO term
enrichment analysis was used (23). GO is a controlled vocabulary that
is structured as a directed acyclic graph, and describes genes and
their products (hereafter referred to simply as 'genes') in any
organism (23). Genes from a
number of organisms have been annotated to GO terms. A widespread
application is the identification of annotation-enriched GO terms
in a list of genes that share biological characteristics, the
so-called study set compared with a larger list of genes, the
population set. These terms are often interpreted as representing
the salient biological features of the genes in the study set
(24). The false discovery rate is
determined. It is a statistical approach used in multiple
hypothesis testing to correct for multiple comparisons. It is
typically used in highthroughput experiments in order to correct
for random events that falsely appear significant (25).
KEGG is a knowledge base for systematic analysis of
gene functions, linking genomic information with higher order
functional information (26). KEGG
pathway enrichment analysis was conducted with P<0.01 based on
Expression Analysis Systematic Explorer (EASE) test applied in The
Database for Annotation, Visualization and Integrated Discovery
(27). The EASE score was used to
detect the significant categories. The threshold of EASE score
<0.01 was considered significant for a category. The principle
of EASE is shown in the following equation:
Where n is the number of background genes; a′ is the
gene number of one gene set in the gene lists; a′+b is the number
of genes in the gene list including at least one gene set; a′+c is
the gene number of one gene list in the background genes; a′ is
replaced with a=a′−1.
Statistical analysis
Differences within groups in all assays were tested
by analysis of variance and Dunnett′s test. P<0.05 was
considered to indicate a statistically significant difference.
Statistical analysis was implemented by SPSS 19.0 software (SPSS
Inc., Chicago, IL, USA). All experiments were repeated three
times.
Results
RT-qPCR
RT-qPCR was used to determine CD44 mRNA expression
in NSCLC and the normal tissue. The electrophoresis results were
recorded by a gel scanning imaging system. Optical density values
were analyzed by Imagemaster VDS image analysis software. The CD44
mRNA expression level was determined by the CD44/β-actin optical
density ratio. CD44/β-actin >0.5 was considered to indicate
positive expression. CD44 expression occurred in AGAR condensed
electrophoresis after 10 cases of the normal tissues was amplified
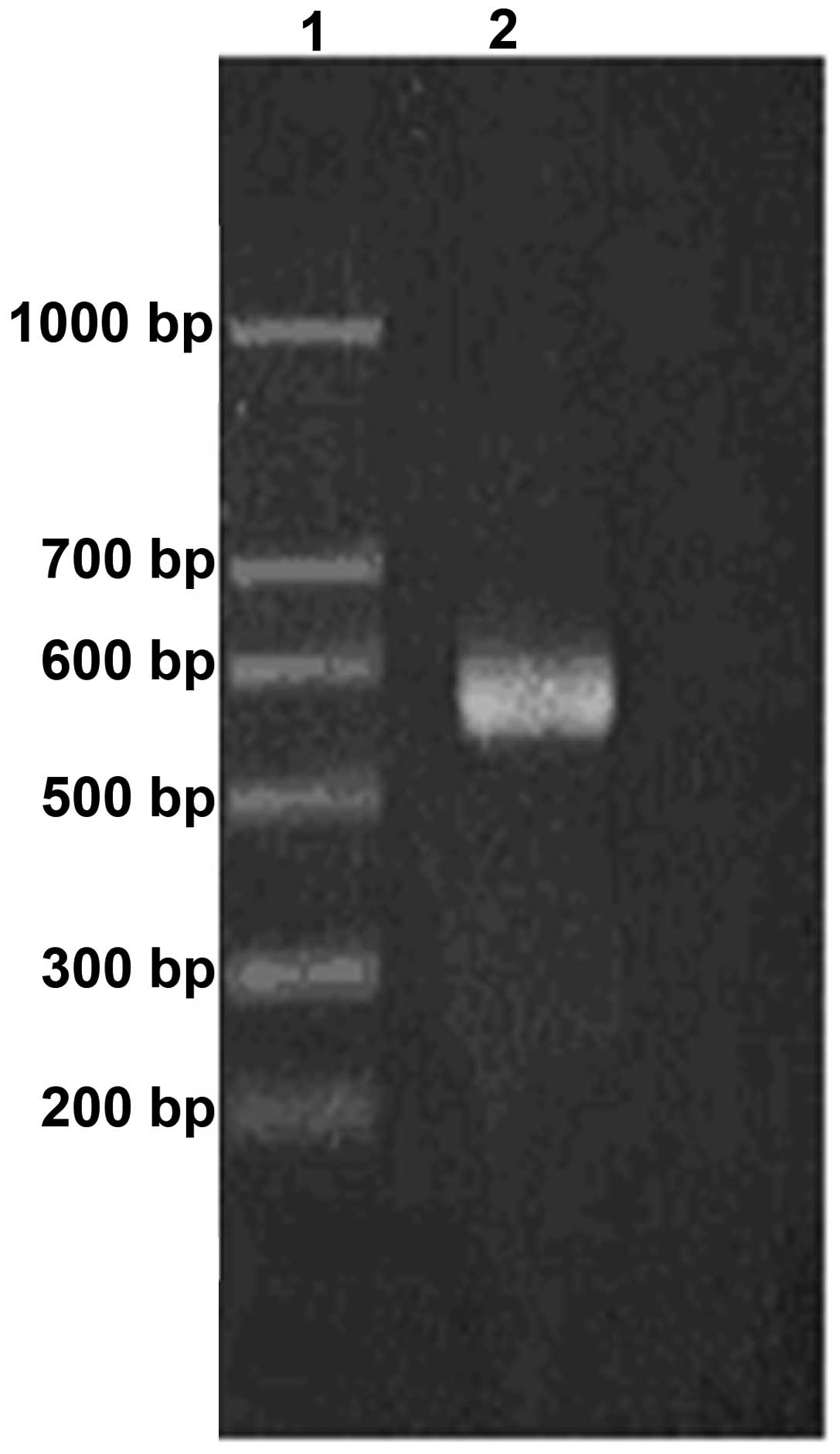
by RT-PCR and the result is shown in Fig. 1. Representative CD44 expression is
indicated in lane 2, compared to the DNA ladder. In the same
conditions, CD34 was overexpressed in 21 of the 36 cases NSCLC
tissues and the overexpression rate was 58.3% (Fig. 2).
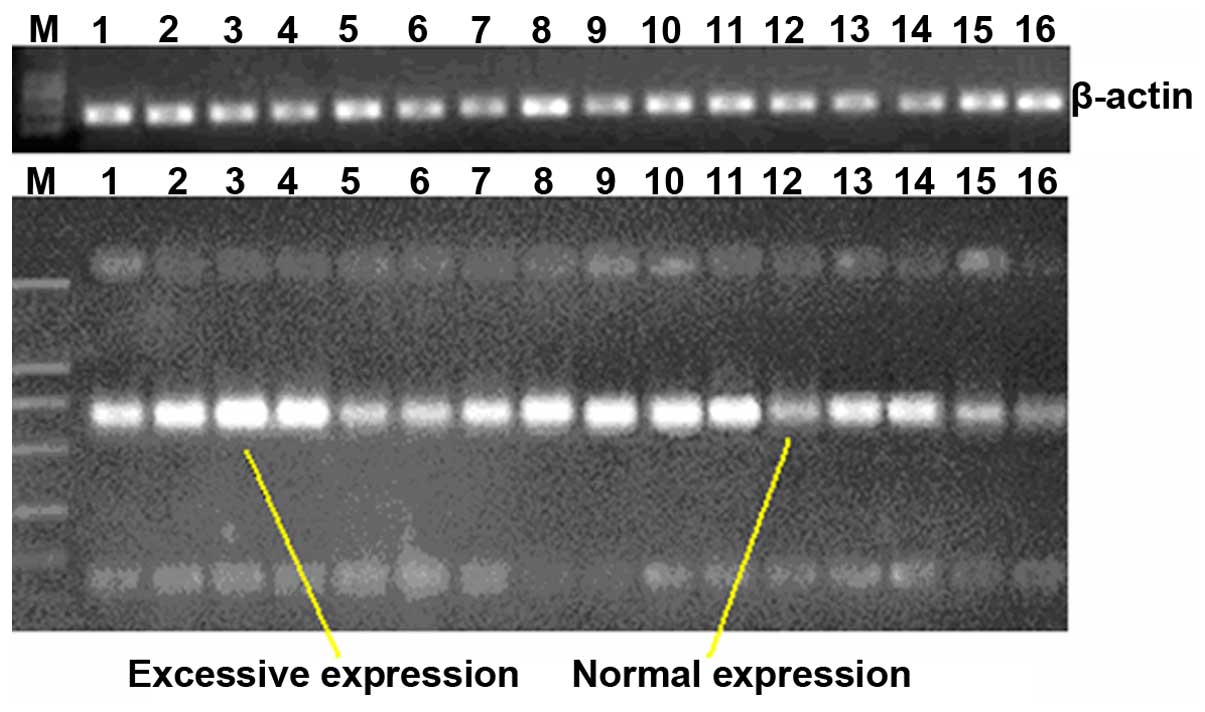 | Figure 2CD44 expression of the 36 cases of
NSCLC after amplification by reverse transcription-quantitative
polymerase chain reaction. M, marker; Lanes 1, 2, 3, 4, 7, 8, 9,
10, 11, 13 and 14, overexpression; and lanes 5, 6, 12, 15 and 16,
normal expression. |
DNA sequencing
In order to further demonstrate that CD34 was
overexpressed, DNA sequencing was used to analyze the CD34
expression conditions in NSCLC and normal tissues. The sequencing
results underwent bioinformatics analysis. In total, 1,375 genes
were found to be differentially expressed between the NSCLC and
normal tissues. Among them, 136 genes were downregulated and 1,239
genes were upregulated. CD44 and all related genes were used to
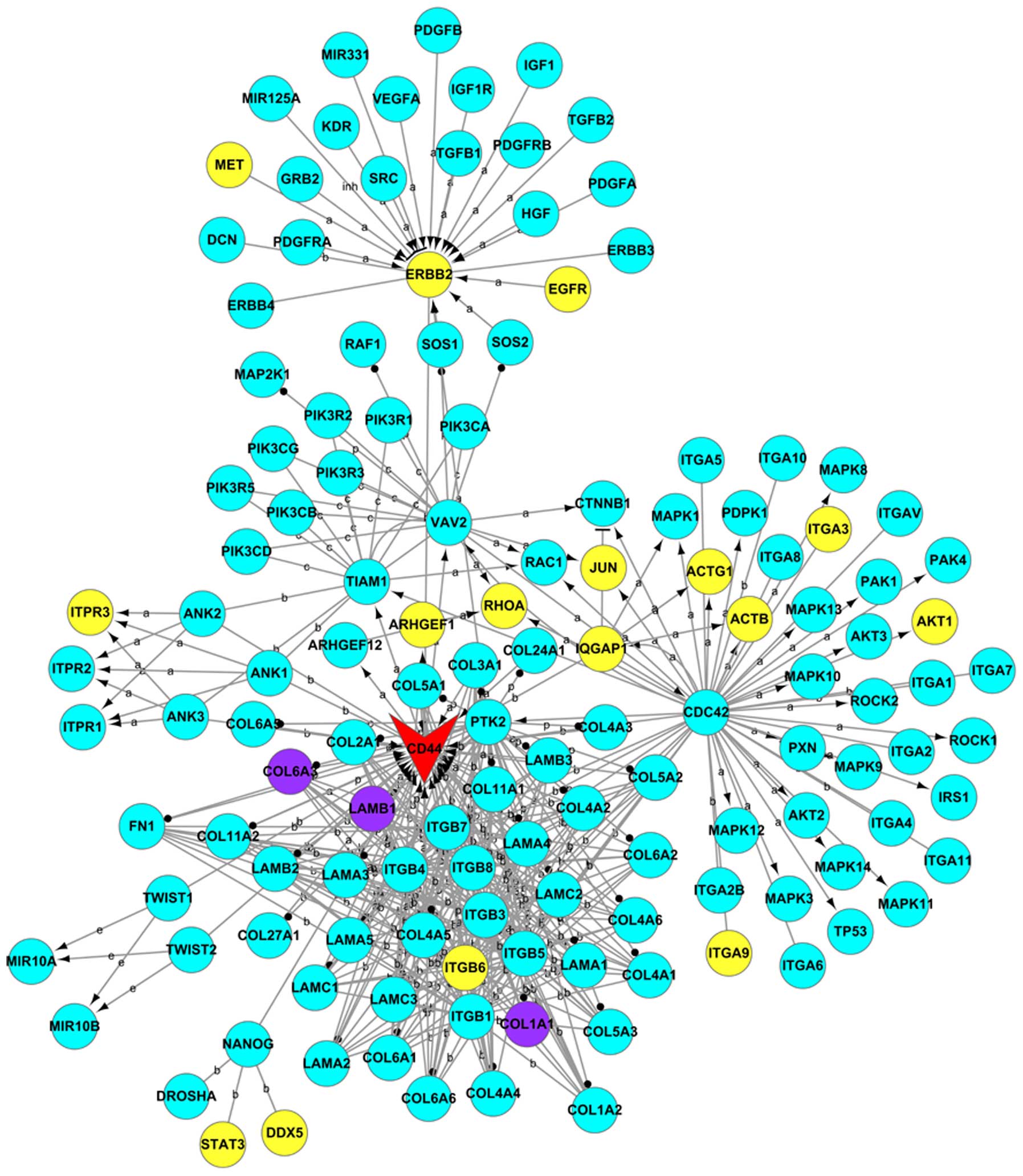
construct Gene_act_net (Fig. 3).
As shown, 20 genes were differentially expressed. Among them, 17
genes were upregulated (MET, ERBB2, EGFR, ITPR3, ARHGEF1, RHOA,
JUN, ACTG1, ITGA3, AKT1, ACTB, IQGAP1, ITGA9, ITGB6, STAT3, DDX5
and CD44) and three genes were downregulated (COL6A3, LAMB1 and
COL1A1).
GO enrichment analysis
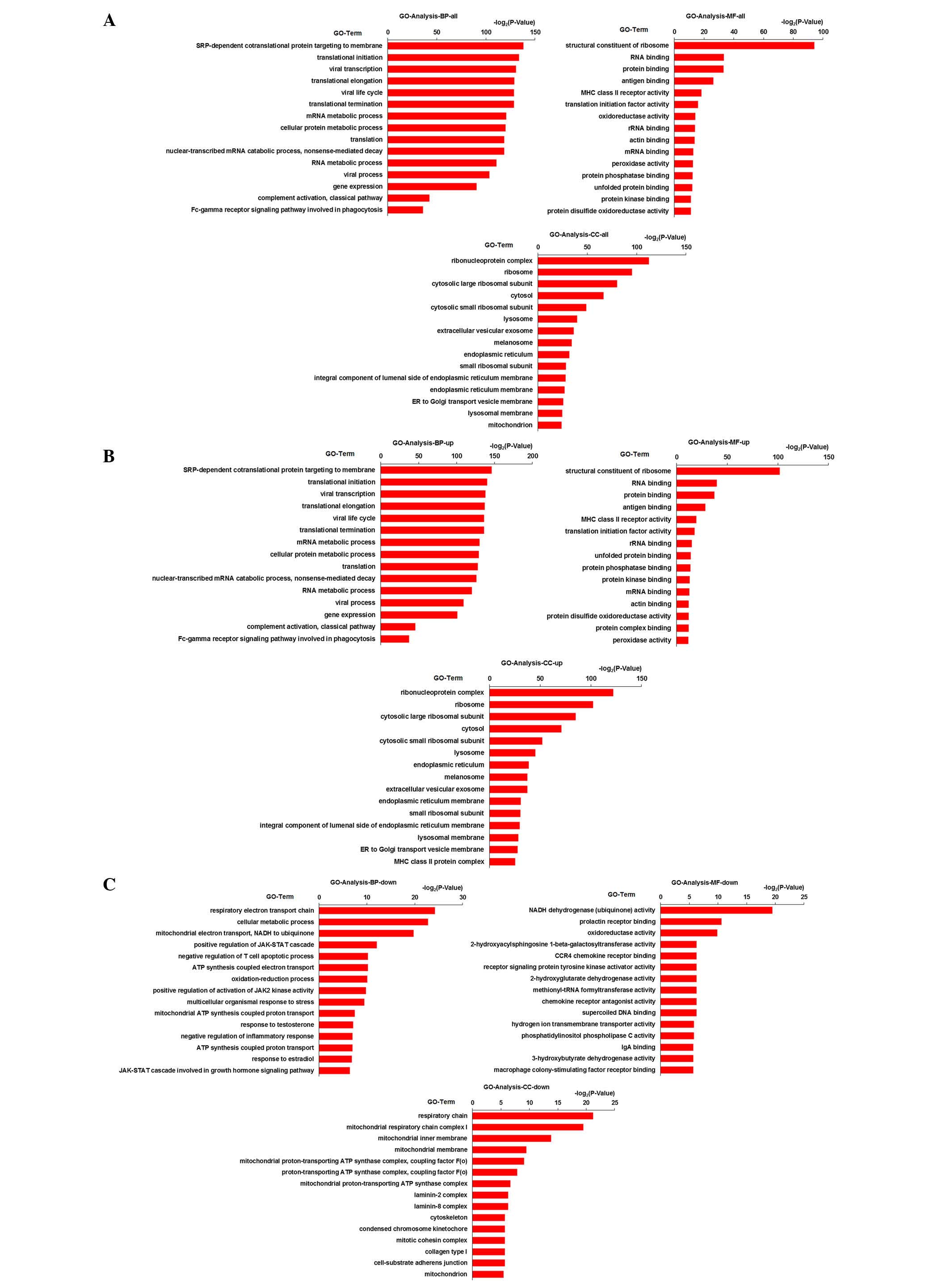
The results of GO enrichment analysis of the
differentially expressed genes are shown in Fig. 4. GO enrichment analysis of total
genes, upregulated genes and downregulated genes is shown in
Fig. 4A–C, respectively. From the
results, it was demonstrated that the following pathways in the
biological process GO term (P<0.05): Respiratory electron
transport chain (5.27E-08), cellular metabolic process (1.40E-07),
mitochondrial electron transport, NADH to ubiquinone (1.16E-06),
SRP-dependent cotranslational protein targeting to membrane
(2.64E-42), translational initiation (5.2E-41) and viral
transcription (5.08E-40). The cellular component GO term
(P<0.05) was associated with respiratory chain (4.11E-07),
mitochondrial respiratory chain complex I (1.39E-06), mitochondrial
inner membrane (6.94E-05), ribonucleoprotein complex (1.33E-34),
ribosome (1.99E-29) and cytosolic large ribosomal subunit
(6.86E-25). Molecular function GO term (P<0.05) was associated
with nicotinamide adenine dinucleotide (NADH) dehydrogenase
(ubiquinone) activity (1.35E-06), prolactin receptor binding
(0.0006), oxidoreductase activity (0.0011), structural constituent
of ribosome (4.75E-29), RNA binding (8.7E-11) and protein binding
(1.04E-10).
Pathway enrichment analysis
The EASE analysis method conducted in the present
study included a number of factors including the statistical
significance of the set of differentially expressed genes in the
pathway, the topology of the signaling pathway and their
interactions.
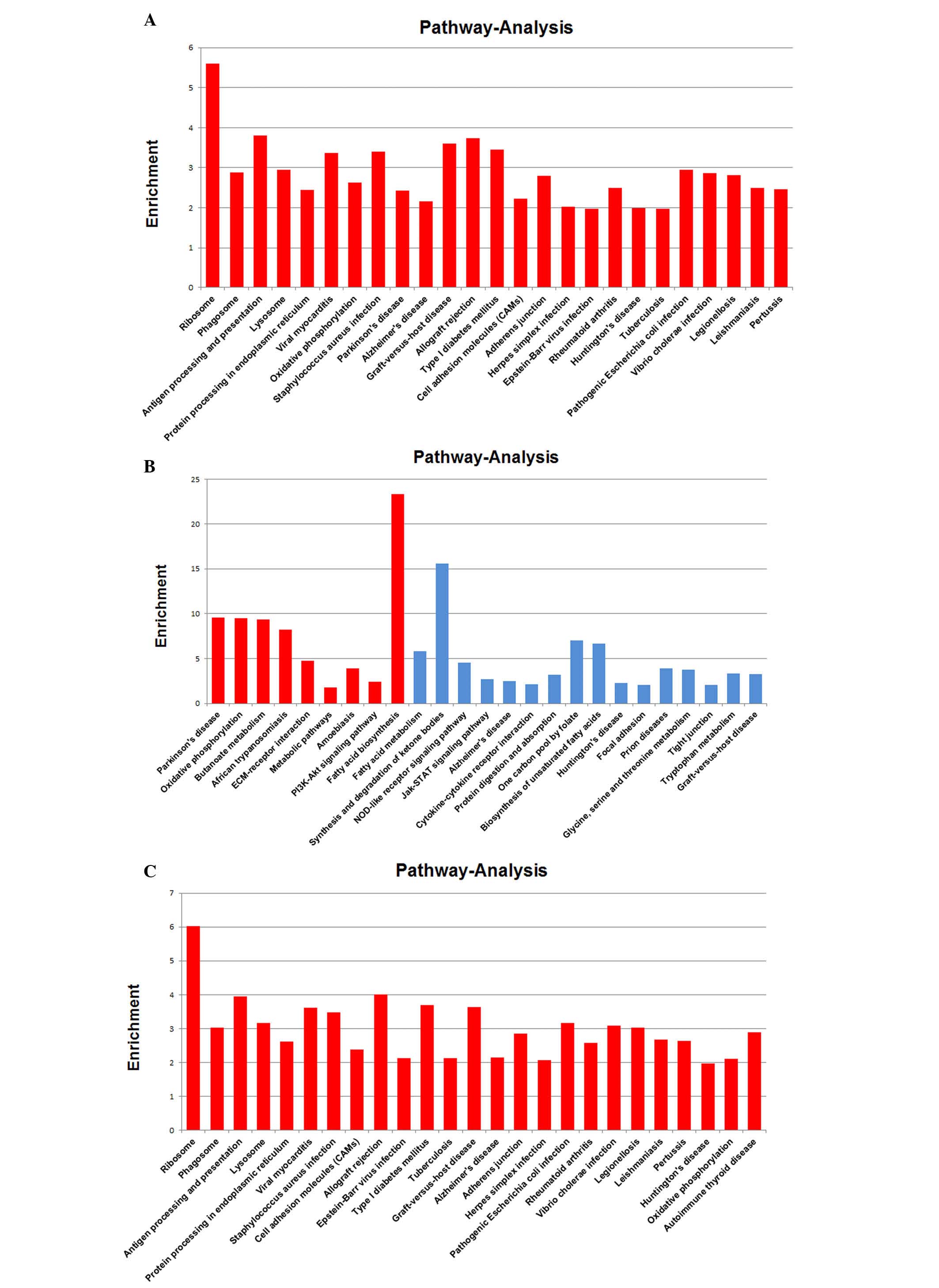
Pathway enrichment analysis of the differentially
expressed genes yielded a number of significant pathways, the
extracellular matrix-receptor interaction and hematopoietic cell
lineage pathways were most affected by overexpression of CD44 and,
thus, important in the development and migration of NSCLC. As shown
in Fig. 5, differentially
expressed genes CD44, SDC3, LAMB1, ITGA9, ITGA3, ITGB6, SDC4,
COL1A1, HSPG2, SDC1 and COL6A3 were involved in the ECM-receptor
interaction pathway. Differentially expressed genes CD44, CD9,
HLA-DRB5, CSF1R, IL1R1, HLA-DRA, ITGA3, CD14, CD55, CD4, CD59 and
HLA-DRB1 were involved in the hematopoietic cell lineage
pathway.
Discussion
Lung cancer is the leading cause of cancer-related
mortality worldwide and its prevalence and mortality is increasing.
NSCLC accounts for 80–85% of lung cancers. Chemotherapy is now
recognized as an important component of treatment for all stages of
NSCLC. However, the majority of patients are not diagnosed until
the disease has spread beyond the primary tumor site. Moreover, the
occurrence rate is increasing annually. Therefore, greater
understanding of NSCLC at the molecular level is required. Defining
new molecular targets may aid the development of more effective
therapeutic strategies. In the present study, RT-qPCR was used to
investigate the expression level of CD44 mRNA and the result showed
that CD44 mRNA was overexpressed. To further prove the
overexpression of CD44, DNA sequencing was used to analyze the
differentially expressed genes between NSCLC tissues and normal
tissues. In total, 1,375 genes were found to be differentially
expressed. Among them, 136 genes were downregulated and 1,239
genes, including CD44, were upregulated. CD44 and all the related
genes were used to construct Gene_act_net. Among them, 17 genes
were upregulated and 3 were downregulated. The 17 upregulated genes
were MET, ERBB2, EGFR, ITPR3, ARHGEF1, RHOA, JUN, ACTG1, ITGA3,
AKT1, ACTB, IQGAP1, ITGA9, ITGB6, STAT3, DDX5 and CD44. The 3
downregulated genes were: COL6A3, LAMB1 and COL1A1.
CD44 is a principle hyaluronate receptor and a
transmembrane glycoprotein (28).
It exists in a standard form (CD44s), and in multiple isoforms
(29). It has been described as
important in various aspects of cancer progression, including cell
growth control, adhesion, migration and invasion (30,31).
A number of primary carcinoma tissues have been shown to express
high levels of CD44 (32). Its
expression was important for NSCLC COX-2-dependent invasion
(33). Miyoshi et al
(15) reported that in NSCLC, a
number of variant forms of CD44 are frequently expressed and that
the expression of CD44v6 is particularly associated with lymph node
metastasis in NSCLC. Yasuda et al (17) found that CD44 was highly expressed
on the surface of lung cancer cells and it stimulated the
downregulation of Fas expression and Fas-mediated apoptosis of lung
cancer cells. Therefore, it was concluded that CD44 was associated
with the occurrence and migration of NSCLC.
MET amplification can activate ERBB3/PI3K/AKT
signaling in EGFR mutant lung cancers and causes resistance to EGFR
kinase inhibitors (34). MET
amplification in NSCLC is likely a primary oncogenic driver and is
a valid clinical target (35). MET
has a prognostic role in surgically resected NSCLC, gene copy
number increased by MET is an independent negative prognostic
factor in surgically resected NSCLC (36). ERBB2 is a 185-Da transmembrane
protein (p185 HER2) tyrosine kinase (37). Mutations in the ERBB2 gene were
recently found in ~2% of primary NSCLC specimens (38). Overexpression of the ERBB2 protein
is observed in a variety of malignancies, including NSCLC (39). EGFR is critical in the control of
cellular proliferation, differentiation and survival (40). Mutations of the EGFR gene have been
identified in specimens from patients with non-small cell lung
cancer respond to anilinoquinazoline EGFR inhibitors (41). EGF is a promising target for
anticancer therapy as it is expressed or highly expressed in a
variety of tumors, including NSCLC (42). AKT1, a downstream mediator of
phosphatidylinositol 3-kinase (PI3K), is a signal transduction
protein that is central in tumorigenesis (43). It has been implicated in lung
tumorigenesis and lung cancer drug resistance, and can be activated
in NSCLC (44,45). IQGAP1 is a scaffold protein whose
function is associated with signal transduction, cell adhesion,
local invasion and distant metastasis of cancer cells (46). Downregulation of IQGAP1 can
decrease proliferation, migration and invasion potential in NSCLC
metastasis (47). Activation of
STAT3 is important in tumorigenesis and tumor progression (48). The overexpression of STAT3 has been
found in various malignancies, including NSCLC (49). Thus, studies have indicated that
MET, ERBB2, EGFR, AKT1, IQGAP1, STAT3 were associated with the
occurrence and migration of NSCLC.
The KEGG pathway of the differentially expressed
genes was investigated. ECM-receptor interaction and hematopoietic
cell lineage pathways were affected by overexpressed CD44. The
ECM-receptor interaction pathway involved CD44, SDC3, LAMB1, ITGA9,
ITGA3, ITGB6, SDC4, COL1A1, HSPG2, SDC1 and COL6A3. ECM-receptor
interactions have a profound influence on major cellular programs
including growth, differentiation, migration and survival (50). Of the 11 genes, CD44, ITGA9, ITGA3
and COL1A1 were associated with NSCLC occurrence. ITGA9 was found
to be downregulated in NSCLC and strongly inhibited colony
formation in renal and lung cancer cell lines (51,52).
Fan et al (53) performed
analysis of expression of integrins in three different lung cancer
cell lines, including A549 adenocarcinoma cells, H1650
bronchioalveolar carcinoma cells and DMS53 small cell carcinoma
cells. Upregulated expression of ITGA4 was detectable in H1650
cells and ITGA3 was observed in all the lung cancer cells (53). High expression of COL1A1 was found
in NSCLC (54). In addition, loss
of expression of SDC1 is known to be associated with biologic
aggressiveness and poor outcome for patients with NSCLC (55). The soluble SDC1 is associated with
shorter survival in chronic lymphocytic leukemia and lung cancer
(56). Overexpression of this
enzyme appears to be crucial to promote tumor growth and metastasis
in a number of types of cancer, including lung cancer (57). CD44, CD9, HLA-DRB5, CSF1R, IL1R1,
HLA-DRA, ITGA3, CD14, CD55, CD4, CD59 and HLA-DRB1 were shown to be
involved in the hematopoietic cell lineage pathway. CD44 was
reported to be important in the occurrence and migration of NSCLC.
CD9 is involved in Schwann cell migration in vitro (58) and is also downregulated in
metastatic tumors (59). ITGA3 is
an important integrin, and integrin α3/β1 was found to be expressed
in 82% of metastatic tumors (60).
It was reported to exhibit a role in MYC-induced liver cancer
(61). CD14 is involved in
hemorrhagic shock-induced alterations of the monocyte tumor
necrosis factor response to endotoxin (62). CD55 is overexpressed in certain
tumor cell lines, and in colorectal carcinomas, it has been shown
to be an indicator of a poor prognosis (63). Therefore, it was concluded that
ECM-receptor interaction and hematopoietic cell lineage pathway are
key in the occurrence and migration of tumor.
Above all, it was concluded that CD44 was
overexpressed in NSCLC and the overexpression was associated with
the occurrence and migration of NSCLC. This may aid in the
development of novel treatment strategies for NSCLC in the
clinic.
Acknowledgments
The current study was supported by the Science and
Technology Development Plan of Jilin Province (grant nos. 20110717
and 20140520019JH), the Key Scientific and Technological Projects
of Jilin Province (grant no. 20150204026YY), the International
Science and Technology Cooperation Project of Jilin Province (grant
no. 20160414047GH), the Strategic Adjustment of Economic Structure
Guiding Funds of Jilin Province (grant no. 2015Y032) and the
Education Department of Jilin Province (grant no. 449).
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Yang P, Allen MS, Aubry MC, Wampfler JA,
Marks RS, Edell ES, Thibodeau S, Adjei AA, Jett J and Deschamps C:
Clinical features of 5,628 primary lung cancer patients: Experience
at Mayo clinic from 1997 to 2003. Chest. 128:452–462. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Pope CA III, Burnett RT, Thun MJ, Calle
EE, Krewski D, Ito K and Thurston GD: Lung cancer, cardiopulmonary
mortality, and long-term exposure to fine particulate air
pollution. JAMA. 287:1132–1141. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Thatcher N, Chang A, Parikh P, Rodrigues
Pereira J, Ciuleanu T, von Pawel J, Thongprasert S, Tan EH,
Pemberton K, Archer V and Carroll K: Gefitinib plus best supportive
care in previously treated patients with refractory advanced
non-small-cell lung cancer: Results from a randomised,
placebo-controlled, multicentre study (Iressa Survival Evaluation
in Lung Cancer). Lancet. 366:1527–1537. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Malvezzi M, Bertuccio P, Levi F, La Vecchi
C and Negri E: European cancer mortality predictions for the year
2012. Ann Oncol. mds024. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Graham MV, Purdy JA, Emami B, Harms W,
Bosch W, Lockett MA and Perez CA: Clinical dose-volume histogram
analysis for pneumonitis after 3D treatment for non-small cell lung
cancer (NSCLC). Int J Radiat Oncol Biol Phys. 45:323–329. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Goldstraw P, Ball D, Jett JR, et al:
Non-small-cell lung cancer. The Lancet. 378:1727–1740. 2011.
View Article : Google Scholar
|
|
8
|
Früh M, Rolland E, Pignon JP, Seymour L,
Ding K, Tribodet H, Winton T, Le Chevalier T, Scagliotti GV,
Douillard JY, et al: Pooled analysis of the effect of age on
adjuvant cisplatin-based chemotherapy for completely resected
non-small-cell lung cancer. J Clin Oncol. 26:3573–3581. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Shepherd F, Pereira J, Ciuleanu T, et al:
A randomized placebo-controlled trial of erlotinib in patients with
advanced non-small cell lung cancer (NSCLC) following failure of
1st line or 2nd line chemotherapy. A National Cancer Institute of
Canada Clinical Trials Group (NCIC CTG) trial. ASCO Annual Meeting
Proceedings. pp. 70222004
|
|
10
|
Yasuda M, Nakano K, Yasumoto K and Tanaka
Y: CD44: Functional relevance to inflammation and malignancy.
Histol Histopathol. 17:945–950. 2002.PubMed/NCBI
|
|
11
|
Mizera-Nyczak E, Dyszkiewicz W, Heider KH
and Zeromski J: Isoform expression of CD44 adhesion molecules,
Bcl-2, p53 and Ki-67 proteins in lung cancer. Tumour Biol.
22:45–53. 2001. View Article : Google Scholar
|
|
12
|
Situ D, Long H, Lin P, Zhu Z, Wang J,
Zhang X, Xie Z and Rong T: Expression and prognostic relevance of
CD44v6 in stage I non-small cell lung carcinoma. J Cancer Res Clin
Oncol. 136:1213–1219. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Aruffo A, Stamenkovic I, Melnick M,
Underhill CB and Seed B: CD44 is the principal cell surface
receptor for hyaluronate. cell. 61:1303–1313. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Weber GF, Ashkar S, Glimcher MJ and Cantor
H: Receptor-ligand interaction between CD44 and osteopontin
(Eta-1). Science. 271:509–512. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Miyoshi T, Kondo K, Hino N, Uyama T and
Monden Y: The expression of the CD44 variant exon 6 is associated
with lymph node metastasis in non-small cell lung cancer. Clin
Cancer Res. 3:1289–1297. 1997.PubMed/NCBI
|
|
16
|
Ariza A, Mate JL, Isamat M, López D, Von
Uexküll-Güldeband C, Rosell R, Fernández-Vasalo A and
Navas-Palacios JJ: Standard and variant CD44 isoforms are commonly
expressed in lung cancer of the non-small cell type but not of the
small cell type. J Pathol. 177:363–368. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yasuda M, Tanaka Y, Fujii K and Yasumoto
K: CD44 stimulation down-regulates Fas expression and Fas-mediated
apoptosis of lung cancer cells. Int Immunol. 13:1309–1319. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Takigawa N, Segawa Y, Mandai K, Takata I
and Fujimoto N: Serum CD44 levels in patients with non-small cell
lung cancer and their relationship with clinicopathological
features. Lung Cancer. 18:147–157. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Szpechcinski A, Rudzinski P, Kupis W,
Langfort R, Orlowski T and Chorostowska-Wynimko J: Plasma cell-free
DNA levels and integrity in patients with chest radiological
findings: NSCLC versus benign lung nodules. Cancer Lett.
374:202–207. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2 (−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
21
|
Shendure J and Ji H: Next-generation DNA
sequencing. Nat Biotechnol. 26:1135–1145. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Guldberg P, Romano V, Ceratto N, Bosco P,
Ciuna M, Indelicato A, Mollica F, Meli C, Giovannini M and Riva E:
Mutational spectrum of phenylalanine hydroxylase deficiency in
sicily: Implications for diagnosis of hyperphenylalaninemia in
southern Europe. Hum Mol Genet. 2:1703–1707. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al: Gene ontology: Tool for the unification of biology. The Gene
Ontology Consortium. Nat Genet. 25:25–29. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Bauer S, Grossmann S, Vingron M and
Robinson PN: Ontologizer 2.0-a multifunctional tool for GO term
enrichment analysis and data exploration. Bioinformatics.
24:1650–1651. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang J: Functional Enrichment Analysis.
Encyclopedia of Systems Biology. Springer; pp. 772. 2013,
View Article : Google Scholar
|
|
26
|
Kanehisa M and Goto S: KEGG: Kyoto
encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30.
2000. View Article : Google Scholar
|
|
27
|
Hosack DA, Dennis G Jr, Sherman BT, Lane
HC and Lempicki RA: Identifying biological themes within lists of
genes with EASE. Genome Biol. 4:R702003. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Banerji S, Ni J, Wang SX, Clasper S, Su J,
Tammi R, Jones M and Jackson DG: LYVE-1, a new homologue of the
CD44 glycoprotein, is a lymph-specific receptor for hyaluronan. J
Cell Biol. 144:789–801. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
De la Torre M, Heldin P and Bergh J:
Expression of the CD44 glycoprotein (lymphocyte-homing receptor) in
untreated human breast cancer and its relationship to prognostic
markers. Anticancer Res. 15:2791–2795. 1995.PubMed/NCBI
|
|
30
|
Kito H, Suzuki H, Ichikawa T, Sekita N,
Kamiya N, Akakura K, Igarashi T, Nakayama T, Watanabe M, Harigaya K
and Ito H: Hypermethylation of the CD44 gene is associated with
progression and metastasis of human prostate cancer. Prostate.
49:110–115. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Pinheiro C, Reis RM, Ricardo S,
Longatto-Filho A, Schmitt F and Baltazar F: Expression of
monocarboxylate transporters 1, 2, and 4 in human tumours and their
association with CD147 and CD44. J Biomed Biotechnol. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Stamenkovic I, Amiot M, Pesando JM and
Seed B: A lymphocyte molecule implicated in lymph node homing is a
member of the cartilage link protein family. Cell. 56:1057–1062.
1989. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Dohadwala M, Batra RK, Luo J, Lin Y,
Krysan K, Pold M, Sharma S and Dubinett SM: Autocrine/paracrine
prostaglandin E2 production by non-small cell lung cancer cells
regulates matrix metalloproteinase-2 and CD44 in
cyclooxygenase-2-dependent invasion. J Biol Chem. 277:50828–50833.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Turke AB, Zejnullahu K, Wu YL, Song Y,
Dias-Santagata D, Lifshits E, Toschi L, Rogers A, Mok T, Sequist L,
et al: Preexistence and clonal selection of MET amplification in
EGFR mutant NSCLC. Cancer cell. 17:77–88. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ou SH, Kwak EL, Siwak Tapp C, Dy J,
Bergethon K, Clark JW, Camidge DR, Solomon BJ, Maki RG, Bang YJ, et
al: Activity of crizotinib (PF02341066), a dual
mesenchymal-epithelial transition (MET) and anaplastic lymphoma
kinase (ALK) inhibitor, in a non-small cell lung cancer patient
with de novo MET amplification. J Thorac Oncol. 6:942–946. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Cappuzzo F, Marchetti A, Skokan M, Rossi
E, Gajapathy S, Felicioni L, Del Grammastro M, Sciarrotta MG,
Buttitta F, Incarbone M, et al: Increased MET gene copy number
negatively affects survival of surgically resected non-small-cell
lung cancer patients. J Clin Oncol. 27:1667–1674. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Nakamura H, Saji H, Ogata A, Hosaka M,
Hagiwara M, Kawasaki N and Kato H: Correlation between encoded
protein overexpression and copy number of the HER2 gene with
survival in non-small cell lung cancer. Int J Cancer. 103:61–66.
2003. View Article : Google Scholar
|
|
38
|
Minami Y, Shimamura T, Shah K, LaFramboise
T, Glatt KA, Liniker E, Borgman CL, Haringsma HJ, Feng W, Weir BA,
et al: The major lung cancer-derived mutants of ERBB2 are oncogenic
and are associated with sensitivity to the irreversible EGFR/ERBB2
inhibitor HKI-272. Oncogene. 26:5023–5027. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Kristiansen G, Yu Y, Petersen S, Kaufmann
O, Schlüns K, Dietel M and Petersen I: Overexpression of c-erbB2
protein correlates with disease-stage and chromosomal gain at the
c-erbB2 locus in non-small cell lung cancer. Eur J Cancer.
37:1089–1095. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Cragg MS, Kuroda J, Puthalakath H, Huang
DC and Strasser A: Gefitinib-induced killing of NSCLC cell lines
expressing mutant EGFR requires BIM and can be enhanced by BH3
mimetics. PLoS Med. 4:1681–1689. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Kobayashi S, Boggon TJ, Dayaram T, Jänne
PA, Kocher O, Meyerson M, Johnson BE, Eck MJ, Tenen DG and Halmos
B: EGFR mutation and resistance of non-small-cell lung cancer to
gefitinib. N Engl J Med. 352:786–792. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Fukuoka M, Yano S, Giaccone G, Tamura T,
Nakagawa K, Douillard JY, Nishiwaki Y, Vansteenkiste J, Kudoh S,
Rischin D, et al: Multi-institutional randomized phase II trial of
gefitinib for previously treated patients with advanced
non-small-cell lung cancer (The IDEAL 1 Trial). J Clin Oncol.
21:2237–2246. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Tang JM, He QY, Guo RX and Chang XJ:
Phosphorylated Akt overexpression and loss of PTEN expression in
non-small cell lung cancer confers poor prognosis. Lung Cancer.
51:181–191. 2006. View Article : Google Scholar
|
|
44
|
Tsurutani J, Fukuoka J, Tsurutani H, Shih
JH, Hewitt SM, Travis WD, Jen J and Dennis PA: Evaluation of two
phosphorylation sites improves the prognostic significance of Akt
activation in non-small-cell lung cancer tumors. J Clin Oncol.
24:306–314. 2006. View Article : Google Scholar
|
|
45
|
Tsao AS, McDonnell T, Lam S, Putnam JB,
Bekele N, Hong WK and Kurie JM: Increased phospho-AKT (Ser473)
expression in bronchial dysplasia: Implications for lung cancer
prevention studies. Cancer Epidemiol Biomarkers Prev. 12:660–664.
2003.PubMed/NCBI
|
|
46
|
Nakamura H, Fujita K, Nakagawa H, Kishi F,
Takeuchi A, Aute I and Kato H: Expression pattern of the scaffold
protein IQGAP1 in lung cancer. Oncol Rep. 13:427–431.
2005.PubMed/NCBI
|
|
47
|
Lele L, Hailin P, Pei Q, et al: The
effect of down regulation of IQGAP1 on the biological behavior of
non-small cell lung cancer PC14/B
|
|
48
|
Gao J, McConnell MJ, Yu B, Li J, Balko JM,
Black EP, Johnson JO, Lloyd MC, Altiok S and Haura EB: MUC1 is a
downstream target of STAT3 and regulates lung cancer cell survival
and invasion. Int J Oncol. 35:337–345. 2009.PubMed/NCBI
|
|
49
|
Yin ZJ, Jin FG, Liu TG, Fu EQ, Xie YH and
Sun RL: Overexpression of STAT3 potentiates growth, survival and
radioresistance of non-small-cell lung cancer (NSCLC) cells. J Surg
Res. 171:675–683. 2011. View Article : Google Scholar
|
|
50
|
Jones FS and Jones PL: The tenascin family
of ECM glycoproteins: Structure, function and regulation during
embryonic development and tissue remodeling. Dev Dyn. 218:235–259.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Dmitriev AA, Kashuba VI, Haraldson K,
Senchenko VN, Pavlova TV, Kudryavtseva AV, Anedchenko EA, Krasnov
GS, Pronina IV, Loginov VI, et al: Genetic and epigenetic analysis
of non-small cell lung cancer with NotI-microarrays. Epigenetics.
7:502–513. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Anedchenko EA, Dmitriev AA, Krasnov GS,
Kondrat'eva TT, Kopantsev EP, Vinogradova TV, Zinov'eva MV,
Zborovskaia IB, Polotskiĭ BE, Sakharova OV, et al: Downregulation
of RBSP3/CTDSPL, NPRL2/G21, RASSF1A, ITGA9, HYAL1 and HYAL2 genes
in non-small cell lung cancer. Mol Biol (Mosk). 42:965–976. 2007.In
Russian.
|
|
53
|
Fan Z, LinLang G and XiangBin M: Analysis
of integrins differential expression in lung cancer cells. Cancer
Research on Prevention and Treatment. 36:734–736. 2009.
|
|
54
|
Hofmann HS, Bartling B, Simm A, Murray R,
Aziz N, Hansen G, Silber RE and Burdach S: Identification and
classification of differentially expressed genes in non-small cell
lung cancer by expression profiling on a global human
59.620-element oligonucleotide array. Oncol Rep. 16:587–595.
2006.PubMed/NCBI
|
|
55
|
Lu Y, Govindan R, Wang L, Liu PY, Goodgame
B, Wen W, Sezhiyan A, Pfeifer J, Li YF, Hua X, et al: MicroRNA
profiling and prediction of recurrence/relapse-free survival in
stage I lung cancer. Carcinogenesis. 33:1046–1054. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Kim YI, Lee A, Lee BH and Kim SY:
Prognostic significance of syndecan-1 expression in cervical
cancers. J Gynecol Oncol. 22:161–167. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Januchowski R, Zawierucha P, Ruciński M,
Nowicki M and Zabel M: Extracellular matrix proteins expression
profiling in chemoresistant variants of the A2780 ovarian cancer
cell line. Biomed Res Int. 3658672014.PubMed/NCBI
|
|
58
|
Anton ES, Hadjiargyrou M, Patterson PH and
Matthew WD: CD9 plays a role in Schwann cell migration in vitro. J
Neurosci. 15:584–595. 1995.PubMed/NCBI
|
|
59
|
Ono M, Handa K, Withers DA and Hakomori S:
Motility inhibition and apoptosis are induced by
metastasis-suppressing gene product CD82 and its analogue CD9, with
concurrent glycosylation. Cancer Res. 59:2335–2339. 1999.PubMed/NCBI
|
|
60
|
Zhu R, Xu R, Jiang X, Cai Y, Zou Y, Du M
and Qin L: Expression profile of cancer-related genes in human
adult bone marrow-derived neural stemlike cells highlights the need
for tumorigenicity study. J Neurosci Res. 85:3064–3070. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Aravalli RN, Talbot NC and Steer CJ: Gene
expression profiling of MYC-driven tumor signatures in porcine
liver stem cells by transcriptome sequencing. World J
Gastroenterol. 21:2011–2029. 2015.PubMed/NCBI
|
|
62
|
Nwariaku F, Sikes P, Lightfoot E, McIntyre
K and Mileski WJ: Role of CD14 in hemorrhagic shock-induced
alterations of the monocyte tumor necrosis factor response to
endotoxin. J Trauma. 40:564–567. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Madjd Z, Durrant LG, Bradley R, Spendlove
I, Ellis IO and Pinder SE: Loss of CD55 is associated with
aggressive breast tumors. Clin Cancer Res. 10:2797–2803. 2004.
View Article : Google Scholar : PubMed/NCBI
|