Introduction
Acanthopanax henryi (Oliv.)
Harms belongs to the Araliaceae family, and may be
used as a traditional Oriental medicine for the treatment of
rheumatism and inflammation (1,2).
Phytochemical studies have identified lignans and other compounds
in the bark of Acanthopanax henryi (Oliv.) Harms, including
syringin, syringaresinol, diglucoside, octacosanoic acid and
beta-sitosterol (3).
Pharmacological studies have reported that the MeOH extract and
fraction of the root bark of Acanthopanax henryi exerts
anti-inflammatory and anticancer effects (3–5). At
present, several studies have been conducted regarding the
pharmacological effects of Acanthopanax henryi root bark;
however, studies on the leaves of Acanthopanax henryi
(Oliv.) Harms are few (5,6). Therefore, the present study aimed to
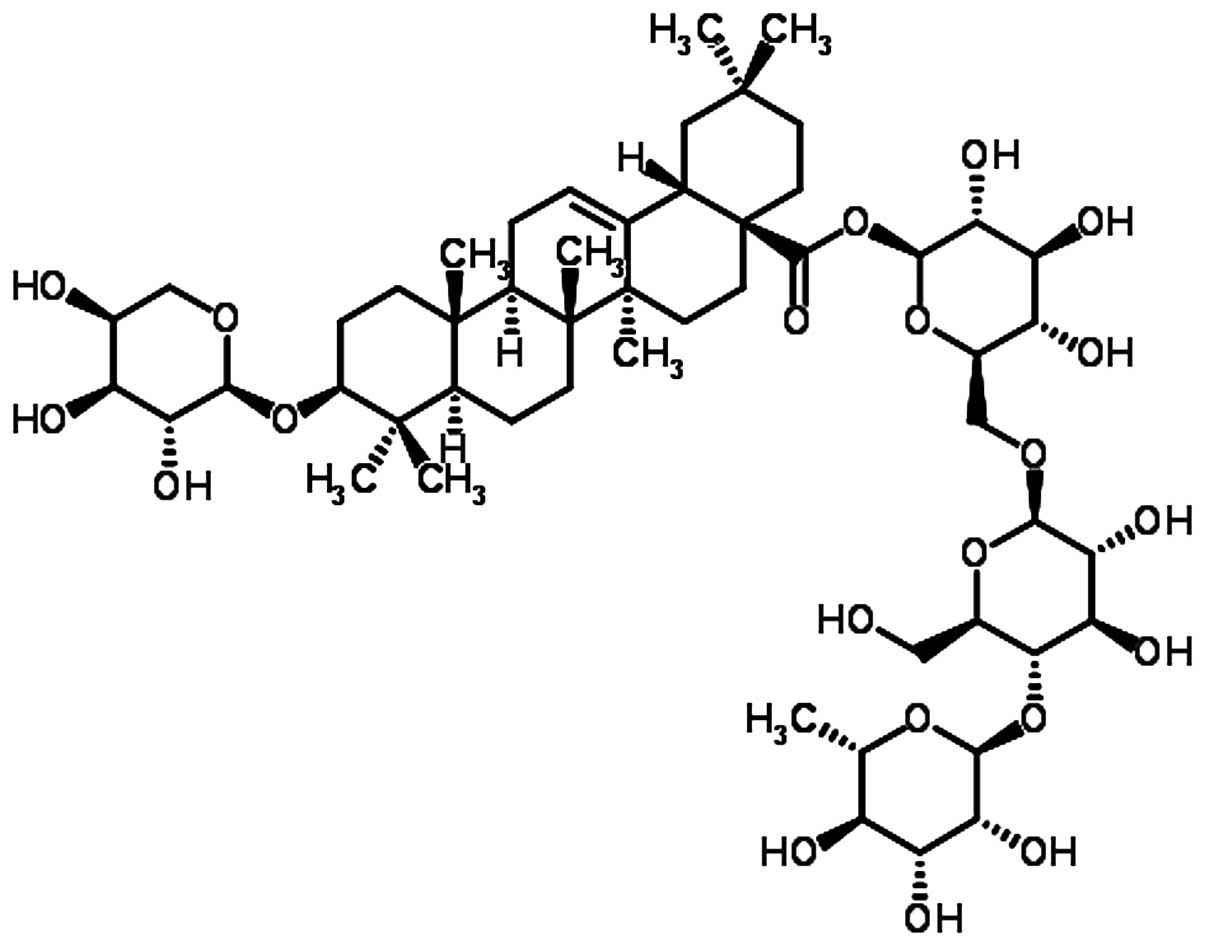
investigate the anti-inflammatory effects of Ciwujianoside C3 (CJS
C3); full name, echinocystic acid
3-O-β-D-glucopyranosyl-(1→3)-O-α-L-arabinopyranoside, isolated from
the leaves of Acanthopanax henryi (Oliv.) Harms (Fig. 1).
Inflammation is a physiological response, and
asthma, obesity and diabetes are common inflammatory diseases
(7). Inflammatory responses
induced by microbial infections stimulate the innate immune system
against foreign components, including lipopolysaccharide (LPS)
(8), which is a cell wall
component of gram-negative bacteria that is detected by Toll-like
receptor 4 (TLR4). Macrophages are able to bind to LPS to induce
the activation of inflammatory signals and the release of
proinflammatory cytokines, chemokines and mediators of the
inflammatory response (9).
Nitric oxide (NO), also known as nitrogen monoxide,
which is synthesized from L-arginine by nitric oxide synthase
(NOS), regulates several physiological functions (10). The NO free radical produced by the
inducible NOS (iNOS) isoform is an essential component of host
innate immunity and the inflammatory response to various pathogens
(11).
The biosynthesis of prostaglandins (PGs) is
initialized by the cyclooxygenase (COX) isoenzymes, COX-1 and
COX-2. COX-2 is an inducible isoform of COX that is present in
inflammatory cells, which generally produces PGs associated with
inflammation, fever and pain (12).
TLRs have important roles in the molecular
mechanisms underlying inflammation (13), particularly TLR4, a protein that in
humans is encoded by the TLR4 gene. TLR4 is able to detect LPS, and
therefore has an important role in activation of the innate immune
system (14,15). It has previously been demonstrated
that LPS-stimulated inflammation is predominantly mediated by TLR4
and cluster of differentiation 14 (16).
The extracellular-signal regulated protein kinases
(ERK) pathway is able to phosphorylate various transcription
factors upon activation, as well as two classes of
mitogen-activated protein kinases (MAPKs): p38 MAPK and c-jun
N-terminal kinases (JNK) (17).
LPS stimulation of RAW 264.7 cells rapidly activates all of these
MAPKs (18).
Nuclear factor (NF)-κB is involved in the cellular
response to various stimuli, including stress, cytokines, free
radicals, ultraviolet light, irradiation, oxidized LDL, and
bacterial or viral antigens. NF-κB has a key role in regulating the
immune response to infection, and is responsible for cytokine
production and cell survival (19–23).
The present study aimed to determine the mechanisms
underlying the anti-inflammatory effects of CJS C3; therefore, its
effects on LPS and TLR4 binding, and on the production of
proinflammatory mediators and cytokines were investigated. The
present study confirmed that the MAPK and NF-κB signaling pathways
were activated in RAW 264.7 macrophages that had been treated with
LPS.
Materials and methods
Plant sample
The leaves of Acanthopanax henryi (Oliv.)
Harms were collected in October 2012 in Xinhua (China). The plant
species was confirmed by Professor Xiang-Qian Liu (Hunan Key
Laboratory of Traditional Chinese Medicine modernization, Hunan
University of Chinese Medicine, Changsha, China), and the voucher
specimen (no. 20121125) was deposited at the School of Pharmacy,
Hunan University of Chinese Medicine (Changsha, China).
Extraction and isolation
The dried leaves of A. henryi (Oliv.) Harms
(10 kg) were cut into small pieces, were extracted three times with
MeOH (3×100 L) at room temperature, and were concentrated to obtain
a dark-green residue (0.8 kg) under reduced vacuum. The residue was
then suspended in H2O and partitioned with petroleum
ether. The water fraction was fractionated using column
chromatography (CC) on macroporous resin eluted with a gradient of
EtOH/H2O (0, 30, 50, 75 and 95%) into five fractions
(1–5). Fraction 4 (75% EtOH, 14.0 g) was
subjected to silica gel CC eluted with
CHCl3/MeOH/H2O (25:1:0/1:1:0.2) to give
fifteen fractions (A–O). Fraction L (0.67 g) was refractionated on
silica gel CC eluted with CHCl3/MeOH/H2O
(6:1:0.1/2:1:0.1) to give six sub-fractions (L1–L6). Sub-fraction
L3 (106.0 mg) was subjected to silica gel CC and was finally
purified by Sephadex LH-20 (MeOH; GE Healthcare Life Sciences,
Little Chalfont, UK) to yield 35.0 mg CJS C3 (24).
The compound structures were identified by mass
spectroscopy, 1D-nuclear magnetic resonance (NMR) and 2D-NMR, and
the spectral data were compared with those reported previously in
the literature (24).
1H NMR and 13C NMR spectra were measured on a
Varian INOVA 400M spectrometer (Agilent Technologies, Inc., Santa
Clara, CA, USA) with chemical shifts reported as ppm
(tetramethylsilane as internal standard). Electrospray ionization
mass spectra were carried out on a Agilent 6530 Accurate-Mass Q-TOF
(Agilent Technologies, Inc.).
High performance liquid chromatography
(HPLC)
The purity of CJS C3 was >98%, as determined
HPLC, as previously described (25). Briefly, CJS C3 was dissolved in
MeOH to a concentration of 0.1 mg/ml, for HPLC analysis with
Kinetex XB-C18 analytical column (100 mm × 4.6 mm × 2.6 µm;
Phenomenex, Inc., Torrance, CA, USA) at 30°C. Elution was conducted
using mobile phase A (water) and mobile phase B (acetonitrile) with
a gradient as follows: 0–2 min, 29–31% B; 2–13 min, 31–35% B; 13–15
min, 35–40% B; 15–23 min, 40–44% B; 23–25 min, 44–46% B; 25–31 min,
46–49% B; 31–38 min, 49–55% B. The flow rate was constant at 1.0
ml/min, and the effluents were monitored at 210 nm using an Agilent
1200 HPLC system with variable wavelength detector (Agilent
Technologies, Inc.). The purity value was found to be >98% by
peak area normalization method. The value of purity was obtained by
calculating the percentage of its peak area to that of the total
peaks in the HPLC chromatogram.
Cell culture
The RAW 264.7 murine macrophage cell line was
obtained from the Korea Research Institute of Bioscience and
Biotechnology (Seoul, South Korea). The cells were cultured in RPMI
1640 medium (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA,
USA) supplemented with 10% fetal bovine serum (FBS; Dako UK Ltd.,
Cambridge, UK) and 100 U/ml penicillin/streptomycin sulfate. Cells
were incubated in a humidified atmosphere containing 5%
CO2 at 37°C. For stimulation, the medium was replaced
with fresh RPMI 1640, and the cells were stimulated with LPS (200
ng/ml; Sigma-Aldrich; Merck Millipore, Darmstadt, Germany) in the
presence or absence of CJS C3 (10, 20 and 40 µM).
Cell viability assay
Cell viability was determined using a
3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethonyphenyl)-2
(4-sulfophenyl)-2H-tetrazolium (MTS) assay. RAW 264.7 cells were
plated at a density of 5×104 cells/ml in 96-well plates
(SPL Life Sciences Co., Ltd., Pocheon, Korea). Each experiment
included a non-treated group as a control. To determine a
concentration that was non-toxic to cells, various concentrations
of CJS C3 (10, 20 and 40 µM) were added to the cells and the
plates were incubated for 24 h at 37°C in a 5% CO2
atmosphere. MTS solution (5 mg/ml) was then added to each well and
the cells were cultured for a further 2 h, after which the optical
density was measured at 490 nm.
Measurement of NO production
NO production was assayed by measuring the nitrite
concentration in the supernatant of cultured RAW 264.7 cells. Cells
were seeded at a density of 7×105 cells/ml in 96-well
culture plates and were cultured for 18 h. The cells were
stimulated with LPS (200 ng/ml) in the absence or presence of test
reagents for 24 h, after which they were briefly centrifuged. The
supernatant was mixed with an equal volume of Griess reagent (1%
sulfanilamide, 0.1% naphthyl ethylenediamine dihydrochloride and
2.5% H3PO4) and was incubated at room
temperature for 5 min. Nitrite concentrations were determined by
measuring the absorbance of the supernatant at 570 nm. Sodium
nitrite (NaNO2) was used to generate a standard
curve.
Enzyme-linked immunosorbent assay
(ELISA)
Cells were seeded at a density of 5×105
cells/ml in 24-well tissue culture plates and were treated with CJS
C3 (10, 20, and 40 µM) for 1 h, prior to stimulation with
LPS (200 ng/ml). Following an incubation at 37°C for 24 h, the
medium was collected in a microcentrifuge tube and centrifuged
(2,000 × g, 5 min, 4°C). Levels of TNF-α and IL-6 in the
culture media were quantified using ELISA kits (cat. nos. 555240
and 555268, respectively), according to the manufacturer's protocol
(BD Biosciences, San Jose, CA, USA). ELISA plates (Falcon; BD
Biosciences) were coated overnight at 4°C with anti-mouse
interleukin (IL)-6 antibody (1:1,000; cat. no. 554402) and TNF-α
antibody (1:250; cat. no. 51-26731E) diluted in coating buffer (0.1
M sodium carbonate; pH 9.5) and were washed three times with
phosphate-buffered saline (PBS) containing 0.05% Tween-20.
Non-specific protein binding sites were subsequently blocked with
assay diluent (PBS containing 10% FBS; pH 7.0) for ≥1 h.
Immediately following this incubation, samples and IL-6 standards
were added to the wells, and the plates were incubated for a
further 2 h. Subsequently, detector solution [biotinylated
anti-mouse IL-6 monoclonal antibody (1:1,000; cat. no. 554402),
TNF-α monoclonal antibody (1:500; cat. no. 51-26732E) and
streptavidin-horseradish peroxidase (HRP) reagent (1:1,000; cat.
no. 554066) all from BD Biosciences] was added to each well and the
plates were incubated for an additional 1 h. Tetramethylbenzidine
was added to each well, and the plates were incubated for 30 min in
the dark prior to reaction termination using stop solution (1M
H3PO4). Absorbance was then measured at 450
nm. All standards and samples were assayed in duplicate.
Measurement of PGE2
production
PGE2 concentrations were determined using
a PGE2 direct Biotrak assay (cat. no. Amersham; GE
Healthcare Life Sciences). Cells were seeded at a density of
5×105 cells/ml in 24-well tissue culture plates. CJS C3
(10, 20 and 40 µM) and LPS (200 ng/ml) were added to the
culture medium and the plates were incubated at 37°C for 24 h. The
medium was then collected in microcentrifuge tubes and centrifuged
(2,000 × g, 5 min, 4°C). The supernatants were decanted into
fresh microcentrifuge tubes and the concentration of
PGE2 was determined using the enzyme immunoassay kit,
according to the manufacturer's protocol.
Western blot analysis
CJS C3-pretreated (10, 20 and 40 µM) RAW
264.7 cells (2×106 cells/ml) were stimulated with LPS
(200 ng/ml) for 24 h and were then washed twice in ice-cold PBS (pH
7.4). The cell pellets were resuspended in lysis buffer on ice for
20 min and cell debris was removed by centrifugation (2,000 ×
g, 5 min, 4°C). The protein concentration in each sample was
determined using the Bio-Rad protein assay reagent (Bio-Rad
Laboratories, Inc., Hercules, CA, USA) according to the
manufacturer's protocol. Equal amounts of protein (20 µg)
were separated by 8% sodium dodecyl sulfate-polyacrylamide gel
electrophoresis and were then transferred onto a polyvinylidene
fluoride membrane (EMD Millipore, Bedford, MA, USA). The membrane
was blocked with 5% non-fat milk in Tris-buffered saline with Tween
20 (150 mmol/l NaCl, 20 mmol/l Tris-HCl and 0.05% Tween 20; pH
7.4). After blocking with 3% bovine serum albumin (Sigma-Aldrich;
Merck Millipore), the membranes were incubated with the following
primary antibodies: Anti-iNOS (1:1,000; cat. no. sc-651),
anti-COX-2 (1:1,000; cat. no. sc-1745),
anti-mouse-phosphorylated-ERK 1/2 MAPK (1:1,000; cat. no. sc-7383),
anti-phosphorylated-JNK (1:1,000; cat. no. sc-6254), anti-ERK
(1:1,000; cat. no. sc-93) and JNK (1:1,000; cat. no. sc-571) all
obtained from Santa Cruz Biotechnology, Inc. (Dallas, TX, USA) for
18 h at 4°C. After washing twice with Tris-buffered saline,
membranes were immunoblotted with HRP-conjugated
anti-immunoglobulin G antibody (1:2,000; cat. no. Z025902; Dako UK
Ltd.) for 1 h at room temperature. Epitopes on proteins were
recognized specifically by the antibodies and were visualized using
an enhanced chemiluminescence kit (Amersham; GE Healthcare Life
Sciences). The membrane was also immunoblotted with anti-β-actin
(1:1,000; cat. no. 47778; Santa Cruz Biotechnology, Inc.) to
demonstrate equal protein loading. The bands were evaluated by
using ImageQuant LAS 4000 Mini Biomolecular Imager (GE Healthcare)
and the quantitative measurement of band intensity was performed
using ImageJ (version 1.45S; National Institutes of Health,
Bethesda, MA, USA)
RNA isolation and cDNA synthesis
Using the Easy Blue total RNA Extraction kit (iNtRon
Biotechnology, Seongnam, South Korea) total RNA was extracted from
RAW 264.7 cells, according to the manufacturer's protocol. Total
RNA was then dissolved in DEPC-treated distilled water. RNA samples
used in the present study had an A260/A280 nm value between 1.6 and
2.0, as determined using a NanoDrop spectrophotometer (Thermo
Fisher Scientific, Inc., Pittsburgh, PA, USA). A Power cDNA
synthesis kit (iNtRon Biotechnology) was used to reverse transcribe
each sample to cDNA. Reverse transcription (RT) conditions were as
follows: 5 min at 75°C, 1 h at 42°C and 5 min at 70°C.
RT-polymerase chain reaction (PCR)
analysis
Synthesized cDNA (0.05 µg) from RAW 264.7
cells was amplified using Sensi 2X PCR premix (Lugen Sci Co., Ltd.,
Bucheon, Korea) and specific primers (Table I). The conditions for amplification
were as follows: Denaturation at 94°C for 3 min for one cycle,
followed by 35 cycles of denaturation at 94°C for 45 sec, annealing
at 56°C for 45 sec (iNOS), 53°C for 45 sec (COX-2) or 57°C for 45
sec (IL-6), and extension at 72°C for 90 sec. A final extension
step was performed at 72°C for 7 min. The PCR products were
separated by 2% agarose gel electrophoresis and were stained with
Dyne Gel Safe Red kit (II) (Dyne Bio, Seongnam, South Korea). The
expression levels were confirmed using a UV detector.
 | Table IGene-specific primers used for
PCR. |
Table I
Gene-specific primers used for
PCR.
| PCR | Gene | Primer
sequence |
|---|
| RT-PCR | β-actina | Forward
5′-CATCCGTAAAGACCTCTATGCCAAC-3′ |
| | Reverse
5′-ATGGAGCCACCGATCCACA-3′ |
| IL-6 | Forward
5′-CATGTTCTCTGGGAAATCGTGG-3′ |
| | Reverse
5′-AACGCACTAGGTTTGCCGAGTA-3′ |
| iNOS | Forward
5′-AGCCCAACAATACAAATGACCCTA-3′ |
| | Reverse
5′-TTCCTGTTGTTTCTATTTCCTTTGT-3′ |
| COX-2 | Forward
5′-CACTCAGTTTGTTGAGTCATTC-3′ |
| | Reverse
5′-GATTAGTACTGTAGGGTTAATG-3′ |
| TNF-α | Forward
5′-ACGGCATGGATCTCAAAGAC-3′ |
| | Reverse
5′-GTGGGTGAGGAGCAGTAGT-3′ |
| RT-qPCR | IL-6 | Forward
5′-TCTATACCACTTCACAAGTCGGA-3′ |
| | Reverse
5′-GAATTGCCATTGCACAACTCTTT-3′ |
| iNOS | Forward
5′-GCAGAGATTGGAGGCCTTGTG-3′ |
| | Reverse
5′-GGGTTGTTGCTGAACTTCCAGTC-3′ |
| COX-2 | Forward
5′-GCCAGGCTGAACTTCGAAACA-3′ |
| | Reverse
5′-GCTCACGAGGCCACTGATACCTA-3′ |
| TNF-α | Forward
5′-ATGAGCACTGAAAGCATGATCC-3′ |
| | Reverse
5′-ATCCGTAAAGACCTCTATGCCAAC-3′ |
RT-quantitative (q)PCR analysis
Synthesized cDNA from RAW 264.7 cells was amplified
using specific primers (Table I)
and the Fast SYBR Green PCR Master Mix (Applied Biosystems; Thermo
Fisher Scientific, Inc., Waltham, MA, USA). PCR products were
measured using a StepOnePlus Real-Time RT-PCR system (Thermo Fisher
Scientific, Inc.). Relative gene expression was calculated based on
the comparative quantification cycle (Cq) method (26), using StepOne software v2.3 (Applied
Biosystems; Thermo Fisher Scientific, Inc.). β-actin mRNA
expression was used as an endogenous control.
Immunofluorescence staining
RAW 264.7 cells were cultured in Nunc™ chambered
coverglass (Thermo Fisher Scientific, Inc.) for 24 h and were
stimulated with LPS in the presence or absence of CJS C3 (10, 20
and 40 µM). The cells were fixed in 4% formaldehyde in PBS
for 15 min at room temperature, and were permeabilized with 100%
MeOH for 10 min at −20°C. Specimens were blocked with blocking
buffer (PBS containing 5% FBS and 0.3% Triton X-100) for 1 h, and
were incubated overnight with anti-NF-κB/p65 antibody (1:200; cat.
no. sc-8008; Santa Cruz Biotechnology, Inc.) at 4°C.
Fluorochrome-conjugated secondary antibodies (1:500; cat. no.
A11029; Thermo Fisher Scientific, Inc.) were then applied for 1 h
at room temperature in the dark. After washing with PBS, nuclei
were counterstained with 4′, 6-diamidino-2-phenylindole, and
fluorescence was visualized under a fluorescence microscope (Carl
Ziess, Oberkochen, Germany).
Cells were stimulated with Alexa Fluor (AF)-LPS
(Sigma-Aldrich; Merck Millipore) for 1 h in the presence or absence
of CJS C3 for the LPS/TLR4 complex formation assay. The cells were
fixed and stained with a rabbit polyclonal anti-TLR4 antibody
(1:200; sc-16240; Santa Cruz Biotechnology, Inc.) overnight at 4°C.
Subsequently, the cells were incubated with AF 488-conjugated
secondary antibodies (1:500; cat. no. A11029; Thermo Fisher
Scientific, Inc.) for 1 h. The stained cells were observed under a
fluorescence microscope.
Statistical analysis
Statistical analysis was performed using one-way
analysis of variance followed by Tukey honest significant
difference test for multiple comparisons. Data are presented as the
mean ± standard deviation of duplicate determinations from three
separate experiments. SPSS 22 software (IBM SPSS, Armonk, NY, USA)
was used to conduct statistical analysis. P<0.05 was considered
to indicate a statistically significant difference.
Results
Effects of CJS C3 on cytotoxicity and NO
production in RAW 264.7 cells
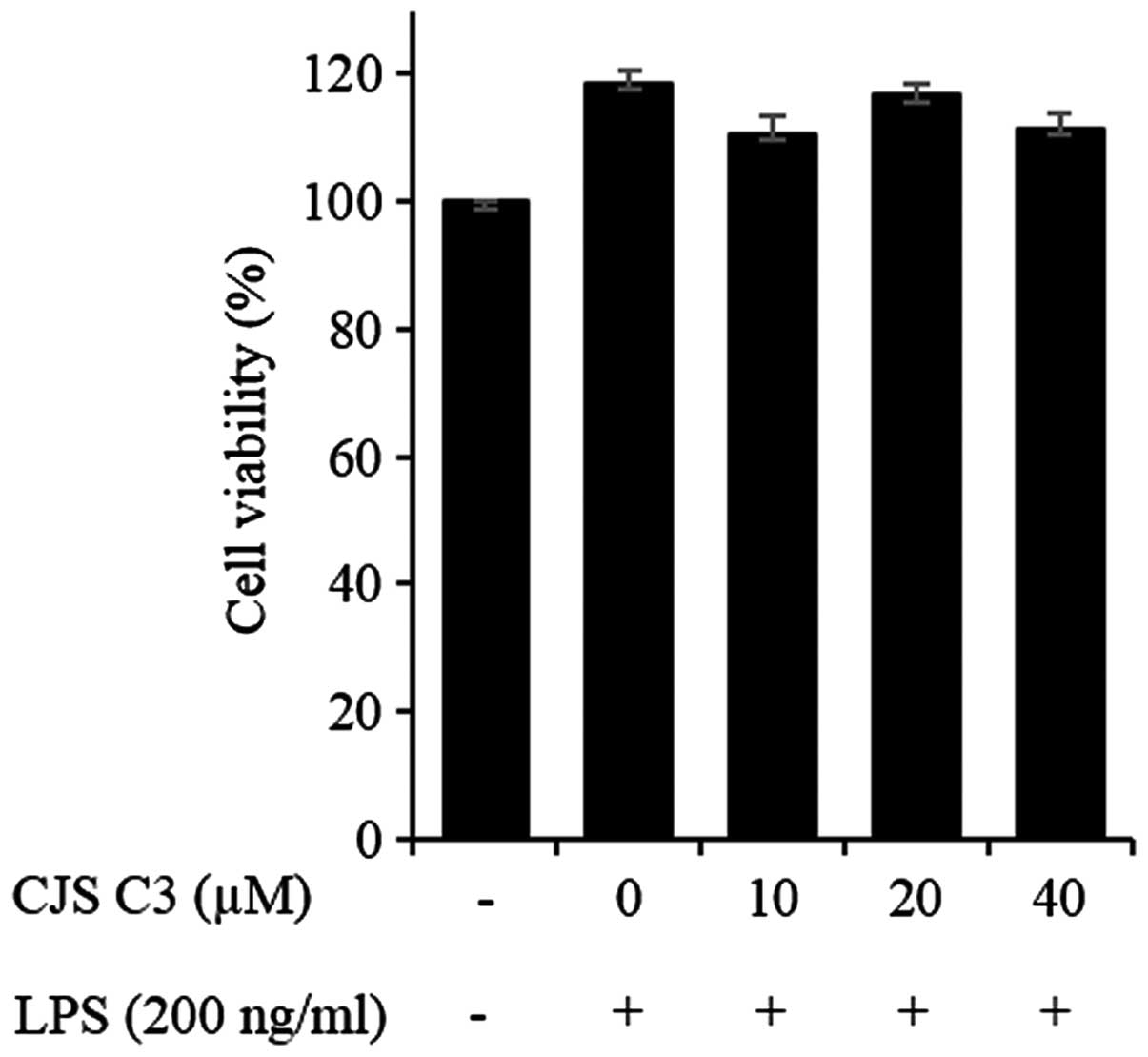
CJS C3 did not affect the viability of RAW 264.7
cells when used at the following concentrations: 10, 20 and 40
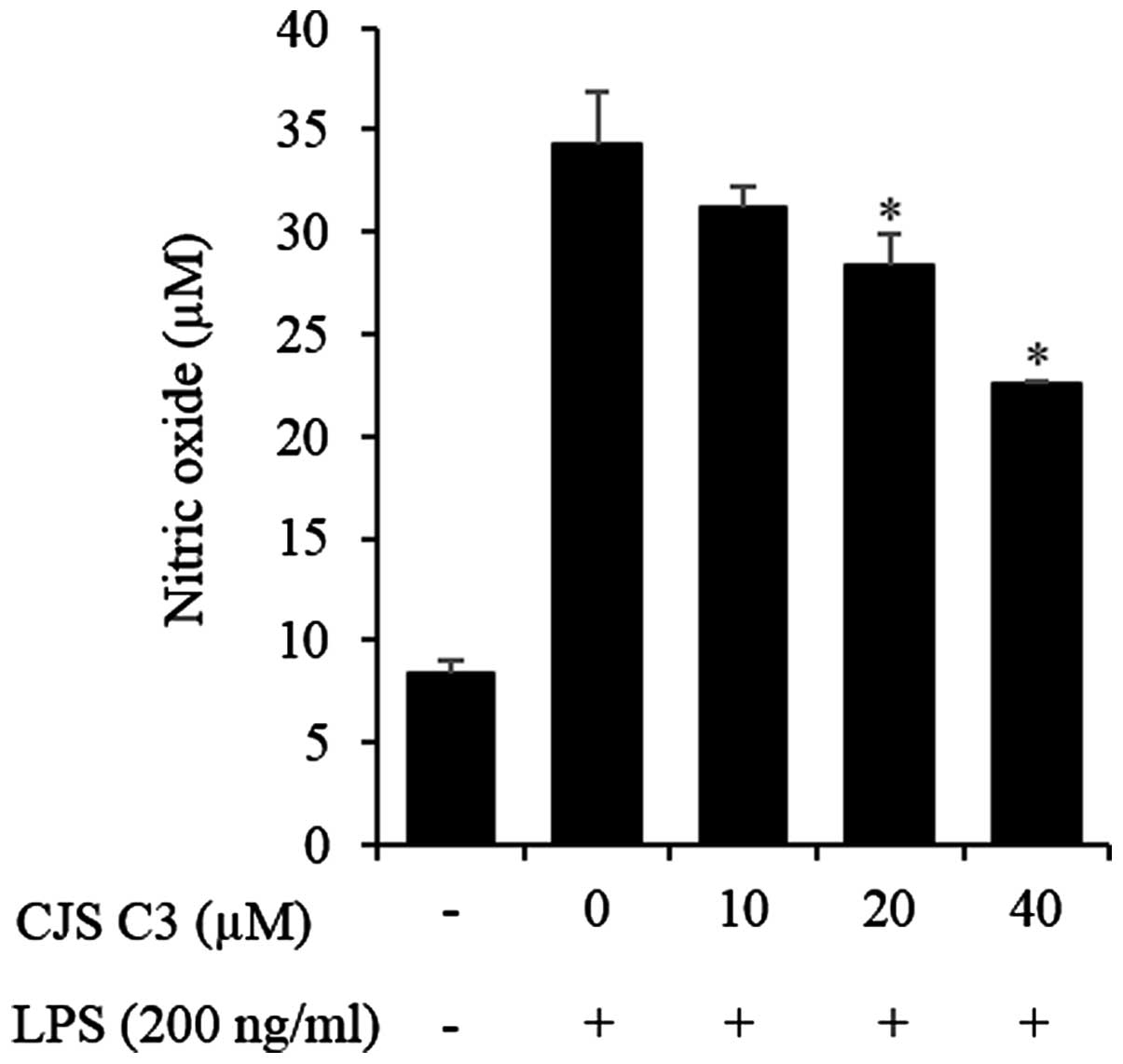
µM (Fig. 2). NO production
was examined in LPS-stimulated RAW 264.7 cells in the presence or
absence of CJS C3 for 24 h by Griess assay. Supernatants from
LPS-stimulated cells had significantly increased nitrite levels
compared with the controls. The effects of LPS were inhibited
following treatment with 20 or 40 µM CJS C3(Fig. 3).
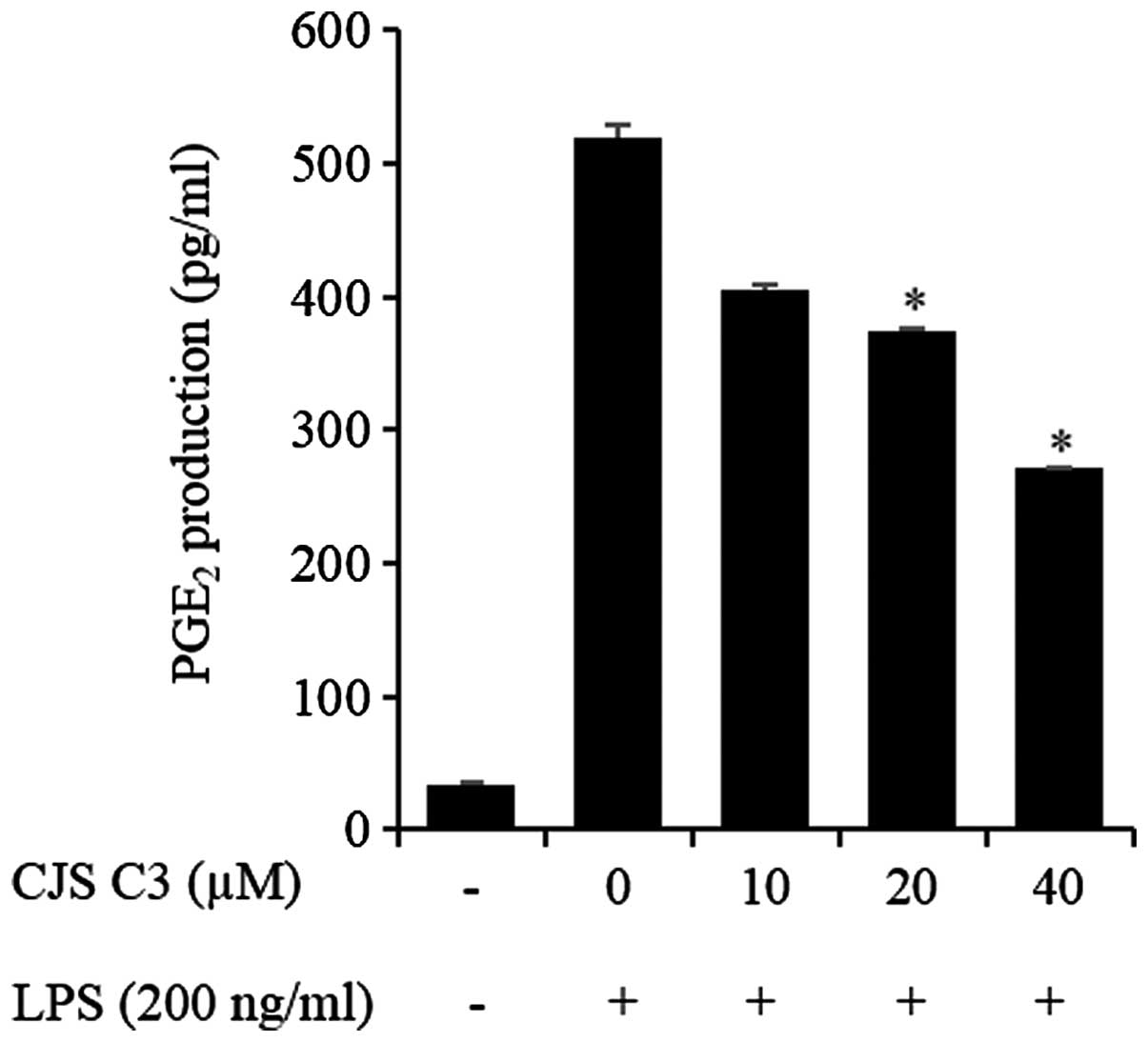
Effects of CJS C3 on PGE2
production in LPS-stimulated RAW 264.7 cells
The effects of CJS C3 on LPS-induced secretion of
PGE2 were examined by ELISA. As presented in Fig. 4, treatment of RAW 264.7 cells with
LPS resulted in a marked increase in PGE2 release
compared with in the untreated control group. However, CJS C3
inhibited LPS-mediated PGE2 production in a
concentration-dependent manner.
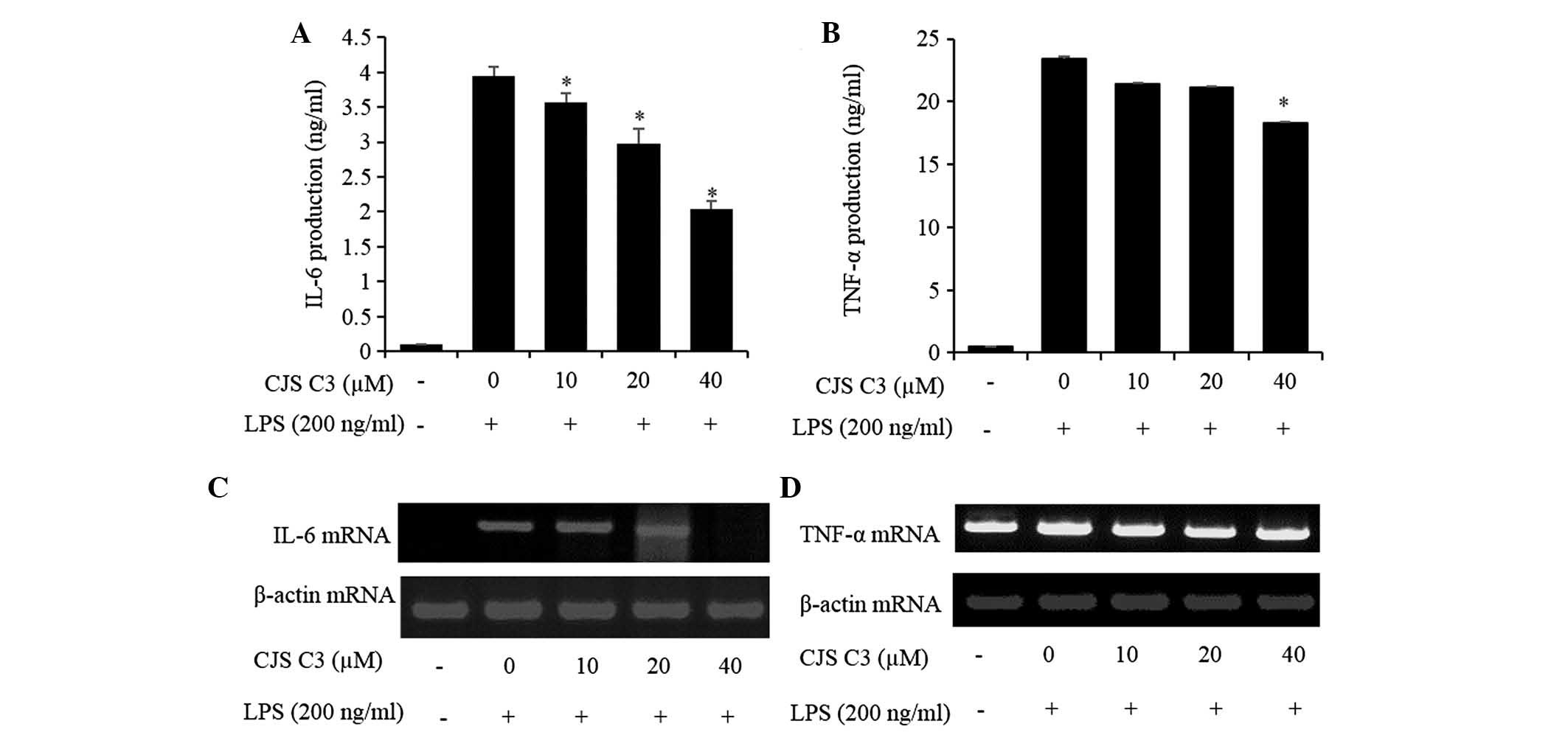
Effects of CJS C3 on IL-6 and tumor
necrosis factor (TNF)-α production in LPS-stimulated RAW 264.7
cells
The effects of CJS C3 on secretion of inflammatory
cytokines, such as IL-6 and TNF-α, were evaluated in LPS-treated
RAW 264.7 cells. IL-6 and TNF-α levels were significantly increased
in the culture medium of LPS-stimulated RAW 264.7 cells. However,
pretreatment with CJS C3 significantly decreased the release of
these cytokines in a concentration-dependent manner (Fig. 5A and B). Furthermore, the results
of an RT-PCR indicated that CJS C3 also markedly suppressed the
mRNA expression levels of these cytokines (Fig. 5C and D).
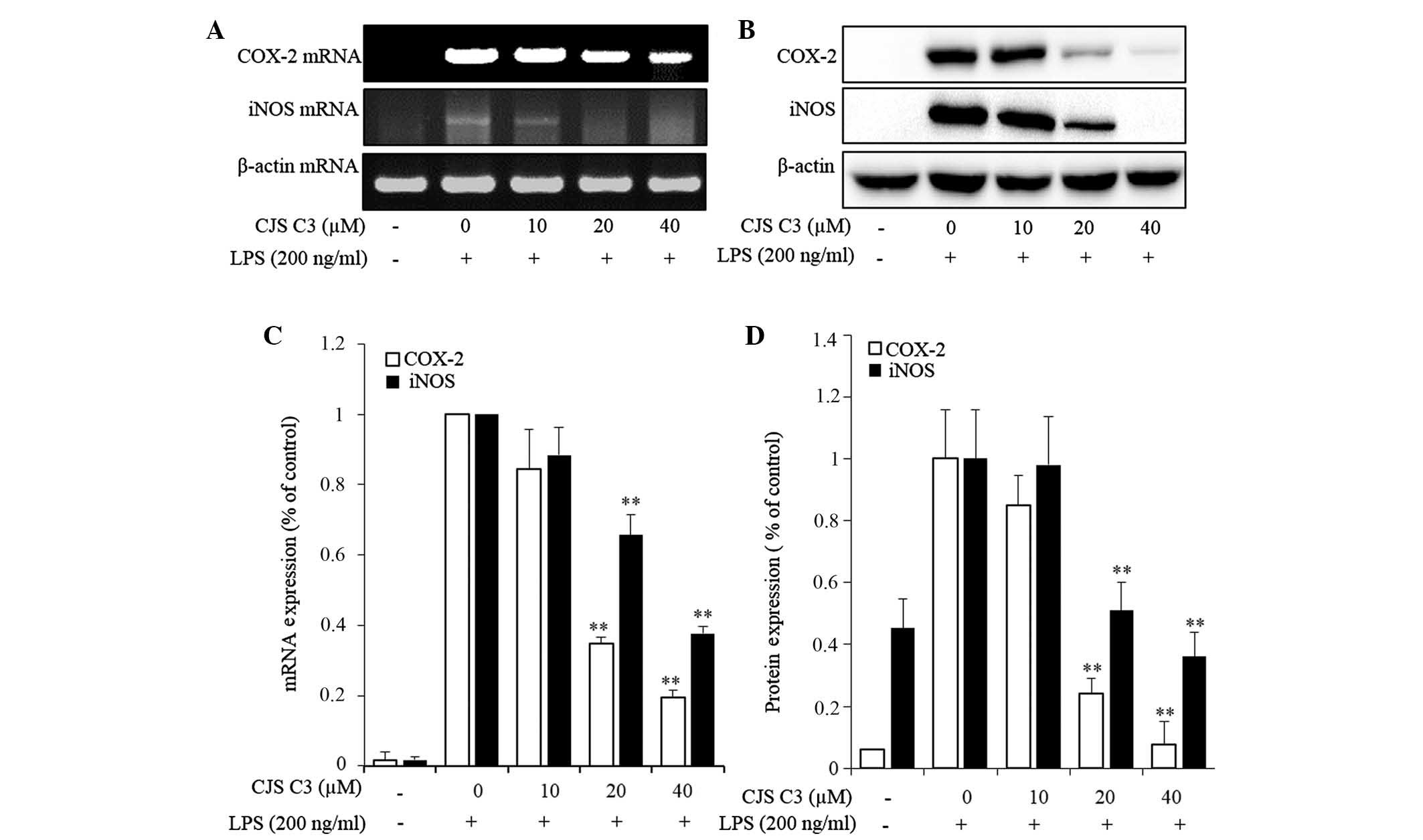
Effects of CJS C3 on LPS-stimulated COX-2
and iNOS expression in RAW 264.7 cells
To elucidate the mechanism underlying decreased
PGE2 levels and NO production in LPS-treated RAW 264.7
cells, the effects of CJS C3 on COX-2 and iNOS mRNA and protein
expression levels were determined by RT-PCR and western blot
analysis. The mRNA and protein expression levels of COX-2 and iNOS
were undetectable in unstimulated murine macrophages. However, the
mRNA and protein expression levels of COX-2 and iNOS were markedly
increased in response to LPS stimulation. In the CJS C3 treatment
group IL-6 and COX-2 expression exhibited a significant
concentration-dependent inhibition (Fig. 6). These results indicate that
decreased COX-2 and iNOS expression may contribute to the
inhibitory effects of CJS C3 on LPS-stimulated NO and
PGE2 production.
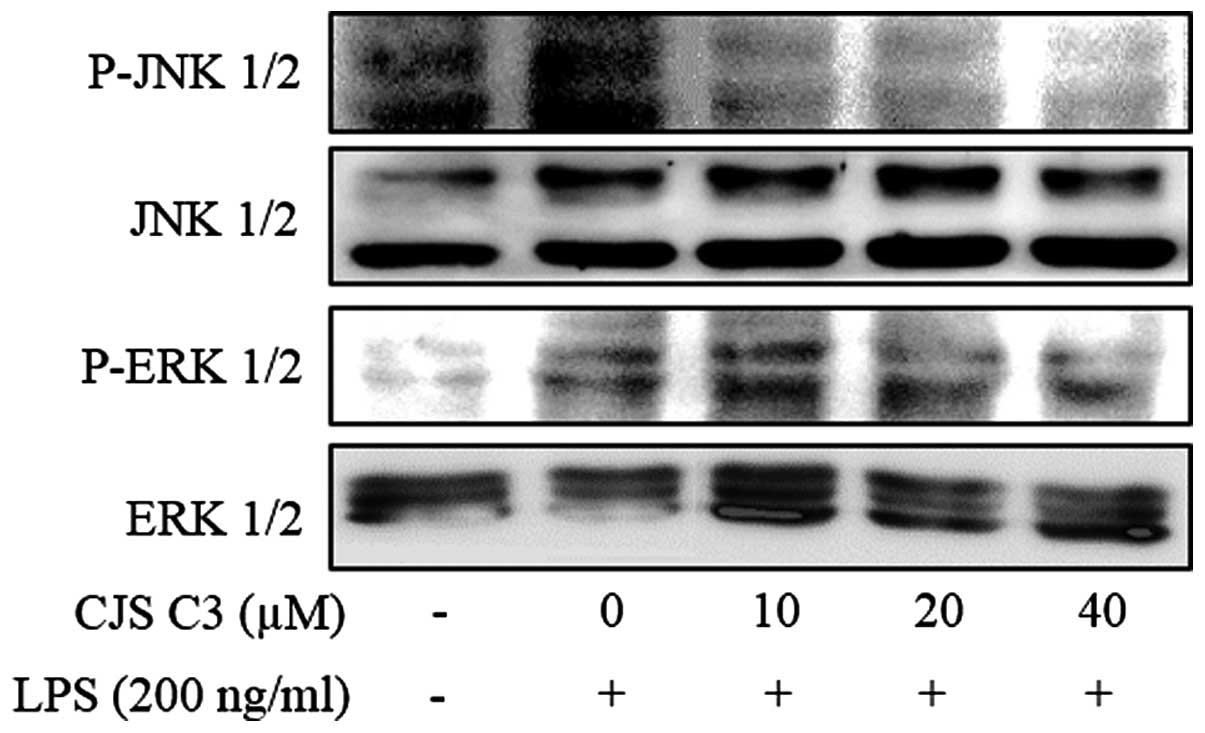
Effects of CJS C3 on the phosphorylation
of MAPKs in LPS-stimulated RAW 264.7 cells
Since MAPK signaling molecules have a crucial role
in regulating the LPS-induced inflammatory process, the present
study analyzed the phosphorylation levels of MAPKs in
LPS-stimulated RAW 264.7 cells by western blotting. In addition,
the effects of CJS C3 on phosphorylation of ERK and JNK MAPKs were
determined in LPS-treated cells. As presented in Fig. 7, ERK and JNK phosphorylation was
effectively suppressed by CJS C3 treatment. These results suggest
that activation of ERK and JNK may be blocked by CJS C3
treatment.
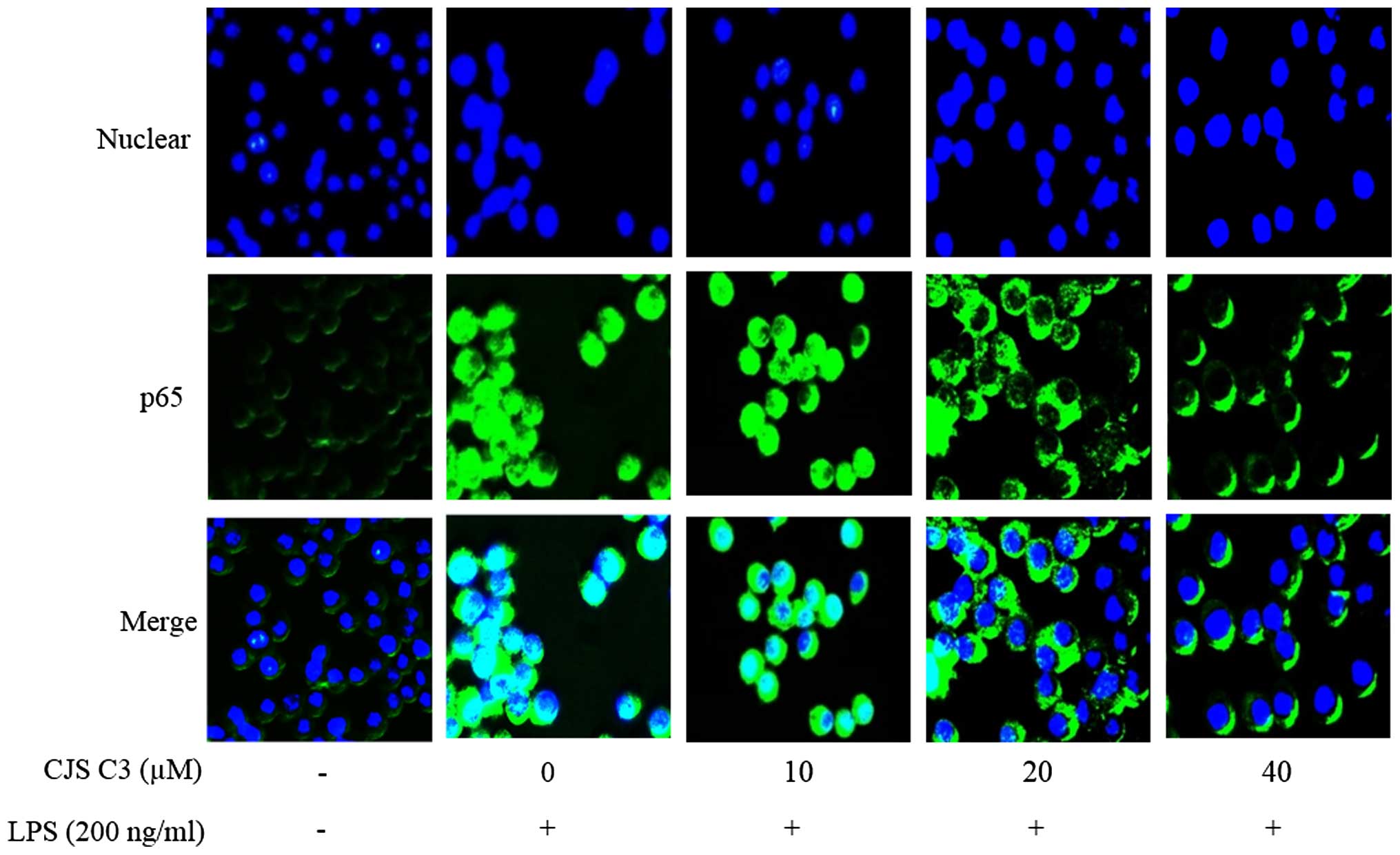
Effects of CJS C3 on LPS-induced nuclear
translocation of NF-κB in RAW 264. 7 macrophages
NF-κB is an important transcription factor that
regulates the expression of iNOS, COX-2 and proinflammatory
cytokines. Therefore, using immunofluorescence staining, the
present study investigated whether CJS C3 could suppress the NF-κB
signaling pathway. The immunofluorescence images revealed that
NF-κB/p65 was normally sequestered in the cytoplasm, and that
nuclear accumulation of NF-κB/p65 was strongly induced following
stimulation of RAW 264.7 cells with LPS. LPS-induced translocation
of NF-κB/p65 was completely inhibited after pretreating the cells
with CJS C3 (Fig. 8, Merge
panel).
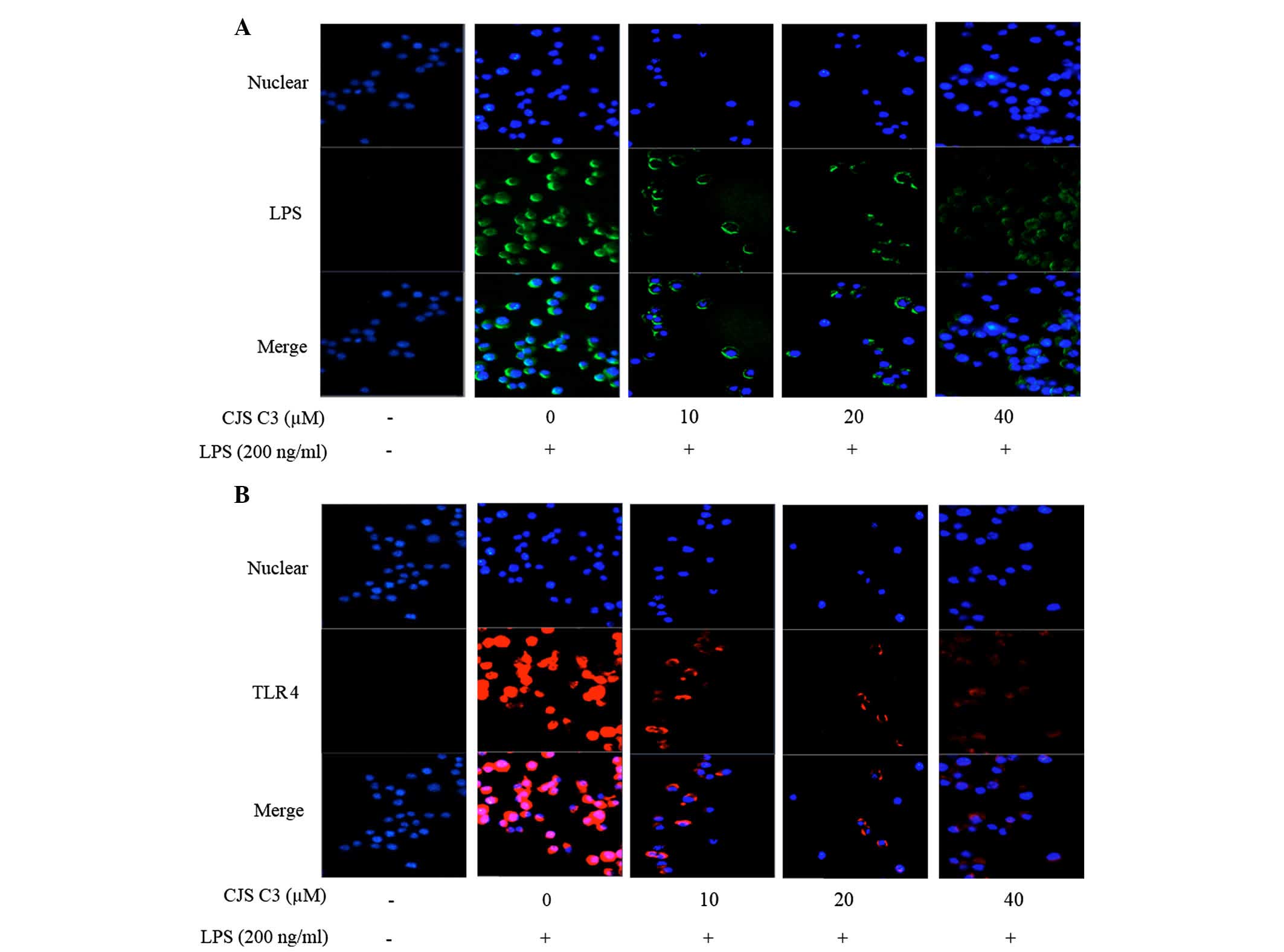
Effects of CJS C3 on LPS binding and TLR4
expression
The present study also investigated the effects of
CJS C3 on the LPS-activated TLR4 signaling pathway, in order to
further determine the mechanisms underlying its anti-inflammatory
effects. AF-LPS was used to determine whether CJS C3 was able to
inhibit the interaction between LPS and TLR4 in murine macrophages.
When the cells were treated with AF-LPS alone, the fluorescence
intensities of LPS and TLR4 were observed outside the cell membrane
by immunofluorescence assay. However, treatment with AF-LPS and CJS
C3 significantly inhibited the fluorescence intensity of TLR4
(Fig. 9A and B). These results
suggest that LPS-stimulated activation of the TLR4 signaling
pathway was potently suppressed by CJS C3.
Discussion
The Acanthopanax spp. belongs to the
Araliaceae family, the stem barks and roots of which have been used
as a tonic and as a prophylactic treatment in Oriental medicine
according to ancient use (26).
Acanthopanax henryi Harms is a member of the
Acanthopanax spp., the stem bark of which was originally
used to treat arthritis, rheumatism, edema and traumatic injuries
in China (27). Recently,
Acanthopanax henryi Harms leaves have started to garner the
attention of researchers. The present study demonstrated that CJS
C3, extracted from the leaves of Acanthopanax henryi Harms,
indicated anti-inflammatory activity in LPS-stimulated RAW 264.7
cells The reported pharmacological effects of active components
extracted from the leaves of Acanthopanax henryi Harms
include inhibition of tryrosinase and acetylcholinesterase
(6,28). To the best of our knowledge, no
other studies have been conducted regarding Acanthopanax
henryi Harms leaves. CJS C3, which was extracted from
Acanthopanax henryi Harms leaves in the present study, has
also been isolated from Acanthopanax senticosus (29). The present study aimed to determine
the anti-inflammatory effects of CJS C3, extracted from
Acanthopanax henryi Harms leaves, on LPS-stimulated RAW
264.7 cells.
Inflammation implies an irregularity in cytokine
levels. Proinflammatory cytokines, such as IL-6 and TNF-α, have
important roles in several inflammatory processes, and recruit
other immune cells involved in the pathogenesis of various
inflammatory conditions. Accordingly, overproduction of IL-6 and
TNF-α is associated with the development of chronic inflammatory
conditions, including septic shock, cachexia, psoriasis, rheumatoid
arthritis and cytotoxicity (30–33).
Therefore, the present study investigated the effects of CJS C3 on
the synthesis of proinflammatory cytokines. The results
demonstrated that CJS C3 effectively suppressed the overproduction
of IL-6 and TNF-α in a concentration-dependent manner (Fig. 5A and B). The suppressing effect
extended to the inhibition of TNF-α and IL-6 transcription
(Fig. 5C and D).
Production of IL-6 and TNF-α is associated with
synergistic activation of NO and PGE2 production in
LPS-activated RAW 264.7 macrophages (32–34).
NO is generated by phagocytes, such as monocytes, macrophages and
neutrophils. Phagocytes contain iNOS, which is activated by
interferon-γ or TNF. In this manner, the immune system regulates
phagocytes, which have a key role in inflammation and immune
responses (35–39).
Among the PGs, PGE2 is the most prominent
mediator in inflammation, fever and pain; it also has physiological
functions in the gastrointestinal tract, kidney, and the immune and
central nervous systems. Increased PGE2 formation during
inflammation predominantly depends on the concomitant induction of
COX-2 (40).
According to the results of the present study, CJS
C3 significantly inhibited LPS-induced NO and PGE2
production in RAW 264.7 cells (Figs.
3 and 4). This suppression was
possibly caused by inhibition of iNOS and COX-2 expression at the
transcriptional level in LPS-stimulated RAW 264.7 cells (Fig. 6).
MAPKs are involved in directing cellular responses
to a diverse array of stimuli, including mitogens, osmotic stress,
heat shock and proinflammatory cytokines. They regulate cell
functions, including proliferation, gene expression,
differentiation, mitosis, cell survival and apoptosis (41). The MAPK family consists of
serine/threonine kinases, such as ERK, p38 MAPK and JNK (42), which control cellular signal
transduction from the cell surface to the nucleus. Furthermore,
phosphorylation and activation of MAPKs has previously been
implicated in signaling pathways relevant to LPS-induced
inflammation, thus suggesting that MAPKs are important targets for
anti-inflammatory molecules (43,44).
NF-κB is a protein complex that controls
transcription of DNA (19,20). NF-κB regulates the expression of
several genes that code for mediators involved in immune and
inflammatory responses, including iNOS, COX-2, TNF-α and IL-6.
Therefore, NF-κB is considered a rational target for novel types of
anti-inflammatory treatment (45,46).
The present data indicated that the effects of CJS C3 appear to
involve inhibition of NF-κB activation by blocking the MAPK
signaling pathway (Fig. 7 and
8).
LPS-activated macrophages, which bind to TLR4,
induce the activation of specific intracellular pathways through
receptor dimerization and recruitment of various adapter molecules,
such as myeloid differentiation primary response 88 (47). These LPS-initiated signaling
cascades lead to activation of the MAPK and NF-κB signaling
pathways (48). The results of the
present study demonstrated that treatment of LPS-stimulated
macrophages with CJS C3 significantly inhibited the fluorescence
intensity of TLR4 (Fig. 9). The
results of the present study suggest that the anti-inflammatory
properties of CJS C3 may result from inhibition of pro-inflammatory
mediators by suppressing the initiation of the inflammatory
response and inhibiting the MAPK-NF-κB signaling pathways.
Acknowledgments
The present study was supported by the Basic Science
Research Program through the National Research Foundation of Korea
(NRF) funded by the Ministry of Education (NRF-2013R1A1A2064673),
and the National Research Foundation of Korea (NRF) grant funded by
the Korean Government (MSIP) (2008-0062484).
References
|
1
|
Park SY, Yook CS, Nohara T, Mizutani T and
Tanaka T: Random amplified polymorphic DNA analysis of genetic
relationships among Acanthopanax species. Arch Pharm Res.
27:1270–1274. 2004. View Article : Google Scholar
|
|
2
|
Park SY: Studies on RAPD analysis and
triterpenoidal constituents of Acanthopanax species. Kumamoto
University Press; 3. pp. 1–3. 2002
|
|
3
|
Phuong NT, Lee KA, Jeong SJ, Fu CX, Choi
JK, Kim YH and Kang JS: Capillary electrophoretic method for the
determination of diterpenoid isomers in Acanthopanax species. J
Pharm Biomed Anal. 40:56–61. 2006. View Article : Google Scholar
|
|
4
|
Kang JS, Linh PT, Cai XF, Kim HS, Lee JJ
and Kim YH: Quantitative determination of eleutheroside B and E
from Acanthopanax species by high performance liquid
chromatography. Arch Pharm Res. 24:407–411. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Li Z, Kim JH, Liu XQ, Lee KT and Yook CS:
Anticancer effects in vitro and chemical compositions of extracts
of Acanthopanax henryi. Journal of Acupuncture and Herbs. 1:49–54.
2014.
|
|
6
|
Li Z, Li XJ, Kwon OX, Wang X, Zou QP, Liu
XQ and Lee HK: Chemical constituents from leaves of Acanthopanax
henryi (II). Nat Prod Sci. 21:196–204. 2015.
|
|
7
|
Lumeng CN and Saltiel AR: Inflammatory
links between obesity and metabolic disease. J Clin Invest.
121:2111–2117. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Balija TM and Lowry SF: Lipopolysaccharide
and sepsis-associated myocardial dysfunction. Cur Opin Infect Dis.
24:248–253. 2011. View Article : Google Scholar
|
|
9
|
Peri F, Piazza M, Calabrese V, Damore G
and Cighetti R: Exploring the LPS/TLR-4 signal pathway with small
molecules. Biochem Soc Trans. 38:1390–1395. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Nathan C: Nitric oxide as a secretory
product of mammalian cells. FASEB J. 6:3051–3064. 1992.PubMed/NCBI
|
|
11
|
Zhao L, Weber PA, Smith JR, Comerford ML
and Elliott GT: Role of inducible nitric oxide synthase in
pharmacological 'preconditioningʼ with monophosphoryl lipid A. J
Mol Cell Cardiol. 29:1567–1576. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hawkey CJ: COX-2 inhibitors. Lancet.
353:307–314. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Mclnturff JE, Modlin RL and Kim J: The
role of toll-like receptors in the pathogenesis and treatment of
dermatological disease. J Invest Dermatol. 125:1–8. 2005.
View Article : Google Scholar
|
|
14
|
Rock FL, Hardiman G, Timans JC, Kastelein
RA and Bazan JF: A family of human receptors structurally related
to Drosophila Toll. Proc Natl Acad Sci USA. 95:588–593. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Medzhitov R, Preston-Hurlburt P and
Janeway CA Jr: A human homologue of the Drosophila Toll protein
signals activation of adaptive immunity. Nature. 388:394–397. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wu J, Zhou J, Chen X, Fortenbery N,
Eksioglu EA, Wei S and Dong J: Attenuation of LPS-induced
inflammation by ICT, a derivate of icariin, bia inhibition of the
CD14/TLR-4 signaling pathway in human monocytes. Int
Immunopharmacol. 12:74–79. 2012. View Article : Google Scholar
|
|
17
|
Chen CC and Wang JK: p38 but not p44/42
mitogen-activated protein kinase is required for nitric oxide
synthase induction mediated by lipopolysaccharide in RAW 264.7
macrophages. Mol Pharmacol. 55:481–488. 1999.PubMed/NCBI
|
|
18
|
Xia Z, Dickens M, Raingeaud J, Davis RJ
and Greenberg ME: Opposing effects of ERK and JNK-p38 MAP kinases
on apoptosis. Science. 270:1326–1331. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Gilmore TD: Introduction to NF-kappaB:
Players, pathways, perspectives. Oncogene. 25:6680–6684. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Brasier AR: The NF-kappaB regulatory
network. Cardiovasc Toxicol. 6:111–130. 2006. View Article : Google Scholar
|
|
21
|
Perkins ND: Integrating cell-signalling
pathways with NF-kappaB and IKK function. Nat Rev Mol Cell Biol.
8:49–62. 2007. View Article : Google Scholar
|
|
22
|
Gilmore TD: The Rel/NF-kappaB signal
transduction pathway: Introduction. Oncogene. 18:6842–6844. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Tian B and Brasier AR: Identification of a
nuclear factor kappa B-dependent gene network. Recent Prog Horm
Res. 58:95–130. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Shao CJ, Kasai Ryoji, Xu JD and Tanaka O:
Saponins from leaves of Acanthopanax senticosus Harms., Ciwujia:
Structures of Ciwujianosides B, C1, C2,
C3, C4, D1, D2 and E.
Chem Pharm Bull. 36:601–608. 1988. View Article : Google Scholar
|
|
25
|
Li Z: Simultaneous determination of
fifteen triterpenoid saponins in different medicinal parts of
Acanthopanax henryi by HPLC-CAD-ESI-MS. Study on chemical
constituents of Acanthopanax henryi (Oliv.) Harms. Hunan University
of Traditional Chinese Medicine; Changsha: pp. 45–66. 2015, In
Chinese.
|
|
26
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
27
|
Hou JP and Jin Y: Herbal painkillers:
Relief of lingering arthritic pain and rheumatism. The Healing
Power of Chinese Herbs and Medicinal Recipes. Russo E: Routledge
Taylor & Francis Group; Oxford: pp. 413–416. 2005
|
|
28
|
Zhang XD, Li Z, Liu GZ, Wang X, Kwon OK,
Lee HK, Whang WK and Liu XQ: Quantitative determination of 15
bioactive triterpenoid saponins in different parts of Acanthopanax
henryi by HPLC with charged aerosol detection and confirmation by
LC-ESI-TOF-MS. J Sep Sci. 39:2252–2262. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Jiang W, Li W, Han L, Liu L, Zhang Q,
Zhang S, Nikaido T and Koike K: Biologically active triterpenoid
saponins from Acanthopanax senticosus. J Nat Prod. 69:1577–1581.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Opal SM and DePalo VA: Anti-inflammatory
cytokines. Chest. 117:1162–1172. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
McInnes IB and Scheett G: Cytokines in the
pathogenesis of rheumatoid arthritis. Nat Rev Immunol. 7:429–442.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Aggarwal BB and Natarajan K: Tumor
necrosis factors: Developments during the last decade. Eur Cytokine
Netw. 7:93–124. 1996.PubMed/NCBI
|
|
33
|
Brennan FM and McInnes IB: Ebidence that
cytokines play a role in rheumatoid arthritis. J Clin Invest.
118:3537–3545. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Jun CD, Choi BM, Kim HM and Chung HT:
Involvement of protein kinase C during taxol-induced activation of
murine peritoneal macrophages. J Immunol. 154:6541–6547.
1995.PubMed/NCBI
|
|
35
|
Green SJ, Mellouk S, Hoffman SL, Meltzer
MS and Nacy CA: Cellular mechanisms of nonspecific immunity to
intracellular infection: Cytokine-induced synthesis of toxic
nitrogen oxides from L-arginine by macrophages and hepatocytes.
Immunol Lett. 25:15–19. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Gorczyniski RM and Stanely J: Clinical
Immunology. Landes Bioscience; Austin, TX: 1999
|
|
37
|
Green SJ, Nacy CA, Schreiber RD, Granger
DL, Crawford RM, Meltzer MS and Fortier AH: Neutralization of gamma
interferon and tumor necrosis factor alpha blocks in vivo synthesis
of nitrogen oxides from L-arginine and protection against
Francisella tularensis infection in Mycobacterium bovis BCG-treated
mice. Infect Immun. 61:689–698. 1993.PubMed/NCBI
|
|
38
|
Kamijo R, Gerecitano J, Shapiro D, Green
SJ, Aguet M, Le J and Vilcek J: Generation of nitric oxide and
clearance of interferon-gamma after BCG infection are impaired in
mice that lack the interferon-gamma receptor. J Inflamm. 46:23–31.
1995.PubMed/NCBI
|
|
39
|
Green SJ, Scheller LF, Marletta MA, Seguin
MC, Klotz FW, Slayter M, Nelson BJ and Nacy CA: Nitric oxide:
Cytokine-regulation of nitric oxide in host resistance to
intracellular pathogens. Immunol Lett. 43:87–94. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Koeberle A and Werz O: Microsomal
prostaglandin E2 synthase-1. Anti-inflammatory Drug Discovery.
Levin JI and Laufer S: Royal Society of Chemistry Publishing;
Cambridge: pp. 8–12. 2012
|
|
41
|
Pearson G, Robinson F, Beers Gibson T, Xu
BE, Karandikar M, Berman K and Cobb MH: Mitogen-activated protein
(MAP) kinase pathways: Regulation and physiological functions.
Endocr Rev. 22:153–183. 2001.PubMed/NCBI
|
|
42
|
Nishida E and Gotoh Y: The MAP kinase
cascade is essential for diverse signal transduction pathways.
Trends Biochem Sci. 18:128–131. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Kirkwood KL and Rossa C Jr: The potential
of p38 MAPK inhibitors to modulate periodontal infections. Curr
Drug Metab. 10:55–67. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Kaminska B: MAPK signaling pathways as
molecular targets for anti-inflammatory therapy-from molecular
mechanisms to therapeutic benefits. Biochim Biophys Acta.
1754:253–262. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Surh YJ, Chun KS, Cha HH, Han SS, Keum YS,
Park KK and Lee SS: Molecular mechanisms underlying chemopreventive
activities of anti-inflammatory phytochemicals: Down-regulation of
COX-2 and iNOS through suppression of NF-kappa B activation. Mutat
Res. 480–481:243–268. 2001. View Article : Google Scholar
|
|
46
|
Makarov SS: NF-kappaB as a therapeutic
target in chronic inflammation: Recent advances. Mol Med Today.
6:441–448. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Tian L, Zhou DS, Wang KZ, Zhang W, Shi ZB,
Fan LH and Sun S: Association of toll-like receptor 4 signaling
pathway with steroid-induced femoral head osteonecrosis in rats. J
Huazhong Univ Sci Technolog Med Sci. 34:679–686. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Kang OH, Chae HS, Choi JG, Oh YC, Lee YS,
Kim JH, Seung MJ, Jang HJ, Bae KH, Lee JH, et al: Ent-pimara-8(14),
15-dien-19-oic acid isolated from the roots of Aralia cordata
inhibits induction of inflammatory mediators by blocking NF-kappaB
activation and mitogen-activated protein kinase pathways. Eur J
Pharmacol. 601:179–185. 2008. View Article : Google Scholar : PubMed/NCBI
|