Introduction
Dermatomyositis (DM) is a type of idiopathic
inflammatory myopathy (IM), which affects the skin, muscle and
other organs, and is associated with significant morbidity and
mortality (1). The incidence of DM
is estimated at 9.63 cases per million individuals, according to a
population-based study (2). DM
affects children and adults, and occurs more commonly in female
patients (3). DM is closely
associated with an increased risk of malignancy (4), including ovarian, lung, pancreatic,
stomach and colorectal cancers, and non-Hodgkin's lymphoma
(5). The rarity of the disease,
the different features of the multiple disease subsets, and the
lethal complications associated with DM make it difficult to cure
(6).
Several factors are thought to contribute to DM,
including environmental factors and genetic susceptibility. The
association between DM and various environmental triggers,
including medication, sunlight and infection, have been examined
(7). In addition, a genetic
component may predispose individuals to DM; the most important
genetic region associated with DM appears to be the major
histocompatibility complex (MHC), and a few candidate genes, such
as signal transducer and activator of transcription (STAT) 4 and
tumor necrosis factor, have been reported to be relevant to the
development of DM (8). The first
case of DM was reported in 1875 (9); however, the underlying molecular
mechanisms of this disease remain incompletely understood.
Pathological studies of muscle tissue in DM have reported
histological abnormalities in capillaries, and the infiltration of
muscle by B cells, cluster of differentiation (CD)4+ T
cells, macrophages and plasma cells, and perifascicular atrophy
(10). Therefore, DM has
classically been considered a humorally mediated autoimmune disease
(10). Recently, a microarray
study reported that type 1 interferon (IFN)-mediated immune
pathogenesis is likely to be involved in DM (11). Identification of the involved
molecular mechanisms may lead to the development of effective
therapeutic approaches for the treatment of patients with DM.
The present study downloaded the GSE48280 dataset,
and identified the differentially expressed genes (DEGs) between
the DM and healthy control (HC) muscle tissue samples, in order to
explore the molecular mechanisms underlying DM. In addition,
functional and pathway enrichment analyses were conducted, and
protein-protein interaction (PPI) networks of the DEGs were
constructed to identify the key genes and signaling pathways in DM.
The findings of the present study may be helpful to improve
understanding regarding the pathogenesis of DM, and provide insight
into the development of a novel therapeutic strategy.
Data and methods
Microarray data
The GSE48280 gene expression profile (12) was downloaded from the Gene
Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE48280).
The profile contained five patients with DM, five patients with
polymyositis, four patients with inclusion body myositis and five
HCs, the muscle samples from which were analyzed based on the
Affymetrix Human Gene 1.0 ST Array platform (Affymetrix, Santa
Clara, CA, USA). In the present study, only DM and HC samples were
analyzed via a series of bioinformatics methods.
Data preprocessing and differential
expression analysis
The original CEL files were transformed to
probe-level data. If several probes mapped to a single gene, the
mean of the probes was used to determine the gene expression value.
The probe-level data were converted to gene symbols using the Perl
procedure (version 5.24.0; www.perl.org).
Following normalization, the limma package (13) in R (version 3.2.2; www.r-project.org) was conducted to identify DEGs
between the DM and HC samples. Only genes with a fold change (FC)
value (|log2FC|)>1 and a P-value <0.05 were regarded as DEGs,
which were considered the signature genes of DM.
Gene Ontology (GO) and Kyoto Encyclopedia
of Genes and Genomes (KEGG) pathway analyses
GO (geneontology.org) and KEGG (http://www.genome.jp/kegg/) pathway enrichment
analyses for the identified DEGs were performed using Cytoscape
software (version 3.2.1) with the ClueGO V2.1.7 plug-in (14). The ClueGO plug-in generates
functionally grouped GO annotation networks for a large number of
genes. GO categories were divided into biological process,
molecular function, cellular component and immune system process
terms. The P-value was calculated by right-sided hypergeometric
tests, and Benjamini-Hochberg adjustment was used for multiple test
correction. GO terms with a P-value <0.01, and KEGG pathways
with a P-value <0.05 were considered significant.
PPI network construction and hub gene
identification
The Search Tool for the Retrieval of Interacting
Genes (STRING; http://string-db.org/) database is an
online tool, which has been designed as pre-computed global
resource to evaluate PPI information (15). In the present study, the STRING
database was applied to construct PPI networks for the screened
DEGs, with a combination score >0.99 as the threshold.
Subsequently, the PPI networks were visualized and analyzed using
Cytoscape software (version 3.2.1) (16) based on the STRING database.
According to a previous study regarding biological
networks, the majority of PPI networks in the present study
exhibited scale-free network properties (17). PPI networks have a small number of
highly connected protein nodes (known as hubs) and many poorly
connected nodes. The connectivity degree was statistically analyzed
by their combined scores based on the database algorithms. The
important nodes in the PPI networks were identified and labeled as
hub genes. Since the majority of the PPI networks obeyed the
scale-free attribution, node degree >20 was selected as the
threshold to obtain hub genes in the present study.
Results
Identification of DEGs between DM and HC
samples
According to the cut-off criteria of P<0.05 and
|log2FC| >1.0, 201 DEGs were identified between the five DM and
five HC samples using the limma package. A total of 180 DEGs were
upregulated and 21 DEGs were downregulated.
GO and KEGG pathway analyses
All DEGs were subjected to GO and KEGG analysis.
Following GO enrichment analysis using ClueGO, a total of 143 GO
terms were associated with the DEGs. The top 20 significantly
enriched GO terms are presented in Table I. The top five significantly
enriched GO terms were as follows: Type I IFN signaling pathway,
defense response, response to cytokine, cytokine-mediated signaling
pathway, and cellular response to cytokine stimulus.
 | Table ITop 20 Gene Ontology (GO) functional
enrichment analysis of differentially expressed genes
(P<0.01). |
Table I
Top 20 Gene Ontology (GO) functional
enrichment analysis of differentially expressed genes
(P<0.01).
| Category | Term | Count | P-value |
|---|
| Immune system
process | GO:0060337: Type I
interferon signaling pathway | 31 | 3.45E-41 |
| Biological
process | GO:0006952: Defense
response | 84 | 1.34E-37 |
| Biological
process | GO:0034097:
Response to cytokine | 57 | 1.95E-37 |
| Biological
process | GO:0019221:
Cytokine-mediated signaling pathway | 47 | 5.39E-35 |
| Biological
process | GO:0071345:
Cellular response to cytokine stimulus | 49 | 3.39E-32 |
| Biological
process | GO:0009615:
Response to virus | 38 | 3.46E-27 |
| Immune system
process | GO:0034341:
Response to interferon-gamma | 26 | 6.14E-26 |
| Biological
process | GO:0051707:
Response to other organism | 48 | 9.91E-25 |
| Biological
process | GO:0098542: Defense
response to other organism | 38 | 2.95E-23 |
| Immune system
process | GO:0071346:
Cellular response to interferon-gamma | 22 | 6.58E-22 |
| Immune system
process | GO:0060333:
Interferon-gamma-mediated signaling pathway | 19 | 3.28E-20 |
| Biological
process | GO:0043901:
Negative regulation of multi-organism process | 20 | 4.95E-19 |
| Biological
process | GO:0019079: Viral
genome replication | 17 | 1.89E-17 |
| Biological
process | GO:0050792:
Regulation of viral process | 20 | 4.17E-17 |
| Biological
process | GO:0043900:
Regulation of multi-organism process | 29 | 1.63E-16 |
| Biological
process | GO:0031347:
Regulation of defense response | 37 | 1.86E-16 |
| Immune system
process | GO:0045088:
Regulation of innate immune response | 27 | 1.31E-15 |
| Immune system
process | GO:0050778:
Positive regulation of immune response | 36 | 4.79E-15 |
| Biological
process | GO:0044403:
Symbiosis, encompassing mutualism through parasitism | 36 | 1.26E-14 |
| Biological
process | GO:0035456:
Response to interferon-beta | 9 | 8.91E-14 |
Following the KEGG analysis, 27 KEGG pathways that
were significantly enriched with DEGs were identified (Table II). The top five significantly
enriched KEGG pathways were as follows: Herpes simplex infection,
influenza A, antigen processing and presentation, measles, and
hepatitis C.
 | Table IIEnriched Kyoto Encyclopedia of Genes
and Genomes (KEGG) pathways of differentially expressed genes
(P<0.05). |
Table II
Enriched Kyoto Encyclopedia of Genes
and Genomes (KEGG) pathways of differentially expressed genes
(P<0.05).
| Term | Description | Count | P-value |
|---|
| KEGG:05168 | Herpes simplex
infection | 21 | 2.31E-13 |
| KEGG:05164 | Influenza A | 18 | 7.66E-11 |
| KEGG:04612 | Antigen processing
and presentation | 13 | 1.30E-10 |
| KEGG:05162 | Measles | 13 | 8.29E-08 |
| KEGG:05160 | Hepatitis C | 11 | 4.99E-06 |
| KEGG:04610 | Complement and
coagulation cascades | 8 | 1.14E-05 |
| KEGG:05133 | Pertussis | 8 | 1.85E-05 |
| KEGG:05150 | Staphylococcus
aureus infection | 6 | 2.96E-04 |
| KEGG:05330 | Allograft
rejection | 5 | 4.14E-04 |
| KEGG:04145 | Phagosome | 9 | 4.41E-04 |
| KEGG:05332 | Graft-versus-host
disease | 5 | 5.44E-04 |
| KEGG:04940 | Type I diabetes
mellitus | 5 | 6.20E-04 |
| KEGG:04622 | RIG-I-like receptor
signaling pathway | 6 | 5.76E-04 |
| KEGG:05142 | Chagas disease
(American trypanosomiasis) | 7 | 7.30E-04 |
| KEGG:05320 | Autoimmune thyroid
disease | 5 | 1.17E-03 |
| KEGG:05416 | Viral
myocarditis | 5 | 1.79E-03 |
| KEGG:04620 | Toll-like receptor
signaling pathway | 6 | 3.97E-03 |
| KEGG:04623 | Cytosolic
DNA-sensing pathway | 4 | 1.47E-02 |
| KEGG:05143 | African
trypanosomiasis | 3 | 1.43E-02 |
| KEGG:04514 | Cell adhesion
molecules (CAMs) | 6 | 1.55E-02 |
| KEGG:05020 | Prion diseases | 3 | 1.52E-02 |
| KEGG:05161 | Hepatitis B | 6 | 1.46E-02 |
| KEGG:04919 | Thyroid hormone
signaling pathway | 5 | 2.32E-02 |
| KEGG:03050 | Proteasome | 3 | 2.29E-02 |
| KEGG:05144 | Malaria | 3 | 2.92E-02 |
| KEGG:04064 | NF-kappa B
signaling pathway | 4 | 3.31E-02 |
| KEGG:00330 | Arginine and
proline metabolism | 3 | 4.74E-02 |
PPI network construction and hub gene
identification
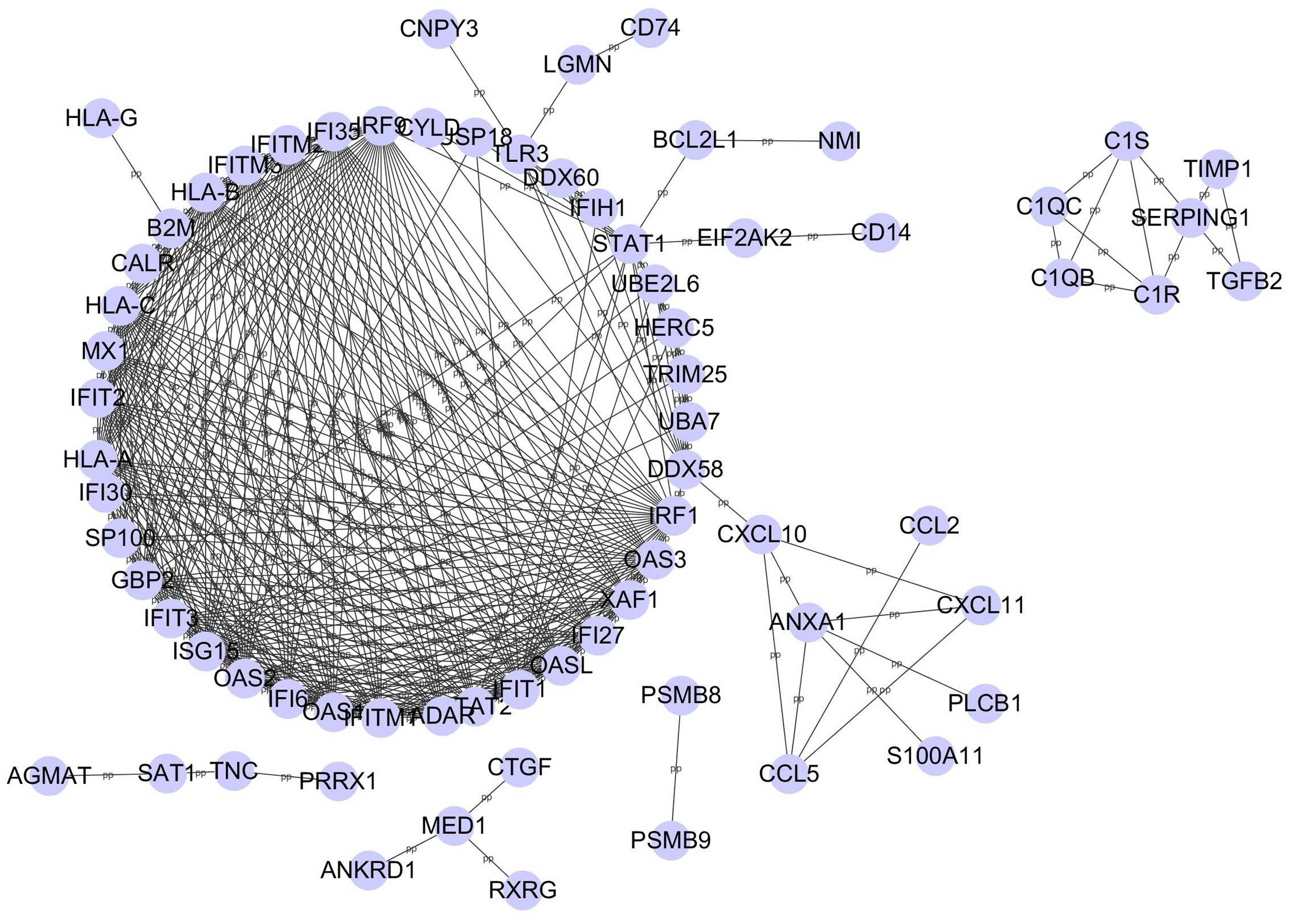
The PPI networks were constructed using Cytoscape
based on the STRING database, and included 71 nodes and 389 edges
with a combined score >0.99 (Fig.
1). The degree of all nodes was analyzed and genes with a node
degree >20 are listed in Table
III. Genes with a high degree of association included: ISG15
ubiquitin-like modifier (ISG15), interferon regulatory factor 1
(IRF1), interferon-induced protein with tetratricopeptide repeats 1
(IFIT1), MHC, class I, A (HLA-A), HLA-B, HLA-C, guanylate binding
protein 2 (GBP2), IRF9, 2′–5′-oligoadenylate synthetase-like
(OASL), 2′–5′-oligoadenylate synthetase 1 (OAS1), OAS2, OAS3,
STAT2, IFIT2, IFIT3, MX dynamin-like GTPase1 (MX1), interferon
induced transmembrane protein 1 (IFITM1), IFITM2, IFITM3,
interferon, alpha-inducible protein 6 (IFI6), IFI27, IFI35, XIAP
associated factor 1 (XAF1) and adenosine deaminase, RNA-specific
(ADAR). Among those hub genes, ISG15 exhibited the highest degree
(degree, 30).
 | Table IIIDegree of each hub node in the
protein-protein interaction networks. |
Table III
Degree of each hub node in the
protein-protein interaction networks.
| Gene symbol | Degree |
|---|
| ISG15 | 30 |
| IRF1 | 29 |
| IFIT1 | 28 |
| HLA-A, HLA-B,
HLA-C, GBP2, IRF9 | 27 |
| OASL, OAS1, OAS2,
OAS3 | 26 |
| STAT2 | 24 |
| IFIT2, IFIT3, MX1,
IFITM1, IFITM2, IFITM3, IFI6, IFI27, IFI35, XAF1, ADAR | 23 |
Discussion
DM is an autoimmune disease, which seriously impacts
the quality of life of affected patients. The underlying
pathogenesis of DM remains elusive. The present study analyzed gene
expression profiles, identified the DEGs associated with DM, and
enabled the identification of potential therapeutic targets. In the
present study, 201 DEGs were identified, of which 180 were
upregulated and 21 were downregulated. Following GO functional
enrichment analysis using ClueGO, the type I IFN signaling pathway
was revealed to be the most significantly enriched GO term within
the DEGs. Previous studies have reported that patients with DM
exhibit an activated type I IFN response, as demonstrated by
increased type I IFN activity, and an IFN signature in the muscle,
blood and skin (1,18,19),
which has also been described in patients with systemic lupus
erythematosus, rheumatoid arthritis, Sjögren's syndrome and
systemic sclerosis (20). In
addition, type 1 IFN-producing plasmacytoid dendritic cells are
abundant in DM muscle and skin (21). When compared with other types of
IM, the overproduction of IFN-I inducible transcripts and proteins
in muscle tissue is a unique feature of DM (22). Furthermore, the iatrogenic
administration of recombinant IFNs has been reported to induce DM
(23). Taken together, these data
indicated that the type I IFN signaling pathway may be important in
the pathogenesis of DM.
A KEGG pathway enrichment analysis of the DEGs was
conducted, and 27 significantly enriched pathways were identified.
The majority of these pathways can be divided into two categories:
Infectious diseases and the immune system. The enriched immune
system-related pathways include antigen processing and
presentation, complement and coagulation cascades pathway,
RIG-I-like receptor (RLR) signaling pathway, Toll-like receptor
(TLR) signaling pathway, and cytosolic DNA-sensing pathway. Among
them, the TLR signaling, RLR signaling and cytosolic DNA-sensing
pathways are responsible for detecting pathogens and generating
innate immune responses. Type I IFNs are primarily induced by the
activation of pattern-recognition receptors, such as TLRs, RLRs and
cytoplasmic DNA sensor receptors (24). In the present study, chemokine (C-C
motif) ligand 5 (CCL5), CD14, C-X-C motif chemokine 10 (CXCL10),
CXCL11, STAT1 and TLR3 DEGs were enriched in the TLR signaling
pathway, whereas CXCL10, CYLD lysine 63 deubiquitinase, DEXD/H-Box
helicase 58 (DDX58), interferon induced with helicase C domain 1
(IFIH1), ISG15 and tripartite motif-containing protein 25 DEGs were
enriched in the RLR signaling pathway. Furthermore, ADAR, CCL5,
CXCL10 and DDX58 DEGs participated in the cytosolic DNA-sensing
pathway. CCL5 is a key proinflammatory chemokine, which may have a
role in driving recruitment of leukocytes, angiogenesis and
fibrosis in chronic inflammatory diseases. Furthermore, CCL5
blockade is able to prevent immunopathology (25). CXCL10 and CXCL11 are members of the
CXC subfamily of chemokines. Serum levels of CXCL10 are elevated in
patients with active DM (26).
CXCL10 has also been reported to be secreted by human skeletal
muscle cells, which promote inflammation (27). A previous study using an animal
model of C-protein-induced myositis demonstrated that suppressing
CXCL10 with monoclonal antibodies was able to inhibit muscular
inflammation (28). TLR3 is a
member of the TLR family, which has a fundamental role in pathogen
recognition and activation of innate immunity. An in vitro
study revealed that TLR3 is involved in the overexpression of MHC-I
in IM (29). Cappelletti et
al reported that TLR3 is localized on vascular endothelial
cells, muscle infiltrating cells and regenerating myofibers in DM
(30). In addition, Li et
al (31) demonstrated that
TLR-3 and RIG-I are implicated in the formation of perifascicular
atrophy in DM.
RLRs comprise DDX58 (RIG-I), IFIH1 [also known as
melanoma differentiation-associated protein 5 (MDA5)] and LGP2, and
are known as cytosolic RNA receptors that induce the expression of
type I IFN genes. Sato et al (32) detected anti-clinically amyopathic
DM (CADM)-140 antibodies encoded by MDA5 in patients with
amyopathic DM; of these patients, 50% had rapidly progressive
interstitial lung disease (RP-ILD). In addition, it has been
reported that anti-MDA5 autoantibodies may be useful for diagnosing
DM, particularly the CADM and RP-ILD subsets among Asian cohorts
(33). Suárez-Calvet et al
(12) reported that RIG-I was
upregulated in pathological muscle fibers from patients with DM
compared with other types of IM, as determined by
immunohistochemistry. In addition, human myotubes were shown to
produce IFN-β in response to RIG-I stimulation, and the autocrine
effects of IFN-β could further amplify the expression of IFN genes.
The results of the present KEGG analysis highlighted the role of
the type I IFN signaling pathway in the pathogenesis of DM, and
suggested that DEGs associated with these pathways, including CCL5,
CXCL10, TLR3, DDX58 and IFIH1, may be used as potential targets for
DM diagnosis and treatment.
Hub nodes have more complex interactions compared
with other genes, thus indicating that they have important roles in
the underlying mechanisms of disease (34). Therefore, identification of hub
genes may facilitate the development of effective therapeutic
approaches for the treatment of patients with DM. The following
DEGs: ISG15, IRF1, IFIT1, HLA-A, HLA-B, HLA-C, GBP2, IRF9, OASL,
OAS1, OAS2, OAS3, STAT2, IFIT2, IFIT3, MX1, IFITM1, IFITM2, IFITM3,
IFI6, IFI27, IFI35, XAF1 and ADAR were identified as hub genes in
the PPI networks, thus indicating that these genes may have pivotal
roles in DM development. Notably, all of these hub genes are
involved in the type I IFN signaling pathway in DM. The ISG15 gene
had the highest degree in the PPI networks (degree, 30). Salajegheh
et al demonstrated that ISG15 is one of the most upregulated
genes in DM muscle compared with normal muscle, and the ISG15
protein localizes to atrophic myofibers, as determined by
immunohistochemistry (23). ISG15
is a ubiquitin-like modifier (35), which is believed to function as a
possible mediator of muscle atrophy via protein conjugation, and
may contribute to disease activity in DM. MX1, IFIT1 and IFIT3 have
been detected in DM muscle, and have previously been identified as
ISG15-conjugated in IFN-β-stimulated HeLa cells (36). Furthermore, MX1 is an IFN-I-induced
protein that provides innate defense against several viruses. A
previous study reported that MX1 is abundant in DM muscle and skin,
and is specifically expressed in DM but not in other types of IM
(37). In addition, MX1 RNA in
juvenile DM peripheral blood mononuclear cells is positively
correlated with muscle involvement (38). Therefore, ISG15 and MX1 may
contribute to disease activity, and may be considered potential
molecular markers and effective treatment targets in DM.
Human leukocyte antigen (HLA-A, HLA-B, HLA-C) is the
human-specific MHC. A previous study reported that MHC-I antigens
were not detected in normal muscle, but were upregulated in IM
muscle (39). In transgenic mice,
overexpression of MHC-I is sufficient to induce myositis (40). The induction of MHC-I in IM may
involve type I IFN and the 'IFN-I signature' is also induced in IMs
(10). These data suggested that
MHC-I may be considered a useful adjunctive diagnostic test for
DM.
Other hub genes associated with the type I IFN
signaling pathway may also have an important role in the
pathogenesis of DM. For example, the OAS family proteins, which are
predominantly induced by IFN type I, consist of OAS1, OAS2, OAS3
and OASL. OAS levels are strongly associated with autoimmune
diseases (41). Sanayama et
al (42) proposed that OASL
may be a biomarker for the response of patients with rheumatoid
arthritis to tocilizumab, which is the most commonly used drug to
treat this disease. Identification of these genes and their precise
roles may clarify the mechanism and offer useful information
regarding the treatment of DM.
In conclusion, the present study attempted to
explore the potential molecular mechanisms underlying DM using
bioinformatics analyses. The findings of the present study may
contribute to our understanding regarding the molecular mechanisms
underlying DM. The identified DEGs, including CCL5, CXCL10, TLR3,
DDX58, IFIH1, ISG15 and MX1, may potentially be used as targets for
DM diagnosis and treatment. However, further studies are required
to further verify this hypothesis.
Acknowledgments
This work was partly supported by the Natural
Science Foundation of China (grant no. 81170562); The abstract of
the current study was partly presented at the 14th
Chinese National Conference of Medical Genetics proceedings.
References
|
1
|
Wong D, Kea B, Pesich R, Higgs BW, Zhu W,
Brown P, Yao Y and Fiorentino D: Interferon and biologic signatures
in dermatomyositis skin: Specificity and heterogeneity across
diseases. PLoS One. 7:e291612012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bendewald MJ, Wetter DA, Li X and Davis
MD: Incidence of dermatomyositis and clinically amyopathic
dermatomyositis: A population-based study in Olmsted County,
Minnesota. Arch Dermatol. 146:26–30. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Mantegazza R, Bernasconi P, Confalonieri P
and Cornelio F: Inflammatory myopathies and systemic disorders: A
review of immunopathogenetic mechanisms and clinical features. J
Neurol. 244:277–287. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Nickoloff BJ and Nestle FO:
Dermatomyositis. Dermatologic Immunity Curr Dir Autoimmun. 10.
Karger AG; Basel: pp. 313–332. 2008, View Article : Google Scholar
|
|
5
|
Hill CL, Zhang Y, Sigurgeirsson B, Pukkala
E, Mellemkjaer L, Airio A, Evans SR and Felson DT: Frequency of
specific cancer types in dermatomyositis and polymyositis: A
population-based study. Lancet. 357:96–100. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Antiga E, Kretz CC, Klembt R, Massi D,
Ruland V, Stumpf C, Baroni G, Hartmann M, Hartschuh W, Volpi W, et
al: Characterization of regulatory T cells in patients with
dermatomyositis. J Autoimmun. 35:342–350. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Reed AM and Ytterberg SR: Genetic and
environmental risk factors for idiopathic inflammatory myopathies.
Rheum Dis Clin North Am. 28:891–916. 2002. View Article : Google Scholar
|
|
8
|
Rothwell S, Cooper RG, Lamb JA and Chinoy
H: Entering a new phase of immunogenetics in the idiopathic
inflammatory myopathies. Curr Opin Rheumatol. 25:735–741. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Dalakas MC: Muscle biopsy findings in
inflammatory myopathies. Rheum Dis Clin North Am. 28:779–798. 2002.
View Article : Google Scholar
|
|
10
|
Hornung T and Wenzel J: Innate
immune-response mechanisms in dermatomyositis: An update on
pathogenesis, diagnosis and treatment. Drugs. 74:981–998. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Greenberg SA: Sustained autoimmune
mechanisms in dermatomyositis. J Pathol. 233:215–216. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Suárez-Calvet X, Gallardo E, Nogales-Gadea
G, Querol L, Navas M, Díaz-Manera J, Rojas-Garcia R and Illa I:
Altered RIG-I/DDX58-mediated innate immunity in dermatomyositis. J
Pathol. 233:258–268. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Diboun I, Wernisch L, Orengo CA and
Koltzenburg M: Microarray analysis after RNA amplification can
detect pronounced differences in gene expression using limma. BMC
Genomics. 7:2522006. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Bindea G, Mlecnik B, Hackl H, Charoentong
P, Tosolini M, Kirilovsky A, Fridman WH, Pagès F, Trajanoski Z and
Galon J: ClueGO: A Cytoscape plug-in to decipher functionally
grouped gene ontology and pathway annotation networks.
Bioinformatics. 25:1091–1093. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
von Mering C, Huynen M, Jaeggi D, Schmidt
S, Bork P and Snel B: STRING: A database of predicted functional
associations between proteins. Nucleic Acids Res. 31:258–261. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lamb J, Crawford ED, Peck D, Modell JW,
Blat IC, Wrobel MJ, Lerner J, Brunet JP, Subramanian A, Ross KN, et
al: The Connectivity Map: Using gene-expression signatures to
connect small molecules, genes, and disease. Science.
313:1929–1935. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Baechler EC, Bauer JW, Slattery CA,
Ortmann WA, Espe KJ, Novitzke J, Ytterberg SR, Gregersen PK,
Behrens TW and Reed AM: An interferon signature in the peripheral
blood of dermatomyositis subjects is associated with disease
activity. Mol Med. 13:59–68. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kao L, Chung L and Fiorentino DF:
Pathogenesis of dermatomyositis: Role of cytokines and interferon.
Curr Rheumatol Rep. 13:225–232. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Greenberg SA: A gene expression approach
to study perturbed pathways in myositis. Curr Opin Rheumatol.
19:536–541. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Walsh RJ, Kong SW, Yao Y, Jallal B, Kiener
PA, Pinkus JL, Beggs AH, Amato AA and Greenberg SA: Type I
interferon-inducible gene expression in blood is present and
reflects disease activity in dermatomyositis and polymyositis.
Arthritis Rheum. 56:3784–3792. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hall JC and Rosen A: Type I interferons:
Crucial participants in disease amplification in autoimmunity. Nat
Rev Rheumatol. 6:40–49. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Salajegheh M, Kong SW, Pinkus JL, Walsh
RJ, Liao A, Nazareno R, Amato AA, Krastins B, Morehouse C, Higgs
BW, et al: Interferon-stimulated gene 15 (ISG15) conjugates
proteins in dermatomyositis muscle with perifascicular atrophy. Ann
Neurol. 67:53–63. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Härtlova A, Erttmann SF, Raffi FA, Schmalz
AM, Resch U, Anugula S, Lienenklaus S, Nilsson LM, Kröger A,
Nilsson JA, et al: DNA damage primes the type I interferon system
via the cytosolic DNA sensor STING to promote anti-microbial innate
immunity. Immunity. 42:332–343. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Marques RE, Guabiraba R, Russo RC and
Teixeira MM: Targeting CCL5 in inflammation. Expert Opin Ther
Targets. 17:1439–1460. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Bilgic H, Ytterberg SR, Amin S, McNallan
KT, Wilson JC, Koeuth T, Ellingson S, Newman B, Bauer JW, Peterson
EJ, et al: Interleukin-6 and type I interferon-regulated genes and
chemokines mark disease activity in dermatomyositis. Arthritis
Rheum. 60:3436–3446. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Crescioli C, Sottili M, Bonini P, Cosmi L,
Chiarugi P, Romagnani P, Vannelli GB, Colletti M, Isidori AM, Serio
M, et al: Inflammatory response in human skeletal muscle cells:
CXCL10 as a potential therapeutic target. Eur J Cell Biol.
91:139–149. 2012. View Article : Google Scholar
|
|
28
|
Kim J, Choi JY, Park SH, Yang SH, Park JA,
Shin K, Lee EY, Kawachi H, Kohsaka H and Song YW: Therapeutic
effect of anti-C-X-C motif chemokine 10 (CXCL10) antibody on C
protein-induced myositis mouse. Arthritis Res Ther. 16:R1262014.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Tournadre A, Lenief V, Eljaafari A and
Miossec P: Immature muscle precursors are a source of interferon-β
in myositis: Role of Toll-like receptor 3 activation and
contribution to HLA class I up-regulation. Arthritis Rheum.
64:533–541. 2012. View Article : Google Scholar
|
|
30
|
Cappelletti C, Baggi F, Zolezzi F,
Biancolini D, Beretta O, Severa M, Coccia EM, Confalonieri P,
Morandi L, Mora M, et al: Type I interferon and Toll-like receptor
expression characterizes inflammatory myopathies. Neurology.
76:2079–2088. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Li L, Dai T, Lv J, Ji K, Liu J, Zhang B
and Yan C: Role of Toll-like receptors and retinoic acid inducible
gene I in endogenous production of type I interferon in
dermatomyositis. J Neuroimmunol. 285:161–168. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Sato S, Hoshino K, Satoh T, Fujita T,
Kawakami Y, Fujita T and Kuwana M: RNA helicase encoded by melanoma
differentiation-associated gene 5 is a major autoantigen in
patients with clinically amyopathic dermatomyositis: Association
with rapidly progressive interstitial lung disease. Arthritis
Rheum. 60:2193–2200. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Lu X, Peng Q and Wang G: Discovery of new
biomarkers of idiopathic inflammatory myopathy. Clin Chim Acta.
444:117–125. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Langfelder P, Mischel PS and Horvath S:
When is hub gene selection better than standard meta-analysis? PloS
One. 8:e615052013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
D'Cunha J, Knight E Jr, Haas AL, Truitt RL
and Borden EC: Immunoregulatory properties of ISG15, an
interferon-induced cytokine. Proc Natl Acad Sci USA. 93:211–215.
1996. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhao C, Denison C, Huibregtse JM, Gygi S
and Krug RM: Human ISG15 conjugation targets both IFN-induced and
constitutively expressed proteins functioning in diverse cellular
pathways. Proc Natl Acad Sci USA. 102:10200–10205. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Arshanapalli A, Shah M, Veerula V and
Somani AK: The role of type I interferons and other cytokines in
dermatomyositis. Cytokine. 73:319–325. 2015. View Article : Google Scholar
|
|
38
|
O'Connor KA, Abbott KA, Sabin B, Kuroda M
and Pachman LM: MxA gene expression in juvenile dermatomyositis
peripheral blood mononuclear cells: Association with muscle
involvement. Clin Immunol. 120:319–325. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Salaroli R, Baldin E, Papa V, Rinaldi R,
Tarantino L, De Giorgi LB, Fusconi M, Malavolta N, Meliconi R,
D'Alessandro R and Cenacchi G: Validity of internal expression of
the major histocompatibility complex class I in the diagnosis of
inflammatory myopathies. J Clin Pathol. 65:14–19. 2012. View Article : Google Scholar
|
|
40
|
van der Pas J, Hengstman GJ, ter Laak HJ,
Borm GF and van Engelen BG: Diagnostic value of MHC class I
staining in idiopathic inflammatory myopathies. J Neurol Neurosurg
Psychiatry. 75:136–139. 2004.PubMed/NCBI
|
|
41
|
Choi UY, Kang JS, Hwang YS and Kim YJ:
Oligoadenylate synthase-like (OASL) proteins: Dual functions and
associations with diseases. Exp Mol Med. 47:e1442015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Sanayama Y, Ikeda K, Saito Y, Kagami S,
Yamagata M, Furuta S, Kashiwakuma D, Iwamoto I, Umibe T, Nawata Y,
et al: Prediction of therapeutic responses to tocilizumab in
patients with rheumatoid arthritis: Biomarkers identified by
analysis of gene expressionin peripheral blood mononuclear cells
using genome-wide DNA microarray. Arthritis Rheumatol.
66:1421–1431. 2014. View Article : Google Scholar : PubMed/NCBI
|