Introduction
Interferon-induced transmembrane protein 3 (IFITM3)
is a member of the gene family encoding IFITM, which can be
upregulated by type I IFN (IFN-α and IFN-β) and type II IFN
(IFN-γ). In humans, the IFITM gene family comprises at least four
members, termed IFITM1, IFITM2, IFITM3 and IFITM5, respectively
(1). The IFITM genes are clustered
on chromosome 11, encoding for proteins upregulated by IFN. With
the exception of IFITM5, the genomic and protein sequence
identities of the IFITM genes are high. Their protein structures
are similar, each containing two transmembrane domains, one
conserved intracellular loop and two extra-cellular terminals. As
one of the IFITM family members, IFITM3 has been demonstrated to be
pivotal in IFN signaling, as downregulating the expression of
IFITM3 using small interfering RNA was observed to reduce the
antiviral activities performed by IFN-γ by 40–70% (2).
Endogenous IFITM3 may have important regulatory
effects intracellularly. IFITM3 is involved in cellular development
and differentiation (3). Previous
studies have identified genetic variations or altered expression of
IFITM3 that may be associated with immune diseases, such as viral
infections (2) schizophrenia
(4), autism (5,6),
inflammatory bowel disease (7) and
cancer (8–10). The human HeLa cell line is one of
the most widely used cell line in molecular biology for
investigating gene functions (11,12),
and the endogenous expression of IFITM3 has been detected in HeLa
cells previously (2). To
investigate the global transcriptional profile when IFITM3 is
downregulated, the present study used a knockdown (KD) approach and
identified a series of altered transcripts when IFITM3 was
downregulated in HeLa cells. Investigating alterations in the
global transcriptional profile may improve current understanding of
the molecular mechanism of the antimicrobial function of
IFITM3.
Materials and methods
KD of endogenous IFITM3
HeLa cells (America Type Culture Collection,
Manassas, VA, USA; cat. no. CRL-2266) at a density of
3–5×106 per T75 flask (Greiner Bio-One, Frickenhausen,
Germany) were cultured in Dulbecco's modified Eagle's medium
(DMEM)/15% fetal bovine serum (FBS)/10 mM HEPES (Invitrogen; Thermo
Fisher Scientific, Inc., Waltham, MA, USA) 1 day prior to
lentiviral infection. Following culture, the IFITM3 short hairpin
(sh)RNA lentivirus (Santa Cruz Biotechnology, Inc.; cat. no.
sc-97053-V) or control shRNA lentivirus (Santa CruzBiotechnology,
Inc.; cat. no. sc-108080), each with a multiplicity of infection of
50%, was added to polybrene with a 8 µg/ml final concentration
(Sigma-Aldrich; Thermo Fisher Scientific, Inc.; cat. no. S2667) to
the cultures and incubated overnight at 37°C. On the third day,
selection medium (DMEM/15%FBS/10 mM HEPES containing 50 µg/ml
puromycin) was added for sorting of the stably infected cells.
After 1 week, the adherent cells were digested with 0.25% trypsin
and 0.02% EDTA, and resuspended for single cell culture at a
density of 1.0×103 cells/well in 96-well plates, with
the same selection medium as that described above, for monoclonal
colony amplification.
Reverse transcription-polymerase chain
reaction (RT-PCR) analysis
Cell selection was performed by adding puromycin to
the cell culture medium, following which the resistant monoclonal
colonies were identified. The efficiency of endogenous IFITM3 KD in
the HeLa cells were confirmed by RT-PCR analysis. The re-cultured
HeLa cells were harvested and RNA was extracted using a standard
TRIzol procedure (Invitrogen; Thermo Fisher Scientific, Inc.; cat.
no. 15596-018). The RNA was subjected to cDNA synthesis using M-MLV
reverse transcriptase (Promega; Madison, WI, USA; cat. no. M170) in
a 20 µl liquid phase reaction, from which 1 µl of cDNA was used for
subsequent PCR amplification. The primers used as an internal
control for RT-PCR to amplify human 18S were as follows: Forward
5′-GGAAGGGCACCACCAGGAGT and reverse 5′-TGCAGCCCCGGACATCTAAG. The
primers used for amplifying the IFITM3 and IFITM3-targeted RNAs via
PCR analysis (Opticon® DNA engine; MJ Research, Waltham,
MA, USA, cat. no. CFD3200) are listed in Table I. SYBR Premix Ex Taq II (Takara
Biotechnology Co., Ltd. Dalian, China) were used for quantitative
(q) PCR in a 25 µl reaction volume with 95°C for 30 sec followed by
40 cycles with each cycle consisting of 95°C for 5 sec and 60°C 30
sec. Relative expression ratio of targeted genes were analyzed
using the 2−∆∆Cq method (13) with 18S as a reference gene.
 | Table I.Primers used for reverse
transcription-quantitative polymerase chain reaction analysis. |
Table I.
Primers used for reverse
transcription-quantitative polymerase chain reaction analysis.
| Gene | Forward primer
(5′-3′) | Reverse primer
(5′-3′) |
|---|
| IFITM3 |
ATGAATCACACTGTCCAAACCTTCT |
CTATCCATAGGCCTGGAAGATCAG |
| VDUP1 |
CGATAGTTTCGGGTCAGG |
GATACATAAGTTCAAGGTCCAA |
| VDR |
CAGGGTGGGATGGAGGAGAAG |
TGGGTGGTGGAGTGAGAATAAGAA |
| UBE2N |
AACTTTATTTAGACGCTGTAGATGG |
AATGTTATTAGTGAGGGCTGTGAT |
| TMED7 |
ATTGGATAGCCATCCTAGTCACT |
GCTGGTCTTCAAACACCGTAA |
| PTGS2 |
TGTCCCTTTACTTCATTCAGTGTTC |
ATGACTCCTTTCTCCGCAACA |
| PLCB4 |
AAGCCTGCTGTAGTTGAGTTGC |
CTTGACGAGTGTTATGCGTGTTT |
| MYD88 |
GAATCCCTGTAGGAAATGGTGAAGC |
AGGAAGTGGAATGGGCGGTGT |
| MAPK13 |
GCCAAATCCTACATCCAGTCCCT |
TCCAGCATCTTCTCCAGCAGGT |
| HIST1H1A1 |
TCCGTGTCAGAGCTGATCGTG |
GCGGCTGTTGTTCTTCTCCAC |
| HIST1H4J |
GATCCGGGACGCCGTGACCTAT |
GGGACGCTCAACCACCGAAACC |
| HIST1H4K |
TCCGGGACGCCGTGACCTATA |
GGACGCTCAACCACCGAAACC |
| HIST1H1B1 |
CTTGCCACCATGTCGGAAACC |
CCAGCTTAATGCGGCTGTTATTCTT |
| HIST1H2AA |
GTGTATTTGGCGGCAGTGTTAGA |
TGCTTTGGGCTTTATGGTGGT |
Analyses of mRNA expression using
oligonucleotide arrays
The RNA KD and control (CT) HeLa cells were cultured
for mRNA extraction three times independently. The three RNA
samples from the KD cells were mixed with equal quality, as were
those from the CT cells. Microarray analysis was performed using a
Human Whole Genome Oligo Microarray (Agilent Technologies, Inc.,
Santa Clara, CA, USA; cat. no. G4112A). According to the protocols
of the Low RNA Input Linear Amplification and Labeling Kit Plus
(Agilent Technologies; cat. no. 5184-3523), double-stranded cDNA
was synthesized and applied as a template to label cRNA. Cy3
(scanned as red) was used to label the cRNA from the KD group from
that of CT group. The ratio of fluorescence intensity for each
probe (KD/CT) was determined to indicate the difference between the
RNA-KD and CT cells. Certain transcripts identified as
IFITM3-targeted were subjected to quantitative (q)PCR analysis to
enable relative quantification. Gene Ontology (GO) enrichment
analysis was performed according to the GO database (www.geneontology.org) to identify significant
functional categories among differentially expressed genes.
Furthermore, pathway analysis was performed using the Kyoto
Encyclopedia of Genes and Genomes (www.genome.jp/kegg) and BioCarta (www.biocarta.com) pathway analysis programs. Each
P-value was calculated with Fisher's exact test using the
R-package, whereas each q-value was calculated with John Storey's
method using the R-package (14).
Results
IFITM3-targeted transcripts
The transcriptional expression profiles of the
endogenous IFITM3-KD and CT HeLa cells were investigated using a
human Agilent GeneChip microarray platform. Using the 2-fold change
as the cut-off, the Agilent microchip revealed 1,011 downregulated
and 615 upregulated transcripts. It was noted that certain
alterations gathered in gene families, including the histone
(HIST), caveolae (CAV), pregnancy-specific β-1-glycoprotein (PSG),
calmodulin (CALM), E twenty-six (ETS) and golgin (GOLG) families
(Fig. 1).
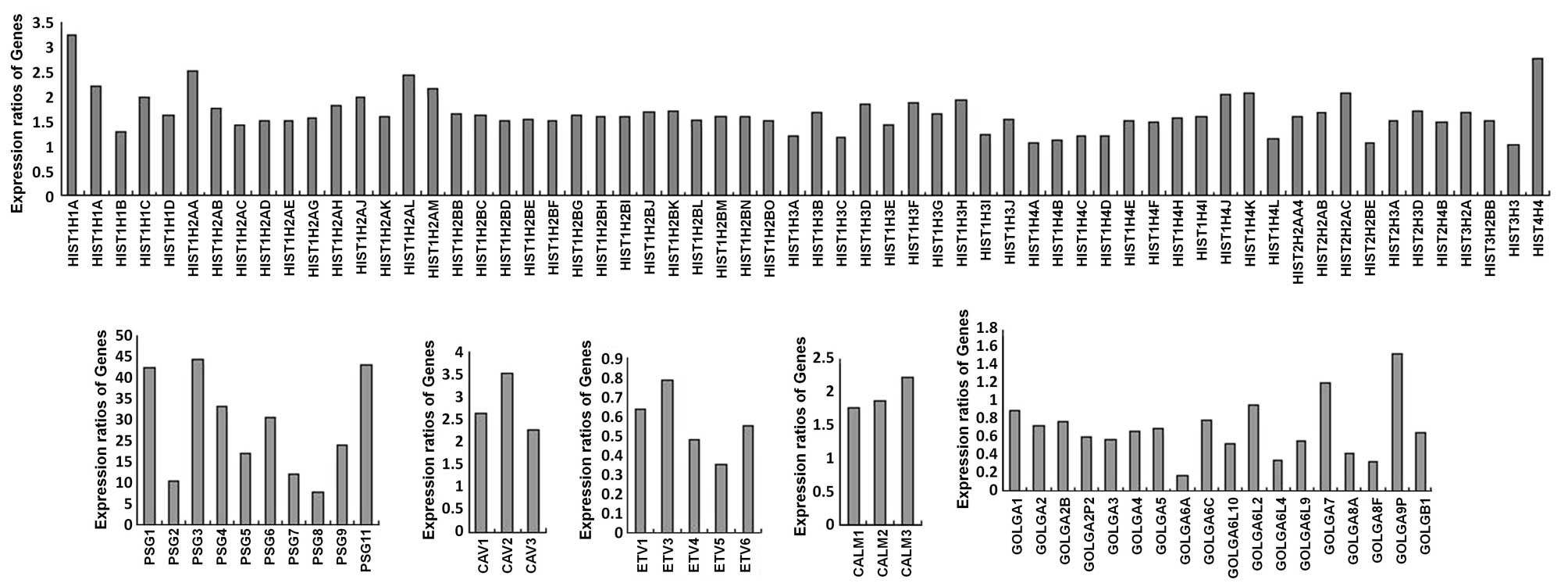 | Figure 1.Gene families altered by interferon
inducible transmembrane protein 3 knockdown. Expression ratios of
altered levels of genes in HIST, CAV, PSG, CALM, ETV and GOLG
families between IFITM3 knocked down cells and control cells were
showed as columns. HIST, histone, CAV, caveolae; PSG,
pregnancy-specific β-1-glycoprotein; CALM, calmodulin; ETV, ets
variant; GOLG, golgin. |
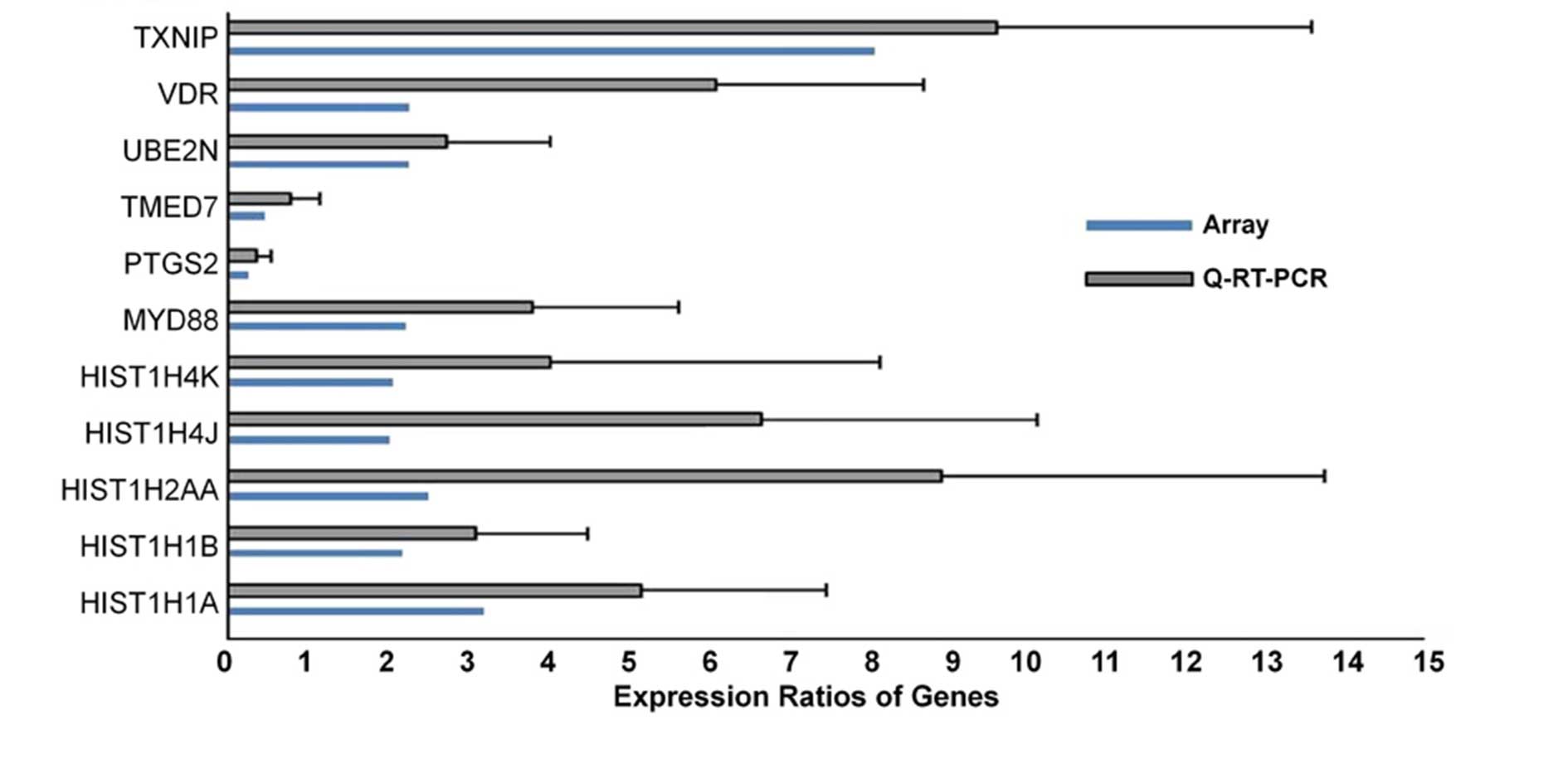
Validation of IFITM3-targeted
transcripts using qPCR analysis
Several IFITM3-targeted transcripts of the HIST
family, including HIST cluster 1 (HIST1) H1a, HIST1 H1b, HIST1
H2aa, HIST1 H4j and HIST1 h4k, were further assessed using RT-qPCR
analysis. In addition, certain additional transcripts, including
mitogen-activated protein kinase 13, myeloid differentiation
primary response 88, prostaglandin-endoperoxide synthase 2,
transmembrane emp24 protein transport domain containing 7,
ubiquitin-conjugating enzyme E2 N, vitamin D receptor and
thioredoxin interacting protein, were randomly selected for
validation. The gene expression levels determined using RT-qPCR
analysis appeared coincident with those of the microarray (Fig. 2).
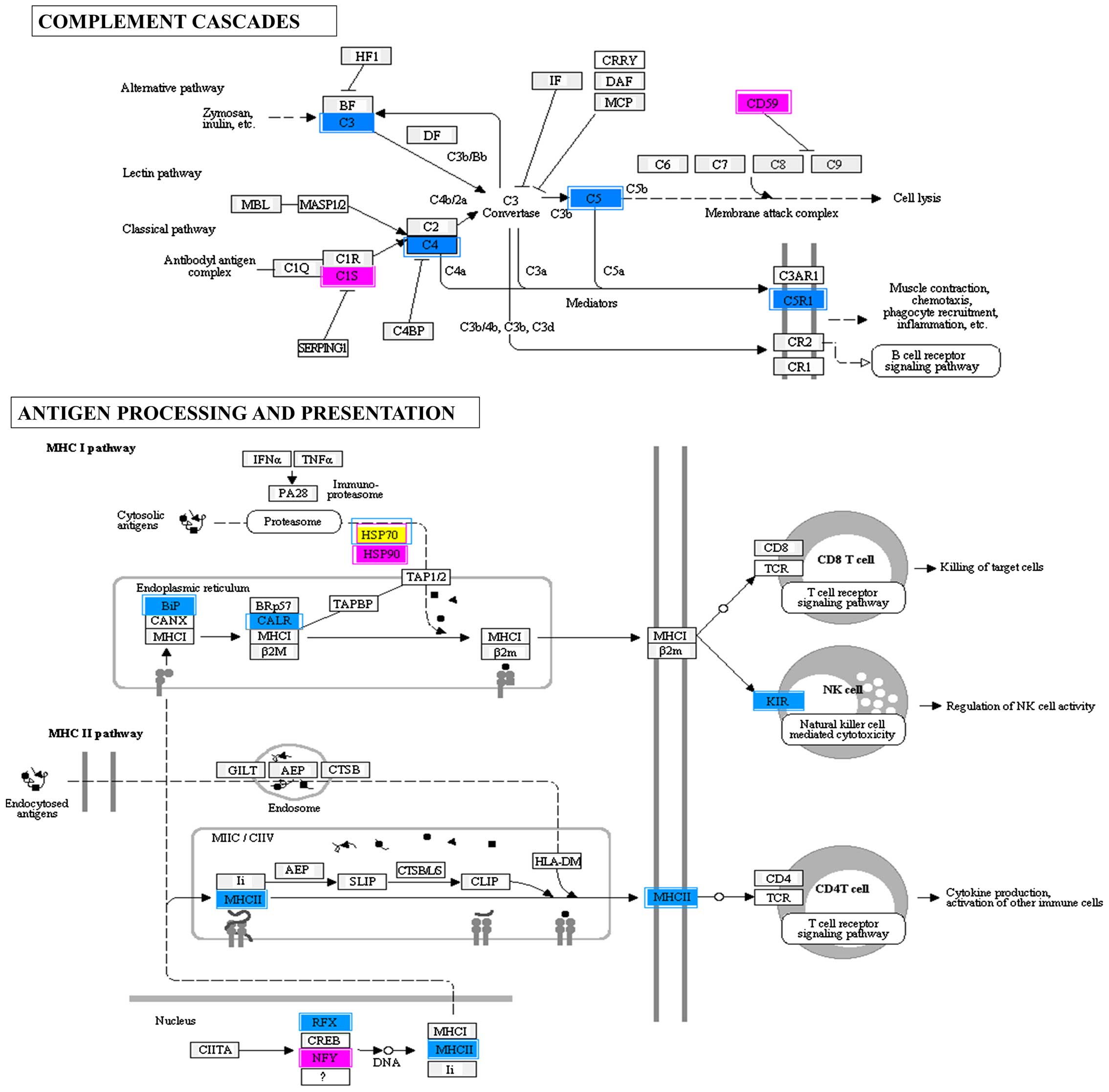
Analysis of altered pathways
In the present study, when endogenous expression of
IFITM3 in the HeLa cells was reduced, several genes in the antigen
processing and presentation pathway appeared to be either
upregulated or downregulated.
The IFITM3-downregulated targets were subjected to
clustering pathway analysis, and the associated gene pathways (21
upregulated and 21 downregulated) are listed in Table II. Alterations of gene expression
involving the complement cascades, antigen processing and
presentation pathways are detailed independently in Fig. 3. It was found that the majority of
the transcripts detected in the complement pathway exhibited a
downregulatory trend, particularly transcripts of complement
component 3 (C3), complement component 4b (C4B) and complement
component 5 (C5). In terms of the antigen processing and
presentation pathway, transcripts of heat shock protein 90 kDa α
class A member 1, heat shock 70 kDa protein 1A, heat shock 70 kDa
protein 1-like, heat shock 70 kDa protein 2, heat shock 70 kDa
protein 8, nuclear transcription factor Y (NFY) α and NFY β were
upregulated, whereas the transcripts of heat shock 70 kDa protein
5, heat shock 70 kDa protein 6, regulatory factor X-associated
protein, killer cell immunoglobulin-like receptor (KIR) three
domains and long cytoplasmic tail 3, KIR two domains and short
cytoplasmic tail 4, calreticulin and major histocompatibility
complex class II, DQ α2, were downregulated.
 | Table II.Associated pathways with altered
regulation. |
Table II.
Associated pathways with altered
regulation.
| Pathway | Count | P-value | q-value |
|---|
| Upregulated |
|
|
|
| Focal
adhesion | 20 | 1.2e-15 | 2.0e-13 |
| Calcium
signaling pathway | 14 | 6.5e-10 | 1.1e-8 |
|
Regulation of actin
cytoskeleton | 13 | 4.9e-8 | 5.1e-7 |
| Gap
junction | 9 | 9.6e-8 | 8.9e-7 |
| MAPK
signaling pathway | 13 | 7.1e-7 | 0.0000048 |
| Wnt
signaling pathway | 10 | 7.1e-7 | 0.0000048 |
|
Glioma | 7 | 0.0000013 | 0.0000078 |
|
Nitrogen metabolism | 5 | 0.0000015 | 0.0000090 |
|
Cytokine-cytokine receptor
interaction | 12 | 0.0000026 | 0.000013 |
|
Melanogenesis | 8 | 0.0000029 | 0.000014 |
|
Pathogenic Escherichia
coli infection | 6 | 0.0000068 | 0.000026 |
|
ECM-receptor interaction | 7 | 0.0000072 | 0.000026 |
| Antigen
processing and presentation | 7 | 0.000011 | 0.000034 |
| Axon
guidance | 8 | 0.000015 | 0.000047 |
|
Complement and coagulation
cascades | 6 | 0.000026 | 0.000074 |
|
Systemic lupus
erythematosus | 8 | 0.000032 | 0.000088 |
|
Long-term potentiation | 6 | 0.000041 | 0.00010 |
| B cell
receptor signaling pathway | 6 | 0.000041 | 0.00010 |
|
Phosphatidylinositol signaling
system | 6 | 0.000045 | 0.00011 |
|
Prostate cancer | 6 | 0.00010 | 0.00023 |
| Downregulated |
|
|
|
| MAPK
signaling pathway | 19 | 4.4e-10 | 9.1e-9 |
|
Arachidonic acid
metabolism | 9 | 1.5e-8 | 1.5e-7 |
|
Glycine, serine and threonine
metabolism | 8 | 2.4e-8 | 2.1e-7 |
|
Long-term depression | 9 | 1.1e-7 | 6.8e-7 |
| Calcium
signaling pathway | 12 | 0.0000013 | 0.0000066 |
|
Cytokine-cytokine receptor
interaction | 14 | 0.0000019 | 0.0000091 |
| VEGF
signaling pathway | 8 | 0.0000023 | 0.000010 |
| Cell
adhesion molecules | 10 | 0.0000031 | 0.000012 |
| GnRH
signaling pathway | 9 | 0.0000036 | 0.000014 |
| Insulin
signaling pathway | 10 | 0.0000040 | 0.000015 |
|
Nitrogen metabolism | 5 | 0.0000062 | 0.000022 |
|
Aminoacyl-tRNA
biosynthesis | 6 | 0.0000062 | 0.000022 |
|
Linoleic acid metabolism | 5 | 0.000020 | 0.000057 |
|
Histidine metabolism | 5 | 0.000023 | 0.000065 |
|
Neuroactive ligand-receptor
interaction | 12 | 0.000038 | 0.00010 |
| Antigen
processing and presentation | 7 | 0.000067 | 0.00016 |
|
Tryptophan metabolism | 5 | 0.00012 | 0.00026 |
|
Complement and coagulation
cascades | 6 | 0.00013 | 0.00028 |
| Type II
diabetes mellitus | 5 | 0.00015 | 0.00031 |
|
Regulation of actin
cytoskeleton | 10 | 0.00020 | 0.00040 |
Discussion
Alterations in expression gathered in
gene families
When IFITM3 was downregulated by shRNA, the HeLa
cells showed an altered gene expression profile. The expression
levels of hundreds of gene transcripts were altered and associated
with multiple gene pathways. The differentially expressed profile
obtained in the present study provided evidence that IFITM3 is
involved in regulating a broad range of transcripts, and this is
likely to result in different consequences in terms of the
biological functions of cells. Of note, similar changes were found
to be gathered in families.
It was found that the majority of gene transcripts
in the HIST, CAV, PSG and CALM families, were upregulated. Histones
are basic nuclear proteins, which are responsible for the
nucleosome structure within the chromosomal fiber in eukaryotes.
The expression of HIST genes is coupled temporally and functionally
with DNA replication, and is controlled at the transcriptional and
post-transcriptional levels (15).
The PSG family belongs to a member of the immunoglobulin
superfamily (16). Evidence shows
that it is involved in modulation of the innate immune system
(17). All the CALM family gene
members (CALM1, CALM2 and CALM3) encode an identical protein,
calmodulin, which modulates a calcium-activated cadherin function.
CALM members are considered to be involved in the intracellular
invasion and colonization of human intestinal epithelial cells by
Campylobacter jejuni in vitro. CALM1 was found to exist
close to a risk locus of C. jejuni colonization in the avian
intestine in a population-based genome-wide association study
(18).
By contrast, the majority of the gene transcripts in
the ETS family and GOLG family were downregulated. The ETS proteins
are transcription factors; they regulate several target genes,
which modulate cellular functions, including growth, apoptosis,
development, differentiation and oncogenic transformation (19). The GOLG family of proteins localize
to the Golgi, and appear to be involved in membrane traffic and
Golgi structure; individual golgins are found in different
locations in the Golgi stack, and are typically anchored to the
membrane at their carboxyl termini by a transmembrane domain or by
binding a small GTPase (20).
Although these altered gene families require further validation to
confirm their association with IFITM3, the results of the present
study provide insight for an improved understanding of the
molecular functions of IFITM3.
Potential role of IFITM against
microorganisms
Previously, IFITM3 has been found to inhibit the
replication of human immunodeficiency virus-1 (21). In addition, IFITMs, particularly
IFITM3, have been confirmed to inhibit viral infections, by
influenza A virus, flaviviruses (dengue virus, West Nile virus and
hepatitis C) and filoviruses (Ebola virus and Marburg virus)
(22–24). In our previous study, a functional
polymorphism of IFITM3 was found to contribute to tuberculosis
susceptibility (25). Thus, IFITM3
may limit microorganism infection by adjusting the host immune
ability. However, the precise mechanisms remain to be fully
elucidated.
As is known, the control of pathogen infections is
critically dependent on the recognition and elimination of infected
cells. As a biological process of the innate immune system, antigen
processing prepares antigens for presentation to specific cells of
the immune system.
According to the clustering pathway analysis
performed in the present study, when the endogenous expression of
IFITM3 in HeLa cells was reduced, 21 upregulated and 21
downregulated pathways were identified. Several genes in the
antigen processing and presentation pathway appeared to be either
upregulated or downregulated. Of note, the majority of the
components in complement signaling were downregulated, particularly
C3, C4b, and C5 (Table III).
Among these altered complement molecules, C5 showed the most marked
reduction. As a feature of the innate immune system, the complement
system assists in clearing pathogens from an organism. The
complement system can be activated by the classical complement
pathway, the alternative complement pathway or the mannose-binding
lectin pathway (26,27). In all three pathways, a
C3-convertase cleaves and activates C3, creating C3a and C3b, and
causing a cascade of further cleavage and activation events. C3b
binds to the surface of pathogens, leading to increased
internalization by phagocytic cells through opsonization.
Individuals with C3 deficiency are susceptible to bacterial
infection (28). Increased levels
of C3a have been found in the bronchoalveolar lavage fluid (BALF)
of mice infected with the pathogenic avian influenza, H5N1
(29). C4b is the basic form of
complement factor 4, which is a member of the classical activation
pathway. Extensive deposition of its fragment has been found in the
lungs in cases of influenza-associated mortality (30). C5 is comprised of α and β
polypeptide chains (C5a and C5b), which are linked by a disulfide
bridge. C5a is an important chemotactic cleavage product, assisting
in the recruitment of inflammatory cells. C5b initiates the
membrane attack pathway, by forming in the membrane attack complex,
which is the cytolytic end product of the complement cascade and
forms a transmembrane channel to cause osmotic lysis of the target
cell. Kupffer cells and other macrophage cell types assist in
clearing complement-coated pathogens. Individuals with C5 mutation
show a propensity for severe recurrent infections. Increased levels
of C5a have been found in the BALF of mice infected with H5N1 and
influenza virus A (29,31).
 | Table III.Genes detected in the complement
pathway. |
Table III.
Genes detected in the complement
pathway.
| Gene ID | Symbol | Full name | Probe ID | Systematic
name | Fold change |
|---|
| 715 | C1R | Complement
component 1, r subcomponent | A_23_P125423 | NM_001733 | 1.32 |
| 716 | C1S | Complement
component 1, s subcomponent | A_23_P2492 | NM_001734 | 2.1 |
| 718 | C3 | Complement
component 3 | A_23_P101407 | NM_000064 | 0.42 |
| 721 | C4B | Complement
component 4B | A_23_P42282 | NM_001002029 | 0.41 |
| 727 | C5 | Complement
component 5 | A_23_P71855 | NM_001735 | 0.3 |
| 3627 | C7 | Complement
component 7 | A_23_P213857 | NM_000587 | 0.83 |
| 733 | C8G | Complement
component 8G | A_23_P20713 | NM_000606 | 0.77 |
| 629 | CFB | Complement factor
B | A_23_P156687 | NM_001710 | 0.54 |
| 8518 | CFD | Complement factor
D | A_23_P119562 | NM_001928 | 0.72 |
| 10747 | MASP2 | Mannan-binding
lectin serine peptidase 2 | A_23_P301971 | NM_139208 | 1.27 |
| 719 | C3AR1 | Complement
component 3a receptor 1 | A_23_P2431 | NM_004054 | 0.64 |
| 728 | C5AR1 | Complement
component 5a receptor 1 | A_23_P153562 | NM_001736 | 0.69 |
| 3689 | ITGB2 | Complement
component 3 receptor 3 and 4 subunit | A_23_P329573 | NM_000211 | 0.64 |
The results of the present study suggested that
sufficient expression of IFITM3 appeared to be pivotal for the
recognition and elimination of infected cells by altering the
transcription of genes involving the complement cascade.
As a member of the IFITM protein family, IFITM3 is
involved in various biological processes. In the present study, the
use of endogenous IFITM3-KD provided information regarding global
gene expression alterations, and assisted in identifying the
IFITM3-targeted transcripts and pathways. The results of the
present study may enable more detailed investigation of the
biological functions of IFITM3 in the future.
Acknowledgements
This study was supported by grants from the National
Natural Science Foundation of China (grant nos. 81301403 and
81071315) and the Basic-Clinical Research Cooperation Grant from
Capital Medical University (grant no. 11JL53).
References
|
1
|
Siegrist F, Ebeling M and Certa U: The
small interferon-induced transmembrane genes and proteins. J
Interferon Cytokine Res. 31:183–197. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Brass AL, Huang IC, Benita Y, John SP,
Krishnan MN, Feeley EM, Ryan BJ, Weyer JL, van der Weyden L, Fikrig
E, et al: The IFITM proteins mediate cellular resistance to
influenza A H1N1 virus, West Nile virus, and dengue virus. Cell.
139:1243–1254. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Tanaka SS, Yamaguchi YL, Tsoi B, Lickert H
and Tam PP: IFITM/Mil/fragilis family proteins IFITM1 and IFITM3
play distinct roles in mouse primordial germ cell homing and
repulsion. Dev Cell. 9:745–756. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Saetre P, Emilsson L, Axelsson E, Kreuger
J, Lindholm E and Jazin E: Inflammation-related genes up-regulated
in schizophrenia brains. BMC Psychiatry. 7:462007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Garbett K, Ebert PJ, Mitchell A, Lintas C,
Manzi B, Mirnics K and Persico AM: Immune transcriptome alterations
in the temporal cortex of subjects with autism. Neurobiol Dis.
30:303–311. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Seno MM Ghahramani, Hu P, Gwadry FG, Pinto
D, Marshall CR, Casallo G and Scherer SW: Gene and miRNA expression
profiles in autism spectrum disorders. Brain Res. 1380:85–97. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Seo GS, Lee JK, Yu JI, Yun KJ, Chae SC and
Choi SC: Identification of the polymorphisms in IFITM3 gene and
their association in a Korean population with ulcerative colitis.
Exp Mol Med. 42:99–104. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hisamatsu T, Watanabe M, Ogata H, Ezaki T,
Hozawa S, Ishii H, Kanai T and Hibi T: Interferon-inducible gene
family 1-8U expression in colitis-associated colon cancer and
severely inflamed mucosa in ulcerative colitis. Cancer Res.
59:5927–5931. 1999.PubMed/NCBI
|
|
9
|
Fan J, Peng Z, Zhou C, Qiu G, Tang H, Sun
Y, Wang X, Li Q, Le X and Xie K: Gene-expression profiling in
Chinese patients with colon cancer by coupling experimental and
bioinformatic genomewide gene-expression analyses: Identification
and validation of IFITM3 as a biomarker of early colon
carcinogenesis. Cancer. 113:266–275. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Andreu P, Colnot S, Godard C, Laurent-Puig
P, Lamarque D, Kahn A, Perret C and Romagnolo B: Identification of
the IFITM family as a new molecular marker in human colorectal
tumors. Cancer Res. 66:1949–1955. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Shen L, Liu Q, Ni J and Hong G: A
proteomic investigation into the human cervical cancer cell line
HeLa treated with dicitratoytterbium (III) complex. Chem Biol
Interact. 181:455–462. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhang J, Timoney PJ, MacLachlan NJ,
McCollum WH and Balasuriya UB: Persistent equine arteritis virus
infection in HeLa cells. J Virol. 82:8456–8464. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Storey JD, Taylor JE and Siegmund D:
Strong control, conservative point estimation and simultaneous
conservative consistency of false discovery rates: A unified
approach. J R Statist Soc B. 66:187–205. 2004. View Article : Google Scholar
|
|
15
|
Happel N and Doenecke D: Histone H1 and
its isoforms: Contribution to chromatin structure and function.
Gene. 431:1–12. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Khan WN, Teglund S, Bremer K and
Hammarström S: The pregnancy-specific glycoprotein family of the
immunoglobulin superfamily: Identification of new members and
estimation of family size. Genomics. 12:780–787. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Snyder SK, Wessner DH, Wessells JL,
Waterhouse RM, Wahl LM, Zimmermann W and Dveksler GS:
Pregnancy-specific glycoproteins function as immunomodulators by
inducing secretion of IL-10, IL-6 and TGF-beta1 by human monocytes.
Am J Reprod Immunol. 45:205–216. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Connell S, Meade KG, Allan B, Lloyd AT,
Downing T, O'Farrelly C and Bradley DG: Genome-wide association
analysis of avian resistance to Campylobacter jejuni colonization
identifies risk locus spanning the CDH13 gene. G3 (Bethesda).
3:881–890. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Oikawa T and Yamada T: Molecular biology
of the Ets family of transcription factors. Gene. 303:11–34. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Munro S: The golgin coiled-coil proteins
of the Golgi apparatus. Cold Spring Harb Perspect Biol. 3(pii):
a0052562011.PubMed/NCBI
|
|
21
|
Lu J, Pan Q, Rong L, He W, Liu SL and
Liang C: The IFITM proteins inhibit HIV-1 infection. J Virol.
85:2126–2137. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yao L, Dong H, Zhu H, Nelson D, Liu C,
Lambiase L and Li X: Identification of the IFITM3 gene as an
inhibitor of hepatitis C viral translation in a stable STAT1 cell
line. J Viral Hepat. 18:e523–e529. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Huang IC, Bailey CC, Weyer JL, Radoshitzky
SR, Becker MM, Chiang JJ, Brass AL, Ahmed AA, Chi X, Dong L, et al:
Distinct patterns of IFITM-mediated restriction of filoviruses,
SARS coronavirus, and influenza A virus. PLoS Pathog.
7:e10012582011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Feeley EM, Sims JS, John SP, Chin CR,
Pertel T, Chen LM, Gaiha GD, Ryan BJ, Donis RO, Elledge SJ and
Brass AL: IFITM3 inhibits influenza A virus infection by preventing
cytosolic entry. PLoS Pathog. 7:e10023372011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Shen C, Wu XR, Jiao WW, Sun L, Feng WX,
Xiao J, Miao Q, Liu F, Yin QQ, Zhang CG, et al: A functional
promoter polymorphism of IFITM3 is associated with susceptibility
to pediatric tuberculosis in Han Chinese population. PLoS One.
8:e678162013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Janeway CA Jr, Travers P, Walport M and
Shlomchik MJ: Immunobiology: The complement system and innate
immunity. 5th. Garland Science; New York, NY: ISBN 0-815321,
2001-3642-X, 2001.
|
|
27
|
Abbas AK, Lichtman AH and Pillai S:
Cellular and Molecular Immunology. 6th. Elsevier; Philadelphia, PA:
pp. 272–288. 2010
|
|
28
|
Miller EC, Chase NM, Densen P, Hintermeyer
MK, Casper JT and Atkinson JP: Autoantibody stabilization of the
classical pathway C3 convertase leading to C3 deficiency and
Neisserial sepsis: C4 nephritic factor revisited. Clin Immunol.
145:241–250. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
O'Brien KB, Morrison TE, Dundore DY, Heise
MT and Schultz-Cherry S: A protective role for complement C3
protein during pandemic 2009 H1N1 and H5N1 influenza A virus
infection. PLoS One. 6:e173772011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Monsalvo AC, Batalle JP, Lopez MF, Krause
JC, Klemenc J, Hernandez JZ, Maskin B, Bugna J, Rubinstein C,
Aguilar L, et al: Severe pandemic 2009 H1N1 influenza disease due
to pathogenic immune complexes. Nat Med. 17:195–199. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Garcia CC, Weston-Davies W, Russo RC,
Tavares LP, Rachid MA, Alves-Filho JC, Machado AV, Ryffel B, Nunn
MA and Teixeira MM: Complement C5 activation during influenza A
infection in mice contributes to neutrophil recruitment and lung
injury. PLoS One. 8:e644432013. View Article : Google Scholar : PubMed/NCBI
|